Abstract
Plant and non-plant species possess cryptochrome (CRY) photoreceptors to mediate blue-light regulation of development or the circadian clock. The blue light-dependent homooligomerization of Arabidopsis CRY2 is a known early photoreaction necessary for its functions, but the photobiochemistry and function of light-dependent homooligomerization and heterooligomerization of cryptochromes, collectively referred to as CRY photooligomerization, have not been well-established. Here we show that photooligomerization is an evolutionarily conserved photoreaction characteristic of the CRY photoreceptors in plant and some non-plant species. Our analyses of the kinetics of the forward and reverse reactions of photooligomerization of Arabidopsis CRY1 and CRY2 provide a previously unrecognized mechanism underlying the different photosensitivity and photoreactivity of these two closely related photoreceptors. We found that photooligomerization is necessary but not sufficient for the functions of CRY2, implying that CRY photooligomerization must accompany with additional function-empowering conformational changes. We further demonstrate that the CRY2-CRY1 heterooligomerization plays roles in regulating functions of Arabidopsis CRYs in vivo. These results are consistent with the hypothesis that photooligomerization is an evolutionary conserved mechanism that determines the photosensitivity and photoreactivity of plant CRYs.
Short summary
Photooligomerization of Arabidopsis cryptochromes is fast, fluence rate-dependent and dark reversible and the photosensitivity of photooligomerization determines the photoreactivity of crytochromes. Photooligomerization is also an evolutionary conserved photoreaction characteristic of the CRY photoreceptors in plant and some non-plant species. Besides, photooligomerization is necessary but not sufficient for CRY2 functions and CRY2-CRY1 heterooligomerization plays roles in regulating functions of Arabidopsis CRYs.
INTRODUCTION
Cryptochromes (CRYs) are photoreceptors that mediate blue light regulation of development in plants and light entrainment of the circadian clock in plant and non-plant species (Cashmore et al., 1999; Sancar, 2000; Wang and Lin, 2019). Most higher plants possess two phylogenetically distinguishable clades of CRYs: CRY1 and CRY2, corresponding to the two CRYs first discovered in Arabidopsis (Ahmad and Cashmore, 1993; Guo et al., 1998). CRYs have two domains: the highly conserved FAD (Flavin Adenine Dinucleotide)-binding PHR (Photolyase Homologous Region) domain and the more divergent CCE (CRY C-terminal Extension, also referred to as CCT) domain of various lengths (Lin and Shalitin, 2003). The PHR domains of Arabidopsis CRY1 (residues 1–494) and CRY2 (residues 1–489) share about 50% amino acid sequence identity; whereas the CCE domains of CRY1 (residues 495–681) and CRY2 (residues 490–612) share less than 13% amino acid sequence identity (Lin and Shalitin, 2003; Lin et al., 1998). Arabidopsis CRY1 and CRY2 have distinct and similar functions (Wang and Lin, 2019). For example, both CRY1 and CRY2 mediate blue-light inhibition of hypocotyl elongation, whereas CRY2 mediates long-day promotion of flowering (Ahmad and Cashmore, 1993; Lin et al., 1998). The blue light-dependent protein-protein interactions are the primary mechanism underlying signal transductions of the CRY photoreceptors (Wang and Lin, 2019). Arabidopsis CRYs physically interact with transcription factors, such as CIBs (Cryptochrome Interacting bHLH transcription factors) and PIFs (Phytochrome Interacting Factors), to regulate transcription directly, and they also interact with the CUL4COP1-SPAs E3 ubiquitin ligase or auxin and brassinosteroid regulators (AUX/IAA, BES1, HBI1), to modulate gene expression (Wang and Lin, 2019; Wang et al., 2018). The PHR domain of CRYs is directly involved in protein-protein interactions of CRYs with most known CRY-signaling proteins, but the CCE domain is also necessary for the functions of plant CRYs (Wang and Lin, 2019).
Two elegant experiments have demonstrated that homodimerization of Arabidopsis CRY1 and CRY2 is required for the functions of plant CRYs (Rosenfeldt et al., 2008; Sang et al., 2005). And it was reported recently that CRY2 homodimerization is a blue light-dependent photoreaction that is necessary for the CRY2 photoactivation in vivo (Wang et al., 2016). Because the photoexcited CRY2 forms microscopically visible homooligomers, also referred to as CRY2 nuclear bodies or photobodies, in the absence of other CRY2-interacting proteins (Mas et al., 2000; Ozkan-Dagliyan et al., 2013; Yu et al., 2009; Zuo et al., 2002), we hypothesize that plant CRYs may undergo not only light-dependent homodimerization but also light-dependent homooligomerization and heterooligomerization, collectively referred as photooligomerization, to exert their cellular functions. Several questions of CRY photooligomerization, which are important for our understanding of the mechanism of CRY functions, have not been investigated. For example, it remained unclear what is the basic kinetics of forward or reverse reactions of CRY photooligomerization, whether photooligomerization is a common photoreaction of plant CRYs, how does CRY photooligomerization associate with CRY photosensitivity, whether photooligomerization is sufficient for CRY function, and whether CRY1 and CRY2 undergo heterooligomerization. In this study, we systematically characterized photooligomerization of plant CRYs to address the above questions. We found that photooligomerization is an evolutionarily conserved photoreaction of plant CRYs, that the oligomerization of CRYs in blue light is much faster than the spontaneous thermal relaxation or monomerization of CRYs in darkness. We further showed that the different kinetics of photooligomerization of CRY1 and CRY2 can explain their respective different photosensitivity. Using various genetics approaches, we also demonstrated that photooligomerization of CRY2 is necessary but not sufficient for its functions, and that blue light-responsive CRY2-CRY1 heterooligomerization may regulate their functions in plants.
RESULTS
CRY photooligomerization is fast, fluence rate-dependent, and dark-reversible
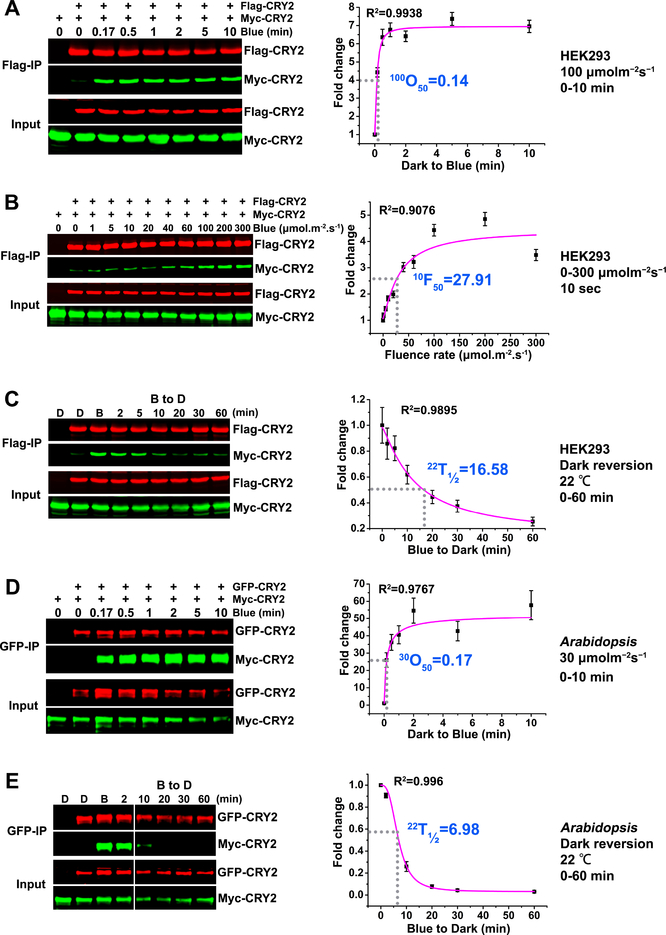
We first investigated the kinetics of photooligomerization of Arabidopsis CRY2, using ex vivo co-IP (co-immunoprecipitation) assays that we had previously established (Wang et al., 2016). In this method, two differentially tagged Arabidopsis CRY2, such as Flag-CRY2 and Myc-CRY2, were co-expressed in HEK293 (Human Embryonic Kidney 293) cells and analyzed for their physical interaction by quantitative co-IP assays, using the near-infrared fluorescence imaging system (Odyssey® CLx, Li-COR). The HEK293 cells contain no plant protein, so the direct protein-protein interaction analyzed by the co-IP assay is not affected by the indirect protein-protein interactions or blue light-induced CRY2 degradation. Results of this experiment shows that the levels of immunoprecipitated Flag-CRY2 stayed unchanged in HEK293 cells over the time before or after light treatment, but the level of Myc-CRY2 co-immunoprecipitated with Flag-CRY2 increased rapidly until it reached saturation in cells irradiated with blue light (Fig. 1A, Fig. S1A). CRY2 photooligomerization occurred fast, reaching 50% saturation in approximately 8 to15 seconds in HEK293 cells irradiated with 100 μmolm−2s−1 blue light (Fig. 1A, 100O50=0.14 min; Table 1, 100O50=0.25 min),or about 50 seconds in HEK293 cells exposed to 30 μmolm−2s−1 blue light (Table 1, 30O50=0.78 min; Fig. S1A, 30O50=0.83 min). The rate of CRY2 photooligomerization reaction increases in response to increased fluence rates of blue light (Fig. 1B) for the defined time of irradiation. For the irradiation time of 10 seconds, the fluence rate of blue-light that renders 50% saturation of CRY2 photooligomerization is approximately 28 μmolm−2s−1 (Fig. 1B, 10F50=27.91 μmolm−2s−1; Table 1, 10F50=28.9μmolm−2s−1). Photoactivated photoreceptors commonly reverse their conformational changes to that of their inactive ground state in darkness by the process of thermal relaxation, which is referred to as dark reversion or thermal relaxation. Indeed, after the blue light-treated HEK293 cells were transferred to darkness, CRY2 photooligomers undergo spontaneous reversion to monomers, which was shown by decreasing amounts of Myc-CRY2 co-immunoprecipitated by Flag-CRY2 (Fig. 1C). In the HEK293 cells exposed to 30 μmolm−2s−1 blue light for 5 min before transferred to darkness, CRY2 monomerization reached 50% saturation in about 17 min the dark (Fig. 1C, 22T1/2=16.58 min; Table 1, 22T1/2 =18.62 min), suggesting that the CRY2 oligomerization reaction under 30 μmolm−2s−1 blue light is about 21 times faster than the reverse reaction or the CRY2 monomerization reaction in the dark (22T1/2≈17 min, 30O50≈0.8 min). As expected for the dark reversion reaction being a thermal relaxation process, the rate of spontaneous dark reversion or monomerization of CRY2 in darkness is faster at higher temperature. In comparison with the rate of dark reversion of CRY2 oligomers at 22°C, the rate of CRY2 monomerization increased about 3 times at 37°C (Fig. S2A, 37T1/2=5.42 min). We concluded that CRY2 photooligomerization in heterologous cells is fast, fluence rate-dependent, and spontaneously but slowly reversible to monomers in the dark.
Figure 1. CRY2 photooligomerization ex vivo or in vivo.
(A-C): CRY2 photooligomerization in HEK293 cells (ex vivo).
(A) The kinetics analyses of CRY2 photooligomerization in response to blue light. HEK293 cells were transfected with equal amount of two plasmids encoding Flag-CRY2 or Myc-CRY2. 40 hours after transfection, cells were exposed to blue light of 100 μmolm−2s−1. After 10 seconds to 10 minutes of light treatment, the light-treated and control (left in the dark) cells were killed by lysis buffer. Flag-CRY2 was immunoprecipitated by Flag-agrose gels. The immunoblots were probed by the anti-Flag and anti-Myc antibody, and quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY2 (co-IP product) were normalized by the respective Flag-CRY2 (IP product), and presented as Fold Change (FC) by the formula FC = [Myc-CRY2/Flag-CRY2]Blue / [Myc-CRY2/Flag-CRY2]Dark. 100O50 indicates the time (minute) required to reach 50% saturation of CRY2 photooligomerization at the fluence rate of 100 μmolm−2s−1 of blue light.
(B) The fluence-rate response analyses of CRY2 photooligomerization. HEK293 cells transfected with Flag-CRY2 or Myc-CRY2 were exposed to blue light of the indicated fluence rates for 10 seconds. The same analyzation was performed as described in (A). 10F50 indicates the fluence rate of blue light required to achieve 50% CRY photooligomerization after light exposure for 10 seconds.
(C) The dark reversion analyses of CRY2 photooligomerization. HEK293 cells transfected with Flag-CRY2 or Myc-CRY2 were irradiated with 30 μmolm−2s−1 blue light for 5 min before transferring to darkness (at 22°C) for the indicated times. The same analyzation was performed as described in (A-B). 22T1/2 indicates the time (minute) required to reverse 50% of CRY2 photooligomers into monomers at the temperature of 22°C.
(D-E): CRY2 photooligomerization in plants (in vivo).
(D) The kinetics analyses of CRY2 photooligomerization in response to blue light in plants. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY2 and Myc-CRY2 were grown on petri dishes in the dark, exposed to 30 μmolm−2s−1 blue light, and tissues were collected at the indicated time (minute). GFP-Trap agarose beads were used for immunoprecipitation. The immunoblots were probed by the anti-GFP antibody to detect IP product and anti-Myc antibody to detect Co-IP product, and quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY2 (co-IP product) were normalized by the respective GFP-CRY2 (IP product), and presented as Fold Change (FC) by the formula FC = [Myc-CRY2/GFP-CRY2]Blue / [Myc-CRY2/GFP-CRY2]Dark. 30O50 indicates the time (minute) required to reach 50% saturation of CRY2 photooligomerization in plants at the fluence rate of 30 μmolm−2s−1 of blue light.
(E) The dark reversion analyses of CRY2 photooligomerization. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY2 and Myc-CRY2 were irradiated with 30 μmolm−2s−1 blue light for 5 min before transferring to darkness for the indicated times. 22T1/2 indicates the time (minute) required to reverse 50% of CRY2 photooligomers into monomers.
The data for curve fitting were derived from three technical repeats by performing three independent western blots. The data were analyzed by Logistic Nonlinear Curve Fit method. R2 stands for coefficient of regression. Standard errors are shown.
Table 1.
Kinetics parameters of CRY photooligomerization.
| Parameters | Experiment #1 | Experiment #2 | Sources | |
|---|---|---|---|---|
| CRY2 | 30O50 (minute) | 0.83 | 0.78 | HEK 293 |
| 100O50 (minute) | 0.14* | 0.25* | HEK 293 | |
| 10F50 (μmolm−2s−1) | 27.91* | 28.9 | HEK 293 | |
| 22T½ (minute) | 16.58* | 18.62* | HEK 293 | |
| 37T½ (minute) | 5.42 | HEK 293 | ||
| 30O50 (minute) | 0.17* | 0.26 | Arabidopsis | |
| 10F50 (μmolm−2s−1) | 16.89 | 13.56 | Arabidopsis | |
| 10T½ (minute) | 8.42* | Arabidopsis | ||
| 22T½ (minute) | 6.98* | Arabidopsis | ||
| 37T½ (minute) | 5.46* | Arabidopsis | ||
| CRY1 | 30O50 (minute) | 1.47 | 1.70 | Arabidopsis |
| 100O50 (minute) | 0.37* | 0.38 | Arabidopsis | |
| 60F50 (μmolm−2s−1) | 14.90 | 16.21 | Arabidopsis | |
| 22T½ (minute) | 2.56 | 1.82 | Arabidopsis |
The table summarizes results of the experiments for the measurement of kinetics parameters of Arabidopsis CRY1 and CRY2 photooligomerization in HEK293 (Human Embryonic Kidney) cells or Arabidopsis seedlings. Symbols of parameters are the following: 30O50 or 100O50: time (minute) required to reach 50% saturation of CRY photooligomerization at the fluence rate of 30 or 100 μmolm−2s−1, respectively; 10F50 or 60F50: the fluence rate of blue light (μmolm−2s−1) required to achieve 50% CRY photooligomerization after light exposure for 10 or 60 seconds, respectively; 10T1/2, 22T1/2, or 37T1/2: time (minute) required to reach 50% saturation of CRY monomerization at the temperature of 10 °C, 22 °C, or 37 °C, respectively. Numbers with asterisk (*) indicates the measurements were means of three technical repeats, and numbers without asterisk indicates the measurements were derived from one technical repeat.
We next examined in vivo CRY2 photooligomerization in transgenic plants co-expressing GFP-CRY2 and Myc-CRY2, using the similar co-IP assay as described above. In contrast to the HEK293 ex vivo system, the kinetics parameters of CRY homooligomerization measured by the co-IP assay in vivo are likely interfered by interactions of CRYs with non-CRY molecules and by light-dependent CRY turnover. Therefore, the kinetics parameters of CRY homooligomerization reaction measured by the co-IP assay in vivo may suffer from over-estimation or under-estimation. As expected, the forward and reverse reactions of CRY2 photooligomerization in plant cells exhibited somewhat different kinetics to that observed in the heterologous HEK293 cells. Similar kinetics of CRY2 photooligomerization in plant cells was observed by the near-infrared fluorescence imaging method and the conventional ECL (enhanced chemiluminescence) method (Fig. 1D, Fig. S1B). For example, when etiolated seedlings were exposed to 30 μmolm−2s−1 blue light, CRY2 photooligomerization reached 50% saturation within approximately 10 to 16 seconds (Fig. 1D, 30O50=0.17 min; Table 1, 30O50=0.26 min), which is about 3 to 5 times faster than that in HEK293 cells (Table 1, 30O50=0.83 and 0.78 min). Consistently, when the reaction reached saturation, the normalized fractions of CRY2 oligomers increased about 40 to 50 folds in plant cells in comparison to about 2.5 fold increased in HEK293 cells under the same light condition (Fig. 1D, Fig. S1A). Furthermore, the CRY2 photooligomerization reaction seems more sensitive to the blue light in plant cells than that in the heterologous HEK293 cells. In plants irradiated for 10 seconds, the fluence rate that renders 50% saturation of CRY2 photooligomerization within 10 seconds is approximately 14 to 17 μmolm−2s−1 (Table 1, 10F50=13.56 μmolm−2s−1; Fig. S1C, 10F =16.89 μmolm−2s−1). Therefore, the total fluence required for CRY2 photooligomerization 50 is about 140–170 μmolm−2 (14–17 μmolm−2s−1 × 10 seconds) in plant cells, or only half that of the 280 μmolm−2 (28 μmolm−2s−1 × 10 seconds) needed to trigger the same extent of CRY2 photooligomerization in HEK293 cells. These observations may be explained by the more favorable native chemical or cellular environment for the CRY2 photooligomerization reaction. But the exact nature of the native environment favoring CRY2 photooligomerization remains unclear. One may speculate that the generally more reduced cellular environment of plant cells may be associated with this phenomenon. For example, the redox potential of plant or mammalian nuclear/cytosol compartments have been estimated to be approximately −320 mV (Meyer et al., 2007; Schwarzlander et al., 2008) or −280 mV (Go and Jones, 2010), respectively. It is intuitive that a more reduced cellular environment may favor photoreduction of CRYs to facilitate the conformational changes associated with CRY photoactivation. Alternatively, existence of CRY2-interacting proteins in plant cells or some plant metabolic reactions may also contribute to the more robust CRY2 photooligomerization in plant cells.
We then compared the dark reversion or monomerization reaction of CRY2 in the heterologous HEK293 and the native plant cells. In these experiments, seedlings irradiated with blue light (30 μmolm−2s−1) for 5 min were transferred to darkness, and the decline of CRY2 oligomers were analyzed for up to 60 min. Similar kinetics of CRY2 monomerization was obtained by the ECL method (Fig. S2B). Similar to that observed in HEK293 cells, the rate of CRY2 dark reversion also increased at higher temperature in Arabidopsis seedlings (Fig.1E, 22T1/2=6.98 min; Fig. S2C, 10T1/2=8.42 min; Fig.S2D, 37T1/2=5.46 min), confirming that CRY dark reversion is a thermal relaxation reaction. Similar to the photooligomerization reaction, the rate of CRY2 monomerization reaction is also favored in the native plant cells. In plants, CRY2 oligomers decreased by 50% within about 7 min in darkness at 22°C (Fig. 1E, 22T1/2=6.98 min), which is about 2.5 folds faster than the rate of CRY2 dark-reversion in the heterologous HEK293 cells at the same temperature of 22°C (Table 1, 22T1/2=16.58 and 18.62 min). In other words, in the absence of light, the half-life of CRY2 oligomers in plants (~7 min) is much shorter than that in HEK293 cells (~18 min). This observation may be explained by the existence of CRY2 inhibitory proteins BICs in plant cells but not HEK293 cells. Although BICs exhibit higher affinity to photoexcited CRY2, they also exhibit low affinity binding to CRY2 in the dark (Wang et al., 2016). This “background” CRY2-BIC interaction in darkness may explain why the half-life of CRY2 oligomer is shorter in Arabidopsis than that in HEK293. The BIC-dependent reduction of the half-life of CRY2 oligomer may act to “desensitize” CRY2 or to increase the amplitude of photoresponses of CRYs and photosensitivity of plants. However, these hypotheses remain to be tested directly.
Photosensitivity of photooligomerization determines photoreactivity of CRY1 and CRY2
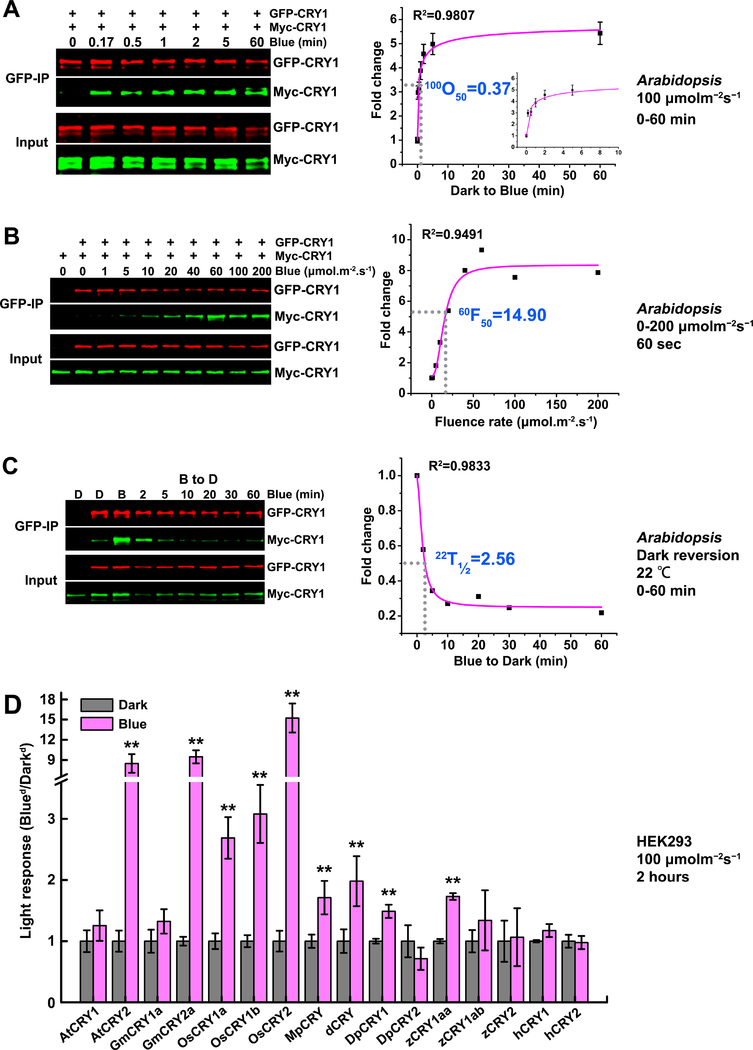
Arabidopsis CRY1 also undergoes blue light-dependent photooligomerization in plants (Fig. 2A–C). We failed to detect a clear light responsiveness of CRY1 oligomerization in the heterologous HEK293 cells (Fig. 2D, Fig. S3A), which may be because CRY1 oligomerization is more dependent on the native cellular environment, such as possible chaperone molecules or specific ionic or redox conditions. Analyses of CRY1 photooligomerization in Arabidopsis plants co-expressing GFP-CRY1 and Myc-CRY1 by co-IP assays indicate that CRY1 is less sensitive to blue light than CRY2. First, in contrast to CRY2 photooligomerization in etiolated seedlings irradiated with 30 μmolm−2s−1 blue light (Table 1, 30O50=0.17 and 0.26 min), it took much longer for CRY1 photooligomerization to reach saturation under the same condition (Table 1, 30O50=1.70 min; Fig. S1D, 30O50=1.47 min). In other words, the CRY1 photooligomerization reaction is about 6 to 8 times slower than CRY2 in plants. This phenomenon is similarly observed in different light conditions. For example, the rate of photooligomerization of CRY1 in plants illuminated with 100 μmolm−2s−1 blue light (100O50≈0.4 min, Fig. 2A, Table 1) is about 2 times slower than the rate of oligomerization of CRY2 (30O5≈0.2 min, Table 1) in plants illuminated with a much lower fluence rate (30 μmolm−2s−1) of blue light. Second, the fluencerate of blue light rendering 50% saturation of CRY1 oligomerization in seedlings irradiated for 60 seconds is about 15 μmolm−2s−1 (Fig. 2B, 60F50 =14.9 μmolm−2s−1;Table 1, 60F50=16.21 μmolm−2s−1). By comparison, 6 times more blue light is needed for CRY1 to reach the same extent of photooligomerization than that for CRY2 under the same condition (Table 1, 10F50=13.56 to 16.89 μmolm−2s−1). In other words, CRY2 photooligomerization is about 6 times more sensitive to blue light than that of CRY1. The photooligomerized CRY1 also undergoes monomerization in darkness. In light-irradiated plants transferred to darkness, it took approximately 2 minutes for CRY1 to reach 50% saturation of monomerization in darkness (Fig. 2C, 22T1/2=2.56 min; Table 1, 22T1/2=1.82 min)at 22°C, indicating that the half-life of CRY1 oligomers in darkness is shorter than that of CRY2 (Fig. 1E, 22T1/2=6.98 min).
Figure 2. The photooligomerization of CRY1 in plants and the conservation of CRY homooligomerization across speices.
(A-C): CRY1 photooligomerization in plants (in vivo).
(A) The kinetics analyses of CRY1 photooligomerization in response to blue light in plants. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY1 and Myc-CRY1 were grown on petri dishes in the dark, exposed to blue light of 100 μmolm−2s−1, and tissues were collected at the indicated time (minute). GFP-CRY1 were co-immunoprecipitated by GFP-trap beads. The immunoblots were probed by the anti-GFP and anti-Myc antibody, and quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY1 (co-IP product) were normalized by the respective GFP-CRY1 (IP product), and presented as Fold Change (FC) by the formula FC = [Myc-CRY1/GFP-CRY1]Blue / [Myc-CRY1/GFP-CRY1]Dark. 100O50 indicates the time (minute) required to reach 50% saturation of CRY1 photooligomerization in plants at the fluence rate of 100 μmolm−2s−1 of blue light. The insert small graph shows the first 10 minutes in the curve. The data for curve fitting were derived from three technical repeats by performing three independent western blots. Standard errors are shown.
(B) The fluence-rate response analyses of CRY1 photooligomerization in plants. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY1 and Myc-CRY1 were exposed to blue light of the indicated fluence rates for 60 seconds. The same analyzation was performed as described in (A). 60F50 indicates the fluence rate of blue light required to achieve 50% CRY1 photooligomerization after light exposure for 60 seconds.
(C) The dark reversion analyses of CRY1 photooligomerization. Seven-day-old Arabidopsis seedlings co-expressing GFP-CRY1 and Myc-CRY1 were irradiated with 30 μmolm−2s−1 blue light for 30 min before transferring to darkness for the indicated times. 22T1/2 indicates the time (minute) required to reverse 50% of CRY2 photooligomers into monomers at the temperature of 22°C.
The data in (A-C) were analyzed by Logistic Nonlinear Curve Fit method. R2 stands for coefficient of regression.
(D): The conservation of CRY homooligomerization across speices.
HEK293 cells were transfected with equal amount of two plasmids encoding Flag-CRY or Myc-CRY from different species: AtCRYs of Arabidopsis, GmCRYs of soybean (Glycine max), OsCRYs of rice (Oryza sativa), MpCRY of liverwort (Marchantia polymorpha), dCRY of Drosophila, DpCRYs of Monarch butterfly (Danaus plexippus), zCRYs of Zebrafish, and hCRYs of human (Homo sapien). After transfection, cells were exposed to 100 μmolm−2s−1 for 2 hours or left in the dark. Flag-CRYs were co-immunoprecipitated by Flag-agrose gels. The immunoblots were probed by the anti-Flag antibody to detect IP product and anti-Myc antibody to detect Co-IP product. Three independent immunoblots were quantified by the Image Studio Lite software in the Odyssey® CLx System (LI-COR). The Myc-CRY2 (co-IP product) were normalized by the respective Flag-CRY2 (IP product), and presented as light response by the formula: light response = [Myc-CRY2/Flag-CRY2]Blue / [Myc-CRY2/Flag-CRY2]Dark. Mean values are shown (n=3, **P≤0.01). The representative immunoblots for quantification were shown in supplemental figure S3.
It has been previously shown that CRY2 is a “low light” photoreceptor that mediates photoresponses primarily in low intensities or fluence rates of blue light, whereas CRY1 is a “high light” photoreceptor that mediate photoresponses primarily in high intensities of blue light (Lin et al., 1998). This phenomenon was previously explained by the fact that CRY2, but not CRY1, is degraded in high light (Lin et al., 1998). However, the blue light-dependent degradation of CRY2 only explains why there is lower amount of active CRY2 than CRY1 in high light, and it was unclear whether CRY2 may be more active than CRY1 in low light. Given that photooligomerization is the early photoreaction necessary for photoactivation of both CRY1 and CRY2, the comparative kinetics of photooligomerization described here provides an additional explanation for the different photosensitivity of CRY1 and CRY2. Based on the new results (Fig. 1–2), we propose that the higher photosensitivity of CRY2 photooligomerization allows CRY2 to be activated in response to low blue light. The relatively longer half-life of CRY2 in darkness is also consistent with its role as a low-light photoreceptor, because the active oligomers of CRY2 may persist for a longer time in plant cells under low or no light. The structural mechanisms underlying different photosensitivity of the photooligomerization reaction of CRY1 and CRY2 remain to be further investigated.
Photooligomerization is necessary but not sufficient for the function of CRY2
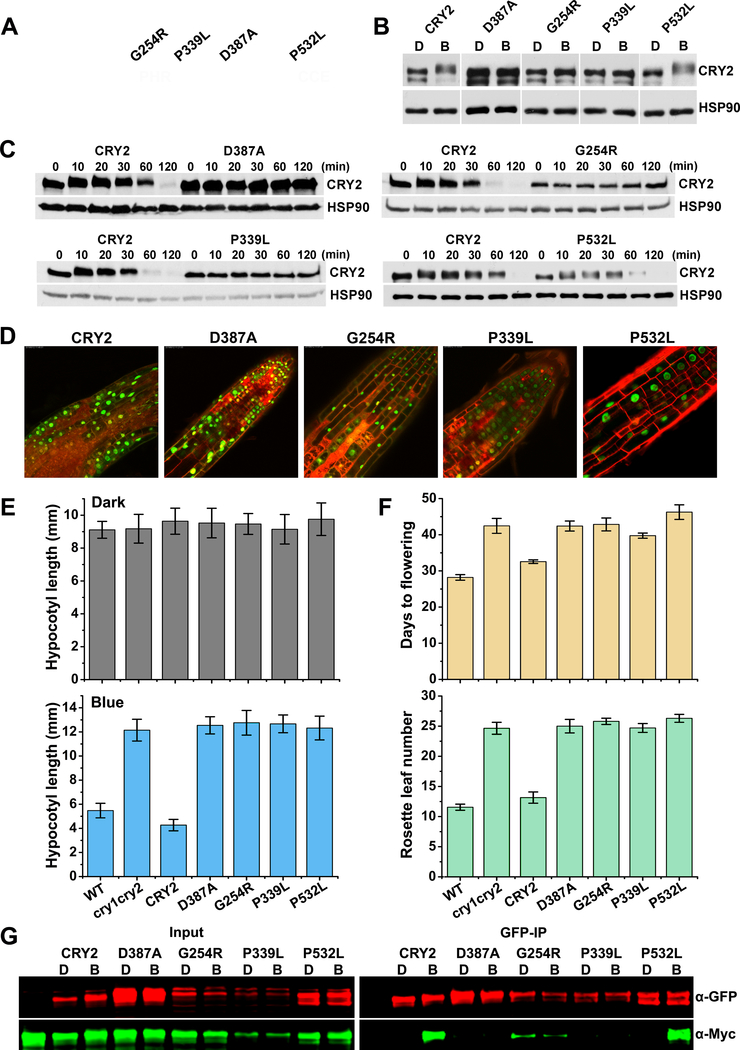
It has been previously demonstrated that photooligomerization is the photoactivation mechanism necessary for the biochemical and physiological functions of Arabidopsis CRY1 and CRY2 (Rosenfeldt et al., 2008; Sang et al., 2005; Wang et al., 2016). However, it was unclear whether CRY photooligomerization is sufficient for CRY function, which is also important to our understanding of the photoactivation mechanism of CRYs. To address this question, we analyzed four loss-of-function mis-sense cry2 mutations identified from 11 cry2 mis-sense mutations that were isolated in our previous genetic studies (Guo, 2001; Guo et al., 1998; Liu et al., 2008), including G254R (CRY2G254R), P339L (CRY2P339L), D387A (CRY2D387A), and P532L (CRY2P532L) (Fig.3). The CRY2D387A and CRY2G254R mutants have been previously reported (Guo et al., 1998; Liu et al., 2008). D387 is required for FAD-binding to CRY2 and it is universally conserved in CRYs in plants and other major evolutionary lineages (Liu et al., 2008). G254 and P339L are also universally conserved in CRYs. P532 is one of the two core residues of the non-canonical VP motif of CRY2 that has been recently shown to interact with COP1 (Lau et al., 2019) and SPA1 (Ponnu et al., 2019). The cry1 mutant (hy4–9, or CRY1P549) that is equivalent to CRY2P532L has been reported previously (Ahmad et al., 1995). The GFP-fusion proteins of these four mutant proteins are located in the nucleus but showed no apparent activity in hypocotyl inhibition or floral promotion (Fig. 3D–F). Surprisingly, although CRY2G254R, CRY2P339L, and CRY2D387A exhibited little blue light-responsive phosphorylation or degradation as expected (Fig. 3B–C), the CRY2P532L mutant protein showed seemingly normal photoresponsive phosphorylation or degradation (Fig. 3B–C). These results suggest that CRY2P532L must retain light responsiveness but fail to transfer the photoactivity to physiological activities. We prepared transgenic plants co-expressing the GFP- and Myc-tagged fusion proteins of these 4 cry2 mutants, and analyzed their photooligomerization activity in plants by the co-IP assay (Fig. 3G). Results of this experiment showed that three mutants, CRY2D387A, CRY2G254R, and CRY2P339L, lost photooligomerization activity (Fig. 3G). The apparently low abundance of CRY2P339L prevented us from quantifying its rate of photooligomerization. CRY2G254R exhibited weak homooligomerization without discernable photoresponse. Importantly, the CRY2P532L mutant protein exhibited seemingly normal level of expression and photooligomerization activity (Fig. 3G). The fact that CRY2P532L lost physiological activity but retained photooligomerization and other aspects of photo-responsiveness argues strongly that photooligomerization is necessary but not sufficient for the biochemical and physiological functions of CRY2. Our observation is consistent with the recent study of the function of the VP motif of CRY2 (Ponnu et al., 2019).
Figure 3. Photooligomerization is necessary but not sufficient for CRY2 function.
(A) Distribution of mutations found in Arabidopsis CRY2 genes. CRY2 is composed of PHR domain (1–489 aa) and the C-terminal extension (CCE, 490–612 aa) as indicated. cry2 mutants were roughly labelled on the top. G254R, the residue glycine at position 254 was changed to arginine; P339L, the residue proline at position 339 was changed to leucine; D387A, the residue aspartic acid at position 387 was changed to alanine; P532L, the residue proline at position 532 was changed to leucine. Green rectangle stands for nuclei localization signals.
(B) Phosphorylation analyses of cry2 mutants. GFP tag was fused to N-terminal of CRY2 and cry2 mutants to generate GFP-CRY2 and GFP-cry2 mutants, and the constructs were transformed into cry1cry2 double mutant background. Six-day-old dark-grown transgenic seedlings were transferred to 50 μmolm−2s−1 blue light for 20 min. Proteins were extracted, fractioned by SDS-PAGE gels, blotted and probed with anti-CRY2 antibody. Membranes were stripped and reprobed with anti-HSP90 for loading controls. The arrowhead indicates the electrophoretic-mobility-shift (phosphorylated) CRY2 and the arrow indicates mostly the unphosphorylated CRY2.
(C) Degradation analyses of cry2 mutant proteins. Six-day-old dark-grown transgenic seedlings were transferred to 50 μmolm−2s−1 blue light for the indicated time (10, 20, 30, 60,120 min). Proteins were extracted, fractioned by SDS-PAGE gels, blotted and probed with anti-CRY2 antibody. Membranes were stripped and reprobed with anti-HSP90 for loading controls.
(D) Subcellular localization analyses of cry2 mutant proteins. Seven-day-old GFP-cry2 mutant seedlings grown in long day were analyzed by confocal microscope. PI stain (red color) was used to show the cell boundaries. Scale bar: 20 μm.
(E) Measurement of hypocotyl length of transgenic cry2 mutants. Seedlings were grown in the dark (upper pannel) or continuous blue (10 μmolm−2s−1, lower pannel) for six days before measurement. Hypocotyl lengths with standard deviations (n≥20) are shown.
(F) Measurement of flowering time of transgenic cry2 mutants. Plants were grown in long day (16 hour light / 8 hour dark) condition. Days to flowering (upper pannel) and rosette leaf numbers (lower pannel) with standard deviations are shown (n≥20).
(G) Photooligomerization analyses of cry2 mutants. Seven days old transgenic Arabidopsis seedlings co-expressing GFP-CRY2 and Myc-CRY2 or GFP-cry2 mutants and Myc-cry2 mutants were grown in the dark, exposed to blue light of 30 μmolm−2s−1 for 5 min or left in the dark. GFP tagged proteins were co-immunoprecipitated by GFP-trap beads. The immunoblots were analyzed by probing with anti-GFP and anti-Myc antibodies.
Photooligomerization is an evolutionarily conserved photoreaction of plant and animal CRYs
We investigated whether CRYs from other plants and animal species may also undergo photooligomerization in HEK293 cells. In this experiment, we co-expressed Flag- and Myc-tagged CRYs in HEK293 cells, including GmCRYs of soybean, OsCRYs of rice, MpCRY of liverwort, DpCRYs of Monarch butterfly, dCRY of Drosophila, zCRYs of Zebrafish, and hCRYs of human, and analyzed their photooligomerization by the co-IP assays. Results of this experiment show that photooligomerization of plant CRYs is conserved evolutionarily from dicot, monocot, to liverwort. For example, most plant CRY2 proteins examined exhibited light-dependent oligomerization (Fig. 2D, Fig. 4, Fig. S3–4). Many CRY1s, including OsCRY1a, OsCRY1b, and GmCRY1c, also exhibited robust photooligomerization in the heterologous HEK293 cells, although Arabidopsis CRY1 and soybean GmCRY1a or GmCRY1b showed light-independent oligomerization in HEK293 cells (Fig. 2D, Fig. 4, Fig. S3–4). On the other hand, most animal CRYs examined, except dCRY, DpCRY1 and zCRY1aa, exhibited no photooligomerization in HEK293 cells (Fig. 2D, Fig. S3). It is interesting that, among the animal CRYs, only those that had been previously reported to be photoactive, such as dCRY of Drosophila, or potentially photoactive, such as DpCRY1 of butterfly and zCRY1aa (Frøland Steindal and Whitmore, 2019; Sancar, 2000), showed photooligomerization (Fig. 2D, Fig. S3). Those animal CRYs that are reported to act primarily as the light-independent transcription repressors (Sancar, 2000), such as hCRY1 and hCRY2 of human, showed no light-dependent oligomerization (Fig. 2D, Fig. S3). Such “coincidence” appears to correlate well with the photoactivity or proposed photoactivity of those animal CRYs. We hypothesize that photooligomerization is a photoreaction evolutionarily conserved in not only plant CRYs but also photoactive animal CRYs, although the photoreceptor activity of most animal CRYs, except dCRY, remain to be experimentally tested.
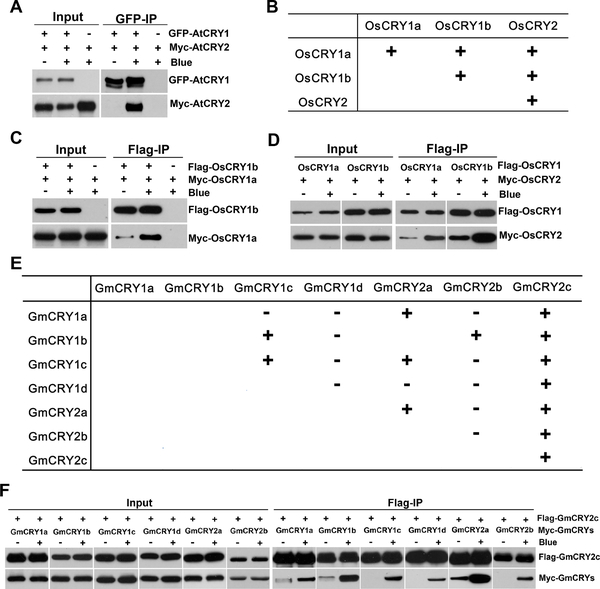
Figure 4. Heterooligomerization of plant CRYs.
(A) Blue-light dependent heterooligomerization of Arabidopsis CRY1 and CRY2. Three-week-old LD-grown Arabidopsis plants co-expressing GFP-CRY1 and Myc-CRY2 were adapted to the dark for 24 hours before sample collection and then treated with blue light (30 μmolm−2s−1) for 10 min or kept in the dark. Total proteins were extracted and immunoprecipitated with GFP-trap beads. The IP (GFP-CRY1) and Co-IP (Myc-CRY2) signals were probed with GFP and Myc antibodies, respectively.
(B-D) Homo- and heterooligomerization analyses of rice CRYs in HEK293. (B) The table is a summary of the immunoblots from (C, D) and supplemental figure S3 (E, F, G), showing the blue-light dependent homo- and heterooligomerization of rice CRYs (OsCRYs). (C-D) HEK293 cells were co-transfected with indicated Flag-CRY and Myc-CRY. Cells were treated with 100 μmol m−2s−1 blue light for 2 hours or kept in the dark at 22°C. Flag-CRYs were co-immunoprecipitated by Flag-agrose gels. The IP signals (Flag-CRY) or the Co-IP signals (Myc-CRY) were detected by immunoblots probed with Flag or Myc antibodies, respectively. (E-F) Homo and heterooligomerization analyses of soybean CRYs in HEK293. (E) The table is a summary of the immunoblots from (F), supplemental figure S3 (C, D) and supplemental figure S4, showing the homo- and heterooligomerization of soybean CRYs (GmCRYs). (F) Heterooligomerization of soybean GmCRYs were analyzed as described in (C-D). “−” represents no oligomerization; “ ” represents blue light-independent oligomerization; “+” represents blue light-dependent oligomerization.
” represents blue light-independent oligomerization; “+” represents blue light-dependent oligomerization.
Blue light-dependent heterooligomerization regulates the activity of CRYs
We finally investigated whether CRYs in Arabidopsis or other plant species may undergo blue light-responsive heterooligomerization and whether CRY heterooligomerization may affect the functions of CRYs. We first examined CRY1-CRY2 heterooligomerization in transgenic Arabidopsis plants co-expressing GFP-CRY1 and Myc-CRY2. Fig. 4A shows robust photooligomerization between Arabidopsis CRY1 and CRY2 in plants. The blue light-dependent oligomerization was also observed among various isoforms of CRY1 and CRY2 from rice and soybean in the heterologous HEK293 system (Fig. 4B–F, Fig. S4). For example, both rice CRY1, OsCRY1a and OsCRY1b, showed light-responsive heterooligomerization with each other and with OsCRY2 (Fig. 4B–D). The seven soybean CRYs showed a more complex pattern of photooligomerization in HEK293 cells. As summarized in Fig. 4E, different CRYs exhibited different photooligomerization characteristics in the heterologous HEK293 cells. For example, soybean GmCRY2c exhibited photoresponsive homooligomerization and photoresponsive heterooligomerization with all soybean GmCRYs tested, whereas other soybean GmCRYs exhibited more complex photoresponsivity and oligomerization characteristics. Although the physiological significance of the photooligomerization activity or lack of such activity shown by rice and soybean CRYs in the heterologous HEK293 cells remain to be verified in vivo, our results clearly demonstrate that plant CRYs are capable of blue light-dependent heterooligomerization.
It is technically difficult to directly test the functional relevance of CRY heterooligomers in vivo. However, the function of CRY oligomerization has been tested by the dominant effect of a truncated CRY fragment, taking advantage of the fact that overexpression of a truncated protein fragment that lacks the native biochemical activity of the holoprotein may retain certain protein-protein interaction activities such that to act as a competitive inhibitor of the endogenous protein in vivo. For example, it has been previously reported the Myc-CNT1 fusion protein, which contains PHR domain of CRY1, referred to as CNT1, causes dominant-negative effects by “homooligomerize” with endogenous CRY1 (Sang et al., 2005). Because the endogenous CRY1 and CRY2 have different activities under different light intensities as described above, we reasoned that overexpression of fusion proteins containing the PHR domain of CRY2 may also cause the dominant-negative effect by interacting with and suppressing the activity of endogenous CRY1 under high light. To test this, we prepared transgenic lines overexpressing 13 different PHR fusion proteins of Arabidopsis CRYs: four contain PHR1 (PHR domain of CRY1) and nine contain PHR2 (PHR domain of CRY2). We used two different length of PHR1 (490 or 520 amino acids), and three different lengths of PHR2 (489, 500, or 520 amino acids). The PHR domains are fused to different affinity tags (GFP or Nanoluc) (Hall et al., 2012), in different orientations, and controlled by two different constitutive promoters to avoid structure-dependent effects (Fig. 5A, 5F). Because nuclear CRY1 and CRY2 are responsible for the hypocotyl inhibition function (Wu and Spalding, 2007; Yu et al., 2007) and that the NLS of at least CRY2 is in the CCE domain (Zuo et al., 2012), the PHR domains are fused to either NLS of CRY2 (NLSc) or NLS of SV40 virus (NLSs). To prevent post-transcriptional transgene silencing, 13 PHR proteins are overexpressed in the rdr6 background (Peragine et al., 2004), which has the wild-type CRY1 and CRY2 genes (so labelled as wild-type). The appropriate expression and nuclear localization of the PHR fusion proteins, and the phenotypic change in the independent transgenic lines were confirmed (Fig 5, Fig. S5, Fig. S6). All 13 PHR-overexpressing transgenic lines showed long hypocotyl phenotype when grown in continuous blue light, demonstrating all the 13 PHR fusion proteins exert dominant negative effects (Fig. 5B–D, 5G–I, Fig. S5). Consistent with the respective activities of CRY1 and CRY2 in hypocotyl inhibition, PHR1-overexpressing seedlings exhibited stronger blue light-specific long hypocotyl phenotype (Fig. 5G–H, Fig. S5) than that of PHR2-overexpressing seedlings (Fig. 5B–C, Fig. S5). The long hypocotyl phenotype of the PHR1- or PHR2-overexpressing plants are wavelength-specific (Fig. 5C, 5H), and fluence rates-dependent (Fig. 5D, 5I), suggesting that the PHR-dependent dominant negative effect is caused by inhibition of the endogenous CRYs. The dominant-negative effect of PHR1 observed in our experiments is consistent with that reported previously (Sang et al., 2005). As previously proposed by those authors, the “homooligomerization” of the PHR1 moiety of PHR1 fusion proteins and endogenous CRY1 can satisfactorily explain the dominant-negative effect of PHR1 fusion proteins. However, “homooligomerization” of the PHR2 moiety of PHR2 fusion proteins and the endogenous CRY2 cannot satisfactorily explain the long hypocotyl phenotype of seedlings overexpressing any of the nine PHR2 fusion proteins grown under high light (>10 μmolm−2s−1) (Fig. 5B–D, Fig. S5). This is because CRY2 does not play a major role in inhibiting hypocotyl in high intensities of blue light and the cry2 monogenic mutants do not normally exhibit long-hypocotyl phenotype under highlight (Lin et al., 1998).
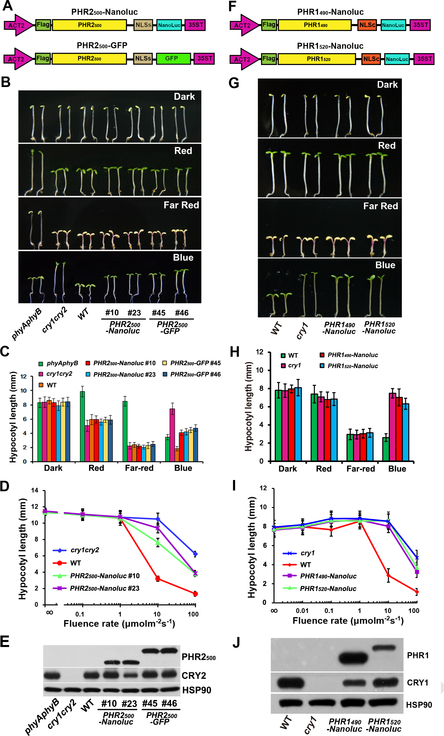
Figure 5. Overexpression of CRY-PHR domain in plants results in reduced sensitivity to blue light.
(A,F) The structures of DNA constructs used for generating CRY2-PHR (A) and CRY1-PHR (F) transgenic lines. PHR2500, CRY2 1–500 aa; PHR1490, CRY1 1–490 aa; PHR1520, CRY1 1–520 aa; NLSs, SV40 nuclear localization signal; NLSc, nuclear localization signal from CRY2 (541–557 aa).
(B,G) Hypocotyl phenotypes of PHR2 (B) and PHR1 (G) transgenic lines. Seedlings with PHR2 or PHR1 overexpressed in wild-type background were grown in the dark, continuous blue light (30 μmol m−2s−1), far-red light (2.5 μmol m−2s−1) and red light (30 μmol m−2s−1) for six days. WT, cry1,cry1cry2 and phyAphyB mutants are used as controls.
(C,H) Hypocotyl measurements of the indicated genotypes shown in (B) and (G). Hypocotyl lengths with standard deviations are shown (n≥20).
(D,I) Fluence rate responses of the hypocotyl inhibition response of PHR2 and PHR1 transgenic lines. Six-day-old seedlings of the indicated genotypes were grown under continuous blue light with fluence rates of 0, 0.1, 1, 10 and 100 μmol m−2s−1. Hypocotyl lengths with standard deviations (n≥20) are shown.
(E,J) Immunoblots showing PHR2 or PHR1 recombinant protein expression levels in transgenic lines. PHR2500, PHR1490 and PHR1520 fusion proteins were detected with anti-Flag antibody. CRY2 and CRY1 proteins were detected by anti-CRY2 or anti-CRY1 antibody, respectively. HSP90 is used as loading control.
Interestingly, no transgenic lines showed late-flowering phenotype, suggesting that none of the 13 PHR-fusion proteins exerts dominant negative effect on the CRY2 function (Fig. S7 A–B). This phenomenon may be explained by previous reports that showed the relatively lower amount of active CRY2 is required to promote floral initiation than that of CRY1 required to inhibit hypocotyl elongation. For example, the cry1 or cry2 mutation is known to be semi-recessive or recessive, respectively (Bruggemann et al., 1996; Guo et al., 1998), demonstrating that half dosage of CRY2 is sufficient for normal flowering but half dosage of CRY1 is insufficient for normal hypocotyl inhibition (Fig. S7C–D). Furthermore, we have recently analyzed the protein concentration-adjusted activity or the relative specific activities of 57 CRY2 mutations altered at residues absolutely conserved between Arabidopsis and human CRYs (Liu, 2019). Among those CRY2 mutants examined, 45.2% retained wild type-like relative specific activity promoting floral initiation, whereas only 9.7% exhibited wild type-like relative specific activity inhibiting hypocotyl elongation.
Our results that the PHR2 fusion proteins inhibit the function of CRY1 led to a proposition that CRY heterooligomerization may regulate the activities of CRYs in plants. This hypothesis predicts that PHR2 fusion proteins would physically interact with the endogenous CRY1 and CRY2, that PHR2 fusion proteins should inhibit light-dependent homooligomerization of CRYs in the dosage-dependent manner, and that PHR2 fusion proteins should inhibit blue light-dependent biochemical reactions of the endogenous CRY2 in plants. We tested these predictions using transgenic Arabidopsis or HEK293 cells co-expressing different proteins (Fig. 6). Different PHR2 fusion proteins were used in different experiments to meet different needs of individual co-IP assays. First, co-IP assays of samples prepared from transgenic plants overexpressing a Flag-PHR2 fusion protein (Flag-PHR2500) shows that the PHR2 fusion protein interacted with both endogenous CRY1 and CRY2 in plants (Fig. 6A). Second, in transgenic plants, the PHR2-HA fusion protein (PHR2500-HA) inhibited blue light-dependent “homooligomerization” of GFP-CRY1 and Myc-CRY1 (Fig. 6B) as well as blue light-dependent “homooligomerization” of GFP-CRY2 and Myc-CRY2 (Fig. 6C). In the third experiment, HEK293 cells were co-transfected by the fixed amount of plasmids encoding Flag-CRY2 and Myc-CRY2 but increasing amount of the plasmid encoding the PHR2-GFP fusion protein. Fig. 6D shows that Flag-CRY2 pulled down increased amount of the PHR2-GFP fusion protein when its expression increased (Fig. 6D). Concomitantly, the amount of Myc-CRY2 co-precipitated by Flag-CRY2 decreased quickly in cells expressing higher level of PHR2-GFP fusion protein (Fig. 6D), indicating that heterooligomerization between PHR2-GFP and Flag-CRY2 competed with and inhibited “homooligomerization” between Flag-CRY2 and Myc-CRY2 (Fig. 6D). Finally, PHR2 fusion proteins (PHR2-NLS-Nanoluc and PHR2-NLS-GFP) show no phosphorylation and degradation under blue light, although CRY2 fusion protein (GFP-CRY2) shows normal blue-light dependent phosphorylation and degradation under the same condition (Fig. 6E–H). The lack of photoreactivity of PHR2 fusion proteins is consistent with our interpretation of the phenotypes of the transgenic plants overexpressing these fusion proteins (Fig. 5). Importantly, overexpression of a PHR2 fusion protein in the wild-type Arabidopsis caused dominant-negative inhibition of blue light-dependent degradation of the endogenous CRY2 (Fig. 6E–H). Taken together, results of these experiments argue that heterooligomerization of one CRY with another CRY may serve roles to regulate the activities of different CRYs in plants exposed to different light conditions in nature.
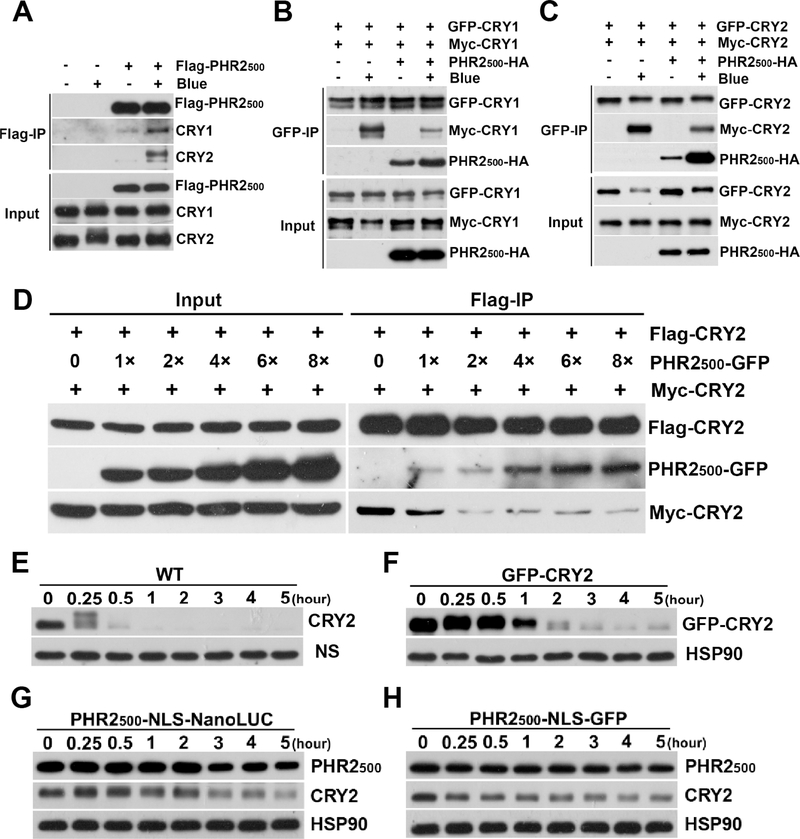
Figure 6. The heterooligomerization of PHR2 with CRY1 and CRY2 results in the PHR2 dominant negative phenotypes of plants.
(A) Blue light-dependent heterooligomerization of PHR2 with endogenous CRY1 and CRY2 in plants. Seven-day-old dark-grown PHR2 transgenic plants were exposed to 30 μmolm−2s−1 blue light for 10min or kept in dark. Total proteins were extracted and immunoprecipitated with Flag-agarose gels. Proteins were analyzed by anti-Flag, anti-CRY1 and anti-CRY2 antibodies.
(B) PHR2 inhibits blue light-dependent homooligomerization of CRY1 in plants. PHR2500-NLS-HA was overexpressed in the transgenic plants co-expressing GFP-CRY1 and Myc-CRY1. Seven-day-old dark-grown seedlings co-expressing PHR2, GFP-CRY1 and Myc-CRY1 were exposed to 30 μmolm−2s−1 blue light for 10 min or kept in dark. Total proteins were extracted and immunoprecipitated with GFP-trap beads. Proteins were analyzed by anti-HA, anti-GFP and anti-Myc antibodies.
(C) PHR2 inhibits blue light-dependent homooligomerization of CRY2 in plants. PHR2500-NLS-HA was overexpressed in the transgenic plants co-expressing GFP-CRY2 and Myc-CRY2. The same analyses were performed as described in (B).
(D) PHR2 inhibits CRY2 homooligomerization in a dosage dependent manner in HEK293. HEK293 cells co-transfected with Flag-CRY2, Myc-CRY2 and different dosages of PHR2 were treated with 100 μmolm−2s−1 blue light for 30 min at 22. Total proteins were extracted and immunoprecipitated with Flag-agarose gels. Proteins were analyzed by anti-Flag, anti-GFP and anti-myc antibodies.
(E-H) PHR2 inhibits blue light-dependent degradation of endogenous CRY2 in plants. Six-day-old dark-grown seedlings of indicated genotype were treated with 100 μmol m−2s−1 blue light for indicated time (hour). Total proteins were extracted, fractioned by 10% SDS–PAGE gels, blotted, probed with anti-CRY2 or anti-Flag antibodies, stripped, and reprobed with anti-HSP90 antibody. HSP90 or NS (non-specific bands) are used as loading controls.
DISCUSSION
In this study, we investigated CRY photooligomerization, including photoresponsive CRY homooligomerization and heterooligomerization. Our study of the kinetics of CRY photooligomerization and dark reversion may affect several aspects of our understanding of the action and regulatory mechanisms of CRYs. First, CRY2 photooligomerization is faster than any presently known photoreaction of CRY2 measured at the ambient temperature, except the electron transfer reactions associated with CRY2 photoreduction (Fig. 1–2). For example, in etiolated seedlings illuminated with similar intensities of blue light (ca. 20–30 μmolm−2s−1), the light-induced CRY2 photobody formation or CRY2 degradation took many minutes to reach 50% saturation (Yu et al., 2007; Yu et al., 2009), in contrast to a few seconds of CRY2 photooligomerization (Fig. 1). The fast speed of the CRY2 photooligomerization supports the hypothesis that CRY2 photooligomerization is one of the earliest photoactivation reaction of this photoreceptor. Second, the half-life of CRY2 measured at ~22°C in seedlings (~7 min) that contain various CRY2-interacting proteins, such as CIBs (CRY-Interacting bHLHs) (Liu et al., 2008), PIFs (Phytochrome Interacting Factors) (Liu et al., 2008; Ma et al., 2016), PPKs (Photoregulatory Protein Kinases) (Liu et al., 2017), the E3 ubiquitin ligase COP1 (Constitutive Photomorphogenesis 1) (Deng et al., 1992; Shalitin et al., 2002), and CRY inhibitors BICs (Blue-light Inhibitor of CRYs) (Wang et al., 2016), appears only one third of the half-life of CRY2 oligomers measured at ~22°C in HEK293 (~18 min) cells that contain no CRY2 regulatory proteins. The shorter half-life of CRY2 oligomers in vivo would suggests that CRY2 monomerization would be faciliated by CRY2-interacting proteins in vivo, but this proposition remains to be tested. Third, the half-life (~7 min) of the photoactivated CRY2 oligomers measured in vivo at ~22°C, whereas the photoreduced CRY2 (neutral semiquinone FADH°) measured in vivo at 0°C has the half-life (~16 min) (Herbel et al., 2013). But due to the different temperatures these two parameters are measured, the biochemical and physiological significance of this difference remains to be investigated. Fourth, given that CRY2 oligomers but not monomers are both active and degraded, it may be interesting to investigate how the equilibrium of CRY2 oligomerization and monomerization in light conditions may affect the total activity and abundance of CRY2 and photomorphogenic responses of plants.
We showed that photooligomerization is evolutionarily conserved in plant CRYs as well as in photoresponsive animal CRYs (Fig. 2). We propose that, like Arabidopsis CRYs, other photoresponsive CRYs may also rely on photooligomerization for their respective cellular functions. Our analyses of the kinetics of forward and reverse reaction of CRY photooligomerization demonstrate that photooligomerization of Arabidopsis CRY2 is more sensitive to blue light than CRY1 and that CRY2 oligomers have longer half-life in darkness than CRY1. These results provide an alternative explanation for why CRY1 and CRY2 act as high-light and low-light receptors, respectively. Moreover, we showed that plant CRY1 and CRY2 undergo not only blue light-dependent homooligomerization but also blue light-dependent heterooligomerization. Our results that the PHR domain of CRY2 can interact with CRY1 to inhibit CRY1 activity support the hypothesis that CRY heterooligomerization may serve a role in the regulation of CRY activity in plants. For example, it is conceivable that CRY2 may heterooligomerize with CRY1 to compete for and weaken the CRY1-CRY1 homooligomerization. However, this effect may be reduced under high light, due to CRY2 degradation, resulting in increased amount of active CRY1 in high light.
We also demonstrate that photooligomerization is necessary but not sufficient for the functions of CRY2. This result is consistent with the current hypothesis of the mechanism of CRY2 photoactivation (Wang et al., 2016). The current hypothesis of CRY2 photoactivation argues that CRY2 exists as inactive monomers in the dark; absorption of blue light triggers conformational changes to facilitate its homooligomerization; the CRY2 homooligomers interact with CRY2-signaling proteins to modulate genome expression and photomorphogenic development. According to this hypothesis, only CRY2 homooligomers have the sufficient affinity and specificity to interact with the CRY2-signaling proteins to accomplish the cellular functions of CRY2. Therefore, CRY2 photooligomerization is necessary for its function. However, the structural elements of CRY2 required for its interaction with CRY2-signaling proteins may take shape primarily after the rapid photooligomerization reaction. Therefore, a lesion of CRY2 could destroy the structural elements required for CRY2 to interact with its signaling proteins without impairment of CRY2 homooligomerization, which would explain why mutations such as P532L is inactive physiologically but active biochemically in photooligomerization. For instance, CRY2P532L mutation substitutes the proline residue of the non-canonical VP motif that forms the structural core for the CRY2-COP1 and CRY2-SPA1 interaction (Lau et al., 2019; Ponnu et al., 2019). The VP motif is apparently not required for photooligomerization but it is required for the protein-protein interaction, such as interaction with COP1 and SPAs. Therefore, the CRY2P532L mutation may reduce its affinity to COP1 without impairing the photooligomerization, which explains why CRY2P532L is defective in function but retains its photooligomerization activity. Because photooligomerization of CRY2 is required for its phosphorylation, ubiquitination, and degradation (Wang et al., 2016), the CRY2P532L mutant protein that continue to photooligomerize could also be phosphorylated and degraded. Additional studies are needed to test this proposition and to define potential structural elements other than the VP motif that enable CRY2 to undergo high affinity interactions with CRY2-signaling proteins in response to blue light.
MATERIALS AND METHODS
Plasmid construction and plant materials
The wild type Arabidopsis in this study is rdr6–11, which suppresses gene silencing (Peragine et al., 2004).
The plant binary vectors used in this paper were modified from pCambia3301 by replacing 35S promoter by Actin2 promoter and replacing GUS gene by 2XFlag-GFP to generate pFGFP vector. To generate p4Myc vector, pCambia3301vector were modified by replacing 35S promoter by Actin2 promoter, replacing GUS gene by 4XMyc and also replacing the Bar gene by HPT gene which converts to hygromycin resistance in plants. To generate the pNanoluc vector, pCambia3301vector was modified by replacing 35S promoter by Actin2 promoter, replacing GUS gene by 2XFlag-Nanoluc. To generate the pHA vector, pCambia3301vector was modified by replacing 35S promoter by Actin2 promoter, replacing GUS gene by 3XHA and also replacing the Bar gene by SUL gene which converts to sulfadiazine resistance in plants.
For generating the plant binary plasmids of CRY2 and its substitution mutants, wild type CRY2 CDS and its substitution mutants were cloned into pFGFP and p4Myc to generate N-terminal fusion FGFP-CRY2 and Myc-CRY2. FGFP-CRY2 and its mutants were tranformed into cry1cry2rdr6 mutant background for phenotype analysis (hypocotyl length and flowering time measurements) and biochemistry study (protein expression, protein degradation, phosphorylation and cellular localization). For photooligomerization analyses, FGFP-CRY2 and Myc-CRY2 were co-transformed into rdr6–11, and screened for co-expression transgenic plants.
For generating the binary plamids of CRY-PHR, different length of CRY1 PHR1 and CRY2 PHR2 coding sequence were cloned into pFGFP vector (driven by Actin2 promoter) or pEGAD vector (driven by 35S promoter) or pNanoluc vector (driven by Actin2 promoter) by In-Fusion cloning method, and the NLS signal sequences were introduced into the constructs by overlapping PCR. The Ti plasmids were transformed into rdr6–11 background.
For generating the binary plasmids of PHR2500-NLS-HA, the coding sequence of CRY2 1 to 500 aa was cloned into pHA by In-Fusion cloning method, and the NLS signal sequences were introduced into the constructs by overlapping PCR. The Ti plasmids were transformed into the transgenic lines co-expressing GFP-CRY1 and Myc-CRY1 or co-expressing GFP-CRY2 and Myc-CRY2.
Human expression pQCMV-FGFP vectors were modified from pEGFP-N1 vectors (Clontech) by inserting a Kozak sequence followed by a Flag epitope tag in front of EGFP. The pCMVmyc plasmid was provided by Dr. Ke Shuai at UCLA. Coding sequences was cloned into pQCMV-FGFP and pCMVmyc by infusion cloning method to generate Flag-, Myc-, or GFP- fusion constructs.
Human cell culture and transfection
Human embryonic kidney (HEK) 293T cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) FBS, 100 IU penicillin and 100 mg/L streptomycin in humidified 5% (v/v) CO2 in air, at 37°C. HEK293T cells were seeded at a density of approximately 8 × 105 cells/ well of a 3-cm plate and transfected using a calcium phosphate precipitation protocol. Briefly, plasmid DNA (2~3 μg) was mixed with 2.5 M CaCl2 (1/20 of total volume) and 2xHeBS [250 mM NaCl, 10 mM KCl, 1.5 mM Na2HPO4, 12 mM Dextrose and 50 mM HEPES pH7.5, adjusted the pH of the final solution to 7.05], and kept at room temperature for 5 min before applying to host cells. The medium was aspirated from each well, the DNA mixture was added slowly into wells without disturbing the cells on the bottom, and the plates were rotated gently to allow the solution to coat the whole well. 3 ml fresh media containing 25 μM chloriquine was added to each well, the plates were placed in a 37°C incubator overnight, and the culture medium wa s changed with 3 ml fresh medium without chloriquine per well. The cells were usually harvested 36~48 hr after transfection.
Co-immunoprecipitation (Co-IP) assays
For HEK293 Co-IP experiments, all the procedures were carried out in the dark. Transfected cells were lysed in 1% Brij buffer [1% Brij-35, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM PMSF and 1x Protease inhibitor cocktail] for 20 min with rotating in the dark at 4°C. After centrifuging at 14,000 g for 10 min at 4 °C, the supernatant was mixed with 20 μl FLAG affinity gels (FLAG M2 gels, sigma, F2426), incubated at 4 °C with rotating for 2 hr. Gels were washed 4 times with ice-cold 1% Brij buffer. The bound proteins were eluted from the beads with 2 x SDS-PAGE Sample Buffer by heating at 100 °C for 4 min.
For Co-IP experiments in Arabidopsis, 2 g transgenic plantswere ground in liquid N2 and homogenized in IP buffer [50 mM Tris-HCl pH7.5, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, 1mM EDTA, 20mM NaF and 1x Protease inhibitor cocktail]. After centrifuging at 16,000 g for 10 min at 4°C, the supernatant for eac h sample was mixed with 30 μL GFP-trap beads and rotated at 4 °C for 2 hours. Then beads w ere pelleted and washed 4 times with ice-cold IP buffer. The bound proteins were eluted from the beads with 2 x SDS-PAGE sample buffer by heating at 100 °C for 4 min, and a nalyzed by immunoblotting.
Protein interaction quantification with Li-COR
To quantify protein abundance, pure nitrocellulose blotting membranes (BioTrace NT, Pall Life Sciences) were used for protein transfer. The rest of the western blot procedures were as followering: protein membranes were blocked with 0.5% Casein in PBS for an hour; primary antibodies, from two different animal sources, for detecting IP signals and co-IP signals were mixed together and diluted in 0.5% Casein in PBST, incubated with membranes for an hour; membranes were washed three times with PBST with shaking; secondary antibodies of Alexa Fluor 680 and Alexa Fluor 790 from corresponding animal sources were mixed together and diluted in 0.5% Casein in PBST, incubated with membranes for one hour; membranes were washed three times with PBST with shaking; The fluorescence signals were detected by Odyssey® CLx System (LI-COR), and the intensity was quantified with Image Studio Lite software (LI-COR). The intensity of each co-IP signal was normalized by its respective IP signal intensity to present as interaction fold change. The interaction fold changes were calculated by the following formula:
Confocal microscopy
Seeds of transgenic Arabidopsis expressing PHR-NL-GFP were surface sterilized and sowed on the Murashige and Skoog (MS) plates supplemented with 3% (w/v) sucrose. After stratification in the dark at 4 °C for 3 days, plates were kept in the growth chamber with long-day conditions for 5 days. The seedlings were incubated in PI (propidium Iodide, Sigma P-4170) solution (10 mg/L) for 2 min, quickly dipped in distilled H2O, followed by mounting in H2O on glass slides and examined by a confocal laser scanning microscope (LSM 880, Carl Zeiss).
Antibodies used for immunoblotting
The primary antibodies used in this study are as following: anti-CRY1 (homemade), anti-CRY2 (homemade), anti-HSP90 (AbM51099-31-PU, Beijing Protein Innovation), anti-HA (3F10, Roche), anti-GFP (HT801, TransGen Biotech), anti-FLAG (F3165, Sigma) and anti-Myc (05–724, Millipore).
The fluorescent secondary antibodies are from ThermoFisher Scientific: goat anti-mouse Alexa Fluor 790 (A11357); goat anti-rabbit IgG-Alexa Fluor 790(A11369); goat anti-mouse IgG-Alexa Fluor 680 (A-21058); goat anti-rabbit IgG-Alexa Fluor 680 (A-21109).
Supplementary Material
ACKNOWLEDGEMENT
This work is supported in part by the National Institute of Health (GM56265 to CL), FAFU-ICE fund (KXGH17011 to QW), FAFU OYIA fund (XJQ201801 to QW), the National Natural Science Foundation of China (31801018to QL, 31600228 to SC) and the Natural Science Foundation of Fujian Province (2019J06014 to QW, 2018J01605 to QL). The authors thank the UCLA-FAFU (Fujian Agriculture and Forestry University) Joint Research Center on Plant Proteomics, Haixia Institute of Science and Technology, and Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology for institutional supports.
Footnotes
Publisher's Disclaimer: This is a PDF file of an article that has undergone enhancements after acceptance, such as the addition of a cover page and metadata, and formatting for readability, but it is not yet the definitive version of record. This version will undergo additional copyediting, typesetting and review before it is published in its final form, but we are providing this version to give early visibility of the article. Please note that, during the production process, errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmad M, and Cashmore AR (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad M, Lin C, and Cashmore AR (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8:653–658. [DOI] [PubMed] [Google Scholar]
- Bruggemann E, Handwerger K, Essex C, and Storz G (1996). Analysis of fast neutron-generated mutants at the Arabidopsis thaliana HY4 locus. Plant J 10:755–760. [DOI] [PubMed] [Google Scholar]
- Cashmore AR, Jarillo JA, Wu YJ, and Liu D (1999). Cryptochromes: blue light receptors for plants and animals. Science 284:760–765. [DOI] [PubMed] [Google Scholar]
- Deng XW, Matsui M, Wei N, Wagner D, Chu AM, Feldmann KA, and Quail PH (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71:791–801. [DOI] [PubMed] [Google Scholar]
- Frøland Steindal IA, and Whitmore D (2019). Circadian Clocks in Fish—What Have We Learned so far? Biology 8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y-M, and Jones DP (2010). Redox Control Systems in the Nucleus: Mechanisms and Functions. Antioxidants & Redox Signaling 13:489–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H (2001). Characterization of an Arabidopsis blue light photoreceptor cryptochrome 2 and blue light signal transduction in de-etiolation responses. Ph.D. Thesis, University of California, Los Angeles. [Google Scholar]
- Guo H, Yang H, Mockler TC, and Lin C (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279:1360–1363. [DOI] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. (2012). Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chemical Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbel V, Orth C, Wenzel R, Ahmad M, Bittl R, and Batschauer A (2013). Lifetimes of Arabidopsis cryptochrome signaling states in vivo. The Plant Journal 74:583–592. [DOI] [PubMed] [Google Scholar]
- Lau K, Podolec R, Chappuis R, Ulm R, and Hothorn M (2019). Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. The EMBO Journal e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, and Shalitin D (2003). Cryptochrome Structure and Signal transduction. Annu. Rev. Plant Biol 54:469–496. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, and Cashmore AR (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A 95:2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Su T, He W, Wang Q, and Lin C (2019). The universally conserved residues are not universally required for stable protein expression or functions of cryptochromes. Molecular Biology and Evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, and Lin C (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–1539. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. (2017). Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat Commun 8:15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, and Liu H (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci U S A 113:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Devlin PF, Panda S, and Kay SA (2000). Functional interaction of phytochrome B and cryptochrome 2. Nature 408:207–211. [DOI] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, and Hell R (2007). Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52:973–986. [DOI] [PubMed] [Google Scholar]
- Ozkan-Dagliyan I, Chiou Y-Y, Ye R, Hassan BH, Ozturk N, and Sancar A (2013). Formation of Arabidopsis Cryptochrome 2 Photobodies in Mammalian Nuclei: APPLICATION AS AN OPTOGENETIC DNA DAMAGE CHECKPOINT SWITCH. Journal of Biological Chemistry 288:23244–23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, and Poethig RS (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18:2368–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J, Riedel T, Penner E, Schrader A, and Hoecker U (2019). Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeldt G, Viana RM, Mootz HD, von Arnim AG, and Batschauer A (2008). Chemically induced and light-independent cryptochrome photoreceptor activation. Mol. Plant 1:4–12. [DOI] [PubMed] [Google Scholar]
- Sancar A (2000). Cryptochrome: the second photoactive pigment in the eye and its role in circadian photoreception. Annu Rev Biochem 69:31–67. [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, and Yang HQ (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzlander M, Fricker MD, Muller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, and Meyer AJ (2008). Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231:299–316. [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, and Lin C (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417:763–767. [DOI] [PubMed] [Google Scholar]
- Wang Q, and Lin C (2019). Mechanisms of CRY-mediated photoresponses in plants. Annu. Rev. Plant Biol. In preparation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han Y-J, et al. (2016). Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354:343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zuo Z, Wang X, Liu Q, Gu L, Oka Y, and Lin C (2018). Beyond the photocycle—how cryptochromes regulate photoresponses in plants? Current Opinion in Plant Biology 45:120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, and Spalding EP (2007). Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci U S A 104:18813–18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, and Lin C (2007). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19:3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, and Lin C (2009). Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Frugis G, and Chua NH (2002). The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30:349–359. [DOI] [PubMed] [Google Scholar]
- Zuo ZC, Meng YY, Yu XH, Zhang ZL, Feng DS, Sun SF, Liu B, and Lin CT (2012). A study of the blue-light-dependent phosphorylation, degradation, and photobody formation of Arabidopsis CRY2. Mol Plant 5:726–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.