Abstract
Cellular immune responses play a fundamental role in controlling viral replication and AIDS progression in human immunodeficiency virus (HIV)-infected subjects and in simian immunodeficiency virus (SIV)-infected macaques. Integrase defective lentiviral vector (IDLV) represents a promising vaccine candidate, inducing functional and durable immune responses in mice and non-human primates. Here, we designed HIV- and SIV-based IDLVs to express the HIVACAT T cell immunogen (HTI), a mosaic antigen designed to cover vulnerable sites in HIV-1 Gag, Pol, Vif, and Nef. We observed that HTI expression during lentiviral vector production interfered profoundly with IDLV particles release because of sequestration of both HIV- and SIV-Gag proteins in the cytoplasm of the vector-producing cells. However, modifications in IDLV design and vector production procedures greatly improved recovery of both HIV- and SIV-based IDLV-HTI. Immunization experiments in BALB/c mice showed that both IDLVs elicited HTI-specific T cell responses. However, immunization with HIV-based IDLV elicited also a T cell response toward exogenous HIV proteins in IDLV particles, suggesting that SIV-based IDLV may be a preferable platform to assess the induction of transgene-specific immune responses against rationally designed HIV structural antigens. These data support the further evaluation of IDLV as an effective platform of T cell immunogens for the development of an effective HIV vaccine.
Keywords: HIV-1, vaccine, lentiviral vector, immunization, IDLV, integrase defective, Gag, Pol, mosaic, HTI
Introduction
Integrase defective lentiviral vectors (IDLVs), based on human or simian immunodeficiency viruses (HIV-1 and SIV), represent an attractive tool for broadly applicable immunization strategies.1, 2, 3 Like their integrating counterparts, IDLVs retain the ability to transduce with high efficiency terminally differentiated non-dividing cells, including antigen-presenting cells (APC), such as dendritic cells (DCs) and macrophages,4 but with a superior safety profile. Wild-type promoter sequences in the U3 region of 3′ long terminal repeat (LTR) have been deleted to generate self-inactivating (SIN) vectors,5 whereas mutations in the catalytic domain of Integrase (IN) gene impair IN protein functionality.6 This hampers integration of vector genome in the target cell and, therefore, avoids the risk for insertional mutagenesis, disruption of host gene expression or functionality, and malignant transformation.7 Consequently, IDLVs are replication defective by definition. The transgene of interest is expressed from the episomal forms of the vector that are transcriptionally active and stable in non-dividing cells,8,9 allowing for efficient and long-term expression of the target gene. These properties and the low pre-existing anti-vector immunity make IDLVs a promising vaccine platform against infectious pathogens for which traditional methods are unsafe or unsuccessful.
We and others previously showed that HIV-based IDLV can be exploited for delivering viral antigens capable of eliciting specific and persistent cellular and humoral immune responses in mice, including influenza nucleoprotein (NP) and hemagglutinin (HA),10,11 human papillomavirus (HPV) E7,12 hepatitis B virus (HBV) surface antigen (HBsAg),13 West Nile virus,14 and HIV-1 envelope proteins.15,16 Subsequent immunization of Rhesus macaques with SIV-based IDLV-expressing HIV Envelope (IDLV-Env) induced persistent and specific antibodies (Abs) and T cells detectable up to 1 year post-injection. Immunization was boosted by a second inoculum with the same vector and was associated with Ab affinity maturation, antigen-specific memory B cell persistence, and development of specific and functional T cell responses.17,18 Several studies have shown that T cell-mediated immune response plays an important role in controlling viral load in HIV-infected individuals and SIV-infected macaques.19, 20, 21, 22, 23 In particular, Rhesus macaques immunized with CMV vector-expressing SIV proteins developed persistent effector memory responses that control SIV infection after mucosal challenge in 50% of vaccinated animals.24,25 Therefore, a T cell vaccine, able to induce strong and specific anti-HIV-1 cellular immune response, could be essential to control viral replication, thus impacting on epidemic spread.26
Among all HIV-1 proteins, Gag is one of the preferred candidates for a T cell vaccine because it is highly conserved across different HIV-1 isolates and highly immunogenic. Anti-Gag cytotoxic T lymphocytes (CTLs) were associated with reduced viral load in both clade B and clade C HIV-1-infected subjects27 and were responsible for control of viral replication after SIV challenge of macaques.28 Nevertheless, immune responses directed toward Gag are not always an indicator of antiviral activity. In fact, full-length wild-type Gag-based vaccine could divert immune responses toward “decoy” epitopes in the Gag protein that are immunodominant, but not protective,29 and exclude favorable epitopes present in other HIV-1 proteins, including Pol or Vif. In this context, Mothe et al.30 have developed the HIVACAT T cell immunogen (HTI), a rationally designed mosaic sequence of close to 500 amino acids (aa) covering 16 regions in gag, pol, vif, and nef HIV-1 genes that are relatively conserved among the different strains of HIV-1. These regions include more than 60 CD4+ and CD8+ T cell beneficial epitopes targeted preferentially by T cells of HIV-1-positive patients with low viral load and independent of beneficial histocompatibility leukocyte antigen (HLA) class I genotypes. Prime-boost immunization of C57BL/6 mice and Indian rhesus macaques with plasmid DNA followed by Modified Vaccinia Ankara (MVA)-expressing HTI induced broad and balanced T cell responses to several segments within Gag, Pol, and Vif.30 Similarly, prime-boost immunization of BALB/c mice with BCG- and ChAdOx1-expressing HTI elicited HTI-specific T cell responses.31
Based on the proven efficiency of IDLVs in inducing strong and durable antigen-specific T cell responses after a single immunization, we exploited IDLV as a platform for delivering the HTI immunogen. To this aim, we had to consider that exogenous Gag and Pol proteins of the HIV-based lentiviral particles may elicit a T cell immunodominant response,32,33 thus skewing the HTI-specific immune response toward decoy epitopes. Also, a dominant-negative effect on multimerization of Gag protein during IDLV assembly can occur when using the HTI immunogen, ultimately leading to cytoplasmic accumulation of Gag protein, as described in similar settings.34,35 To avoid interference of HTI with IDLV assembling, we optimized design and production strategy of both HIV- and SIV-based IDLVs expressing HTI (hIDLV-HTI and sIDLV-HTI, respectively) and evaluated their immunogenicity in BALB/c mice. Results indicate that both IDLVs induced a broad and robust HTI-specific response. However, SIV-based IDLV induced a specific immune response directed only to the HTI transgene, whereas HIV-based IDLV induced also an immune response toward exogenous major histocompatibility complex (MHC) class I-restricted T cell epitopes in IDLV particles, which may distract the T cell response from the most critical T cell targets present in HTI. Overall, these results support the development of IDLV-vectored vaccines expressing rationally designed HIV-1 T cell epitopes for clinical application.
Results
HTI Transgene Interferes with IDLV Production
Previous work using HIV-1 Gag mutants demonstrated that they interfere with Gag oligomerization and HIV-1 particle assembly, whereas non-myristoylated Gag protein accumulates in the cytosolic complex.34, 35, 36, 37, 38, 39 To address whether HTI mosaic affected IDLV production, we compared hIDLV-HTI and sIDLV-HTI vector titers with those of corresponding IDLV-expressing GFP (hIDLV-GFP and sIDLV-GFP, respectively) (Figure 1A). We observed 1 log reduction in IDLV-HTI vector titers, as measured by reverse transcriptase (RT) activity assay, compared with IDLV-GFP, suggesting that the HTI mosaic interfered with the membrane clustering of the Gag expressed by the packaging plasmid. To address the interference of HTI on membrane clustering of Gag, we co-transfected 293T Lenti-X cells with plasmids expressing HIV- or SIV-Gag fused to GFP (pHIVGag-GFP and pSIVGag-GFP, respectively) and HTI fused to mCherry (pHTI-mCherry) for confocal laser scanning microscopy (CLSM) analysis, using a high 3:2 HTI/Gag plasmid ratio, corresponding to the ratio of HTI/Gag used for producing the IDLV in Figure 1A. When transfected alone, HIV- and SIV-Gag were membrane associated, whereas HTI, in the absence of a myristoylation site, localized within the cytoplasm (Figures 1Ba–1Bc). However, in co-transfection experiments, HTI retained most of the Gag proteins into the cytoplasm of transfected cells, preventing membrane association of Gag (Figures 1Bd and 1Be), revealing a dominant-negative effect of HTI on membrane clustering of wild-type Gag.
Figure 1.
Interference of HTI on Vector Release
(A) Recovery of HIV- and SIV-based IDLV-HTI (hIDLV-HTI and sIDLV-HTI, respectively) expressed as percentage of reverse transcriptase (RT) activity compared with the corresponding control IDLVs expressing GFP (100% RT activity). Data are expressed as mean with range of four independent experiments. (B) Confocal laser scanning microscopy (CLSM) of 293T Lenti-X cells after transfection with pHIVGag-GFP (a), pSIVGag-GFP (b), and pHTI-mCherry (c) plasmids alone and after co-transfection with pHTI-mCherry and pHIVGag-GFP (d) or pHTI-mCherry and pSIVGag-GFP (e) plasmids (high 3:2 HTI/Gag plasmid ratio). Nuclei are stained in blue by DAPI. Scale bars are indicated for each image. Results from one representative experiment are shown for each analysis.
To reduce this interference and overcome the low efficiency in IDLV-HTI production, we evaluated whether lowering the HTI/Gag plasmid ratio would improve membrane tethering of Gag. In 293T Lenti-X cells co-transfected with a lower amount of HTI-expressing plasmid compared with Gag-expressing plasmid (corresponding to a low HTI/Gag plasmid ratio of 1:3), the majority of Gag localized on the cell membrane, whereas a smaller amount of HIV- or SIV-Gag proteins was still sequestered within the cytoplasm of the cells by HTI (Figure 2A). This suggests that HTI interferes with membrane clustering of Gag, and that this effect can be mitigated by using higher amounts of Gag.
Figure 2.
Low Levels of HTI Facilitate Release of IDLV
(A) CLSM analysis of 293T Lenti-X cells co-transfected with plasmids pHTI-mCherry and pHIVGag-GFP (a) or pHTI-mCherry and pSIVGag-GFP (b) using low HTI/Gag plasmid ratio (1:3). Nuclei are stained in blue by DAPI. Scale bars are indicated for each image. Results from one representative experiment are shown for each analysis. (B) Flow cytometry analysis (counts versus FL1/GFP) of 293T Lenti-X cells transduced with HIV-based or SIV-based lentiviral vectors expressing GFP (hLV-GFP and sLV-GFP) produced including decreasing amounts of HTI-expressing plasmid during transfection procedure. Shown are data from one representative experiment. (C) RT activity of hIDLV-HTI and sIDLV-HTI produced with high (3:2) or low (1:3) HTI/Gag plasmid ratio. Data are expressed as percentage of RT activity compared with the corresponding control IDLVs expressing GFP (100% RT activity). Mean with range of four independent experiments is shown.
To assess whether lentiviral vector production in the presence of a decreasing amount of HTI-expressing plasmid resulted in increased functional titers, we used lentiviral vectors expressing GFP in both the HIV and SIV backgrounds. Flow cytometry analysis confirmed that transduction efficiency (GFP+ cells) increased when using a decreasing amount of HTI-expressing plasmid (Figure 2B). Based on these results, we selected a 1:3 HTI/Gag plasmid ratio for the production of IDLV-HTI to be used in the in vivo experiments. As shown in Figure 2C, the use of this plasmid ratio resulted in 6- to 8-fold higher recovery of IDLV-HTI in the SIV and HIV background, respectively.
HIV-Based IDLV Particles Induce a T Cell Response against the Immunodominant Epitope of the HIV-Gag Transgene
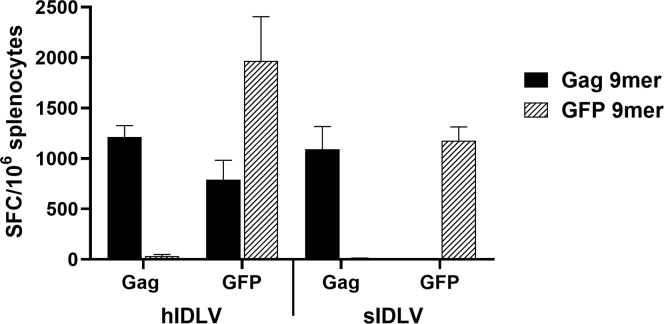
Previous work has shown exogenous MHC class I-restricted T cell responses toward proteins of HIV-1 particles in the absence of viral protein synthesis.40 Also, HIV virus-like particles (VLPs) pseudotyped with vesicular stomatitis virus envelope glycoprotein G (VSV.G) showed higher exogenous MHC class I-restricted T cell responses toward Gag protein than VLP that lacked VSV.G.32,33 In mice, this was associated with immunodominant H-2Kd-restricted HIV-Gag epitope AMQMLKETI,33,41 which is not present in SIV-Gag protein (Figure S1A). This may influence the immune response elicited after immunization with HIV-based and SIV-based IDLV-expressing HIV structural antigens such as Gag. To address the impact of exogenous HIV epitopes derived from IDLV particles in inducing a MHC class I-restricted T cell response, we immunized BALB/c mice with HIV- and SIV-based IDLV expressing a codon-optimized HIV-Gag protein. Corresponding IDLVs expressing GFP were used as controls of immunization. Four weeks after injection, T cell-specific responses were evaluated by interferon γ (IFNγ) ELISpot assay using the immunodominant H-2Kd-restricted HIV-Gag-9-mer AMQMLKETI33,41 and GFP-9-mer HYLSTQSA.42 As expected, mice immunized with either IDLV-GFP or IDLV-Gag elicited transgene-specific immunity, against GFP and HIV-Gag, respectively (Figure 3). In addition, Gag-specific IFNγ-producing T cells were induced also in mice immunized with hIDLV-GFP (Figure 3, left). This exogenous MHC class I-restricted T cell response was directed to the HIV-Gag immunodominant epitope present in the HIV-derived IDLV particle, originated from the packaging plasmid used for hIDLV production, whereas immunization with sIDLV-GFP did not induce a HIV-Gag-specific response (Figure 3, right).
Figure 3.
Exogenous H-2Kd-Restricted T Cell Responses against Gag Protein of IDLV Particles
IFNγ ELISpot assay at 30 days after a single immunization in BALB/c mice either with HIV- or SIV-based IDLVs (hIDLV or sIDLV) expressing HIV-Gag or GFP. Splenocytes were stimulated overnight with the H-2Kd-restricted Gag (black bars) or GFP (hatched bars) epitopes. Data are expressed as specific spot-forming cells (SFCs) per 106 splenocytes. Error bars represent the standard error among animals from the same group.
HTI-Specific Responses in Mice Immunized with sIDLV-HTI, but Not hIDLV-HTI
BALB/c mice were immunized once intramuscularly (i.m.) with 1 × 107 transducing units (TUs)/mouse of hIDLV or sIDLV expressing HTI or GFP. A group of naive mice was included in the study and kept as control for the immunological assays. Antigen-specific T cell responses were assessed 5 weeks from IDLV injection by IFNγ ELISpot on fresh splenocytes using 10 overlapping peptide pools spanning the entire HTI sequence (Table S1), as well as the immunodominant H-2Kd-restricted HIV-Gag epitope. The HIV-Gag-9-mer peptide, not present in the HTI sequence (Figure S1B), was included to evaluate the impact of HIV-Gag protein from hIDLV particles in the induction of HTI-specific responses. H-2Kd-restricted GFP peptide was used as positive control of immunization and as unrelated peptide in IDLV-GFP- and IDLV-HTI-injected mice, respectively (Figure 4).
Figure 4.
HTI-Specific T Cell Response in Mice Immunized with HIV- and SIV-Based IDLVs
IFNγ ELISpot assay at 5 weeks after a single immunization with HIV- or SIV-based IDLV-HTI (hIDLV-HTI or sIDLV-HTI, respectively) and corresponding controls IDLV-GFP (hIDLV-GFP or sIDLV-GFP) in BALB/c mice. Splenocytes were stimulated overnight with the H-2Kd-restricted GFP (hatched bars) or Gag (gray bars) epitopes and 10 pools of peptides spanning the HTI sequence (P1–P10, black bars) (Table S1). Data are expressed as specific spot-forming cells (SFCs) per 106 splenocytes. Error bars represent the standard deviation among animals from the same group.
As expected, GFP-specific T cells were elicited only in mice immunized with hIDLV-GFP and sIDLV-GFP (Figure 4, right panels), whereas no specific responses were detectable in naive mice (data not shown). Similarly, a specific response to the HIV-Gag-9-mer was present only in mice immunized with the HIV-based IDLVs (Figure 4, top panels). Because this epitope is absent in the HTI sequence, this response was due to the HIV-Gag protein in the HIV-based IDLV particles. Some peptide pools were reactive with cells derived from mice immunized with both HTI-expressing IDLVs. In particular, pool 6 spanning the HTI protease (PR) region and pool 7 spanning the HTI RT region elicited the most specific responses because they were induced after immunization with sIDLV-HTI, but not sIDLV-GFP (Figure 4, bottom panels). Interestingly, hIDLV-GFP induced immune responses to pool 4 spanning the HTI p24 (CA) region and pool 7, suggesting that these responses were elicited by structural proteins present in the HIV-based IDLV particles. To evaluate whether in addition to the immunodominant H-2Kd-restricted HIV-Gag epitope AMQMLKETI, also other epitopes are responsible for eliciting strong immunogenic response demonstrated for hIDLV, we characterized the responsive epitopes using a panel of H-2d-restricted peptides included in HTI pools 4, 6, and 7 (Table S1; Figures S1 and S2) described as target of CTL in BALB/c mice immunized with HIV structural antigens.43, 44, 45 Specific responses to peptides #80 and #81 (pool 6, PR), containing the LVGPTPVNI epitope,43,44 and to peptides #111 and #112 (pool 7, RT), containing the YYDPSKDLI epitope,43, 44, 45 were detected in mice immunized with either SIV-based or HIV-based IDLV-HTI (Figure S3). Moreover, splenocytes from mice immunized with hIDLVs were reactive against peptides #46 and #47 (pool 4, p24 CA), containing the IYKRWIILGL epitope,46,47 included in the HTI pool 4 and in the HIV Gag protein, and to peptides #111 and #112 (pool 7, RT), containing the YYDPSKDLI epitope (Figure S3). No specific responses against any of the tested HTI peptide pools were detectable in mice immunized with sIDLV-GFP. These data indicate that both HTI-expressing IDLVs elicited HTI-specific T cell-mediated immune response after administration in BALB/c mice. Importantly, SIV-based IDLV induced a specific immune response directed only to the HTI transgene, whereas HIV-based IDLV induced also exogenous MHC class I-restricted T cell responses toward HIV proteins in IDLV particles. Overall, these data suggest that SIV-based IDLV may be a preferable platform to assess the induction of a transgene-specific immune response against rationally designed HIV antigens, such as HTI.
Inhibition of HTI Synthesis during IDLV Production Attenuates the HTI Dominant-Negative Effect
In an attempt to decrease further the dominant-negative effect of HTI on Gag protein and to improve the IDLV-HTI production, we modified the IDLV design by exploiting the repression of HTI translation from transfer vector during IDLV-HTI production. In particular, we harnessed the Bacillus subtilis Tryptophan RNA-binding Attenuation Protein (TRAP) and a TRAP-binding sequence (tbs), inserted upstream of the HTI sequence, which acts to repress translation of HTI.48,49 To validate this system, we constructed a plasmid containing the tbs sequence upstream the HTI-mCherry fusion protein (ptbsHTI-mCherry). The plasmid was co-transfected in 293T Lenti-X cells with plasmid expressing SIV-Gag, with or without pTRAP, for CLSM analysis (Figure 5A). The presence of tbs sequence did not interfere with mCherry expression and localization in the cellular cytoplasm, whereas addition of pTRAP inhibited mCherry expression (Figures 5Aa and 5Ab), indicating that TRAP protein successfully repressed HTI-mCherry translation from ptbsHTI-mCherry plasmid. When ptbsHTI-mCherry was co-transfected with pSIVGag-GFP at low HTI/Gag molar ratio, HTI trapped the SIV-Gag protein into the cytoplasm of transfected cells, preventing Gag to associate fully with the cellular membrane (Figure 5Ac), similar to what was previously shown (Figure 2Ab). Importantly, when TRAP-expressing plasmid was co-transfected together with pSIVGag-GFP and ptbsHTI-mCherry, SIV-Gag protein fully localized at the cellular membrane (Figure 5Ad). These results were confirmed by western blot (WB) analysis on lysates of 293T Lenti-X cells co-transfected with transfer vector expressing HTI containing upstream the tbs sequence (pGAE-tbsHTI) in the presence of pTRAP plasmid. The parental transfer vector pGAE-HTI devoid of tbs sequence was used as control. As shown in Figure 5B, a band corresponding to HTI was present in cells transfected with HTI- or tbsHTI-expressing plasmids, whereas HTI was undetectable in cells co-transfected with tbsHTI/TRAP, but it was detected in cells transfected with HTI/TRAP, confirming that TRAP protein was able to repress HTI expression only from pGAE-tbsHTI transfer vector. Importantly, addition of pTRAP during vector production in the presence of ptbsHTI improved greatly transduction efficiency (GFP+ cells), suggesting that TRAP protein can counteract the HTI dominant-negative effect on IDLV production (Figure 6A). Indeed, production of sIDLV-tbsHTI using the tbs/TRAP system allowed for a further increase of recovered vector compared with sIDLV-tbsHTI produced using a low HTI/Gag ratio (Figure 6B).
Figure 5.
Inhibition of HTI Synthesis Attenuates the HTI Dominant-Negative Effect
(A) CLSM of 293T Lenti-X cells transfected with ptbsHTI-mCherry alone (a) and in combination with pTRAP (b), pSIVGag-GFP (c), or both (d). Cells were transfected with tbsHTI/TRAP plasmid ratio of 1:1 (b and d) and low tbsHTI/Gag plasmid ratio (1:3) (c and d). Nuclei are stained in blue by DAPI. Scale bars are indicated for each image. Results from one representative experiment are shown. (B) Western blot (WB) on 293T Lenti-X cells transfected with SIV-based transfer vectors expressing HTI or tbsHTI, with or without pTRAP. Cells transfected with GFP-expressing plasmid were used as negative control.
Figure 6.
Inhibition of HTI Synthesis Improves Release of IDLV
(A) A representative experiment of flow cytometry analysis of 293T Lenti-X cells transduced with SIV-based lentiviral vectors expressing GFP produced including ptbsHTI or ptbsHTI + pTRAP during transfection procedure. Mock plasmid pcDNA3 was used as a control. (B) RT activity of SIV-based IDLV-tbsHTI produced with or without addition of pTRAP during the transfection. Results are expressed as percentage of RT activity compared with IDLV-GFP used as control, defined as 100%. Dotted line with cross (×) symbol represents the mean of four independent experiments.
IDLV-HTI Immunogenicity Is Not Impaired by the tbs/TRAP System
To verify that the improved production of IDLV-tbsHTI using the tbs/TRAP system does not affect the HTI-induced immunity, we used SIV-based IDLV-HTI, IDLV-tbsHTI+pTRAP, and IDLV-GFP to immunize BALB/c mice. IFNγ-producing T cells measured by ELISpot assay at 4 weeks after immunization showed no differences between mice immunized with IDLV-HTI and IDLV-tbsHTI produced, including pTRAP during transfection procedure (Figure 7). In particular, in both groups of animals, specific T cell responses against pool 6 (HTI PR) and pool 7 (HTI RT) were confirmed, suggesting that the tbs/TRAP system is an effective tool to increase IDLV-HTI production without affecting its immunogenicity.
Figure 7.
Immunization with IDLV-tbsHTI Does Not Affect the HTI-Specific Immune Response
BALB/c mice were immunized with SIV-based sIDLV-tbsHTI (black bar), sIDLV-HTI (gray bar), or sIDLV-GFP (hatched bar). The specific T cell response was evaluated by IFNγ ELISpot at 30 days after immunization. Cells were stimulated overnight with the H-2d-restricted epitopes Gag-9-mer and GFP-9-mer or 10 pools of peptides spanning the whole HTI sequence (P1–P10) (Table S1). Data are expressed as specific spot-forming cells (SFCs) per 106 splenocytes. Error bars represent the standard deviation among animals from the same group.
Discussion
In this study, we used an optimized IDLV-based platform to deliver the HIV-1-derived HTI. Previous work has shown that HIV-1-positive individuals with high reactivity against sub-dominant epitopes were able to control viral replication compared with individuals with immune response directed against only immunodominant epitopes.50 Im et al.46 demonstrated that inclusion of such sub-dominant epitopes in the design of a T cell immunogen induced robust immune responses and protection in BALB/c mice against replicating vaccinia virus expressing the HIVA immunogen (WR.HIVA), used as a surrogate challenge virus. The HTI immunogen was developed based on the hypothesis that a focused T cell immunogen mainly guided by human immunogenicity data could avoid the induction of dominant and ineffective decoy responses and instead (re)-focus the T cell response on the most critical T cell targets from which the virus cannot escape or if it can escape, it pays a high viral fitness cost.46,50, 51, 52, 53, 54 HTI design has been based on functional data from more than a thousand HIV-1-infected individuals in which only regions in Gag, Pol, Vif, and Nef have been identified as the main target of cellular immune response and are preferentially targeted by T cells in individuals with low viremia, whereas immunodominant epitopes with no beneficial role in viral replication control were excluded from HTI design.30,55 Therefore, after the screening of the whole HIV-1 genome for correlation between T cell response and low viral load, segments in Env, Tat, and Vpu have been excluded from the HTI sequence because there was no evidence of their protective role.
Our results indicate that the HTI protein interfered with production of both HIV- and SIV-based IDLVs expressing HTI (hIDLV-HTI and sIDLV-HTI, respectively). This dominant-negative effect of HTI is likely due to the absence of the membrane-tethering myristoylation signal in the Gag portion contained in HTI and its negative effect on Gag protein multimerization, as previously described in similar settings.34,35 In particular, the myristoylation domain in the p17 matrix (MA) region is crucial not only for Gag-Gag interactions,56 but especially for anchoring Gag to the inner layer of cellular membrane.57 Importantly, it has been shown that non-myristoylated Gag accumulates in the cellular cytoplasm.36,37 Indeed, the absence of myristoylation signal in the portion of Gag protein codified by HTI led to its cytosolic accumulation, whereas the presence of Gag multimerization domains in the same regions resulted in cytoplasmic sequestration of Gag proteins by HTI, as shown by CLSM observation of cells co-transfected with plasmids expressing HIV- or SIV-Gag fused to GFP together with pHTI-mCherry.
In order to mitigate the HTI interference on IDLV release, we increased the amount of packaging plasmids expressing Gag proteins and decreased the transfer vector expressing HTI during vector production. The resulting low HTI/Gag ratio (1:3) weakened the HTI negative effect and allowed for higher IDLV-HTI recovery. A further increase in IDLV-HTI production was obtained by using the tbs/TRAP system,48,49 originally described by Nie and Htun,48 who provided evidence that Bacillus subtilis TRAP may repress the translation of mRNA containing the tbs in transiently transfected mammalian cells. Cloning of tbs upstream the HTI open reading frame (ORF) in the transfer vector and subsequent production of IDLV-tbsHTI using the tbs/TRAP system allowed for a further increase of recovered vector compared with IDLV produced using a lower HTI/Gag ratio. This is in line with data showing enhanced titers of lentiviral vectors using the TRAP system in a different setting.49
In murine immunogenicity experiments, all mice immunized with either IDLV-expressing HTI (sIDLV-HTI, hIDLV-HTI, and sIDLV-tbsHTI) were reactive against HTI peptide pools covering PR (pool 6) and RT (pool 7) regions, which contain previously described H-2d-restricted epitopes.43, 44, 45 Splenocytes from mice immunized with hIDLV-HTI were reactive also against the HTI pool covering Gag p24 (pool 4) and the immunodominant Gag-9-mer AMQMLKETI, which contains the H-2kd-restricted epitope.41,46,47 No responses against these peptides were detected in mice immunized with sIDLV-HTI. This was expected, because SIV-Gag protein has only 54% of sequence identity with HIV-Gag (Figure S1A), and a large portion of HTI-epitopes is therefore absent, in line with previous work showing Gag-specific T cell responses in mice immunized with HIV-1 VLPs.32 This was also confirmed in immunization experiments using hIDLV- or sIDLV-expressing codon-optimized HIV-Gag or GFP. Splenocytes from mice vaccinated with hIDLV-GFP, but not sIDLV-GFP, were also responsive to the H-2d-restricted peptides AMQMLKETI (immunodominant Gag-9-mer), IYKRWIILGL (pool 4, p24 CA), and YYDPSKDLI (pool 7, RT), confirming that HIV particles contributed to the HIV-specific immune response. Indeed, we did not detect any cross-reactivity between HTI or HIV-Gag with the SIV particles from sIDLV. The strong T cell responses against the immunodominant Gag-9-mer epitope (AMQMLKETI) in the vector particle may be detrimental for the HTI vaccine efficacy because they may act as a decoy and divert the immune response from the beneficial epitopes encoded by rationally designed immunogens.58,59 Also, these data suggest that sIDLV may be more suitable than hIDLV as a delivery platform to achieve specific immune responses against rationally designed HIV structural proteins, maximizing the induction of effective anti-HIV cellular immune response and avoiding the induction of T cell response to dominant decoy targets.
Although the validation of the IDLV-HTI system in non-human primates (NHPs) is out of the scope of this manuscript, it will be interesting the evaluation of IDLV expressing HTI in the NHP immunogenicity model, whereas previous work has shown that prime-boost immunization of Indian rhesus macaques with three injections of plasmid DNA followed by two MVA boosts expressing HTI induced robust induction of HTI-specific IFN-γ+ T cells in all macaques starting after the second DNA vaccination. Taken together, these findings support the use of SIV-based IDLV as a platform for delivering improved rationally designed HIV-1 antigens for the development of HIV-1 vaccines and have the additional advantage related to the presence of the SIV-Vpx in the sIDLV, which significantly improves the transduction efficiency of human and simian APCs, favoring the induction of a strong and functional immune response.60,61
Materials and Methods
Vector Construction
Schematic representations of plasmids used in this report are shown in Figure S4. The HIV- and SIV-based SIN lentiviral transfer vectors expressing GFP (pTY2-GFP and pGAE-GFP, respectively), the HIV- and SIV-based IN defective packaging plasmids (pcHelp/IN and pAdSIVD64V, respectively), and the phCMV-VSV.G plasmid producing the pseudotyping VSV.G have been already described.15,62,63 For the construction of pTY2-Gag and pGAE-Gag, the codon-optimized HIV-Gag ORF was cloned into pTY2-GFP or pGAE-GFP using SnaBI/XmaI restriction enzymes by replacing the GFP coding sequence.
To construct pTY2-HTI and pGAE-HTI, the HTI coding sequence was excised from expression plasmid30 using SnaBI/XbaI or ClaI/SalI and cloned into the corresponding sites of pTY2-GFP and pGAE-GFP, respectively. pHIVGag-GFP, expressing the codon-optimized HIV-Gag ORF fused to the GFP sequence, has been previously described.64 The mCherry coding sequence was amplified by using the forward primer mCherryFOR 5′-CCACCGGTATGGATCCATTACCACCATGGTGAGCAAGG-3′ and the reverse primer mCherryREV 5′-GCTCTAGATCTCGAGAGTTACTTGTACAGCTCGTCCATG-3′ to generate a 711-bp fragment digested with AgeI and XbaI, and ligated with SnaBI/AgeI HTI fragment into pCDNA3 (Thermo Fisher Scientific, Waltham, MA, USA) after digestion with XbaI/SnaBI, obtaining the pHTI-mCherry plasmid. To construct pSIVGag-GFP, expressing the SIV-Gag sequence fused to the GFP sequence, codon-optimized SIV-Gag ORF was obtained after pTY2-SIVGagDX65 digestion with NdeI and MscI, ligation with MscI/NotI GFP fragment, and insertion into NotI/NdeI digested pCDNA3 (Thermo Fisher Scientific). The plasmid expressing the TRAP, pMN-cFKT (henceforth referred to as pTRAP), was a gift from Han Htun (Addgene plasmid #18785). To construct pGAE-tbsHTI, CMVtbs sequence was obtained from plasmid ptbs-5Y-YIC (a gift from Han Htun, Addgene plasmid #18786) by PCR with the forward primer CMVS 5′-ACATCGATGGAGTTCCGCGTTACATAAC-3′ and the reverse primer tbsAS 5′-CTCCTAGGAGCTCCACTCGTCTCTGCT-3′ adding a unique restriction site ClaI at the 5′ end and AvrII at the 3′ end. The PCR product was digested with ClaI and AvrII, and ligated into AvrII/ClaI digested pGAE-HTI. For the construction of ptbsHTI-mCherry, the tbsHTI DNA fragment was excised from pGAE-tbsHTI by digestion with SnaBI/HindIII and ligated into HindIII/SnaBI digested pHTI-mCherry.
Production of Lentiviral Vectors
293T Lenti-X human embryonic kidney cell line (Clontech, Mountain View, CA, USA) was maintained in Dulbecco’s modified Eagle’s medium (DMEM), high glucose 4.5 g/L (GIBCO, Life Technologies Italia, Monza, Italy) supplemented with 10% fetal calf serum (Corning, Mediatech, Manassas, VA, USA), and 100 U/mL penicillin/streptomycin (GIBCO). For production of IDLV, 3.5 × 106 293T Lenti-X cells were seeded on 10-cm Petri dishes (Corning Incorporated, Life Sciences, Oneonta, NY, USA) and transiently transfected with lentiviral transfer vector, IN defective packaging plasmid, and VSV.G-envelope plasmid using the Profection Mammalian Transfection System (Promega Corporation, Madison, WI, USA) as previously described using a 3:2:1 ratio (transfer vector/packaging plasmid/VSV.G plasmid).15,17,63 For production of IDLV with a lower HTI/Gag molar amount, the protocol was modified to a 1:3:1 ratio (transfer vector/packaging plasmid/VSV.G plasmid).
pTRAP was included to inhibit HTI translation during the production of IDLV-tbsHTI (tbs:TRAP plasmids molar ratio of 1:1). At 48 h from transfections, supernatants were collected and ultracentrifuged (Beckman Coulter, Fullerton, CA, USA) on a 20% sucrose cushion (Sigma Chemical, St. Louis, MO, USA) at 23,000 rpm for 2.5 h at 4°C using an SW28 swinging bucket rotor (Beckman). Pelleted vector particles were resuspended in 1X phosphate-buffered saline (PBS; GIBCO) and stored at −80°C until use. Each IDLV stock was titered by the RT activity assay, and the corresponding TUs were calculated by comparing the RT activity with one of the IDLV-GFP virions with known infectious titers, thus allowing for the determination of their infectious titer units.66
CLSM
293T Lenti-X cells (2 × 104/well) were seeded in 24-well cluster plates onto 12-mm cover glasses previously treated with l-polylysine (Sigma) and transfected with plasmids using the Profection Mammalian Transfection System (Promega). Twenty-four hours post-transfection, cells were washed and fixed with 4% paraformaldehyde (PFA) for 30 min at +4°C, and the coverslips were mounted with Vectashield antifade mounting medium-containing DAPI (Vector Labs, Burlingame, CA, USA) on the microscope slides. CLSM observations were performed on a Leica TCS SP2 AOBS apparatus (Leica Microsystems, Wetzlar, Germany) using excitation spectral laser lines at 488 and 546 nm (for GFP) or at 586 and 610 nm (for mCherry), and using the confocal software (Leica, Wetzlar, Germany) and Photoshop CS5 (Adobe Systems, San Jose, CA, USA). Signals from different fluorescent probes were taken in sequential scanning mode, several fields were analyzed for each labeling condition, and representative results are shown. Images represented a single central optical section taken in the center of each cell nucleus.
Flow Cytometry
293T Lenti-X cells were seeded in 12-well microplates and transduced with 500 μL of HIV-based or SIV-based IDLV-GFP produced including a scalar amount of pHTI plasmid (from 6 to 0.5 μg), ptbsHTI (6 μg or 4 μg), or ptbsHTI + pTRAP (tbsHTI/TRAP ratio of 1:1). Control plasmid pCDNA3 (6 μg) was used in mock co-transfected cells. Seventy-two hours after transduction, cells were collected, washed with PBS 1X (GIBCO), and fixed with 1% PFA. GFP expression was evaluated by measuring fluorescence using the FACSCalibur (BD Biosciences, Milan, Italy), and data were analyzed using Kaluza Analysis Software (Beckman).
WB
293T LentiX cells were transiently transfected with SIV-based transfer vector expressing HTI or tbsHTI (pGAE-HTI and pGAE-tbsHTI, respectively), with or without TRAP-expressing plasmid (molar ratio of 1:1) using the Profection Mammalian Transfection System (Promega Corporation) as described earlier. SIV-based transfer vector expressing GFP (pGAE-GFP) was used as negative control. Twenty-four hours post-transfections, cells were lysed and protein concentrations were determined by Bradford’s protein assay (Bio-Rad Laboratories, Hercules, CA, USA), and 30 μg of total lysates was resolved by SDS-PAGE (10% polyacrylamide) under reducing conditions. To evaluate protein expression, filters were blotted with mAb anti-HTI n63 Ab, kindly provided by Aelix Therapeutics (Barcelona) (dilution 1:2,000), followed by goat anti-mouse horseradish peroxidase (HRP)-conjugated Abs (dilution 1:3,000; Bio-Rad Laboratories). Immobilion Western reagents (Merck Millipore, Billerica, MA, USA) were used as chemoluminescent substrates. Images were acquired and elaborated by a Multispectral Imaging system UVP (Biospectrum, Upland, CA, USA).
Mice Immunization
Four- to six-week-old BALB/c female mice (Charles River, Calco, Como, Italy) were housed under specific pathogen-free conditions in the animal facility of the Istituto Superiore di Sanità (ISS, Rome, Italy). All animal procedures have been performed in accordance with European Union guidelines and Italian legislation for animal care. All animal studies were authorized by the Italian Ministry of Healthy and reviewed by the Service for Animal Welfare at ISS (Authorization n. 314/2015-PR of 30/04/2015). A single i.m. injection with 1 × 107 TUs/mouse of the indicated IDLVs was administered. In all of the experiments, naive non-immunized mice were kept as negative control for parallel analysis. At 4 or 5 weeks after the immunization, mice were euthanized by CO2 inhalation using approved chambers. At sacrifice, single-cell suspension from spleen was prepared by mechanical disruption, passage through 0.45-μm cell strainers (BD Biosciences, Milan, Italy), and treatment with ammonium chloride potassium (ACK). Splenocytes were maintained in RPMI (GIBCO) supplemented with 10% fetal bovine serum (FBS; Lonza, Treviglio, Milan, Italy), penicillin-streptomycin-glutamine (100 U/mL; GIBCO), 2-mercaptoethanol 50 mM (Sigma Chemicals), HEPES buffer solution 25 mM (GIBCO), sodium pyruvate 1 mM (GIBCO), and non-essential amino acids (GIBCO); remaining cells were stored in liquid nitrogen for further experiments.
IFNγ ELISpot Assay
The IFNγ ELISpot assay was performed using BD ELISpot kit reagents (BD Biosciences) according to the manufacturer’s instructions. 2.5 × 105 splenocytes/well were seeded in 96-well plates previously coated with 5 μg/mL anti-mouse IFNγ. Cells were stimulated overnight with either 5 μg/mL of the H-2Kd-restricted HIV-Gag p24 epitope (AMQMLKETI; NIH AIDS Reagent Program, catalog #11876) or 2 μg/mL of GFP epitope (HYLSTQSAL; PRIMM, Milan, Italy). To assess the immune responses to HTI epitopes, 10 pools of 111 peptides of 15 aa in length (overlapping by 11 residues) spanning the entire HTI sequence, including the linkers regions (Table S1), were used at 2 μg/mL each. Medium alone and concanavalin A (ConA; 5 μg/mL; Sigma Chemicals) were used as negative and positive controls, respectively. Spot-forming cells (SFCs) were counted with an automated ELISpot reader (A.EL.VIS, Hannover, Germany). Values obtained from the medium-treated cells (background) were subtracted from values obtained from cells stimulated with specific peptides, and results were expressed as IFNγ-secreting cells (SFCs) per 106 cells. Samples were scored positive when a minimum of 50 spots per 106 cells was present after background subtraction and 1.5-fold higher than the unstimulated sample.
Author Contributions
A.G. participated in study design, performed the majority of the experiments, analyzed the data, and wrote the manuscript. M.B. performed the RT assay and contributed to performing the ELISpot assays. M.F.P. contributed to vector design, construction, and preparation. S.C. performed the CLSM and WB experiments. R.B. and A. Canitano contributed to vector design and construction. Z.M. contributed to performing the ELISPOT assays. A.D.V. performed the animal experiments. A.O. and C.B. contributed to study design and data analysis. D.N. contributed to study design, data analysis, and editing of the manuscript, and oversaw the animal experiments. A. Cara oversaw the planning and direction of the project, including analysis and interpretation of the data and editing of the manuscript.
Conflicts of Interests
The authors declare no competing interests.
Acknowledgments
The authors are grateful to Maria Blasi for critically discussing the manuscript, Stefania Donnini for secretarial assistance, and Ferdinando Costa and Patrizia Cocco for technical support. This work was supported in part by grants from the National Institute of Allergy and Infectious Diseases (NIAID; 1P01AI110485-01A1) and Italian Ministry of Health Ricerca Finalizzata (PE-2011-02347035 to A. Cara). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 681137 (EAVI2020) and the European Union’s Seventh Programme for Research, Technological Development and Demonstration under grant agreement no. 280873 (ADITEC; to A. Cara and D.N.). The following reagent was obtained through the NIH AIDS Reagent Program, AIDS Program, NIAID, NIH: HIV-1 Gag p24 Peptide from NIAID, DAIDS (catalog #11876).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.01.013.
Supplemental Information
References
- 1.Negri D.R.M., Michelini Z., Cara A. Toward integrase defective lentiviral vectors for genetic immunization. Curr. HIV Res. 2010;8:274–281. doi: 10.2174/157016210791208622. [DOI] [PubMed] [Google Scholar]
- 2.Wanisch K., Yáñez-Muñoz R.J. Integration-deficient lentiviral vectors: a slow coming of age. Mol. Ther. 2009;17:1316–1332. doi: 10.1038/mt.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Negri D.R., Michelini Z., Bona R., Blasi M., Filati P., Leone P., Rossi A., Franco M., Cara A. Integrase-defective lentiviral-vector-based vaccine: a new vector for induction of T cell immunity. Expert Opin. Biol. Ther. 2011;11:739–750. doi: 10.1517/14712598.2011.571670. [DOI] [PubMed] [Google Scholar]
- 4.Negri D.R.M., Bona R., Michelini Z., Leone P., Macchia I., Klotman M.E., Salvatore M., Cara A. Transduction of human antigen-presenting cells with integrase-defective lentiviral vector enables functional expansion of primed antigen-specific CD8(+) T cells. Hum. Gene Ther. 2010;21:1029–1035. doi: 10.1089/hum.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwakuma T., Cui Y., Chang L.J. Self-inactivating lentiviral vectors with U3 and U5 modifications. Virology. 1999;261:120–132. doi: 10.1006/viro.1999.9850. [DOI] [PubMed] [Google Scholar]
- 6.Vargas J., Jr., Gusella G.L., Najfeld V., Klotman M.E., Cara A. Novel integrase-defective lentiviral episomal vectors for gene transfer. Hum. Gene Ther. 2004;15:361–372. doi: 10.1089/104303404322959515. [DOI] [PubMed] [Google Scholar]
- 7.Cesana D., Ranzani M., Volpin M., Bartholomae C., Duros C., Artus A., Merella S., Benedicenti F., Sergi Sergi L., Sanvito F. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol. Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillim-Ross L., Cara A., Klotman M.E. HIV-1 extrachromosomal 2-LTR circular DNA is long-lived in human macrophages. Viral Immunol. 2005;18:190–196. doi: 10.1089/vim.2005.18.190. [DOI] [PubMed] [Google Scholar]
- 9.Cara A., Klotman M.E. Retroviral E-DNA: persistence and gene expression in nondividing immune cells. J. Leukoc. Biol. 2006;80:1013–1017. doi: 10.1189/jlb.0306151. [DOI] [PubMed] [Google Scholar]
- 10.Gallinaro A., Borghi M., Bona R., Grasso F., Calzoletti L., Palladino L., Cecchetti S., Vescio M.F., Macchia D., Morante V. Integrase defective lentiviral vector as a vaccine platform for delivering influenza antigens. Front. Immunol. 2018;9:171. doi: 10.3389/fimmu.2018.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fontana J.M., Christos P.J., Michelini Z., Negri D., Cara A., Salvatore M. Mucosal immunization with integrase-defective lentiviral vectors protects against influenza virus challenge in mice. PLoS ONE. 2014;9:e97270. doi: 10.1371/journal.pone.0097270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grasso F., Negri D.R.M., Mochi S., Rossi A., Cesolini A., Giovannelli A., Chiantore M.V., Leone P., Giorgi C., Cara A. Successful therapeutic vaccination with integrase defective lentiviral vector expressing nononcogenic human papillomavirus E7 protein. Int. J. Cancer. 2013;132:335–344. doi: 10.1002/ijc.27676. [DOI] [PubMed] [Google Scholar]
- 13.Karwacz K., Mukherjee S., Apolonia L., Blundell M.P., Bouma G., Escors D., Collins M.K., Thrasher A.J. Nonintegrating lentivector vaccines stimulate prolonged T-cell and antibody responses and are effective in tumor therapy. J. Virol. 2009;83:3094–3103. doi: 10.1128/JVI.02519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutant F., Frenkiel M.P., Despres P., Charneau P. Protective antiviral immunity conferred by a nonintegrative lentiviral vector-based vaccine. PLoS ONE. 2008;3:e3973. doi: 10.1371/journal.pone.0003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Negri D.R.M., Michelini Z., Baroncelli S., Spada M., Vendetti S., Buffa V., Bona R., Leone P., Klotman M.E., Cara A. Successful immunization with a single injection of non-integrating lentiviral vector. Mol. Ther. 2007;15:1716–1723. doi: 10.1038/sj.mt.6300241. [DOI] [PubMed] [Google Scholar]
- 16.Negri D.R.M., Michelini Z., Baroncelli S., Spada M., Vendetti S., Bona R., Leone P., Klotman M.E., Cara A. Nonintegrating lentiviral vector-based vaccine efficiently induces functional and persistent CD8+ T cell responses in mice. J. Biomed. Biotechnol. 2010;2010:534501. doi: 10.1155/2010/534501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Negri D., Blasi M., LaBranche C., Parks R., Balachandran H., Lifton M., Shen X., Denny T., Ferrari G., Vescio M.F. Immunization with an SIV-based IDLV expressing HIV-1 Env 1086 Clade C elicits durable humoral and cellular responses in rhesus macaques. Mol. Ther. 2016;24:2021–2032. doi: 10.1038/mt.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blasi M., Negri D., LaBranche C., Alam S.M., Baker E.J., Brunner E.C., Gladden M.A., Michelini Z., Zandergrift N.A., Wiehe K.J. IDLV-HIV-1 Env vaccination in non-human primates induces affinity maturation of antigen-specific memory B cells. Commun. Biol. 2018;1:134. doi: 10.1038/s42003-018-0131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds M.R., Weiler A.M., Weisgrau K.L., Piaskowski S.M., Furlott J.R., Weinfurter J.T., Kaizu M., Soma T., León E.J., MacNair C. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J. Exp. Med. 2008;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Bao R., Haigwood N.L., Persidsky Y., Ho W.Z. SIV infection of rhesus macaques of Chinese origin: a suitable model for HIV infection in humans. Retrovirology. 2013;10:89. doi: 10.1186/1742-4690-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts M.R., Nason M.C., West S.M., De Rosa S.C., Migueles S.A., Abraham J., Lederman M.M., Benito J.M., Goepfert P.A., Connors M. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDermott A.B., Koup R.A. CD8(+) T cells in preventing HIV infection and disease. AIDS. 2012;26:1281–1292. doi: 10.1097/QAD.0b013e328353bcaf. [DOI] [PubMed] [Google Scholar]
- 23.Hu X., Valentin A., Dayton F., Kulkarni V., Alicea C., Rosati M., Chowdhury B., Gautam R., Broderick K.E., Sardesai N.Y. DNA prime-boost vaccine regimen to increase breadth, magnitude, and cytotoxicity of the cellular immune responses to subdominant Gag epitopes of Simian Immunodeficiency Virus and HIV. J. Immunol. 2016;197:3999–4013. doi: 10.4049/jimmunol.1600697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen S.G., Ford J.C., Lewis M.S., Ventura A.B., Hughes C.M., Coyne-Johnson L., Whizin N., Oswald K., Shoemaker R., Swanson T. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen S.G., Piatak M., Jr., Ventura A.B., Hughes C.M., Gilbride R.M., Ford J.C., Oswald K., Shoemaker R., Li Y., Lewis M.S. Immune clearance of highly pathogenic SIV infection. Nature. 2013;502:100–104. doi: 10.1038/nature12519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korber B.T., Letvin N.L., Haynes B.F. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 2009;83:8300–8314. doi: 10.1128/JVI.00114-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuñiga R., Lucchetti A., Galvan P., Sanchez S., Sanchez C., Hernandez A., Sanchez H., Frahm N., Linde C.H., Hewitt H.S. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 2006;80:3122–3125. doi: 10.1128/JVI.80.6.3122-3125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephenson K.E., Li H., Walker B.D., Michael N.L., Barouch D.H. Gag-specific cellular immunity determines in vitro viral inhibition and in vivo virologic control following simian immunodeficiency virus challenges of vaccinated rhesus monkeys. J. Virol. 2012;86:9583–9589. doi: 10.1128/JVI.00996-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulkarni V., Valentin A., Rosati M., Alicea C., Singh A.K., Jalah R., Broderick K.E., Sardesai N.Y., Le Gall S., Mothe B. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. PLoS ONE. 2014;9:e86254. doi: 10.1371/journal.pone.0086254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mothe B., Hu X., Llano A., Rosati M., Olvera A., Kulkarni V., Valentin A., Alicea C., Pilkington G.R., Sardesai N.Y. A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J. Transl. Med. 2015;13:60. doi: 10.1186/s12967-015-0392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilpeläinen A., Saubi N., Guitart N., Olvera A., Hanke T., Brander C., Joseph J. Recombinant BCG expressing HTI prime and recombinant ChAdOx1 boost is safe and elicits HIV-1-specific T cell responses in BALB/c mice. Vaccines (Basel) 2019;7:78. doi: 10.3390/vaccines7030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuate S., Stahl-Hennig C., Stoiber H., Nchinda G., Floto A., Franz M., Sauermann U., Bredl S., Deml L., Ignatius R. Immunogenicity and efficacy of immunodeficiency virus-like particles pseudotyped with the G protein of vesicular stomatitis virus. Virology. 2006;351:133–144. doi: 10.1016/j.virol.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Marsac D., Loirat D., Petit C., Schwartz O., Michel M.-L. Enhanced presentation of major histocompatibility complex class I-restricted human immunodeficiency virus type 1 (HIV-1) Gag-specific epitopes after DNA immunization with vectors coding for vesicular stomatitis virus glycoprotein-pseudotyped HIV-1 Gag particles. J. Virol. 2002;76:7544–7553. doi: 10.1128/JVI.76.15.7544-7553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trono D., Feinberg M.B., Baltimore D. HIV-1 Gag mutants can dominantly interfere with the replication of the wild-type virus. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- 35.Cara A., Rybak S.M., Newton D.L., Crowley R., Rottschafer S.E., Reitz M.S., Jr., Gusella G.L. Inhibition of HIV-1 replication by combined expression of gag dominant negative mutant and a human ribonuclease in a tightly controlled HIV-1 inducible vector. Gene Ther. 1998;5:65–75. doi: 10.1038/sj.gt.3300545. [DOI] [PubMed] [Google Scholar]
- 36.Bryant M., Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pal R., Reitz M.S., Jr., Tschachler E., Gallo R.C., Sarngadharan M.G., Veronese F.D. Myristoylation of gag proteins of HIV-1 plays an important role in virus assembly. AIDS Res. Hum. Retroviruses. 1990;6:721–730. doi: 10.1089/aid.1990.6.721. [DOI] [PubMed] [Google Scholar]
- 38.Checkley M.A., Luttge B.G., Soheilian F., Nagashima K., Freed E.O. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology. 2010;400:137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson-Daniels J., Singh P.K., Sowd G.A., Li W., Engelman A.N., Aiken C. Dominant negative MA-CA fusion protein is incorporated into HIV-1 cores and inhibits nuclear entry of viral preintegration complexes. J. Virol. 2019;93 doi: 10.1128/JVI.01118-19. e01118-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buseyne F., Le Gall S., Boccaccio C., Abastado J.-P., Lifson J.D., Arthur L.O., Rivière Y., Heard J.M., Schwartz O. MHC-I-restricted presentation of HIV-1 virion antigens without viral replication. Nat. Med. 2001;7:344–349. doi: 10.1038/85493. [DOI] [PubMed] [Google Scholar]
- 41.Doe B., Walker C.M. HIV-1 p24 Gag-specific cytotoxic T-lymphocyte responses in mice. AIDS. 1996;10:793–794. doi: 10.1097/00002030-199606001-00015. [DOI] [PubMed] [Google Scholar]
- 42.Gambotto A., Dworacki G., Cicinnati V., Kenniston T., Steitz J., Tüting T., Robbins P.D., DeLeo A.B. Immunogenicity of enhanced green fluorescent protein (EGFP) in BALB/c mice: identification of an H2-Kd-restricted CTL epitope. Gene Ther. 2000;7:2036–2040. doi: 10.1038/sj.gt.3301335. [DOI] [PubMed] [Google Scholar]
- 43.Wild J., Bieler K., Köstler J., Frachette M.-J., Jeffs S., Vieira S., Esteban M., Liljeström P., Pantaleo G., Wolf H., Wagner R. Preclinical evaluation of the immunogenicity of C-type HIV-1-based DNA and NYVAC vaccines in the Balb/C mouse model. Viral Immunol. 2009;22:309–319. doi: 10.1089/vim.2009.0038. [DOI] [PubMed] [Google Scholar]
- 44.Ondondo B., Abdul-Jawad S., Bridgeman A., Hanke T. Characterization of T-cell responses to conserved regions of the HIV-1 proteome in BALB/c mice. Clin. Vaccine Immunol. 2014;21:1565–1572. doi: 10.1128/CVI.00587-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larke N., Im E.J., Wagner R., Williamson C., Williamson A.L., McMichael A.J., Hanke T. Combined single-clade candidate HIV-1 vaccines induce T cell responses limited by multiple forms of in vivo immune interference. Eur. J. Immunol. 2007;37:566–577. doi: 10.1002/eji.200636711. [DOI] [PubMed] [Google Scholar]
- 46.Im E.J., Hong J.P., Roshorm Y., Bridgeman A., Létourneau S., Liljeström P., Potash M.J., Volsky D.J., McMichael A.J., Hanke T. Protective efficacy of serially up-ranked subdominant CD8+ T cell epitopes against virus challenges. PLoS Pathog. 2011;7:e1002041. doi: 10.1371/journal.ppat.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cellini S., Fortini C., Gallerani E., Destro F., Cofano E.B., Caputo A., Gavioli R. Identification of new HIV-1 Gag-specific cytotoxic T lymphocyte responses in BALB/c mice. Virol. J. 2008;5:81. doi: 10.1186/1743-422X-5-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nie M., Htun H. Different modes and potencies of translational repression by sequence-specific RNA-protein interaction at the 5′-UTR. Nucleic Acids Res. 2006;34:5528–5540. doi: 10.1093/nar/gkl584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maunder H.E., Wright J., Kolli B.R., Vieira C.R., Mkandawire T.T., Tatoris S., Kennedy V., Iqball S., Devarajan G., Ellis S. Enhancing titres of therapeutic viral vectors using the transgene repression in vector production (TRiP) system. Nat. Commun. 2017;8:14834. doi: 10.1038/ncomms14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frahm N., Kiepiela P., Adams S., Linde C.H., Hewitt H.S., Sango K., Feeney M.E., Addo M.M., Lichterfeld M., Lahaie M.P. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 2006;7:173–178. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 51.Goulder P.J.R., Phillips R.E., Colbert R.A., McAdam S., Ogg G., Nowak M.A., Giangrande P., Luzzi G., Morgan B., Edwards A. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 52.Allen T.M., O’Connor D.H., Jing P., Dzuris J.L., Mothé B.R., Vogel T.U., Dunphy E., Liebl M.E., Emerson C., Wilson N. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 53.Cao J., McNevin J., Malhotra U., McElrath M.J. Evolution of CD8+ T cell immunity and viral escape following acute HIV-1 infection. J. Immunol. 2003;171:3837–3846. doi: 10.4049/jimmunol.171.7.3837. [DOI] [PubMed] [Google Scholar]
- 54.Goonetilleke N., Liu M.K.P., Salazar-Gonzalez J.F., Ferrari G., Giorgi E., Ganusov V.V., Keele B.F., Learn G.H., Turnbull E.L., Salazar M.G., CHAVI Clinical Core B The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 2009;206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mothe B., Llano A., Ibarrondo J., Daniels M., Miranda C., Zamarreño J., Bach V., Zuniga R., Pérez-Álvarez S., Berger C.T. Definition of the viral targets of protective HIV-1-specific T cell responses. J. Transl. Med. 2011;9:208. doi: 10.1186/1479-5876-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou J., Wang J.-J., Chen X., Li H., Ding L., Spearman P. Characterization of a myristoylated, monomeric HIV Gag protein. Virology. 2009;387:341–352. doi: 10.1016/j.virol.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundquist W.I., Krausslich H. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012;2:43. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu X., Lu Z., Valentin A., Rosati M., Broderick K.E., Sardesai N.Y., Marx P.A., Mullins J.I., Pavlakis G.N., Felber B.K. Gag and env conserved element CE DNA vaccines elicit broad cytotoxic T cell responses targeting subdominant epitopes of HIV and SIV Able to recognize virus-infected cells in macaques. Hum. Vaccin. Immunother. 2018;14:2163–2177. doi: 10.1080/21645515.2018.1489949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munson P., Liu Y., Bratt D., Fuller J.T., Hu X., Pavlakis G.N., Felber B.K., Mullins J.I., Fuller D.H. Therapeutic conserved elements (CE) DNA vaccine induces strong T-cell responses against highly conserved viral sequences during simian-human immunodeficiency virus infection. Hum. Vaccin. Immunother. 2018;14:1820–1831. doi: 10.1080/21645515.2018.1448328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negri D.R.M., Rossi A., Blasi M., Michelini Z., Leone P., Chiantore M.V., Baroncelli S., Perretta G., Cimarelli A., Klotman M.E., Cara A. Simian immunodeficiency virus-Vpx for improving integrase defective lentiviral vector-based vaccines. Retrovirology. 2012;9:69. doi: 10.1186/1742-4690-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mochizuki H., Schwartz J.P., Tanaka K., Brady R.O., Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J. Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Michelini Z., Negri D.R.M., Baroncelli S., Spada M., Leone P., Bona R., Klotman M.E., Cara A. Development and use of SIV-based Integrase defective lentiviral vector for immunization. Vaccine. 2009;27:4622–4629. doi: 10.1016/j.vaccine.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hermida-Matsumoto L., Resh M.D. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 2000;74:8670–8679. doi: 10.1128/jvi.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buffa V., Negri D.R.M., Leone P., Borghi M., Bona R., Michelini Z., Compagnoni D., Sgadari C., Ensoli B., Cara A. Evaluation of a self-inactivating lentiviral vector expressing simian immunodeficiency virus gag for induction of specific immune responses in vitro and in vivo. Viral Immunol. 2006;19:690–701. doi: 10.1089/vim.2006.19.690. [DOI] [PubMed] [Google Scholar]
- 66.Berger G., Durand S., Goujon C., Nguyen X.N., Cordeil S., Darlix J.L., Cimarelli A. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat. Protoc. 2011;6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.