Abstract
Drug resistance, including adriamycin (ADR)-based therapeutic resistance, is a crucial cause of chemotherapy failure in breast cancer treatment. Acquired chemoresistance has been identified to be closely associated with the overexpression of P-glycoprotein (P-gp/ABCB1). Long non-coding RNA (lncRNA) growth arrest-specific 5 (GAS5) can be involved in carcinogenesis; however, its roles in ABCB1-mediated ADR resistance are poorly understood. In this study, we identified a panel of differentially expressed lncRNAs, mRNAs, and microRNAs (miRNAs) in MCF-7 and MCF-7/ADR cell lines through RNA sequencing (RNA-seq) technologies. GAS5 level was downregulated whereas ABCB1 level was upregulated in the resistant breast cancer tissues and cells. Overexpression of GAS5 significantly enhanced the ADR sensitivity and apoptosis, and it inhibited the efflux function and expression of ABCB1 in vitro, while knockdown of GAS5 had the opposite effects. Further mechanism-related investigations indicated that GAS5 acted as an endogenous “sponge” by competing for miR-221-3p binding to regulate its target dickkopf 2 (DKK2), and then it inhibited the activation of the Wnt/β-catenin pathway. Functionally, GAS5 enhanced the anti-tumor effect of ADR in vivo. Collectively, our findings reveal that GAS5 exerted regulatory function in ADR resistance possibly through the miR-221-3p/DKK2 axis, providing a novel approach to develop promising therapeutic strategy for overcoming chemoresistance in breast cancer patients.
Keywords: lncRNA GAS5, miR-221-3p, ABCB1, drug resistance, breast cancer
Graphical Abstract
Introduction
Breast cancer is the most common cancer in women and one of the most common causes of cancer death worldwide, accounting for 30% in diagnosed new cases and 15% in estimated deaths.1 Anthracycline genotoxic drug epirubicin or adriamycin (ADR) is a major chemotherapeutic drug that has been widely applied for breast cancer patients; however, the acquired drug resistance frequently restricted its curative effect and thus led to failure in the clinic.2, 3, 4 Among many factors and causes of acquired drug resistance, overexpression of P-glycoprotein (P-gp/ABCB1) encoding by the multidrug resistance 1 (MDR1) gene plays a significant role in drug resistance due to the increased energy-dependent efflux of cytotoxic drugs from cancer cells.5,6 So far, however, the detailed mechanisms of drug resistance in cancer cells are complex and have not been fully elucidated. Therefore, a better understanding of the molecular mechanisms of drug resistance in breast cancer is necessary in order to identify the promising therapeutic targets and improve the efficiency of chemotherapy for breast cancer patients.
Long non-coding RNAs (lncRNAs) are a class of transcripts more than 200 nt in length and limited protein-coding ability, which have been confirmed to be involved in many cellular and genomic processes associated with carcinogenesis and drug resistance/sensitivity.3,7 Moreover, the importance of lncRNAs in the resistance of breast cancer to multiple drugs has been discussed. For instance, lncRNA TINCR (terminal differentiation-induced non-coding RNA) enhanced trastuzumab resistance and migration, invasion, and the accompanied epithelial-mesenchymal transition (EMT) process in breast cancer by competitive binding to miR-125b, upregulating HER-2 (human epidermal growth factor receptor-2) and Snail-1.8 Knockdown of H19 restored chemosensitivity in a paclitaxel-resistant triple-negative breast cancer cell line by regulating the Akt-mediated apoptotic signaling pathway.9 Moreover, knockdown of H19 repressed the expression of transcription factors related to the Wnt pathway and EMT, and it improved tamoxifen sensitivity for promoting cell growth and inhibiting apoptosis in tamoxifen-resistant breast cancer cells.10 lncRNA SNHG14 contributed to trastuzumab resistance and tumorigenesis in breast cancer cells through regulating PABPC1 expression by H3K27 acetylation.11 Furthermore, extracellular lncRNA-SNHG14 was able to be incorporated into exosomes and transmitted to sensitive breast cancer cells, thus inducing trastuzumab resistance.12 lncRNA growth arrest-specific 5 (GAS5) is an lncRNA 650 bases in length, originally isolated from NIH 3T3 cells using subtraction hybridization in 1988,13 which was downregulated in breast cancer samples and cells.14,15 A recent report demonstrated that upregulation of GAS5 alleviated tamoxifen resistance by acting as a molecular sponge of miR-222, leading to the de-repression of its endogenous target phosphatase and tensin homologs (PTENs) in breast cancer.16 In addition, GAS5 can sensitize tumor cells to UV irradiation and doxorubicin, as well as suppress cell invasion by regulating PTENs and PDCD4 through miR-21.17 Although GAS5 has been suggested to play roles in chemoresistance, the underlying mechanism of GAS5-mediated gene expression having an impact on drug resistance is still elusive.
In this study, we attempted to explore the contributions of GAS5 to the ADR resistance in breast cancer and explore the potential mechanisms. We found that GAS5 expression was decreased whereas ABCB1 was increased in breast cancer-resistant patients and cell lines (Table 1). Furthermore, our results show, for the first time, that GAS5 modulates ABCB1-mediated ADR resistance by targeting the miR-221-3p/dickkopf 2 (DKK2)/β-catenin pathway in breast cancer cells. Thus, GAS5 maybe a promising therapeutic biomarker and target for the treatment of breast cancer patients with ADR resistance.
Table 1.
Clinical Characteristics of Breast Cancer Patients
| Clinicopathological Data | n | Percentage (%) |
|---|---|---|
| Age (years) | ||
| ≤50 | 18 | 69.2 |
| >50 | 8 | 30.8 |
| Primary tumor size (cm) | ||
| ≤5 | 16 | 61.5 |
| >5 | 10 | 38.5 |
| Lymph node metastasis | ||
| Present | 21 | 80.8 |
| Absent | 5 | 19.2 |
| TNM stage | ||
| I–II | 11 | 42.3 |
| III–IV | 15 | 57.7 |
| Histopathological grade | ||
| I–II | 15 | 57.7 |
| III | 11 | 42.3 |
| ER status | ||
| Positive | 13 | 50.0 |
| Negative | 13 | 50.0 |
| PR status | ||
| Positive | 11 | 42.3 |
| Negative | 15 | 57.7 |
| HER-2 status | ||
| Positive | 12 | 46.2 |
| Negative | 14 | 53.8 |
| Molecular subtype | ||
| Luminal A | 5 | 19.2 |
| Luminal B | 9 | 34.6 |
| Her-2 overexpressing | 8 | 30.8 |
| Triple negative | 4 | 15.4 |
TNM, tumor, node, metastasis; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor-2.
Results
GAS5 Is Downregulated while ABCB1 Is Upregulated in Breast Cancer Tissues and Cell Lines
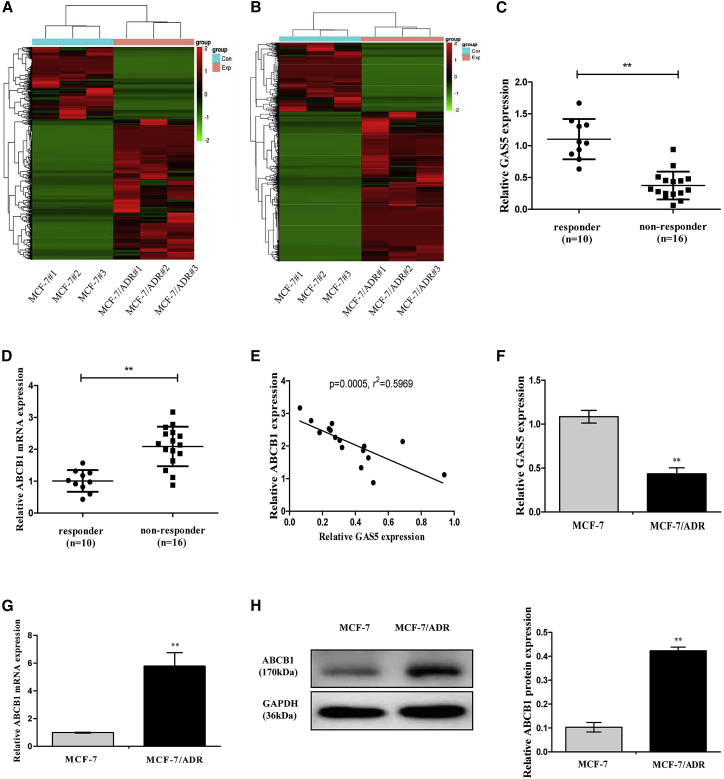
The RNA expression profiles were obtained from MCF-7 (human breast cell line) and ADR-resistant MCF-7 cells (MCF-7/ADR) by RNA sequencing (RNA-seq). A total of 3,811 lncRNAs and 4,088 mRNAs were detected with the criteria of absolute false discovery rate (FDR) value <0.05 and p value <0.01. Among them, 1,264 lncRNAs and 1,943 mRNAs were significantly downregulated, whereas 2,547 lncRNAs and 2,145 mRNAs were significantly upregulated in the MCF-7/ADR cells than that in MCF-7 cells (Figures 1A and 1B, all p < 0.01; >3.0-fold). The 10 lncRNAs and mRNAs with the largest fold changes are shown in Tables 2 and 3. Of particular interest, we noticed that GAS5 was one of the most prominently downregulated lncRNAs and ABCB1 was the most upregulated mRNA in the ADR-resistant breast cancer cells. This suggested that low GAS5 expression and high ABCB1 expression were associated with ADR resistance. Next, we examined the expressions of GAS5 and ABCB1 in clinical samples and cells.
Figure 1.
GAS5 and ABCB1 Expression Levels in Breast Cancer Tissues and Cells
(A) Heatmap of differentially expressed lncRNAs in MCF-7 and MCF-7/ADR cells (n = 3). (B) Heatmap of differentially expressed mRNAs in MCF-7 and MCF-7/ADR cells (n = 3). (C and D) GAS5 expression (C) and ABCB1 mRNA expression (D) in breast cancer tissues from responder and non-responder patients were detected by quantitative real-time PCR. (E) Negative correlation between GAS5 and ABCB1 expressions in breast cancer tissues. (F and G) GAS5 expression (F) and ABCB1 mRNA expression (G) in MCF-7 and MCF-7/ADR cells were detected by quantitative real-time PCR. (H) The ABCB1 protein expression in MCF-7 and MCF-7/ADR cells was detected by western blot analysis. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
Table 2.
Characteristics of lncRNAs with the Largest Fold Change
| lncRNA | Gene ID | Fold Change | Regulation | p Value | FDR |
|---|---|---|---|---|---|
| CYTOR | ENSG00000222041.10 | 5.092170527 | up | 1.0821E−159 | 9.3006E−157 |
| C10orf25 | ENSG00000165511.6 | 7.153890466 | up | 1.904E−132 | 1.1255E−129 |
| TPRG1-AS1 | ENSG00000234076.1 | 5.672774218 | up | 7.442E−108 | 3.1891E−105 |
| ADGRL2 | ENSG00000117114.19 | 5.005623404 | up | 1.63691E−87 | 5.04765E−85 |
| LINC00707 | ENSG00000238266.1 | 12.8054899 | up | 1.71569E−80 | 4.69952E−78 |
| LINC00886 | ENSG00000240875.5 | 5.154727315 | down | 5.2902E−173 | 5.775E−170 |
| AL035661.1 | ENSG00000274173.1 | 8.484605987 | down | 7.65708E−94 | 2.6819E−91 |
| GAS5 | ENSG00000234741.7 | 5.038222071 | down | 1.91332E−89 | 6.15376E−87 |
| AC010503.4 | ENSG00000275234.1 | 6.181951912 | down | 5.82009E−76 | 1.45558E−73 |
| FP325330.3 | ENSG00000279712.1 | 7.88445797 | down | 2.33947E−70 | 5.10034E−68 |
Table 3.
Characteristics of mRNAs with the Largest Fold Change
| Gene Name | Gene ID | Fold Change | Regulation |
|---|---|---|---|
| ABCB1 | ENSG00000085563.14 | 12.48764005 | up |
| GALC | ENSG00000054983.16 | 10.31859095 | up |
| CALD1 | ENSG00000122786.19 | 9.684997032 | up |
| COL12A1 | ENSG00000111799.20 | 7.649996364 | up |
| AEBP1 | ENSG00000106624.10 | 6.849429416 | up |
| GRHL2 | ENSG00000083307.10 | 11.78960888 | down |
| SPDEF | ENSG00000124664.10 | 10.53368044 | down |
| PRLR | ENSG00000113494.16 | 9.023956295 | down |
| ADAMTS19 | ENSG00000145808.9 | 8.368612293 | down |
| MAP7 | ENSG00000135525.18 | 7.322561697 | down |
According to RECIST criteria, we evaluated the clinical responses to neoadjuvant chemotherapy (NAC) in 26 patients. In our study, a total of 6 patients (23%) had a partial response (PR) and 4 patients (15%) had complete responses (CRs), which were regarded as clinical responders, whereas 10 patients (38%) had stable disease (SD) and 6 patients (23%) had progressive disease (PD), which were regarded as non-responders. Therefore, in our study 10 patients were responders and 16 patients were non-responders. To investigate the potential roles of GAS5 and ABCB1 for response to chemotherapy in breast cancer patients, GAS5 and ABCB1 expression were examined by quantitative real-time PCR in breast cancer tissues of clinical responders (n = 10) and non-responders (n = 16). As shown in Figure 1C, GAS5 expression was considerably downregulated in non-responders compared with that in the responders. On the contrary, ABCB1 expression was upregulated in non-responders compared with that in the responders (Figure 1D). Moreover, the mRNA expression level of ABCB1 was positively correlated with GAS5 level in breast cancer tissues (Figure 1E). Due to the relevance between GAS5 and ABCB1 in ADR resistance, we further evaluated the GAS5 and ABCB1 expressions in MCF-7 and MCF-7/ADR cells. The relative expression level of GAS5 was strongly decreased (Figure 1F) whereas ABCB1 was significantly increased (Figures 1G and 1H) by more than 2-fold in the MCF-7/ADR cells compared to the parental MCF-7 cells, indicating that GAS5 and ABCB1 might play key roles in the development of ADR resistance in breast cancer.
GAS5 Influenced Chemoresistance in Breast Cancer Cells
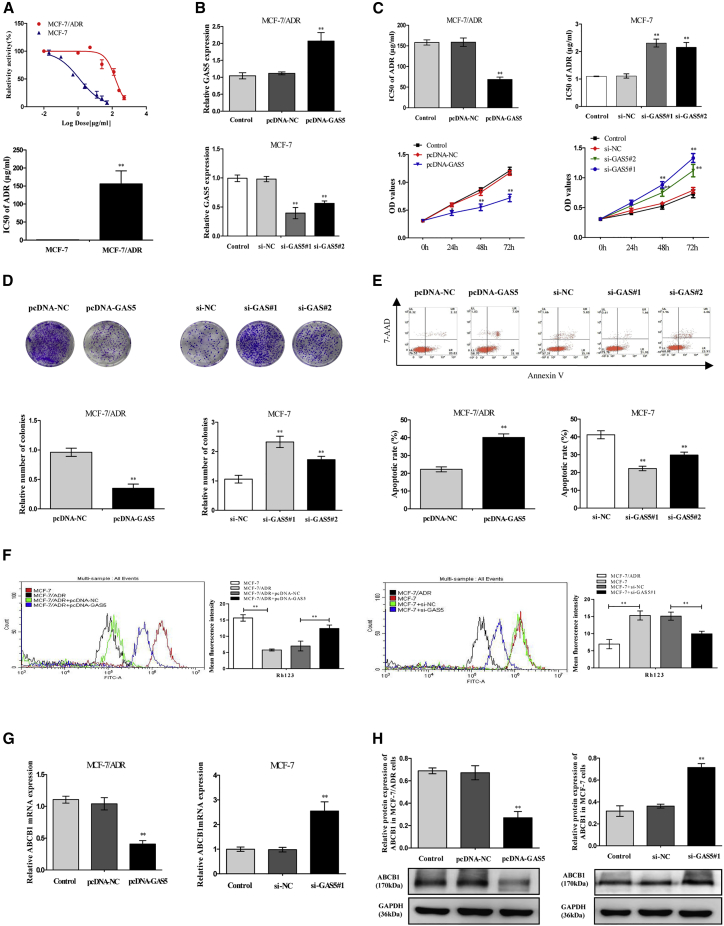
Afterward, we investigated whether GAS5 is associated with the ADR resistance of breast cancer cells. The 50% inhibition concentration (IC50) of ADR was measured to confirm the ADR resistance of MCF-7/ADR cells in comparison with its parental MCF-7 cells. As shown in Figure 2A, the IC50 value of ADR in MCF-7/ADR cells was almost 160-fold compared to that in MCF-7 cells. To further uncover the function of GAS5 on the sensitivity of ADR, the breast cancer cells were transfected with GAS5 overexpression plasmid (pcDNA-GAS5) or two specific GAS5 small interfering RNAs (siRNAs) (si-GAS5#1 and si-GAS5#2). As expected, GAS5 expression was markedly upregulated in MCF-7/ADR cells when transfected with pcDNA-GAS5, whereas GAS5 expression was downregulated by 64% by si-GAS5#1 and 41% by si-GAS5#2 compared with their counterparts, respectively (Figure 2B). Additionally, the value of IC50 of ADR and cell proliferation capacity were all strongly decreased in MCF-7/ADR cells by overexpression of GAS5, while introduction with si-GAS5#1 or si-GAS5#2 in MCF-7 cells showed an inverse effect (Figures 2C and 2D). Furthermore, flow cytometry analysis observed that ADR-induced apoptosis was remarkably increased in pcDNA-GAS5-transfected MCF-7/ADR cells and decreased in si-GAS5#1- or si-GAS5#2-transfected MCF-7 cells (Figure 2E). Overall, all of the data concluded that GAS5 reversed the breast cancer cell chemoresistance to ADR-based chemotherapy in vitro; here, we further examined the effect of GAS5 on the functions and expressions of ABCB1.
Figure 2.
GAS5 Was Associated with ADR Resistance of Breast Cancer Cells
(A) Cells were exposed to a series dose of ADR (0.02, 0.1, 0.5, 2.5, 12.5, and 25 μg/mL for MCF-7, and 0.5, 1, 5, 25, 125, 250 μg/mL for MCF-7/ADR) for 48 h; the cell viability of cells and the 50% inhibitory concentration (IC50) of ADR were then detected using a CCK-8 assay. (B) MCF-7/ADR cells were transfected with pcDNA-GAS5, and MCF-7 cells were introduced with two synthesized individual GAS5 siRNAs (si-GAS5#1 and si-GAS5#2). The GAS5 expression was detected by quantitative real-time PCR. (C) pcDNA-GAS5-transfected MCF-7/ADR cells and si-GAS5#1- or si-GAS5#2-transfected MCF-7 cells were treated with various concentrations of ADR for 48 h, and IC50s of ADR and cell proliferation capacity were measured by a CCK-8 assay. (D) The colony-forming ability was measured by a colony-forming assay. (E) Cell apoptosis was evaluated by flow cytometry. (F) Representative fluorescence-activated cell sorting (FACS) histograms of Rh123 accumulation and intensity of Rh123 fluorescence in pcDNA-GAS5-transfected MCF-7/ADR cells and in si-GAS5#1-transfected MCF-7 cells. (G) The ABCB1 mRNA expression was examined by quantitative real-time PCR. (H) The ABCB1 protein expression was examined by western blot analysis. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
Rhodamine-123 (Rh123), which is an ABCB1-specific fluorescent dye, was used to measure the efflux function of ABCB1.18 As described in Figure 2F, compared with sensitive MCF-7 cells, lesser accumulation of Rh123 was significantly observed in resistant MCF-7/ADR cells. When the GAS5 expression was overexpressed by pcDNA-GAS5, the Rh123 accumulation in MCF-7/ADR cells was increased 1.78-fold compared to pcDNA-negative control (NC) (empty pcDNA3.1 vector) group cell levels. Consistently, the Rh123 accumulation was decreased almost 1.52-fold in MCF-7 cells transfected with specific si-GAS5#1 than in the NC group siRNA (si-NC), indicating that GAS5 inhibited the cellular efflux function of ABCB1. Furthermore, overexpression of GAS5 dramatically decreased ABCB1 mRNA and protein levels by almost 60% in MCF-7/ADR cells, and knockdown of GAS5 by transferring si-GAS5#1 increased the levels by more than 50% in MCF-7 cells (Figures 2G and 2H). In total, our results exhibited that GAS5 might restrain ABCB1 function and expression, thus influencing the chemoresistance of breast cancer cells.
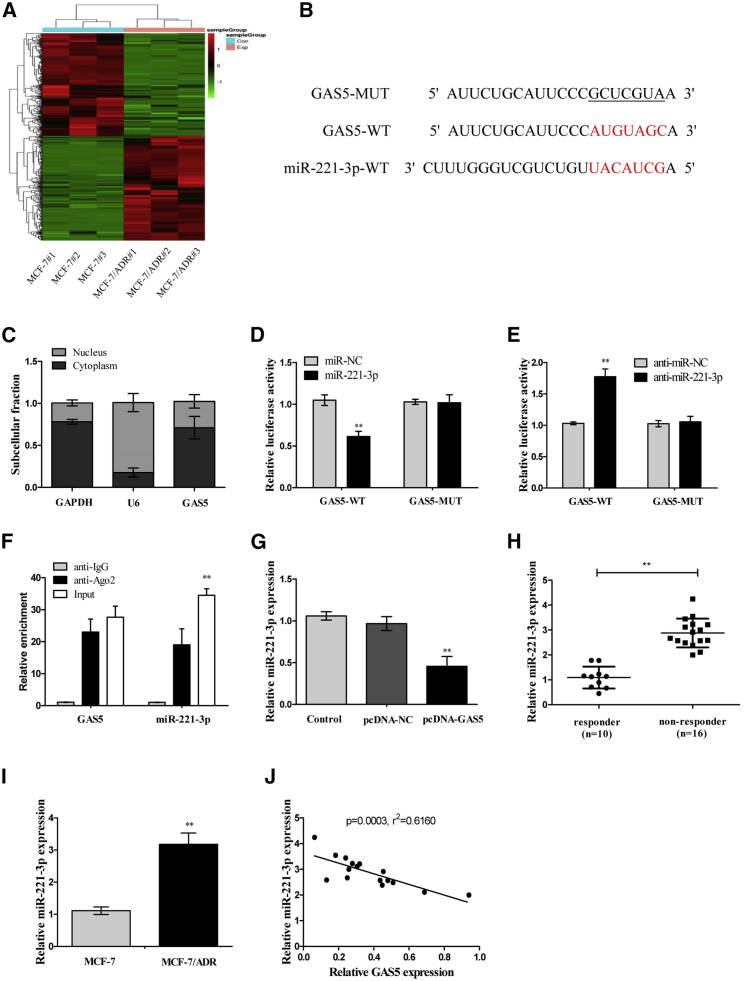
GAS5 Inhibited miR-221-3p Expression by Direct Interaction
In order to explore the potential mechanism of GAS5 involved in ADR sensitivity in breast cancer cells, the online databases DIANA-LncBase, miRcode, and RNA-seq were applied to predict and confirm the most underlying microRNA (miRNA) that can be interacted with GAS5. We selected and identified the 456 miRNAs that were significantly differentially expressed by RNA-seq (Figure 3A, all p < 0.01; >3.0-fold). Of all of the 456 miRNAs, we found that miR-221-3p was the most upregulated miRNA in the MCF-7/ADR cells (Table 4). Interestingly, the results of online bioinformatics analysis indicated that complementary sites exist between miR-221-3p and GAS5 (Figure 3B). Moreover, the subcellular location of GAS5 was determined by quantitative real-time PCR. The results showed that GAS5 was mainly localized in the cytoplasmic fraction, indicating that GAS5 had a chance to interact with cytoplasmic miRNAs (Figure 3C). The further dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay validated the direct binding between miR-221-3p and GAS5 in MCF-7/ADR cells. In the dual-luciferase reporter assay, co-transfection of MCF-7/ADR cells with GAS5 wild-type (WT) reporter vector (GAS5-WT) and miR-221-3p mimics remarkably decreased the luciferase activity of GAS5 compared to those transfected with GAS5 mutant (MUT) type reporter vector (GAS5-MUT) (Figure 3D), and downregulation of miR-221-3p by introducing anti-miR-221-3p clearly promoted the luciferase activity (Figure 3E). However, the mutant reporter activities in all transfected cells had no evident effects (Figures 3D and 3E). Moreover, RIP data revealed that GAS5 and miR-221-3p were highly enriched in the complex precipitated by Argonaute2 (Ago2) antibody relative to nonspecific immunoglobulin G (IgG) control antibody (Figure 3F). Then, the effect of GAS5 on miR-221-3p expression was detected in MCF-7/ADR cells transfected with pcDNA-GAS5. As shown in Figure 3G, miR-221-3p was downregulated by ectopic GAS5. Subsequently, we detected the expression of miR-221-3p in breast tumor tissues and cells. It was shown that miR-221-3p was strongly significantly upregulated in non-responders compared with that in the responders (Figure 3H), and miR-221-3p expression in MCF-7/ADR cells was about 3-fold greater than that in corresponding controls (Figure 3I). Additionally, Pearson’s correlation analysis demonstrated that miR-221-3p expression was inversely correlated with GAS5 expression in breast tumor tissues (Figure 3J). All of these results suggested that GAS5 could directly bind to miR-221-3p and regulate its expression.
Figure 3.
GAS5 Directly Suppressed miR-221-3p Expression in Breast Cancer Cells
(A) The differently expressed miRNAs between MCF-7 and MCF-7/ADR cells were elucidated by a heatmap (n = 3). (B) Sequence alignment of miR-221-3p with the putative binding sites within the wild-type regions of GAS5. (C) The subcellular location of GAS5 in MCF-7 cells was identified by a subcellular fractionation assay. GAPDH and U6 were used as internal references. (D) Luciferase activity in MCF-7/ADR cells co-transfected with the wipe-type or mutant GAS5 reporters (GAS5-WT or GAS5-MUT) and miR-221-3p mimics or miR-control (Con). (E) Luciferase activity in MCF-7/ADR cells co-transfected with GAS5-WT or GAS5-MUT and anti-miR-221-3p or anti-miR-Con. (F) The GAS5 and miR-221-3p levels isolated from Ago2 and IgG immunoprecipitates derived from MCF-7/ADR cells were examined by quantitative real-time PCR. (G) The miR-221-3p expression in MCF-7/ADR cells transfected with pcDNA-GAS5 was examined by quantitative real-time PCR. (H) The miR-221-3p expression in breast cancer tissues was examined by quantitative real-time PCR. (I) The miR-221-3p expression in MCF-7 and MCF-7/ADR cells was examined by quantitative real-time PCR. (J) Pearson’s analysis shows a negative correlation between GAS5 and miR-221-3p expressions in breast cancer tissues. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
Table 4.
Characteristics of miRNAs with the Largest Fold Change
| miRNA | Chromosome | Fold Change | Regulation | p Value | FDR |
|---|---|---|---|---|---|
| hsa-miR-221-3p | chrX | 7.287296594 | up | 1.5134E−250 | 2.8823E−248 |
| hsa-miR-222-3p | chrX | 7.285851132 | up | 1.6712E−248 | 2.8644E−246 |
| hsa-miR-363-3p | chrX | 6.407126736 | up | 9.8099E−243 | 1.5286E−240 |
| hsa-miR-29a-3p | chr7 | 5.79567643 | up | 6.2988E−232 | 8.9968E−230 |
| hsa-miR-10a-5p | chr17 | 5.595258884 | up | 1.6657E−228 | 2.0393E−226 |
| hsa-miR-125b-5p | chr11;chr21 | 3.515215771 | up | 7.7306E−227 | 8.8335E−225 |
| hsa-miR-100-5p | chr11 | 4.749079454 | up | 2.4896E−216 | 2.667E−214 |
| hsa-miR-489-3p | chr7 | 6.667962520 | down | 3.8488E−231 | 5.0746E−229 |
| hsa-miR-200b-3p | chr1 | 3.960448865 | down | 8.5356E−209 | 8.6059E−207 |
| hsa-miR-200c-3p | chr12 | 6.365063854 | down | 1.3053E−203 | 1.2429E−201 |
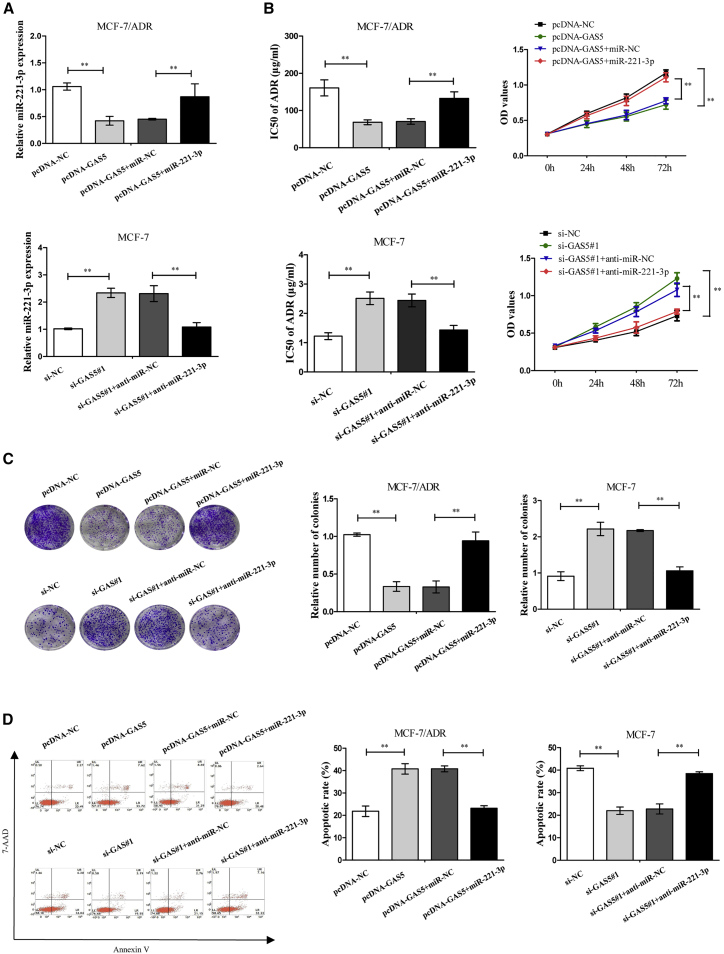
miR-221-3p Mediates the Effects of GAS5 on the ADR Resistance of Breast Cancer Cells
Next, we attempted to explore whether miR-221-3p mediated the effect of GAS5 on the chemoresistance of breast cancer cells. The results indicated that GAS5-triggered reduction of miR-221-3p levels was significantly reversed by miR-221-3p overexpression in MCF-7/ADR cells (Figure 4A). Consistently, si-GAS5#1-induced promotion of miR-221-3p expression was markedly recovered after depletion of miR-221-3p in MCF-7 cells in comparison to homologous controls (Figure 4A). In the Cell Counting Kit-8 (CCK-8) and colony formation assays, GAS5 decreased the IC50 of ADR and cell proliferation, which were strikingly reversed by introduction with miR-221-3p mimics in MCF-7/ADR cells, whereas si-GAS5#1 showed the opposite effects by introduction with anti-miR-221-3p (Figures 4B and 4C). The data from flow cytometry indicated that GAS5-elicited apoptosis was significantly lowered after upregulation of miR-221-3p in MCF-7/ADR cells, while si-GAS5#1-induced reduction on apoptosis was greatly improved following downregulation of miR-221-3p in MCF-7 cells (Figure 4D). Collectively, these findings demonstrated that GAS5 increased the sensitivity of breast cancer cells to ADR through repressing miR-221-3p.
Figure 4.
GAS5-Induced ADR Sensitivity of Breast Cancer Cells Was Abated by miR-221-3p Upregulation
MCF-7/ADR cells were transfected with pcDNA-GAS5 alone or together with miR-221-3p mimics, and MCF-7 cells were transfected with si-GAS5#1 alone or together with anti-miR-221-3p. (A) The miR-221-3p expression in MCF-7/ADR and MCF-7 cells was examined by quantitative real-time PCR. (B) IC50 of ADR and cell proliferation capacity were measured by a CCK-8 assay in MCF-7/ADR and MCF-7 cells. (C) The colony-forming ability was measured by a colony-forming assay. (D) Cell apoptosis was evaluated by flow cytometry. The data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
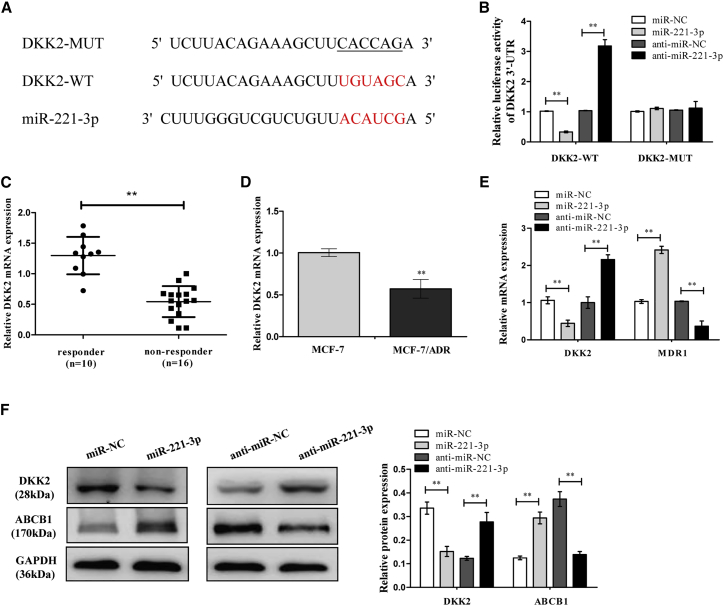
miR-221-3p Directly Targets DKK2, Regulating ABCB1 Expression
Bioinformatics databases (TargetScan, starBase, and miRDB) analysis revealed that DKK2, which is a critical receptor for activation of the Wnt/β-catenin signaling pathway, was identified as a potential target of miR-221-3p (Figure 5A). In the meantime, it has been reported that DKK2 is a target of miR-221 in osteosarcoma.19 Following the above findings, we attempted to explore whether DKK2 was a target of miR-221-3p in breast cancer cells. Later, a luciferase reporter assay was performed to confirm the binding site of miR-221-3p. It indicated that miR-221-3p negatively regulated the luciferase activity of DKK2-WT-3′ UTR, rather than that of DKK2-MUT-3′ UTR (Figure 5B). Subsequently, DKK2 was distinctly downregulated in non-responders compared with responders with respect to breast cancer tissues (Figure 5C), and DKK2 mRNA expression was decreased more in ADR-resistant MCF-7/ADR cells than that in the MCF-7 control group (Figure 5D). Furthermore, Figures 5E and 5F suggested that mRNA and protein expression levels of DKK2 in MCF-7/ADR cells were inactively regulated by miR-221-3p, while the mRNA and protein expression levels of ABCB1 displayed the contrary effect. Therefore, we concluded that miR-221-3p directly targets DKK2 and induces ABCB1 in breast cancer cells.
Figure 5.
miR-221-3p Targets the 3′ UTR of DKK2 and Regulates the ABCB1 Expression
(A) Putative binding sites between DKK2 and miR-221-3p predicted by the TargetScan online website together with the mutant sites of the DKK2-3′ UTR region in the MUT-DKK2-3′ UTR reporter vector. (B) The luciferase activity was detected in MCF-7/ADR cells transfected with DKK2-WT or DKK2-MUT and miR-221-3p mimics or anti-miR-221-3p. (C) The DKK2 mRNA expression in breast cancer tissues was examined by quantitative real-time PCR. (D) The DKK2 mRNA expression in MCF-7 and MCF-7/ADR cells was examined by quantitative real-time PCR. (E) The MCF-7/ADR cells were transfected with miR-221-3p mimics or anti-miR-221-3p. The DKK2 and ABCB1 mRNA expression levels were examined by quantitative real-time PCR. (F) The DKK2 and ABCB1 protein expression levels were examined by western blot analysis. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
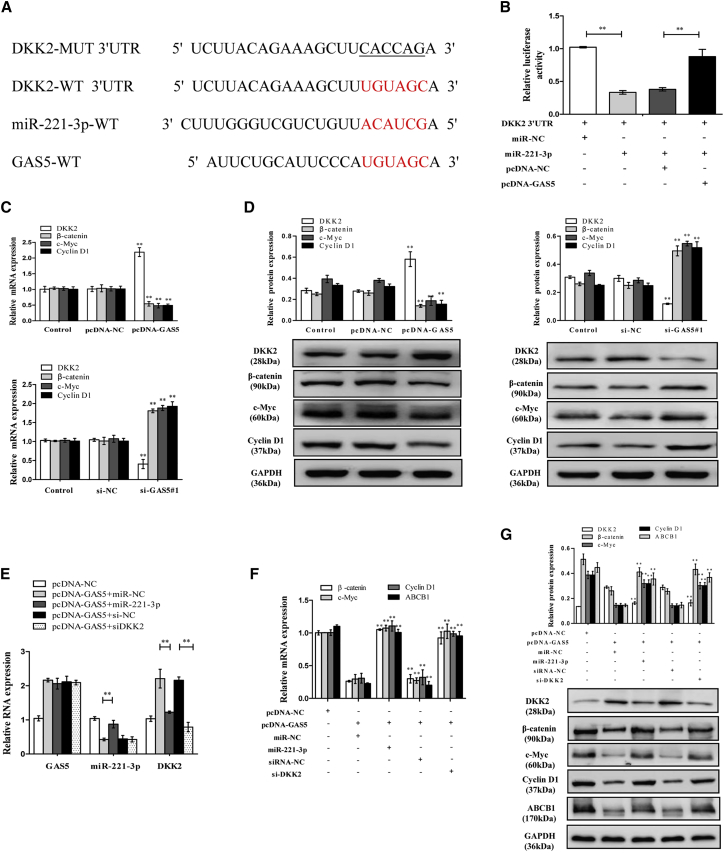
GAS5 Inhibits ABCB1 Expression by Activating the Wnt/β-Catenin Signaling Pathway through the GAS5/miR-221-3p/DKK2 Axis
To determine whether specific crosstalk existed between GAS5 and DKK2 through competition for miR-221-3p binding (Figure 6A), we conducted luciferase report assays by transfecting DKK2 WT reporter vector (DKK2-WT) into cells together with miR-221-3p mimics or miR-221-3p mimics+pcDNA-GAS5. These data indicated that the fluorescence intensity of the DKK2-WT vector was remarkably inhibited by co-transfection with miR-221-3p mimics compared to the corresponding controls. Additionally, miR-221-3p triggered a decrease in luciferase activity of the DKK2-WT vector, which was substantially counteracted by GAS5 upregulation (Figure 6B). In addition, we found that GAS5 increased the DKK2 expression and decreased the expression of the Wnt/β-catenin pathway downstream targets c-Myc and cyclin D1 (Figures 6C and 6D). Then, we attempted to explore whether GAS5 regulated ABCB1 expression via activating the Wnt/β-catenin signaling pathway by the GAS5/miR-221-3p/DKK2 axis. Real-time PCR for rescue experiments revealed that GAS5 positively regulated DKK2 mRNA expression, which was mediated by miR-221-3p. It showed that pcDNA-GAS5 not only increased the DKK2 expression but also decreased the β-catenin, c-Myc, cyclin D1, and ABCB1 expressions, as previously mentioned, while when combined with miR-221-3p mimics or si-DKK2 it reversed the effects of pcDNA-GAS5 (Figures 6E–6G). When combining the preceding findings, the data demonstrated that GAS5 inhibits ABCB1 expression by activating the Wnt/β-catenin signaling pathway through the GAS5/miR-221-3p/DKK2 axis.
Figure 6.
GAS5 Inhibits ABCB1 Expression by Activating the Wnt/β-Catenin Signaling Pathway through the GAS5/miR-221-3p/DKK2 Axis
(A) Putative miR-221-3p binding sites on GAS5 and DKK2. (B) The dual-luciferase reporter assay was performed by transfecting DKK2-WT vector into MCF-7/ADR cells together with miR-221-3p mimics or miR-221-3p mimics+pcDNA-GAS5. (C) The DKK2, β-catenin, c-Myc, and cyclin D1 mRNA expressions in MCF-7/ADR cells transfected with pcDNA-NC or pcDNA-GAS5 and in MCF-7 cells transfected with si-NC or si-GAS5#1 were detected by quantitative real-time PCR. (D) The DKK2, β-catenin, c-Myc, and cyclin D1 protein expressions in MCF-7/ADR cells transfected with pcDNA-NC or pcDNA-GAS5 and in MCF-7 cells transfected with si-NC or si-GAS5#1 were detected by western blot analysis. (E) MCF-7/ADR cells were transfected with pcDNA-GAS5+miR-NC, pcDNA-GAS5+miR-221-3p mimics, pcDNA-GAS5+siRNA-NC, or pcDNA-GAS5+si-DKK2. The GAS5, miR-221-3p, and DKK2 expressions were examined by quantitative real-time PCR. (F) The DKK2, β-catenin, c-Myc, and cyclin D1 mRNA expressions were examined by quantitative real-time PCR. (G) The DKK2, β-catenin, c-Myc, and cyclin D1 protein expressions were examined by western blot analysis. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
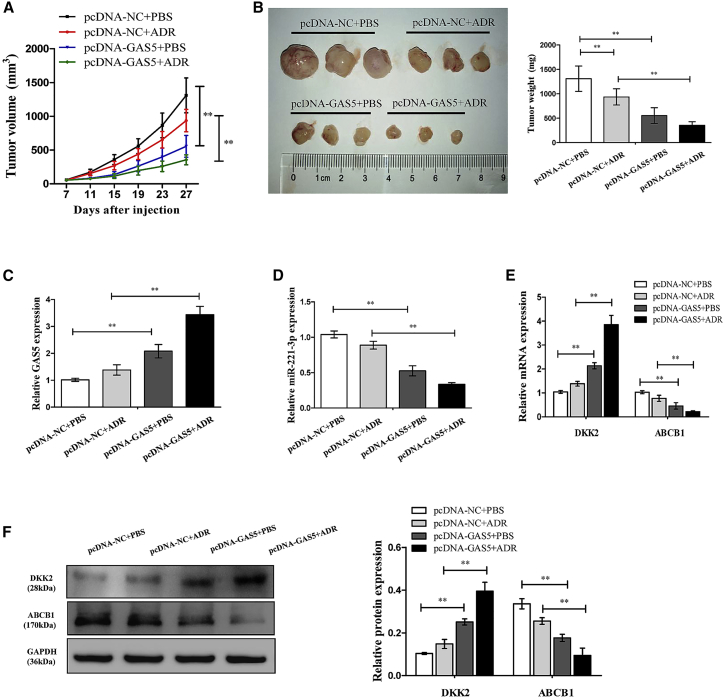
GAS5 Enhances ADR Sensitivity in Breast Cancer In Vivo
To further verify the underlying effects and mechanisms of GAS5 in increasing the sensitivity of breast cancer MCF-7/ADR cells to ADR in vivo, cells infected with pcDNA-GAS5 or pcDNA-NC were subcutaneously injected into each nude mouse to generate tumor xenograft, followed by administration with ADR or PBS. As shown in Figures 7A and 7B, GAS5 overexpression or ADR treatment dramatically decreased the tumor volume and weight compared with the control group. Furthermore, a more distinct inhibition on tumor growth was caused by simultaneous GAS5 overexpression together with ADR treatment, suggesting the vital role of GAS5 on ADR sensitivity in vivo (Figures 7A and 7B). Consistent with previous findings, we found that the expressions of GAS5 (Figure 7C) and DKK2 (Figure 7E) were increased, but the expressions of miR-221-3p (Figure 7D) and ABCB1 (Figure 7E) were decreased, in tumor tissues derived from GAS5-overexpressed MCF-7/ADR cells with or without ADR treatment. A Western blot assay confirmed that the DKK2 expression level was clearly increased while the ABCB1 level was decreased followed by GAS5 overexpression in tumor xenografts with or without ADR treatment (Figure 7F). Overall, these results suggested that overexpression of GAS5 enhances ADR sensitivity in breast cancer in vivo.
Figure 7.
Overexpression of GAS5-Sensitized MCF-7/ADR Cells to ADR In Vivo
MCF-7/ADR cells stably infected pcDNA-GAS5 or pcDNA-NC were subcutaneously injected into nude mice, followed by administration with ADR (4 mg/kg) every 4 days from the 7th day after cell inoculation. (A) Tumor volumes were examined every 4 days for 27 days. (B) Representative photographs and average weights of dissected tumors. (C) The GAS5 expression in resected tumor masses was detected by quantitative real-time PCR. (D) The miR-221-3p expression in resected tumor masses was detected by quantitative real-time PCR. (E) The mRNA expression levels of DKK2 and ABCB1 in resected tumor masses were detected by quantitative real-time PCR. (F) The protein expression levels of DKK2 and ABCB1 in resected tumor masses were detected by western blot analysis. Data are shown as the mean ± SD (n = 3). *p < 0.05, **p < 0.01 versus respective control.
Discussion
During the past decades, the overall prognosis of breast cancer patients diagnosed at the early stage has been remarkably improved through standard chemotherapy after surgical resection. Largely due to drug resistance, however, the 5-year survival rates of breast cancer patients remain dismal.20, 21, 22 It has been proposed that drug resistance has frequently been associated with elevated expression levels of ABCB1 in breast cancer cells.6,23 Overcoming resistance mediated by ABCB1 is still a challenge in breast cancer chemotherapy. Recently, emerging evidence has demonstrated that aberrant lncRNA expression is strongly implicated in the development of drug resistance in breast cancer; however, the molecular mechanisms for regulation of ABCB1 by lncRNA in drug resistance have not been well elucidated.
In our present study, based on the analysis results of RNA-seq, we focused on lncRNA GAS5, which significantly contributes to various aspects of cancer biology and has been identified as a critical player of drug resistance in cancer therapy. Liu et al.24 found that GAS5 was downregulated in both pancreatic cancer tissues and cell lines. Overexpression of GAS5 suppressed cell proliferation, cell invasion, and migration and promoted gemcitabine-induced apoptosis by regulating the miR-221/SOCS3 pathway, mediating EMT and tumor stem cell self-renewal. Similarly, GAS5 upregulation could decrease the malignancy and cisplatin resistance of cervical cancer HeLa cells via the miR-21/STAT3 signaling pathway.25 Furthermore, GAS5 improved the development of multidrug resistance of pancreatic cancer cells by regulating the miR-181c-5p/ABCB1/Hippo pathway. A recent study indicated that GAS5 was significantly downregulated in tamoxifen-resistant MCF-7R cells, and overexpression of GAS5 could enhance the cell sensitivity to tamoxifen in MCF-7R cells both in vitro and in vivo.16 However, it is still unclear whether GAS5 could also affect ADR resistance in breast cancer cells. ADR is an ABCB1 substrate, which limits its effectiveness for current use in treating breast cancer patients.26 Based on the above, we sought to determine the potential mechanisms between GAS5 and ABCB1-mediated ADR resistance in breast cancer. Our results found an inverse correlation of expression patterns between GAS5 and ABCB1. The expression level of GAS5 was significantly low in non-responder breast cancer tissues and in MCF-7/ADR cells, whereas expression level of ABCB1 was very high. Overexpression of GAS5 greatly increased the sensitivity and apoptosis of MCF-7/ADR cells to ADR. Conversely, ADR sensitivity and apoptosis were reduced by treatment with si-GAS5. Moreover, we assessed the impact of GAS5 on the efflux function of ABCB1 by Rh123 transport assay. GAS5 upregulation distinctly increased the intracellular accumulation of the substrate drug Rh123, whereas downregulation decreased compared to the cells transfected with NC. We further found that GAS5 upregulation completely suppressed ABCB1 expression. These findings highlight the importance of GAS5 dysfunction in the development of drug resistance.
Considerable research has shown that one of lncRNA’s functional mechanisms is the competing endogenous RNA (ceRNA)-miRNA-mRNA regulatory network, which maintains the activity and functional balance of gene circuitry in a cancer cell. Any disturbance of this system may trigger a series of cellar pathological processes. Hence, we used RNA-seq and online software (e.g., DIANA-LncBase, miRcode) to search for the potential target miRNAs of GAS5. miR-221-3p is regarded as a tumor suppressor in human cancers, including breast cancer.27,28 Interestingly, miR-221-3p is the top target miRNA when using RNA-seq analysis. Additionally, several conserved cognate sites between GAS5 and miR-221-3p existed, predicting that GAS5 might serve as a ceRNA of miR-221-3p. Li et al.29 revealed that the ceRNA network normally occurs in the cytoplasm. Our study verified that GAS5 was substantially enriched in the cytoplasmic fraction, showing that GAS5 had a chance to interact with cytoplasm miRNAs. Additionally, our data also indicated that the negative correlation between GAS5 and miR-221-3p was confirmed in ADR-resistant breast cancer tissues. In the subsequent luciferase reporter experiments, the results of RIP and quantitative real-time PCR assays indicated that GAS5 could suppress miR-221-3p expression through direct interaction. GAS5 overexpression could suppress the expression of miR-221-3p. Moreover, induced GAS5 increased the sensitivity and apoptosis of MCF-7/ADR cells to ADR, which was antagonized following the restoration of miR-221-3p expression. Knockdown of GAS5 by si-GAS5#1 decreased the sensitivity and apoptosis of MCF-7 cells to ADR, which was fortified after transfection with miR-221-3p inhibitors. All of these data provided evidence that GAS5 sensitized breast cancer cells to ADR via restraining miR-221-3p expression.
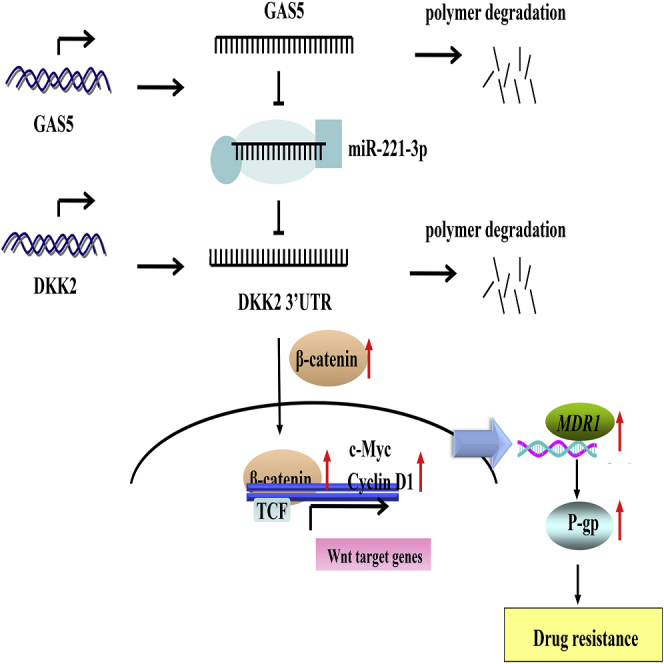
Next, we investigated the potential targets of miR-221-3p, and DKK2 was identified as a functional target in ADR-resistant breast cancer MCF-7/ADR cells. DKK2 is a member of the dickkopf family, which is identified as a secreted modulator of Wnt-β-catenin signaling by binding to the lipoprotein receptor-related protein 5/6 (LRP5/6) component of the Wnt receptor complex.30 DKK2 has been reported to be involved in drug resistance and promote tumor cell survival, proliferation, metastasis, and invasion.30, 31, 32 Remarkably, previous research found that DKK2 could stimulate tumor metastasis and angiogenesis via accelerating the aerobic glycolysis of human colorectal cancer cells, through a novel VEGF-independent, but energy metabolism-related pathway.30 Wang et al.31 showed that miR-221 functions as an oncomiR and controls 5-fluorouracil (5-FU) resistance in esophageal adenocarcinoma cancer by direct targeting of DKK2 expression via modulation of the Wnt/β-catenin-EMT pathway. In our study, DKK2 expression was decreased in non-responder breast cancer tissues and in MCF-7/ADR cells. In addition, GAS5 upregulation increased DKK2 expression whereas GAS5 knockdown repressed DKK2 expression. Additionally, GAS5 inhibited activation of the Wnt/β-catenin pathway. Moreover, the correlation between GAS5 and the miR-221-3p/DKK2 axis was determined by a dual-luciferase reporter assay. In the subsequent rescue experiments, we showed that GAS5 upregulation increased the expression of DKK2 and decreased the expressions of ABCB1 and the Wnt/β-catenin pathway, while these effects were attenuated following miR-221-3p overexpression or DKK2 inhibition. All of these results implied that GAS5 could reverse the ADR resistance by the miR-221-3p/DKK2 axis via the Wnt/β-catenin pathway in ADR-resistant breast cancer cells. In vivo experiments also confirmed that overexpression of GAS5 led to decreases in miR-221-3p and ABCB1 expression, as well as an increase in DKK2 expression, in resected tumors derived from MCF-7/ADR cells with or without ADR treatment. Taken together, all of these results led us to the conclusion that GAS5 could enhance ADR sensitivity in breast cancer cells via the miR-221-3p/DKK2 axis, at least in part through the Wnt/β-catenin pathway.
In conclusion, our study demonstrates the existence of a dual role played by GAS5 in ABCB1-mediated drug resistance of breast cancer. In particular, we highlight a novel ceRNA-miRNA-mRNA regulation mechanism in which GAS5 positively reverses the ABCB1-mediated ADR resistance via the miR-221-3p/DKK2 axis by repressing the Wnt/β-catenin pathway. Our present data provide strong evidence that the GAS5/miR-221-3p/DKK2 axis may be a promising chemosensitizing strategy for the treatment of breast cancer.
Materials and Methods
Patients and Specimens
Twenty-six biopsy-proven patients with invasive primary breast cancer treated with NAC at the First Affiliated Hospital of the University of Science and Technology of China from January 2016 to December 2018 were enrolled in this study. The diagnosis of each case was confirmed by pathologists based on World Health Organization (WHO) classification.
Patients were treated with four cycles of epirubicin (90 or 100 mg/m2) combined with cyclophosphamide (600 mg/m2), followed by four cycles of docetaxel (90 or 100 mg/m2). Each chemotherapy cycle is 3 weeks. All patients received modified radical mastectomy after NAC. The data, including age, primary tumor size, lymph nodes metastasis, TNM (tumor, node, metastasis) stage, histological grade, estrogen receptor (ER) status, progesterone receptor (PR) status, HER-2 status, and subtype, were collected from clinical and pathological records. The clinicopathological characteristics of the patients are summarized in Table 1. The clinical response was assessed by the decrease in tumor size and classified according to RECIST criteria.33 Thus, patients with complete response (CR) or partial response (PR) were regarded as clinical responders, while patients with stable disease (SD) or progressive disease (PD) were regarded as non-responders for further analysis.34,35 Fresh tumor samples were immediately frozen in liquid nitrogen and preserved at −80°C. The present study was approved by the Ethics Committee of Anhui Medical University, and all patients signed an informed consent.
Cell Culture and Transfection
The parental-sensitive human breast cell line MCF-7 and ADR-resistant cell line MCF-7/ADR were kind gifts from Prof. Tao Zhu (University of Science and Technology of China). The cells were cultured in RPMI 1640 medium (Biological Industries, Israel) containing 10% fetal calf serum (Biological Industries, Israel) and 1% penicillin-streptomycin solution at 37°C under a humidified atmosphere of 5% CO2. The MCF-7/ADR cells were cultured in the above-mentioned media that were additionally supplemented with 1 μg/mL ADR (KeyGen Biotech, Nanjing, China) to maintain the drug-resistant phenotype. The GAS5 overexpression plasmid pcDNA3.1-GAS5 (pcDNA-GAS5) was generated by inserting the full-length GAS5 sequence into the NheI and BamHI sites of pcDNA (Invitrogen, USA). All miRNA mimics (miR-221-3p NC [miR-NC]), miRNA inhibitors (anti-miR-221-3p, anti-miR-NC), and si-RNAs (si-NC, si-GAS5#1, and si-GAS5#2) were designed and synthesized by GenePharma (Generay, Shanghai, China). Transfection was performed using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Cells were harvested for further evaluation at 48 h after transfection. The sequences of oligonucleotides used are as follows: miR-221-3p mimics, 5′-AGCUACAUUGUCUGCUGGGUUUC-3′, 5′-AACCCAGCAGACAAUGUAGUCUUU-3′; miR-221-3p mimic NCs, 5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′; miR-221-3p antagomirs, 5′-CAAACCCAGCAGACAAUGUAGCU-3′; miR-221-3p antagomir NCs, 5′-CAGUACUUUUGUAGUACAA-3′.
RNA-Seq Analysis
lncRNAs, miRNAs, and mRNAs in the RNA samples were profiled using human RNA-seq analysis (Smartquerier Biomedicine, Shanghai, China). Differentially expressed lncRNAs, miRNAs, and mRNAs were identified by volcano plot filtering. Upregulated or downregulated lncRNAs were selected based on changes ≥3-fold thresholds and p <0.01.
Drug Sensitivity Assay
The ADR sensitivity of cells was detected using a CCK-8 (Sigma-Aldrich) assay by measuring the IC50 value (ADR concentration causing 50% decrease in absorbance compared with control). Briefly, cells were seeded into each well of a 96-well plate at 2 × 103 per well overnight and incubated with various concentrations of ADR (0.02, 0.1, 0.5, 2.5, 12.5, and 25 μg/mL for MCF-7, and 0.5, 1, 5, 25, 125, and 250 μg/mL for MCF-7/ADR). After incubation with ADR treatment, the cell viability was evaluated using a CCK-8 assay according to the manufacturer’s instructions.
Cell Proliferation Assay
Cell proliferation capacity was determined by using CCK-8 and colony formation assays. At 0, 24, 48, and 72 h after transfection, the CCK-8 solution was added to each well and incubated for 4 h. The absorbance was recorded at a wavelength of 450 nm using a SpectraMax i3x microplate reader (Molecular Devices, USA). With respect to the colony formation assay, cells were seeded in six-well plates and then cultured for 14 days in culture medium with 10% fetal bovine serum (FBS) as previously performed.36 Each assay was repeated three times.
Apoptosis Analysis
Twenty-four hours after the transfection as described above, ADR was added, with the final concentration of 80 μg/mL for MCF-7/ADR and 1 μg/mL for MCF-7. Forty-eight hours later, cell apoptosis was measured by an annexin V-allophycocyanin (APC)/7-aminoactinomycin D (7-AAD) apoptosis detection kit (KeyGen Biotech, China) according to the manufacturer’s protocols. Analyses were conducted using a BD LSR flow cytometer (BD Biosciences).
Intracellular Accumulation of Rh123
Accumulation of Rh123 by flow cytometry was carried out following modified methods as previously described.18 Briefly, the cells were transfected with GAS5 overexpression plasmid (pcDNA-GAS5) or GAS5 siRNA (si-GAS5#1) for 48 h, and then cells were collected and incubated with 5 μg/mL Rh123 at 37°C, 5% CO2 for 30 min. After incubation, the cells were gathered and washed three times with ice-cold PBS and then resuspended in 500 μL of PBS. Immediately, the mean fluorescence intensity of intracellular Rh123 was determined by a CytoFLEX flow cytometer (Beckman Coulter) with excitation/emission wavelengths of 488/525 nm.
Quantitative Real-Time PCR
Total RNA was extracted from breast cancer tissue samples or cell lines by using a RISO RNA isolation reagent (Biomics Biotechnologies, USA). The expression of miRNA was measured using an EzOmics miRNA qPCR detection primer set (Biomics Biotechnologies, USA) and EzOmics one-step qPCR kit (Biomics Biotechnologies, USA) in an Applied Biosystems 7500 instrument.
Total RNA was extracted by using TRIzol reagent (Invitrogen, USA). Reverse transcription was performed using a first-strand cDNA synthesis kit (Fermentas, USA) according to the manufacturer’s protocol. Quantitative real-time PCR analysis was conducted by using SYBR Green PCR master mix (QIAGEN, Germany). The relative expression of target genes was calculated by using the 2−ΔΔCT method and expressed as fold change relative to the respective control. The CT values were normalized using U6 or GAPDH as an internal control. Primer sequences are listed in Table S1. PCR was performed in triplicate.
Separation of Nuclear and Cytoplasmic Fractions
The cytoplasmic and nuclear fractions were isolated from cells using a cytoplasmic and nuclear RNA purification kit (Norgen Biotek, Thorold, ON, Canada) as previously described.37 Thus, the expression level of GAS5 in nuclear and cytoplasm fractions was detected by quantitative real-time PCR assays.
Western Blot Analysis
Cells were harvested and suspended in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, China) and quantified with a bicinchoninic acid (BCA) protein assay kit (Boster, Wuhan, China). After boiling in sample buffer, total proteins were separated by denaturing 8% or 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Subsequently, the membranes were then blocked and incubated with the following antibodies overnight at 4°C: anti-ABCB1 (Millipore, Merck, Germany), anti-β-catenin, anti-c-Myc, and anti-cyclin D1 (Cell Signaling Technology, USA), anti-DKK2 (Abcam, England), and anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) as a control. Horseradish peroxidase-conjugated anti-mouse, anti-rabbit antibodies were used as secondary antibodies correspondingly. The bands were visualized by chemiluminescence reagents (ChemiQ 4600, Bioshine).
Luciferase Reporter Assay
Putative WT and MUT miR-221-3p-binding sites in the 3ʹ UTR of GAS5 or DKK2 mRNA, termed GAS5-WT or GAS5-MUT and DKK2-WT or DKK2-MUT, were cloned into a pmirGLO-report luciferase vector (Promega, Fitchburg, WI, USA). The reporter plasmid was transiently transfected into MCF-7/ADR cells in the presence of miR-221-3p mimics, anti-miR-221-3p, and/or pcDNA-GAS5, si-GAS5#1. Forty-eight hours after transfection, relative luciferase activity was measured with the Dual-Luciferase reporter assay system (Promega, USA).
RIP Assay
RIP assays were performed using the EZ-Magna RIP kit (Millipore, Billerica, MA, USA) following the manufacturer’s protocol. Briefly, the cells were lysed with RIP lysis buffer and then the cell extract was incubated with RIP buffer containing magnetic beads, which were conjugated with human anti-Ago2 antibody (Millipore) and NC normal mouse IgG (Millipore). Samples were then incubated with proteinase K buffer with shaking. Finally, the RNAs in the immunoprecipitates were extracted, purified, and detected by quantitative real-time PCR.
In Vivo Chemosensitivity Assay
The protocol was approved by the Animal Care and Ethics Committee of Anhui Medical University. All experimental procedures were performed in accordance with the National Institutes of Health animal use guidelines. Female BALB/c nude mice (4 weeks old, 18–20 g) were purchased from Slake Jingda Laboratory Animal Company (Hunan, China) and housed under specific pathogen-free conditions. Approximately 2.0 × 107 MCF-7/ADR cells stably infected with pcDNA-GAS5 or pcDNA-control (NC) were subcutaneously injected into each mouse to form xenografts. When the tumor volume reached 50 mm3, ADR was intravenously injected via the tail at a dose of 5 mg/kg every 4 days for five times according to the indicated groups (n = 6 for each group): pcDNA-NC+PBS, pcDNA-NC+ADR, pcDNA-GAS5+PBS, and pcDNA-GAS5+ADR. Tumor growth was examined every 4 days, and tumor volumes were calculated with the following equation: volume = 0.5 × width2 × length. After 27 days, the mice were all sacrificed and the tumor masses were dissected, photographed, and subjected to statistical analyses.
Statistical Analysis
Data were expressed as the mean ± SD and analyzed using SPSS 19.0 statistics software (SPSS, USA). Significant differences among groups were determined by a Student’s t test or one-way ANOVA. Correlations between GAS5 and miR-221-3p were analyzed by Spearman rank correlation. The statistical significance was defined as *p < 0.05 or **p < 0.01. All experiments were performed at least three times.
Author Contributions
Study Design, A.S. and L.Z.; Drafting the Manuscript, Z.C.; Critical Revision of the Manuscript, D.J. and L.J.; Performance of Experiments, Z.C., T.P., and D.J.; Preparation and Assistant Work, Y.G. and X.F.; Necessary Technical Guides, L.Z. and X.F.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Prof. Tao Zhu for offering MCF-7 and MCF-7/ADR cell lines. This research was supported by the National Natural Science Foundation of China (81602344, 81603339, and 81803774); the China Postdoctoral Science Foundation (2019M662207); the Anhui Natural Science Fund Project (1908085QH363); and the Anhui Postdoctoral Science Foundation (2019B375).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.01.030.
Contributor Information
Aizong Shen, Email: anhuiaizongs@163.com.
Lei Zhang, Email: 76zhanglei@126.com.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Li Y., Gao X., Yu Z., Liu B., Pan W., Li N., Tang B. Reversing multidrug resistance by multiplexed gene silencing for enhanced breast cancer chemotherapy. ACS Appl. Mater. Interfaces. 2018;10:15461–15466. doi: 10.1021/acsami.8b02800. [DOI] [PubMed] [Google Scholar]
- 3.Chang L., Hu Z., Zhou Z., Zhang H. Linc00518 contributes to multidrug resistance through regulating the mir-199a/MRP1 axis in breast cancer. Cell. Physiol. Biochem. 2018;48:16–28. doi: 10.1159/000491659. [DOI] [PubMed] [Google Scholar]
- 4.Yao N., Fu Y., Chen L., Liu Z., He J., Zhu Y., Xia T., Wang S. Long non-coding RNA NONHSAT101069 promotes epirubicin resistance, migration, and invasion of breast cancer cells through NONHSAT101069/miR-129-5p/Twist1 axis. Oncogene. 2019;38:7216–7233. doi: 10.1038/s41388-019-0904-5. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z., Ma T., Huang C., Zhang L., Lv X., Xu T., Hu T., Li J. miR-27a modulates the MDR1/P-glycoprotein expression by inhibiting FZD7/β-catenin pathway in hepatocellular carcinoma cells. Cell. Signal. 2013;25:2693–2701. doi: 10.1016/j.cellsig.2013.08.032. [DOI] [PubMed] [Google Scholar]
- 6.Tian F., Dahmani F.Z., Qiao J., Ni J., Xiong H., Liu T., Zhou J., Yao J. A targeted nanoplatform co-delivering chemotherapeutic and antiangiogenic drugs as a tool to reverse multidrug resistance in breast cancer. Acta Biomater. 2018;75:398–412. doi: 10.1016/j.actbio.2018.05.050. [DOI] [PubMed] [Google Scholar]
- 7.Hu J., Wang Z., Shan Y., Pan Y., Ma J., Jia L. Long non-coding RNA HOTAIR promotes osteoarthritis progression via miR-17-5p/FUT2/β-catenin axis. Cell Death Dis. 2018;9:711. doi: 10.1038/s41419-018-0746-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong H., Hu J., Zou K., Ye M., Chen Y., Wu C., Chen X., Han M. Activation of lncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting microRNA-125b in breast cancer. Mol. Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Han J., Han B., Wu X., Hao J., Dong X., Shen Q., Pang H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol. Appl. Pharmacol. 2018;359:55–61. doi: 10.1016/j.taap.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Gao H., Hao G., Sun Y., Li L., Wang Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. OncoTargets Ther. 2018;11:8001–8012. doi: 10.2147/OTT.S172379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong H., Wang W., Mo S., Liu Q., Chen X., Chen R., Zhang Y., Zou K., Ye M., He X. Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J. Cell. Mol. Med. 2018;22:4935–4947. doi: 10.1111/jcmm.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong H., Wang W., Chen R., Zhang Y., Zou K., Ye M., He X., Zhang F., Han J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018;53:1013–1026. doi: 10.3892/ijo.2018.4467. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Schneider C., King R.M., Philipson L. Genes specifically expressed at growth arrest of mammalian cells. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 14.Arshi A., Sharifi F.S., Khorramian Ghahfarokhi M., Faghih Z., Doosti A., Ostovari S., Mahmoudi Maymand E., Ghahramani Seno M.M. Expression analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in breast cancer tissues from young women and women over 45 years of age. Mol. Ther. Nucleic Acids. 2018;12:751–757. doi: 10.1016/j.omtn.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mourtada-Maarabouni M., Pickard M.R., Hedge V.L., Farzaneh F., Williams G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 16.Gu J., Wang Y., Wang X., Zhou D., Shao C., Zhou M., He Z. Downregulation of lncRNA GAS5 confers tamoxifen resistance by activating miR-222 in breast cancer. Cancer Lett. 2018;434:1–10. doi: 10.1016/j.canlet.2018.06.039. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z., Zhu Z., Watabe K., Zhang X., Bai C., Xu M., Wu F., Mo Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z., Huang C., Ma T., Jiang L., Tang L., Shi T., Zhang S., Zhang L., Zhu P., Li J. Reversal effect of quercetin on multidrug resistance via FZD7/β-catenin pathway in hepatocellular carcinoma cells. Phytomedicine. 2018;43:37–45. doi: 10.1016/j.phymed.2018.03.040. [DOI] [PubMed] [Google Scholar]
- 19.Ye K., Wang S., Zhang H., Han H., Ma B., Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J. Cell. Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 20.Xiang S., Dauchy R.T., Hoffman A.E., Pointer D., Frasch T., Blask D.E., Hill S.M. Epigenetic inhibition of the tumor suppressor ARHI by light at night-induced circadian melatonin disruption mediates STAT3-driven paclitaxel resistance in breast cancer. J. Pineal Res. 2019;67:e12586. doi: 10.1111/jpi.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perone Y., Farrugia A.J., Rodríguez-Meira A., Győrffy B., Ion C., Uggetti A., Chronopoulos A., Marrazzo P., Faronato M., Shousha S. SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ERα breast cancer. Nat. Commun. 2019;10:2115. doi: 10.1038/s41467-019-09676-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Díaz-Rodríguez E., Pérez-Peña J., Ríos-Luci C., Arribas J., Ocaña A., Pandiella A. TRAIL receptor activation overcomes resistance to trastuzumab in HER2 positive breast cancer cells. Cancer Lett. 2019;453:34–44. doi: 10.1016/j.canlet.2019.03.042. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Du Z., Pan S., Shi M., Li J., Yang C., Hu H., Qiao M., Chen D., Zhao X. Overcoming multidrug resistance by codelivery of MDR1-targeting siRNA and doxorubicin using EphA10-mediated pH-sensitive lipoplexes: in vitro and in vivo evaluation. ACS Appl. Mater. Interfaces. 2018;10:21590–21600. doi: 10.1021/acsami.8b01806. [DOI] [PubMed] [Google Scholar]
- 24.Liu B., Wu S., Ma J., Yan S., Xiao Z., Wan L., Zhang F., Shang M., Mao A. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol. Ther. Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao T., Lu R., Zhang J., Fang X., Fan L., Huang C., Lin R., Lin Z. Growth arrest-specific 5 attenuates cisplatin-induced apoptosis in cervical cancer by regulating STAT3 signaling via miR-21. J. Cell. Physiol. 2019;234:9605–9615. doi: 10.1002/jcp.27647. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z., Shi T., Zhang L., Zhu P., Deng M., Huang C., Hu T., Jiang L., Li J. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett. 2016;370:153–164. doi: 10.1016/j.canlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Deng L., Lei Q., Wang Y., Wang Z., Xie G., Zhong X., Wang Y., Chen N., Qiu Y., Pu T. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017;8:108712–108725. doi: 10.18632/oncotarget.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ergun S., Tayeb T.S., Arslan A., Temiz E., Arman K., Safdar M., Dağlı H., Korkmaz M., Nacarkahya G., Kırkbeş S., Oztuzcu S. The investigation of miR-221-3p and PAK1 gene expressions in breast cancer cell lines. Gene. 2015;555:377–381. doi: 10.1016/j.gene.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Li G.Y., Wang W., Sun J.Y., Xin B., Zhang X., Wang T., Zhang Q.F., Yao L.B., Han H., Fan D.M. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8:2846–2861. doi: 10.7150/thno.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng F., Zhou R., Lin C., Yang S., Wang H., Li W., Zheng K., Lin W., Li X., Yao X. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics. 2019;9:1001–1014. doi: 10.7150/thno.30056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Zhao Y., Herbst A., Kalinski T., Qin J., Wang X., Jiang Z., Benedix F., Franke S., Wartman T. miR-221 mediates chemoresistance of esophageal adenocarcinoma by direct targeting of DKK2 expression. Ann. Surg. 2016;264:804–814. doi: 10.1097/SLA.0000000000001928. [DOI] [PubMed] [Google Scholar]
- 32.Xiao Q., Wu J., Wang W.J., Chen S., Zheng Y., Yu X., Meeth K., Sahraei M., Bothwell A.L.M., Chen L. DKK2 imparts tumor immunity evasion through β-catenin-independent suppression of cytotoxic immune-cell activation. Nat. Med. 2018;24:262–270. doi: 10.1038/nm.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Therasse P., Arbuck S.G., Eisenhauer E.A., Wanders J., Kaplan R.S., Rubinstein L., Verweij J., Van Glabbeke M., van Oosterom A.T., Christian M.C., Gwyther S.G. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J. Natl. Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 34.Paik J.H., Jang J.Y., Jeon Y.K., Kim W.Y., Kim T.M., Heo D.S., Kim C.W. MicroRNA-146a downregulates NFκB activity via targeting TRAF6 and functions as a tumor suppressor having strong prognostic implications in NK/T cell lymphoma. Clin. Cancer Res. 2011;17:4761–4771. doi: 10.1158/1078-0432.CCR-11-0494. [DOI] [PubMed] [Google Scholar]
- 35.Chintamani S., Singh J.P., Mittal M.K., Saxena S., Bansal A., Bhatia A., Kulshreshtha P. Role of p-glycoprotein expression in predicting response to neoadjuvant chemotherapy in breast cancer—a prospective clinical study. World J. Surg. Oncol. 2005;3:61. doi: 10.1186/1477-7819-3-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song X., Zhang X., Wang X., Chen L., Jiang L., Zheng A., Zhang M., Zhao L., Wei M. lncRNA SPRY4-IT1 regulates breast cancer cell stemness through competitively binding miR-6882-3p with TCF7L2. J. Cell. Mol. Med. 2020;24:772–784. doi: 10.1111/jcmm.14786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaghlool A., Ameur A., Nyberg L., Halvardson J., Grabherr M., Cavelier L., Feuk L. Efficient cellular fractionation improves RNA sequencing analysis of mature and nascent transcripts from human tissues. BMC Biotechnol. 2013;13:99. doi: 10.1186/1472-6750-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.