Abstract
Quantitative structure–activity relationships (QSAR) provides a model that link biological activities of compounds to thier chemical stuctures and molecular docking study reveals the interaction between drug and its target enzyme. These studies were conducted on 1,3-dioxoisoindoline-4-aminoquinolines with the aim of producing a model that could be used to design highly potent antiplasmodium. The compounds were first optimized using Density Functional Theory (DFT) with basis set B3LYP/6-31G∗ then their descriptors calculated. Genetic Function Algorithm (GFA) was used to select descriptors and build the model. One of the four models generated was found to be the best having internal and external squared correlation coefficient (R2) of 0.9459 and 0.7015 respectively, adjusted squared correlation coefficient (Radj) of 0.9278, leave-one-out (LOO) cross-validation coefficient (Q2cv) of 0.8882. The model shows that antiplasmodial activities of 1,3-dioxoisoindoline-4-aminoquinolines depend on ATSC5i, GATS8p, minHBint3, minHBint5, MLFER_A and topoShape descriptors. The model was validated to be predictive, robust and reliable. Hence, it can predict the antiplasmodium activities of new 1,3-dioxoisoindoline-4-aminoquinolines.The docking result indicates strong binding between 1,3-dioxoisoindoline-4-aminoquinolines and Plasmodium falciparum lactate dehydrogenase (pfLDH), and revealed the important of the morpholinyl substituent and amide linker in inhibiting pfLDH. These results could serve as a model for designing novel 1,3-dioxoisoindoline-4-aminoquinolines as inhibitors of PfLDH with higher antiplasmodial activities.
Keywords: Physical chemistry, Theoretical chemistry, Pharmaceutical chemistry, Antiplasmodium, QSAR, Molecular docking, Plasmodium falciparum lactate dehydrogenase, Hybrid compounds
Physical chemistry; Theoretical chemistry; Pharmaceutical chemistry, Antiplasmodium; QSAR; Molecular docking; Plasmodium falciparum lactate dehydrogenase; Hybrid compounds.
1. Introduction
Malaria is an infection with high morbidity and mortality burden all over world which is endemic in African region. Data from the World Health Organization malaria report 2019 revealed the occurrence of an estimate of 228 million cases and 405 000 deaths from the infection worldwide in 2018. Nigeria has 25% and 19% of the global cases and deaths respectively, follow by Democratic Republic of Congo with 11% each of the global cases and deaths. The most vulnerable are children below 5 years of age responsible for 67% (585 000) of the global malaria deaths in 2018 (WHO, 2019). Plasmodium falciparum is the most deadly of the five species of malaria parasite known to infect human (Cohen et al., 2012; WHO, 2019).
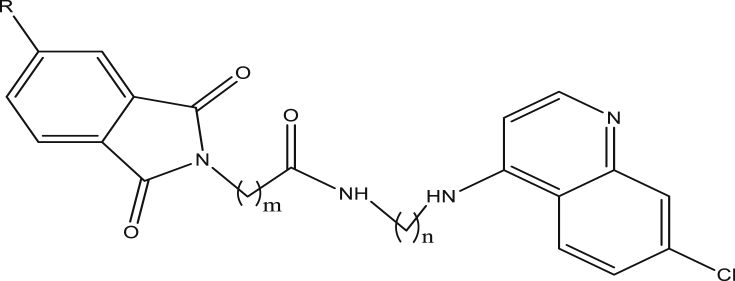
Quinoline present in several antimalarial drugs such as Chloroquine, Amodiaquine, Mefloquine, Primaquine, Ferroquine, etc. has been considered as the most important moiety that impact antimalarial action out of many heterocycle pharmacophores known for treatment of malaria (Kumar et al., 2016; Ilhami et al., 2010). Chloroquine has been the most effective antimalaria for decades, but the wide spread of its resistance led to the development of Artemisinin-based Combination Therapy (ACTs), the WHO recommended drug for treatment of uncomplicated malaria (Beteck et al., 2014). ACTs are drug combinations containing artemisinin derivatives and other antimalarials such as quinoline compounds. Resistance to ACTs was reported in some part of the world which jeopardized their future effectiveness hence, the need for promising antimalaria that will meet the challenge of eradicating the disease (Burrows et al., 2011; Aguiar et al., 2012). Alternative to ACTs are hybrid compounds which have advantage of hitting two or more molecular targets (Oliveira et al., 2015). Earlier this year, Rania and coworkers reported the synthesis of 1,3-dioxoisoindoline-4-aminoquinolines (Figure 1) as potent and noncytotoxic hybrid compounds having excellent antiplasmodial activities against W2 strain of P. falciparum (Rania et al., 2019).
Figure 1.
1,3-dioxoisoindoline-4-aminoquinolines.
Conventional drug discovery methods are expensive and time-consuming, hence, the need for more effective methods in terms of time and resources (Jitender et al., 2010). Discovery of novel drug candidates require efficient and vigorous methods that can screen chemical databases against molecules with known biological activities (Tropsha, 2010). QSAR and molecular docking studies have been successfully deployed in the discovery and design of many drugs for their time and cost effectiveness (Talele et al., 2010). This work aim at applying these techniques to reveal the connection between chemical structures of 1,3-dioxoisoindoline-4-aminoquinolines and their antiplasmodial activities so as to come up with a model that could be used to design highly potent antimalaria.
2. Materials and method
2.1. Data collection
36 compounds of 1,3-dioxoisoindoline-4-aminoquinolines (Figure 1) and their antiplasmodial activities against W2 strain of Plasmodium falciparum were obtained from the work of Rania and co-workers (Rania et al., 2019) and used in this research. The antiplasmodial activities of the compounds were gotten as IC50 (nM) and converted to pIC50 {pIC50 = -logIC50 (M)} for the purpose of this work. The compounds with their respective activities were presented in Table 1.
Table 1.
Compounds of Figure 1 with their antiplasmodial activities.
| Compounds | R | m | N | IC50(nM) |
|---|---|---|---|---|
| 1 | H | 1 | 2 | 5.65 |
| 2 | H | 1 | 4 | 3.66 |
| 3 | H | 1 | 6 | 3.35 |
| 4 | H | 1 | 8 | 0.24 |
| 5 | H | 2 | 2 | 0.30 |
| 6 | H | 2 | 4 | 0.41 |
| 7 | H | 2 | 6 | 0.29 |
| 8 | H | 2 | 8 | 0.15 |
| 9 | H | 3 | 2 | 0.28 |
| 10 | H | 3 | 4 | 0.84 |
| 11 | H | 3 | 6 | 1.57 |
| 12 | H | 3 | 8 | 0.58 |
| 13 | F | 1 | 2 | 1.99 |
| 14 | F | 1 | 4 | 1.05 |
| 15 | F | 1 | 6 | 1.49 |
| 16 | F | 2 | 2 | 0.36 |
| 17 | F | 2 | 4 | 0.22 |
| 18 | F | 2 | 6 | 0.66 |
| 19 | Morpholinyl | 1 | 2 | 3.21 |
| 20 | Morpholinyl | 1 | 4 | 1.28 |
| 21 | Morpholinyl | 1 | 6 | 1.78 |
| 22 | Morpholinyl | 2 | 2 | 0.13 |
| 23 | Morpholinyl | 2 | 4 | 0.22 |
| 24 | Morpholinyl | 2 | 6 | 0.64 |
| 25 | Diethylamino | 1 | 2 | 1.14 |
| 26 | Diethylamino | 1 | 4 | 1.17 |
| 27 | diethylamino | 1 | 6 | 1.22 |
| 28 | diethylamino | 2 | 2 | 0.097 |
| 29 | diethylamino | 2 | 4 | 0.26 |
| 30 | diethylamino | 2 | 6 | 0.53 |
| 31 | 2-(piperazin-1-yl)ethan-1-ol | 1 | 2 | 1.04 |
| 32 | 2-(piperazin-1-yl)ethan-1-ol | 1 | 4 | 0.86 |
| 33 | 2-(piperazin-1-yl)ethan-1-ol | 1 | 6 | 0.46 |
| 34 | 2-(piperazin-1-yl)ethan-1-ol | 2 | 2 | 0.28 |
| 35 | 2-(piperazin-1-yl)ethan-1-ol | 2 | 4 | 0.14 |
| 36 | 2-(piperazin-1-yl)ethan-1-ol | 2 | 6 | 0.18 |
2.2. Geometric optimization
The 2D structures of the molecules shown in Table 1 were drawn using Chemdraw version 12.0.2 software (Li et al., 2004), converted to 3D and optimized using Spartan 14 Version 1.1.4 software (using B3LYP functional and 6-31G basis set) (Becke, 1993) (see Supplementary file Table 1 for 2D and 3D structures).
2.3. Molecular descriptors calculation
A total of 1875 molecular descriptors of the optimized molecules of 1,3-dioxoisoindoline-4-aminoquinolines derivatives were computed with PaDEL-Descriptor software version 2.20 (Yap, 2011).
2.4. Normalization and data pretreatment
Using Eq. (1), the computed descriptors were normalized so that each variable will have equal opportunity in construction of the model (Singh, 2013).
| (1) |
where X is the normalized descriptors, Xi is the descriptor's value for each molecule, Xmin and Xmax are minimum and maximum values for each descriptor. Data Pretreatment software of Drug Theoretical and Cheminformatics Laboratory (DTC Lab) was used to eliminate redundancy in the normalized data.
2.5. Data Division
Kennard-Stone's algorithm was employed to divide the data into training set and test set (Kenard and Stone, 1969) using Data Division software of DTC Lab. Training set (70%) was used in building the model while test set (30%) in validating the model.
2.6. Model generation
Regression analysis of the training set was carried out using Genetic Function Approximation (GFA) technique in Material Studio software to generate the model. Activities in pIC50 were taken as the dependent variable and the descriptors independent variables.
2.7. Internal validation of the model generated
The model generated was assessed using Friedman formula (Friedman, 1991) defined as;
| (2) |
LOF is the Friedman's Lack of fit which is a measure of fitness of a model, SEE is the standard error of estimation, p is the total number of descriptors in the model, d is the user-defined smoothing parameter, c is the number of terms in the model and M is the number of compounds in the training set.
SEE is defined as;
| (3) |
which is the same as the standard deviation of the model and its value has to be low for a model to be good.
Correlation coefficient, R2 of a built model is another parameter considered and the closer it is to 1.0, the better the model built. R2 is expressed as;
| (4) |
where Yprd, Yexp and Ymtrn are the predicted, experimental and mean experimental activities in the training set, respectively.
The stability of a model is not reliable on the value of R2 as it is directly proportional to the number of descriptors in the model. Thus, for a reliable and stable model, R2 is adjusted according to the expression:
| (5) |
where p is the number of descriptors in the model and n number of compounds used in training set.
The cross-validation coefficient, Q2cv expressed as:
| (6) |
where Yprd, Yexp and Ymtrn are the predicted, experimental and average experimental activity in the training set, respectively.
2.8. External validation of the model generated
Test set was used in validating the generated model externally by assessing the value of R2pred expressed as;
| (7) |
where Yprd and Yexp are respectively the predicted and experimental activities of the test set and Ymtrn the mean experimental activity of the training set. The closer the value is to 1.0, the better the model generated (Tropsha et al., 2003).
2.9. Y-Randomization test
Random Multi-Linear regression models are generated (using training set) in Y-randomization test whose R2 and Q2 values have to be low for the QSAR model to be robust (Tropsha et al., 2003). Coefficient of determination, cR2p, whose value has to be greater than 0.5 for passing this test is also calculated in the Y-randomization test and is expressed as;
| (8) |
where R is the correlation coefficient for Y-randomization and R2r is the average ‘R’ of the random models.
2.10. Applicability domain of the generated model
Leverage (hi) method was used in describing the applicability domain of the QSAR models (Veerasamy et al., 2011) and for a chemical compound is expressed as;
| (9) |
where Xi is training compounds matrix of i. X is the n x k descriptor matrix of the training set compound and XT is the transpose matrix of X used to generate the model. The warning leverage, h∗ is the maximum value for X and is expressed as;
| (10) |
where n is the number of training compounds and p is the number of descriptors in the model.
2.11. Descriptors analyses
Correlation analysis was carried out to check the presence of correlation among the descriptors in the built model. This was further evaluated by computing the variance inflation factor (VIF) for each descriptor using the equation below (see supplementary file Table 2 for the values of descriptors used in the model):
| (11) |
VIFi is the variance inflation factor for a given descriptor i in the model R2ij is the correlation coefficient of the multiple regression between the descriptor i and the remaining j descriptors in the model (Beheshti et al., 2016).
The relative significance and influence of every descriptor in the model was estimated by its mean effect (MF) value. The MFj for a given descriptor j in a model is given as;
| (12) |
where bj is the coefficient of descriptor j in the model, dij is the value of the descriptor j in the descriptor matrix for each molecule in the training set, m is the number of descriptors that appear in the model, and n is the number of molecule that made up the training set (Habibi-Yangjeh and Danandeh-Jenagharad, 2009).
2.12. Docking study
Plasmodium falciparum lactate dehydrogenase (PfLDH) enzyme is regarded as a potential molecular target for antimalarials because of the parasite's reliance on glycolysis to produce energy (Penna-Coutinho et al., 2011). Therefore, molecular docking study was conducted between PfLDH and 1,3-dioxoisoindoline-4-aminoquinolines to investigate their interaction. Crystal structure of PfLDH was obtained from protein data bank (PDB ID: 1CET). Ten compounds with best activities were prepared as the ligands for this study. PfLDH was prepared as the receptor with the aid of Discovery Studio software. The prepared receptor and ligands were docked using Autodock Vina in Pyrx software (Trott and Olson, 2010). Discovery Studio Visualizer was use to analyze the results.
3. Result and discussion
Four QSAR models were built using GFA in material studio software to study how the biological activities of 1,3-dioxoisoindoline-4-aminoquinolines as potent antiplasmodium relate with their chemical structures. One of the QSAR models generated was selected for its statistical significance and reported herein as follow:
Table 2 presents the validation parameters of the model which passed the thresholds required for a QSAR model to be accepted (Veerasamy et al., 2011; Tropsha, 2010).
Table 2.
Validation parameters for the selected model.
| S/N | Parameter | Value |
|---|---|---|
| 1 | Friedman LOF | 0.083 |
| 2 | R2train | 0.946 |
| 3 | Adjusted R-squared | 0.928 |
| 4 | Cross-validated R-squared (Q2cv) | 0.888 |
| 5 | Significant regression | Yes |
| 6 | Significance-of-regression F-value | 48.26 |
| 7 | Critical SOR F-value (95%) | 2.672 |
| 8 | Replicate points | 0 |
| 9 | Computed experimental error | 0 |
| 10 | Lack-of-fit points | 18 |
| 11 | Min expt. error for nonsignificant LOF (95%) | 0.103 |
| 12 | R2test | 0.702 |
The model contained 2D Autocorrelation descriptors ATSC5i and GATS8p which described how the considered properties (first ionization potential and atomic polarizabilities) are distributed along the topological structures of the molecules. ATSC5i is defined as centered Broto-Moreau autocorrelation of lag 5 weighted by first ionization potential and it present in the model related the first ionization potential of pairs atoms that are separated by five bonds (lag 5) with the antiplasmodial activities of 1,3-dioxoisoindoline-4-aminoquinolines, while GATS8p is defined as Geary autocorrelation of lag 8 weighted by atomic polarizabilities and it present in the model associated the presence of polarizable pairs of atoms eight bonds apart on the antiplasmodial activities the compounds. The model also contained 2D atom type electro-topological state descriptors minHBint3 and minHBint5 which are defined as Minimum E-State descriptors of strength for potential Hydrogen Bonds of path length 3 (minHBint3) and of path length 5 (minHBint5). These descriptors indicated the importance of hydrogen bonds of path length 3 and 5 (Arthur et al., 2016). Other descriptors in the model were Overall or summation solute hydrogen bond acidity, MLFER_A which is related with molecular linear free energy relations (Platts et al., 1999) and Petitjean topological shape index, topoShape.
Table 3 shows the residual values between the experimental and predicted activities of 1,3-dioxoisoindoline-4-aminoquinolines as potent Plasmodium falciparum inhibitors. The low residual values were indicative of the high predictability of the model.
Table 3.
Experimental and predicted pIC50 of 1,3-dioxoisoindoline-4-aminoquinolines with their residuals.
| Compounds | Experimental pIC50 | Predicted pIC50 | Residual |
|---|---|---|---|
| 1 | 5.248 | 5.331 | -0.083 |
| 2a | 5.437 | 5.523 | -0.087 |
| 3 | 5.475 | 5.523 | -0.048 |
| 4a | 6.62 | 6.355 | 0.2644 |
| 5 | 6.523 | 6.716 | -0.193 |
| 6a | 6.387 | 6.777 | -0.39 |
| 7 | 6.538 | 6.434 | 0.1035 |
| 8 | 6.824 | 6.914 | -0.09 |
| 9 | 6.553 | 6.518 | 0.0352 |
| 10a | 6.076 | 5.909 | 0.1669 |
| 11 | 5.804 | 5.746 | 0.0581 |
| 12a | 6.237 | 6.236 | 0.0007 |
| 13 | 5.701 | 5.655 | 0.0459 |
| 14 | 5.979 | 5.8 | 0.179 |
| 15 | 5.827 | 6.006 | -0.179 |
| 16 | 6.444 | 6.447 | -0.003 |
| 17a | 6.658 | 6.526 | 0.1317 |
| 18 | 6.18 | 6.204 | -0.024 |
| 19 | 5.493 | 5.57 | -0.077 |
| 20a | 5.893 | 5.53 | 0.363 |
| 21a | 5.75 | 5.796 | -0.046 |
| 22 | 6.886 | 6.793 | 0.0932 |
| 23 | 6.658 | 6.634 | 0.0238 |
| 24 | 6.194 | 6.342 | -0.148 |
| 25 | 5.943 | 5.998 | -0.055 |
| 26a | 5.932 | 5.912 | 0.0194 |
| 27a | 5.914 | 6.147 | -0.233 |
| 28 | 7.013 | 6.726 | 0.287 |
| 29 | 6.585 | 6.566 | 0.0193 |
| 30 | 6.276 | 6.286 | -0.011 |
| 31 | 5.983 | 5.867 | 0.1161 |
| 32a | 6.066 | 5.858 | 0.2075 |
| 33 | 6.337 | 6.202 | 0.1356 |
| 34 | 6.553 | 6.644 | -0.091 |
| 35 | 6.854 | 6.942 | -0.088 |
| 36 | 6.745 | 6.751 | -0.006 |
Test set.
Table 4 presents the result of statistical analysis of the descriptors. The low values of the inter-correlation between any pair of descriptors as shown by the Pearson's correlation matrix suggest no significant inter-correlation among the descriptors used in building the model. This was confirmed by the low value of the Variance Inflation Factor which was less than 10 for every descriptor. Hence, the descriptors used in building the model were good. The mean effects of the descriptors gave the relative influences of the descriptors on the antiplasmodial activities of the compounds where the signs and the magnitudes indicated the directions and extent of the influences respectively. The MF values suggested that the descriptors ATSC5i, minHBint3, minHBint5 and MLFER_A decrease the antimalarial activities of 1,3-dioxoisoindoline-4-aminoquinolines with increase in their values while the descriptors GATS8p and topoShape increase the antimalarial activities of 1,3-dioxoisoindoline-4-aminoquinolines with increase in their values. The magnitude of the MF values shows that the descriptor GATS8p has greatest influence on the antiplasmodial activities of the compounds and should be highly considered when designing more potent 1,3-dioxoisoindoline-4-aminoquinolines. The descriptor ATSC5i had the least influence on the antiplasmodial activities of the compounds as shown by the magnitude of its MF.
Table 4.
Pearson's correlation, Variance Inflation Factor (VIF) and Mean Effect (MF) of descriptors used in the selected model.
| Descriptor |
Inter-correlation |
VIF | MF | |||||
|---|---|---|---|---|---|---|---|---|
| ATSC5i | GATS8p | minHBint3 | minHBint5 | MLFER_A | topoShape | |||
| ATSC5i | 1 | 1.391 | -0.005 | |||||
| GATS8p | -0.1181 | 1 | 2.994 | 0.983 | ||||
| minHBint3 | -0.3422 | 0.3350 | 1 | 1.500 | -0.010 | |||
| minHBint5 | -0.4024 | 0.6021 | 0.3330 | 1 | 5.148 | -0.020 | ||
| MLFER_A | 0.1375 | 0.1420 | 0.1317 | -0.5053 | 1 | 2.704 | -0.110 | |
| topoShape | -0.0275 | -0.0451 | 0.2254 | -0.2468 | 0.2461 | 1 | 1.221 | 0.166 |
The reliability, robustness and stability of the built QSAR model were confirmed by Y- Randomization test. This was true for the low R2 and Q2 values for several trials in the result presented in Table 5. The cR2p value which was greater than 0.5 also shows the model is good and not gotten by chance.
Table 5.
Y-Randomization test result.
| Model | R | R2 | Q2 |
|---|---|---|---|
| Original | 0.9726 | 0.9459 | 0.8883 |
| Random 1 | 0.4172 | 0.1741 | -0.7131 |
| Random 2 | 0.4080 | 0.1665 | -0.6273 |
| Random 3 | 0.6915 | 0.4782 | 0.0379 |
| Random 4 | 0.4669 | 0.2180 | -0.4743 |
| Random 5 | 0.6025 | 0.3631 | -0.2769 |
| Random 6 | 0.3653 | 0.1335 | -0.6688 |
| Random 7 | 0.4835 | 0.2338 | -0.6283 |
| Random 8 | 0.5267 | 0.2774 | -0.4792 |
| Random 9 | 0.2620 | 0.0686 | -0.8088 |
| Random 10 | 0.6430 | 0.4135 | -0.3519 |
| Random Models Parameters | |||
| Average r: | 0.4867 | ||
| Average r2: | 0.2526 | ||
| Average Q2: | -0.4990 | ||
| cRp2: | 0.8189 | ||
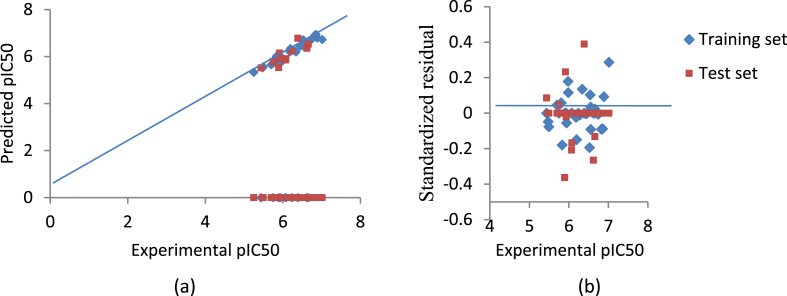
The linearity of the plot of predicted activities against experimental activities of the compounds (Figure 2a) shows the strength of the model in predicting antiplasmodial activities of the 1,3-dioxoisoindoline-4-aminoquinolines. The dispersal of standardized residuals of both training and test set on both sides of zero (Figure 2b) suggested no systematic error in the generated model (Jalali-Heravi and Kyani, 2004).
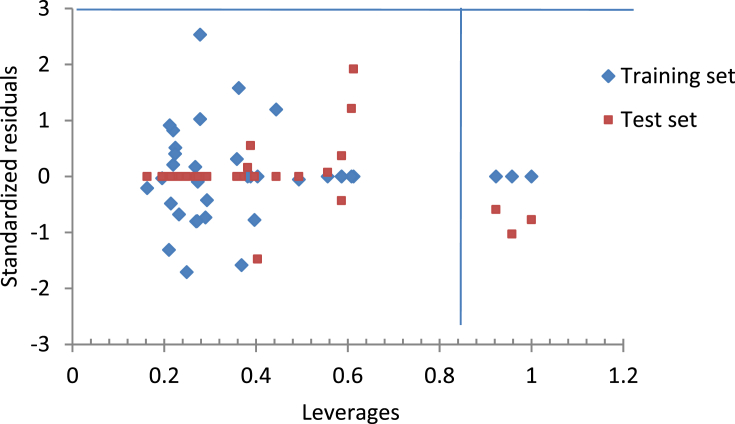
The Williams plot presented by Figure 3 shows no outliers in the compounds but three influential compounds because their leverages were greater than the warning leverage (h∗ = 0.84) which might arose from their structural feature. All the compounds were within the applicability domain except the influential compounds which were all from test set (compound 4, 10 and 32). These compounds should not be considered when designing novel 1,3-dioxoisoindoline-4-aminoquinolines.
Figure 2.
(a) Plot of predicted activities against experimental activities (b) Plot of Standardized residuals against experimental activities of the compounds.
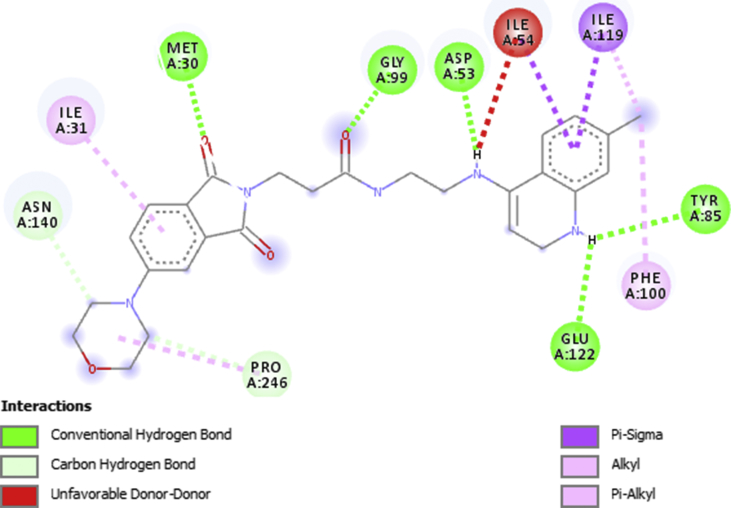
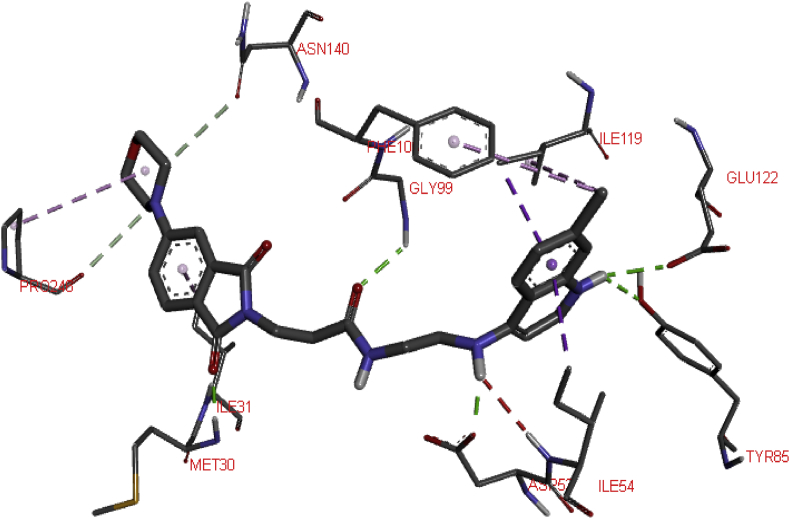
The result of the molecular docking study conducted between Plasmodium falciparum lactate dehydrogenase, PfLDH (receptor) and ten 1,3-dioxoisoindoline-4-aminoquinolines with highest activities (ligands) is presented in Table 3 of the supplementary file. All the docked ligands interacted strongly with the active sites of the receptor with binding affinity ranging from -6.7 to -9.2 kcal/mol. They all exhibited significant hydrogen bonding and hydrophobic interaction with the amino acids of the protein. Figures 4and 5 presented respectively the 3D and 2D 22-pfLDH interactions which had the best binding affinity (-9.2 kcal/mol). Five conventional hydrogen bond were formed between one of the C=O of the 1,3-dioxoisoindoline moiety and residue MET30, NH of the quinoline moiety and residue GLU122 and TYR85, C=O of the amide linker and residue GLY99 and NH of the amino group and residue ASP53. Two Carbon hydrogen bonds type were formed between two carbons of the morpholinyl substituent on the 1,3-dioxoisoindoline moiety and C=O of PRO246 and ASN140 of the target. The ligand also formed Hydrophobic interactions with the receptor via Pi-Sigma type with ILE54 and ILE119, Alkyl-Alkyl type with PRO246 and ILE119 and Pi-Alkyl type with PHE100 and ILE31 of the target. Unfavorable donor-donor interaction was also observed between the H of the amino group of the ligand H of the ILE54 of the receptor. The docked compounds have better binding affinity and interactions than chloroquine (a well known inhibitor of pfLDH).
Figure 5.
2D interaction between Ligand 22 and pfLDH.
Figure 3.
Plot of the standardized residuals against the leverages (Williams plot).
Figure 4.
3D interaction between Ligand 22 and pfLDH.
4. Conclusion
QSAR and molecular docking techniques were successfully applied on 1,3-dioxoisoindoline-4-aminoquinolines as potent antiplasmodial in order to produce a model that relates the chemical structures of the molecules and their antiplasmodial activities. A predictable, reliable and robust model was generated using Genetic Function Approximation (GFA) technique in Material Studio software. The model has internal and external validation R2 values of 0.9459 and 0.701544 respectively. The model revealed that the antiplasmodial activities of 1,3-dioxoisoindoline-4-aminoquinolines is influence by ATSC5i, GATS8p, minHBint3, minHBint5, MLFER_A and topoShape descriptors with greatest and least effect from GATS8p and ATSC5i respectively. The docking study revealed the important of some strategic point on the structural features of the molecules in inhibiting Plasmodium falciparum lactate dehydrogenase (PfLDH). This information could be used together with the built model to design novel 1,3-dioxoisoindoline-4-aminoquinolines inhibitors of PfLDH with higher antiplasmodial activities.
Declarations
Author contribution statement
Aliyu Wappah Mahmud: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Gideon Adamu Shallangwa: Conceived and designed the experiments; Performed the experiments; Wrote the paper.
Adamu Uzairu: Conceived and designed the experiments; Performed the experiments.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors are thankful to Muhammad Tukur Ibrahim for his help.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- Aguiar A.C.C., Rocha E.M., Souza N.B., França T.C., Krettli A.U. New approaches in antimalarial drug discovery and development: a review. Mem. Inst. Oswaldo Cruz. 2012;107:831−–845. doi: 10.1590/s0074-02762012000700001. [DOI] [PubMed] [Google Scholar]
- Arthur D.E., Uzairu A., Mamza P., Abechi S. Quantitative structure–activity relationship study on potent anticancer compounds against MOLT-4 and P388 leukemia cell lines. J. Adv. Res. 2016;7:823–837. [Google Scholar]
- Becke A.D. Becke’s three parameter hybrid method using the LYP correlation functional. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- Beheshti A., Pourbasheer E., Nekoei M., Vahdani S. QSAR modeling of antimalarial activity of urea derivatives using genetic algorithm–multiple linear regressions. J. Saudi Chem. Soc. 2016;20:282–290. [Google Scholar]
- Beteck R.M., Smit F.J., Haynes R.K., N’Da D.D. Recent progress in the development of anti- malarial quinolones. Malar. J. 2014;13:339. doi: 10.1186/1475-2875-13-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows J.N., Leroy D., Lotharius J., Waterson D. Challenges in antimalarial drug discovery. Future Med. Chem. 2011;3:1401–1412. doi: 10.4155/fmc.11.91. [DOI] [PubMed] [Google Scholar]
- Cohen J.M., Smith D.L., Cotter C., Ward A., Yamey G., Sabot O.J., Moonen B. Malaria resurgence: a systematic review and assessment of its causes. Malar. J. 2012;11:122. doi: 10.1186/1475-2875-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J.H. Multivariate adaptive regression splines. Ann. Stat. 1991;1–67 [Google Scholar]
- Habibi-Yangjeh A., Danandeh-Jenagharad M. Application of a genetic algorithm and an artificial neural network for global prediction of the toxicity of phenols to Tetrahymena pyriformis. Monatsh. Chem.-Chem Mon. 2009;140:1279–1288. doi: 10.1007/s00706-009-0185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilhami G., Riad E., Akcahan G., Aun C., Fevzi T. Antioxidant activity of bisbenzylisoquinoline alkaloids from Stephania rotunda: cepharanthine and fangchinoline. J. Enzym. Inhib. Med. Chem. 2010;25(1):44–53. doi: 10.3109/14756360902932792. [DOI] [PubMed] [Google Scholar]
- Jalali-Heravi M., Kyani A. Use of computer-assisted methods for the modeling of the retention time of a variety of volatile organic compounds: a PCA-MLR-ANN approach. J. Chem. Inf. Comput. Sci. 2004;44:1328–1335. doi: 10.1021/ci0342270. [DOI] [PubMed] [Google Scholar]
- Jitender V., Vijay M.K., Evans C.C. 3D-QSAR in drug design: a review. Curr. Top. Med. Chem. 2010;10:95–115. doi: 10.2174/156802610790232260. [DOI] [PubMed] [Google Scholar]
- Kenard R.W., Stone L.A. Computer aided design of experiments. Technometrics. 1969;11(1):137–148. [Google Scholar]
- Kumar S., Singh R.K., Patial B., Goyal S., Bhardwaj T.R. Recent advances in novel heterocyclic scaffolds for treatment of drug-resistant malaria. J. Enzym. Inhib. Med. Chem. 2016;31:173. doi: 10.3109/14756366.2015.1016513. [DOI] [PubMed] [Google Scholar]
- Li Z., Wan H., Shi Y., Ouyang P. Personal experience with four kinds of chemical structure drawing software: review on ChemDraw, ChemWindow, ISIS/Draw, and ChemSketch. J. Chem. Inf. Comput. Sci. 2004;44:1886–1890. doi: 10.1021/ci049794h. [DOI] [PubMed] [Google Scholar]
- Oliveira R., Miranda D., Magalhaes J., Capela R., Perry M.J., O'Neill P.M., Moreira R., Lopes F. From hybrid compounds to targeted drug delivery in antimalarial therapy. Bioorg. Med. Chem. 2015;23 doi: 10.1016/j.bmc.2015.04.017. [DOI] [PubMed] [Google Scholar]
- Penna-Coutinho J., Cortopassi W.A., Oliveira A.A., Franc¸a T.C.C., Krettli A.U. Antimalarial activity of potential inhibitors of plasmodium falciparum lactate dehydrogenase enzyme selected by docking studies. PloS One. 2011;6(7) doi: 10.1371/journal.pone.0021237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts J.A., Butina D., Abraham M.H., Hersey A. Estimation of molecular linear free energy relation descriptors using a group contribution approach. J. Chem. Inf. Comput. Sci. 1999;39(5):835–845. doi: 10.1021/ci990427t. [DOI] [PubMed] [Google Scholar]
- Rania A., Legacb J., Rosenthalb P.J., Kumara V. Substituted 1,3-dioxoisoindoline-4-aminoquinolines coupled via amide linkers: synthesis, antiplasmodial and cytotoxic evaluation. Bioorg. Chem. 2019;88:102–912. doi: 10.1016/j.bioorg.2019.04.006. [DOI] [PubMed] [Google Scholar]
- Singh P. Quantitative structure-activity relationship study of substituted-[1, 2, 4] oxadiazoles as S1P1 agonists. J. Curr. Chem. Pharm, Sci. 2013;3 [Google Scholar]
- Talele T.T., Santosh A.K., Alan C.R. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr. Top. Med. Chem. 2010;10:127–141. doi: 10.2174/156802610790232251. [DOI] [PubMed] [Google Scholar]
- Tropsha A. Best practices for qsar model development, validation, and exploitation. Mol. Inf. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- Tropsha A., Gramatica P., Gombar V.K. The importance of being earnest: validation is the absolute essential for successful application and interpretation of QSPR models. Mol. Inform. 2003;22:69–77. [Google Scholar]
- Trott O., Olson A.J. Autodock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010;22:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veerasamy R., Rajak H., Jain A., Sivadasan S., Varghese C.P., Agrawal R.K. Validation of QSAR models-strategies and importance. Int. J. Drug Des. Discov. 2011;3:511–519. [Google Scholar]
- World Health Organization . 2019. World Malaria Report.http://apps.who.int/malaria/media/world-malaria-report-2019 [Google Scholar]
- Yap C.W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.