Abstract
Discovery of biomarkers is critical to understand tumor heterogeneity and microenvironment. To determine differently expressed makers on cancer tissue for comprehensive profiling, the multiplexed tissue imaging mass cytometer (IMC) which uniquely combines time-of-flight mass spectrometry with metal-labeling technology to enable breakthrough discovery on single cell level was employed to investigate the expression of seven markers related to the epithelial-to-mesenchymal transition [α-smooth muscle actin (α-SMA), vimentin, collagen I, cytokeratin 7, pan-keratin], tumor proliferation (Ki-67), and human leucocyte antigen (HLA-DR) on human pancreatic cancer tissue. The difference was analyzed using bioinformatic tools. We observed the high expression of α-SMA, vimentin, collagen I, and Ki-67 on grade I but not on grade III. HLA-DR was highly expressed on grade I/III but not on grade II. Overall, the expression of markers has elucidated the heterogeneity intratumors. Additionally, to identify biomarkers on pancreatic cancer cells by blind systematic evolution of ligands by exponential enrichment (SELEX), aptamer pull-down assay and liquid chromatography–tandem mass spectrometry were used. Mortalin was identified as a potential a prognostic marker of pancreatic cancer. Our studies demonstrate that the IMC and blind SELEX might be implemented to discover biomarkers which can be used to better understand tumor biology and biomedical research applications.
Introduction
Cancer is a very dynamic disease that can evolve, thereby resulting in heterogeneity [1]. To understand the heterogeneity within tumor tissue, the role of the microenvironment, and the impact of immune cells on cancer development, sophisticated tools are needed for quantitative analysis of a large number of biomarkers while retaining spatial resolution of cells and tissue architecture. Pathological assessment of tissue provides prognostic evaluation and helps to select optimum treatment of regimens based on expression of biomarkers on cancer tissue [2]. To understand heterogeneity of phenotypes and to evaluate the expression of markers on the single cell level, imaging mass cytometry (IMC) is a right approach with multiparametric analyses.
Mass cytometry is an advanced technology that uses a time-of-flight ICP-MA instrument (CyTOF) that can detect dozens of makers simultaneously at a high mass-spectrum acquisition by high-frequency laser ablation [3]. It is a highly multiparametric and quantitative approach for the identification of phenotypes and proteins on cells within tissue architecture. The high purity of metal isotopes affords minimal background and yields high signal-to-noise ratios. Therefore, it provides 135 available detection channels to measure more than 100 markers per cell simultaneously [4]. These unique advantages allow to study the functional complexity of biological systems at the single-cell level. Indeed, multiplexed imaging of human breast tissues by CyTOF enabled the detection of 32 proteins at a cellular resolution of 1 μm [5].
Metastasis is a leading cause of cancer-associated death. Pancreatic ductal adenocarcinoma (PDAC) is among the most fatal cancers due to extensive invasion into surrounding tissues even at an early stage [6]. During malignant progression and dissemination of tumor cells, carcinoma cells undergo an epithelial-to-mesenchymal transition (EMT), in which they acquire invasive stem cell–like features [7]. Rhim et al. showed that the EMT is an early event in tumor malignancy and that inflammation promotes EMT, invasiveness, and tumor cell dissemination [8]. Expression of intermediate filaments such as vimentin and cytokeratins (CKs) is regulated during development and differentiation. Therefore, intermediate filaments have been used to differentiate normal pancreas and pancreatic cancer [9]. In that study, it indicated that CK7 is homogeneously expressed in all pancreatic cancers. Another study also showed that CK7 is positive with high intensity over 3+ in the majority of carcinomas [10]. Thus, CK7 is a good biomarker to evaluate the level of expression and to differentiate the stage of PDAC. Vimentin is present in mesenchymal cells and is used to distinguish them from epithelial cells [11]. Vimentin is associated with a migratory phenotype, and overexpression of vimentin is observed in EMT of cancer [12]. α-Smooth muscle actin (α-SMA) has been used as a marker for activated fibrogenic cells and myofibroblasts that are effector cells of tissue fibrogenesis [13]. Cells expressing α-SMA contribute to EMT in embryogenesis and wound healing in normal epithelial cells [11]. Thus, both vimentin and α-SMA are considered biomarkers of EMT [11]. The desmoplastic component of PDAC has long been appreciated by a massive deposition of extracellular matrix, containing an abundance of collagen I [14], [15]. Collagen I also promotes metastasis in pancreatic cancer by activating c-Jun and N-cadherin [16]. Therefore, vimentin, α-SMA, CK7, and collagen I are good markers to differentiate tumor grades and to determine the prevalence of EMT in human PDAC.
Since the identification of Ki-67 as an antigen in Hodgkin lymphoma cell nuclei [17], it has been widely used as a proliferation marker in tumor cells [18]. Thus, Ki-67 is evaluated as a prognostic marker of disease progression in pancreatic neuroendocrine neoplasms [19]. In this study, Ki-67 is included to differentiate tumor grades of PDAC.
The antigen-presenting molecule human leukocyte antigen (HLA-DR) is part of the major histocompatibility complex class II family of antigen presentation molecules, expressed on antigen-presenting cells [20], [21] and some tumor cells in response to inflammatory reactions compared with normal [22], [23]. For example, the expression of HLA-DR in esophageal epithelium is increased in inflammatory states and early dysplasia upon progression from inflammations to cancer [22]. Also, the HLA-DR in cytotoxic T lymphocytes (CTLs) is investigated to predict the responses of neoadjuvant chemotherapy in cancer [24]. It has been shown that HLA-DR–expressing CTLs are mainly located in intraepithelial tumor structures rather than in the normal region. Interestingly, it also shows that only HLA-DR–positive CTL cells have shown high expression of IFN-γ and granzyme B, suggesting that high expression of HLA-DR+ CTLs are indicative of effector lymphocytes that are utilized for antitumor effects. Reportedly, the expression of HLA-DR in tumors correlates with the presence of immune cells such as CD16+ myeloid cells and T cells [23]. As it has been shown that inflammation promotes EMT in cancer [8], the expression of HLA-DR on PDAC is worth investigating.
Using a multiplexed IMC platform (M-IMC), we determined the level of expression of 7 markers which are five EMT markers (α-SMA, vimentin, CK7, pan-keratin, and collagen I), one proliferation marker (Ki-67), and one leukocyte antigen marker (HLA-DR), in human formalin-fixed paraffin-embedded (FFPE) PDAC samples. Analysis of single-cell phenotypes revealed substantial intratumoral heterogeneity in PDAC. We also identified a potential biomarker on PDAC using blind systematic evolution of ligands by exponential enrichment (SELEX).
Materials and Methods
Reagents
Iridium DNA Intercalator (Fluidigm, 201192A) and metal-labeled antibodies pan-keratin (Nd 148, Fluidigm 3148020D), cytokeratin 7 (Dy 164, Fluidigm 3164028D), Ki-67 (Er 168, Fluidigm 3168022D), HLA-DR (Yb 174, Fluidigm 3174023D), α-SMA (Pr 141, Fluidigm 3141017D), vimentin (Nd 143, Fluidigm 3143029D), and collagen I (Tm 169, Fluidigm 3169023D) were purchased from Fluidigm. Tissue microarray (TMA) samples were purchased from BioMax (PA807). Recombinant mortalin was purchased from Novus Biologicals (NBC1-18380).
IMC Staining
The routine IHC protocols were used for staining. Briefly, slides were dewaxed and hydrated in descending grades of ethanol (100%, 95%, 80%, 70%), rinsed, and inserted into preheated antigen retrieval solution. Then, the slides were incubated with the antibody cocktail overnight at 4°C. Afterward, tissues were stained with Ir-Intercalator (1:400) for nuclear counterstaining. Finally, slides were washed in Distilled water and dried at room temperature.
Imaging Acquisition by the Hyperion
An image of the slide was acquired to be used as a reference. The resulting image was used as a visual reference to select regions of interest (ROIs) for data acquisition in the Fluidigm software. The TMA slide was inserted into the Hyperion Imaging Cytometer (Fluidigm Corporation, San Francisco, CA) for laser ablation. A panel was designed from the metals/antibodies of interest and applied. The ROIs of 1000 μm × 1000 μm were then laser ablated. The total acquisition time for the slide was approximately 160 hours.
The results were stored in Fluidigm's MCD format and were exported as images in 16-bit OME tiff format for downstream quantification. Cell segmentation masks were created to identify single cells by establishing the nuclear and cellular boundaries using Cellprofiler v2.2.0 [25] and exported as text image TIFF format files. These files were imported into histoCAT (histology topography Cytometry Analysis Toolbox) [26] for intensity measurements of each marker and data visualization including cellular spatial distribution images, box plots, heat maps, and t-SNE plots.
Data Analysis
Both Hyperion CyTOF image files and cell segmentation mask files were imported into histoCAT to measure the intensity of each marker at the single-cell level. The markers' intensities were arcsine transformed before cluster analysis and t-SNE analysis. All ROIs of the cells were partitioned to 28 subpopulations automatically using unsupervised PhenoGraph [27] algorithms implemented in histoCAT. The subpopulations were visualized and color coded on the t-SNE plot. Detailed TMA source compositions of each subpopulation cluster and the detailed subpopulation composite of each TMA were visualized in the histogram diagrams. The expressions of the 7 markers in the 28 subpopulations were presented in the heat map to identify the marker specific cells. The count of subpopulation cells in each TMA was clustered to identify the phenotype specific subpopulations. A total of eight marker specific subpopulations were identified. These marker specific subpopulations and their expression status were highlighted on the t-SNE plots using heat maps. The detailed quantitative expression was presented in the box plots. The composition of those marker specific subpopulations of each TMA was visualized in the histogram plots. These marker specific cells were highlighted on each ROI to show their spatial distribution.
Competition Assay on Live Cells
For the aptamer competition assay, 1 × 105 PANC-1 cells were seeded in 35-mm glass-bottom dishes (MatTek, Ashland, MA) and grown in medium for 24 hours. For the competition assays, 200 nM of Cy3-labeled P19 aptamer was used to compete with unlabeled either the P19 or P1 aptamer (1 μM). The fluorescence intensity was quantified in the presence of increasing amounts of competitor using confocal microscopy using a Zeiss LSM 510 Meta Inverted 2 photon confocal microscope system with a C-Apo 40x/1.2NA water immersion objective and analyzed using one-way analysis of variance (ANOVA).
Aptamer Pull-Down Assay
To retrieve the aptamer binding ligand, cell membrane proteins were isolated from PANC-1 cells. To identify the target epitope of the aptamer, biotinylated aptamers at 3′ ends were immobilized using a pull-down process. Plasma membrane proteins were prepared using procedures described by Daniels et al. [28]. In-gel digestion was used for protein purification and analyzed by mass spectrometry for peptide fingerprinting. After sodium dodecyl sulfate polyacrylamide gel electrophoresis, aptamer-retrieved protein bands were excised and in-gel digested [29].
Q-TOF Liquid Chromatography–Tandem Mass Spectrometry (LC-MS/MS)
An Agilent 6520 Q-TOF mass spectrometer equipped with a Chip Cube source was used for LC-MS/MS analyses. A C18 chip with a 43-mm analytical column and a 40-nl trapping column (ProtID-Chip-43, Agilent G4240-62005) was used. Digested samples (10-15 μl) were loaded onto the column at 6 μl/min in 99% buffer A (0.1% formic acid in water)/1% buffer B (0.1% formic acid in acetonitrile) with an extra 8 μl wash volume. The X! Tandem search engine (http://www.thegpm.org/TANDEM/index.html) was used to search the peptide MS/MS spectrum. The dataset was then processed using the Scaffold program (http://www.proteomesoftware.com) to visualize the results. SWISS Prot or NCBI was used to obtain the detailed protein annotation.
Validation of Ligand Binding by the Surface Plasmon Resonance (SPR)
To validate the binding of ligand, Biacore T100 (GE Healthcare, Uppsala, Sweden) was used to monitor label-free the aptamer-target interactions. Biotinylated aptamers were coupled to a streptavidin-coated Biocore chip (SensorChip SA, BR-1003-98, General Electric Company) by an injection in binding buffer. To measure binding kinetics, five different concentrations of mortalin protein were injected at a flow rate of 10 μl/min. BIAevaluation software (GE Healthcare) was used to fit to a 1:1 binding analysis.
Statistical Analysis
Statistically significant differences were determined by one-way ANOVA test using Graph Pad Prism software (GraphPad Software, La Jolla, CA). A P value ≤.05 was considered significant; *P ≤ .05, **P ≤ .01.
Results
Expression of Each Maker Is Acquired by IMC
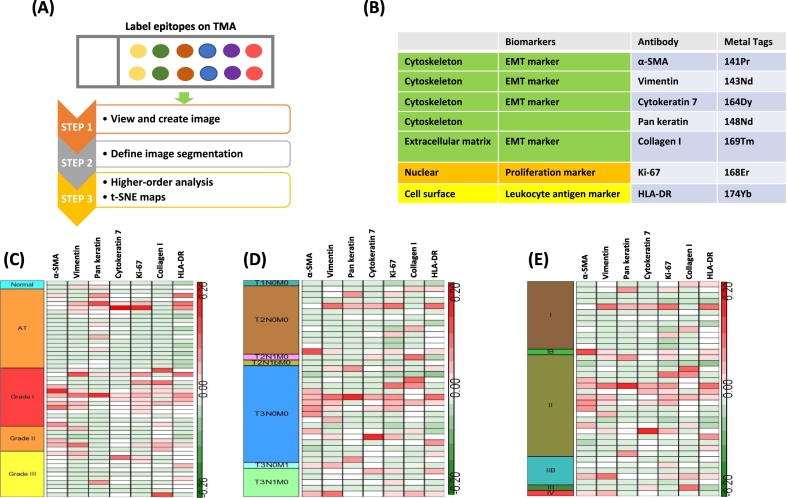
To determine multiparametric expression on pancreatic cancer specimens, TMAs containing FFPE pancreatic cancer tissues of mainly PDAC, adjacent, and normal tissues were used. For multiplexed imaging analysis of CyTOF-based mass cytometry, the workflow of M-IMC was depicted in Figure 1A. Firstly, tissue section was stained with metal-tagged antibodies labeling using routine IHC protocols. Antibodies were selected to relevant targets. In this study, seven markers were employed to stain FFPE pancreatic cancer specimens. For data analysis on single cells, acquired images were defined for segmentation, and high-order analysis was performed using t-SNE plot. The list of metal tags of each antibodies was shown in Figure 1B.
Figure 1.
Workflow of M-IMC. (A) After tissue staining with metal tagged antibodies on TMA, first step was to create images. After single-cell segmentation of tissue images, higher-order analysis was performed to visualize results such as t-SNE maps. (B) The element of each metal tagged antibody is listed. Five EMT markers: α-SMA, vimentin, cytokeratin 7, pan-keratin, and collagen I. One tumor proliferation marker: Ki-67. One immune cell marker: HLA-DR. Two nuclear counterstainings. (C, D, and E) The expression of each target was generated by heat map; heat map of tumor grade (C), TNM stage (D), tumor stage (E).
The Distinct Expression of Each Marker Shows on Tumor Grades of Human Pancreatic Cancer
Utilizing the median value of each of seven markers, the expression was measured and represented depending on tumor grades, tumor-node-metastasis (TNM), and stages with heat map (Figure 1, C, D, and E). In normal pancreas tissue, all seven markers showed a low expression. Some of the adjacent tumor regions (AT) showed high intensity of pan-keratin (7/19), CK7 (2/19), Ki-67 (2/19), vimentin (3/19), and HLA-DR (3/19), but it showed low expression of collagen I (0/19). In grade I, α-SMA (6/14), vimentin (6/14), Ki-67 (8/14), collagen I (7/14), and HLA-DR (5/14) showed a high expression. In grade II, α-SMA (3/6), vimentin (2/6), and collagen I (3/6) were highly expressed, but CK7 (0/6), pan-keratin (0/6), Ki-67 (1/6), and HLA-DR (1/6) showed low expression. The seven markers—α-SMA (1/11), vimentin (2/11), pan-keratin (1/11), CK7 (1/11), Ki-67 (2/11), collagen I (1/11), and HLA-DR (3/11)—showed low expression on higher tumor grade III (Figure 1C).
With TNM classification, all seven markers showed low expression in T1. The high expression of α-SMA (8/23), vimentin (8/23), pan-keratin (3/23), CK7 (4/23), Ki-67 (9/23), collagen I (5/23), and HLA-DR (7/23) was observed in T3 (Figure 1D). The high expression of both α-SMA and vimentin was observed on one metastasized tissue specimen.
With tumor staging classification, α-SMA (8/23), vimentin (8/23), pan-keratin (3/23), CK7 (5/23), Ki-67 (9/23), collagen I (6/23), and HLA-DR (7/23) were highly expressed in stage II (Figure 1E). The expression pattern of these makers was closely related between T3 and stage II.
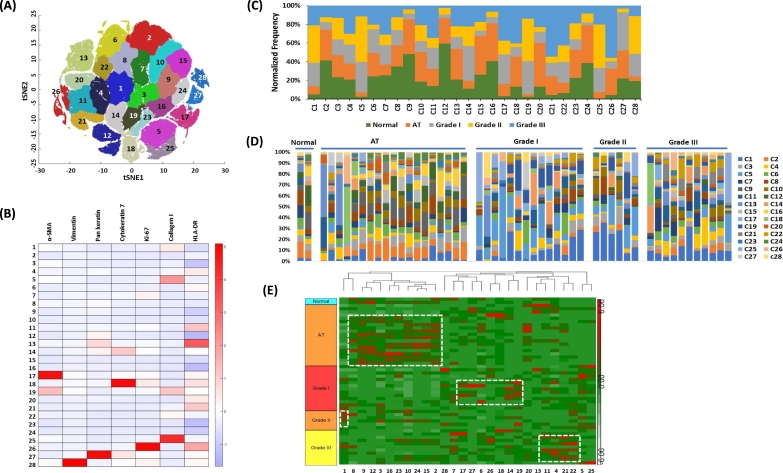
t-SNE Analysis Reveals 28 Clusters with Differently Expressed Patterns of Seven Markers
Given the high dimensionality of the mass cytometry data, t-SNE maps were employed to visualize and investigate high-dimensional cell phenotypes that differentiate these groups (Figure 2A). The expression of each marker on clusters is depicted in the heat maps (Figure 2B). Cluster 17/19 showed high expression of α-SMA. The high expression of vimentin, pan-keratin, CK7, Ki-67, and collagen I was distributed to cluster 28, 27, 18, 26, and 5/25, respectively (Figure 2B). The normalized frequency of tumor grades on each cluster was depicted in Figure 2C. The normalized frequency of each cluster on tumor grades was depicted in Figure 2D. As shown in Figure 2D, it showed the heterogeneity on tumor grades. Hierarchical clustering within the group of tumor grades revealed that clusters 8, 9, 12, 3, 16, 23, 10, 24,15, and 2 were segregated to AT. Clusters 17, 27, 6, 26, 18, 14, and 19 were segregated to grade I. Cluster 1 was located to grade II. Clusters 11, 4, 21, and 22 were segregated to grade III (Figure 2E).
Figure 2.
High-dimensional analysis reveals differently expressed marker on tumor grade. (A) Gating was performed on total t-SNE clusters after arbitrary assignment of cluster numbers. (B) The expression of each marker on each cluster was generated via heat map. (B) The heat map of each cluster was generated to classify on tumor grades. (D) The stacked bar chart was aligned to the heat map that shows the normalized frequencies of each marker composition (colored legends) within the gated clusters. (E) Hierarchical clustering analysis was depicted in each tumor grade.
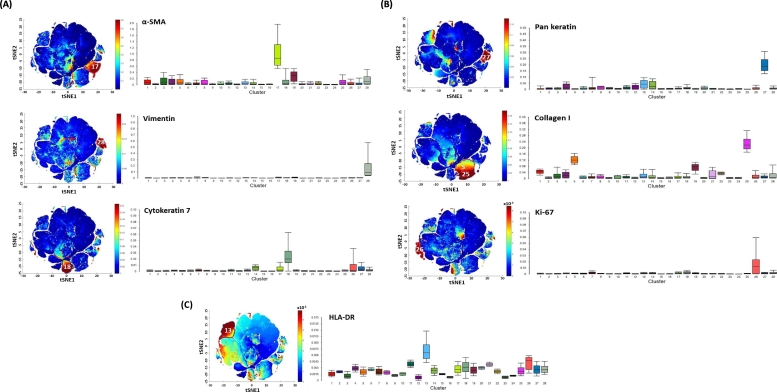
Individual t-SNE Analysis Reveals the Differential Expression of Each Cluster
t-SNE analysis was performed individually to determine expression of each marker on clusters. For EMT markers, the high expression of α-SMA, vimentin, and CK7 was shown in cluster 17, 28, and 18, respectively (Figure 3A). Pan-keratin and collagen I were highly expressed in cluster 27 and 25, respectively (Figure 3B). Ki-67 was upregulated in cluster 26 (Figure 3B). The expression of HLA-DR was shown highly in cluster 13 (Figure 3C).
Figure 3.
Individual t-SNE map shows different expression of pattern in each marker. (A) The t-SNE was performed on three EMT markers (left). Bar graph was generated to show the level of expression (right): α-SMA, vimentin, and cytokeratin 7. (B) The t-SNE was performed on two EMT makers and one tumor proliferation marker (left). Bar graph was generated to show the level of expression (right); pan-keratin, Collagen I, and Ki-67. (C) The t-SNE was performed on an leukocyte antigen marker (left), Bar graph was generated to show the level of expression (right); HLA-DR. (D) The t-SNE was performed on nuclear staining (left), Bar graph was generated to show the level of expression (right); HLA-DR.
The Heterogeneity on Human Pancreatic Cancer Is Revealed
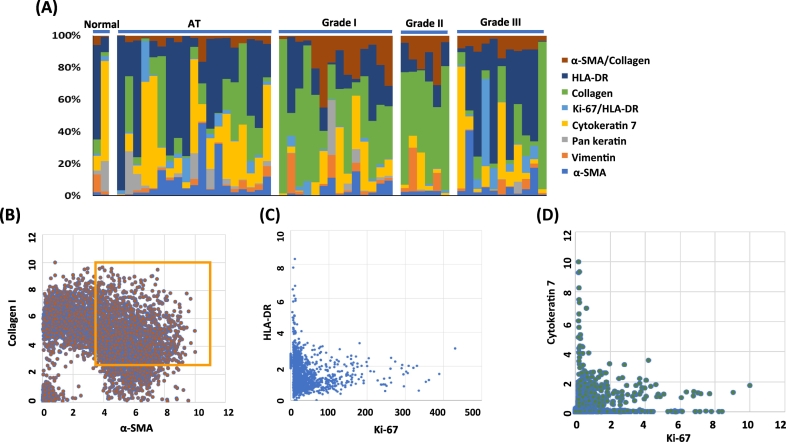
The expression of each maker was analyzed on each TMA sample, and the expression was measured and displayed depending on tumor grades (Figure 4A). It clearly showed heterogeneity in the same tumor grades. The expression of HLA-DR was high in AT and grade III. Collagen I was high in grade I and II. Vimentin was highly expressed in grade I. When we investigated the correlation of each marker, it demonstrated a strong pairwise correlation between α-SMA and collagen I in higher tumor grades (Figure 4B). However, there was minimal correlation between HLA-DR and Ki-67 (Figure 4C) or between CK7 and Ki-67 (Figure 4D).
Figure 4.
The stacked bar graph shows the heterogeneity. (A) The expression of each of the seven makers was stacked in bar graph. It shows the heterogenous expression pattern in the same tumor grade. (B) The expression of collagen and α-SMA was closely correlated on single-cell level in cluster 19. (C) The expression of Ki-67 and HLA-DR was not related on single-cell level in cluster 26. (D) The expression of Ki-67 and Cytokeratin 7 was not related on single-cell level in cluster 26.
Image Segmentation Is Validated with Hematoxylin and Eosin (H&E)–Stained TME Samples
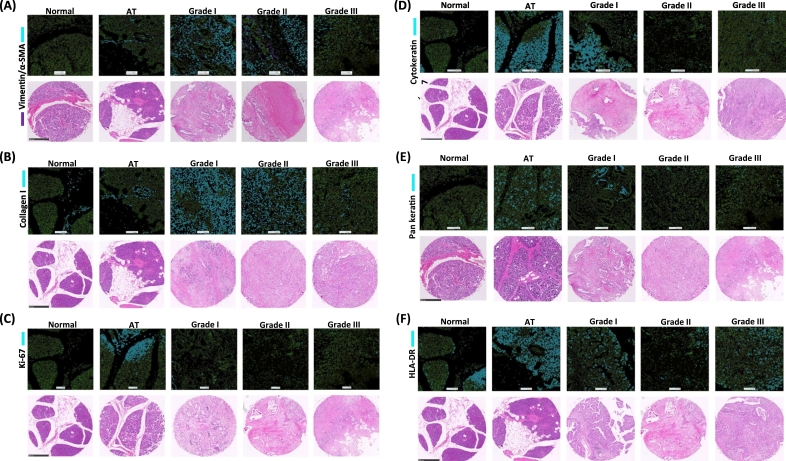
The image segmentation acquired by CyTOF was visualized and compared to an array of a representative TMA which was stained with H&E (Figure 5). All corresponding H&E-stained TMA images were displayed below. An upregulation of both vimentin and α-SMA was observed on grades I and II (Figure 5A). Collagen I was observed in connective tissue regions on AT. The expression of collagen I was excessive on grade I and II because of desmoplasia of PDAC (Figure 5B). The high expression of Ki-67 was observed on some specific regions of tumor cells on AT and grade I (Figure 5C). The expression of CK7 was high on tumor cells of AT and grade I (Figure 5D). Low expression of pan-keratin was observed on tumor grade II and III (Figure 5E). High expression of HLA-DR was observed on tumor cells on AT and tumor grades I and III (Figure 5F).
Figure 5.
The corresponding histoCAT images to H&E staining. (A) Vimentin in purple and α-SMA in blue were shown in top, and corresponding H&E staining is in the bottom. No expression of α-SMA and vimentin was observed on normal region. The high expression on tumor grade I and II and low expression on grade III were observed. (B) The expression of collagen I was dominantly expressed on tumor grades I and II. Collagen I was observed in only connective tissues on normal and adjacent region (AT). (C) The high expression of Ki-67 was observed some of AT and tumor grades I and II. No expression was observed on normal region and AT. (D) The expression of cytokeratin 7 was observed highly on tumor grade I and AT. (E) The high expression of pan-keratin was observed only on tumor grade I and AT. The low expression was observed on tumor grades II and III. (F) The dominant expression of HLA-DR was observed on tumor grades I and III and AT. Low expression of HLA-DR on tumor grade II was observed. Scale bar: 500 μm.
Mortalin Is a Potential Biomarker in Pancreatic Cancer
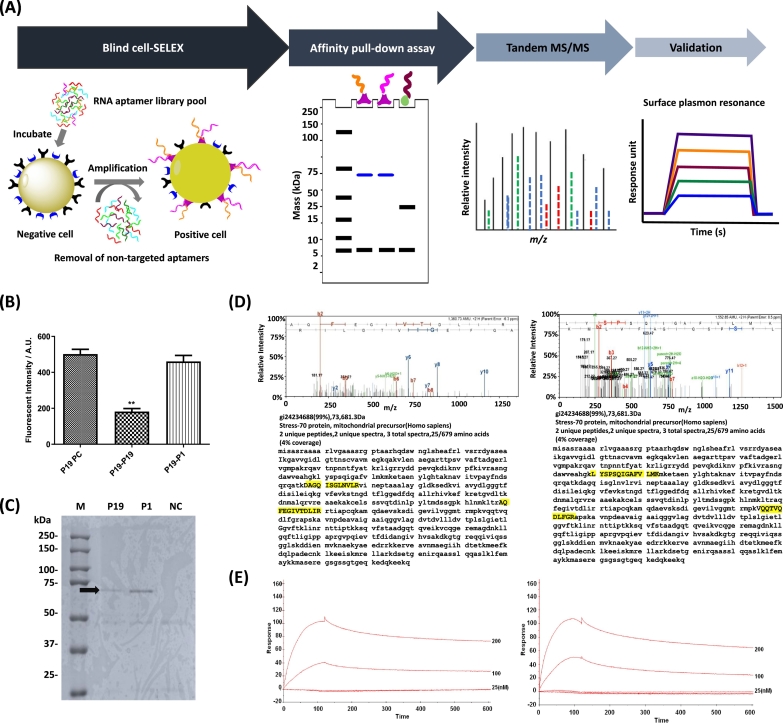
To identify biomarkers on pancreatic cancer, blind SELEX was developed on naive cells [30], [31]. The schematic workflow was depicted in Figure 6A. In previous studies, two pancreatic cancer specific RNA aptamers (P19 and P1) have been identified [30]. However, the aptamer binding ligand has not been determined. To verify whether these aptamers bind to the same cell surface epitope, a competition assay was performed with each Cy3-labeled aptamer against its unlabeled one. It showed a significant decrease of intensity in Cy3-P19 in the presence of its unlabeled one. However, no significant decrease in Cy3-P19 was observed in the presence of unlabeled P1 aptamer (Figure 6B), suggesting that P19 and P1 might recognize distinct epitopes on the target cell surface. To confirm our observation and to identify the target epitope of the aptamer, the cell membrane proteins were extracted and retrieved by biotinylated aptamers using a pull-down process. The retrieved proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis followed by Coomassie staining to visualize the resolved protein bands (Figure 6C). The resolved bands were extracted for protein purification and analyzed by mass spectrometry for peptide fingerprinting. The highest matching peak matched a known peak for HSPA9B which is also known as mitochondrial Hsp70 or mortalin (Figure 6D); P19 (left) and P1 (right). The identified target epitopes of P19 and P1 highlighted showed different peptides which are consistent with results of competition assays. To validate the mass spectrometry results, SPR measurements was performed to confirm the aptamer/epitope interaction (Figure 6E); P19 (left) and P1(right). When complexed to mortalin, it showed a 100–response unit effect at 200 nM as measured by SPR.
Figure 6.
Blind SELEX to identify potential biomarkers on pancreatic cancer. (A) The schematic workflow of blind SELEX is depicted. After untargeted SELEX to identify differently expressed markers on plasma membrane of PANC-1 cells, aptamers are enriched in vitro. Target ligands were retrieved via affinity pull-down assays using biotinylated aptamers. After affinity pull-down assays, MS/MS is used to achieve peptide fingerprinting. Finally, the binding of aptamer with the target ligand is validated by surface plasmon resonance. (B) Competition assay was performed to determine whether P19 and P1 bind to the same epitope. The fluorescence intensity was quantified using confocal microscopy. *One-way ANOVA test; *P ≤ .05, **P ≤ .01. (C) Ten percent of polyacrylamide gel electrophoresis was used to separate immobilized protein samples after pull-down with biotinylated P19, P1, and negative control RNAs. Coomassie-stained gels: M (marker), P19 (lane 1), P1 (lane 2), and NC (irrelevant RNA, lane 3). Arrow indicates target antigens. (D) MS/MS spectra of aptamer binding ligand. Peptide matching and MS/MS spectra of P19 (left) and P1 (right) affinity-purified peptides. Inset: amino acid sequence of the parent peptide showing b- and y-ion series coverage. Target epitopes are highlighted in yellow. (E) Biosensor analysis of the mortalin-aptamer interaction. Binding of mortalin to the P19 (left) and P1 (right) was shown. Biotinylated P19 or P1 was bound to a streptavidin-coupled carboxyl methyl dextran surface, and binding was measured using the SPR technique. The increased response unit was shown in a dose-dependent manner.
Discussion
IMC is an emerging and transformative technique in the histopathology field which can be applied to complex tissue sections [3] [4]. As the Hyperion Imaging System (Fluidigm) can multiplex up to 50 parameters simultaneously in FFPE and frozen tissue sections with subcellular resolution [5], it provides comprehensive and quantitative biological information at subcellular resolution. Immunohistochemistry (IHC) has been used to identify IHC markers on tumor differentiation [32]. However, the approaches of IHC have limitations such as false negatives on low abundant markers and difficulty in multiplexing and in the precise quantification of intensities. Thus, in this study, we developed a workflow of multiplexed IMC and identified potential biomarkers for tumor differentiation on PDAC with quantifiable results.
Determination of the expression of biomarkers is critical to understanding tumor biology and to anticipate clinical outcomes. Invasion into surrounding tissues and metastasis to distant organs in PDAC are initiated at an early stage by EMT [6]. Also, they change their polarity by modulating the organization of their cytoskeletal systems and switching expression from keratin- to vimentin-type intermediate filaments [33]. Therefore, we determined the level of expression of EMT markers in this study. We observed the high expression of EMT markers on PDAC.
The expression of vimentin is positive in 52.8% of PDAC cases [34]. The overexpression of vimentin is closely correlated with accelerated tumor growth, suggesting that vimentin can be used to determine the degree of tumor differentiation [35]. In evaluation of EMT markers on tumor progression and metastasis, it has been reported that the expression of vimentin was transcriptionally upregulated in early states [36]. Thus, we evaluated the feasibility of vimentin as a marker to differentiate the degrees of tumor. Consistent with results reported previously [36], we observed its high expression on grade I, but the low expression of vimentin was observed on higher tumor grade III.
Reportedly, the expression of Ki-67 in PDAC correlates with tumor grade and positive nodal status [34], [37]. Our study showed different results: high expression on grade I and low expression on higher tumor grade II/III.
The abundant desmoplastic stroma reaction in PDAC is considered to be an important reason for its aggressiveness [38]. It also suggests a prognostic role for α-SMA expression and stromal density in the peritumoral stroma [38]. In relationship of tumor differentiation with α-SMA, it has been reported that stromal α-SMA has no significant association with tumor grades [39]. But our study showed different results: high expression on grade I/II but low expression on higher grade III.
The deposition of extracellular matrix collagen in the stromal compartment promotes PDAC survival [40] and malignant phenotype [41]. The expression of collagen I is elevated in PDAC tissues compared to normal [42]. No significant difference between collagen composition and tumor grade was reported [42]. But our data showed different results: high expression on grade I/II but low on higher grade III.
The high expression of CK7 has been shown in PDAC [10]. But there are no reports to differentiate tumor grades with CK7. Our data showed the low expression of CK7 on tumor grade II/III. But interestingly, high expression of pan-keratin on tumor adjacent region (AT) was observed, suggesting that AT cells are transformed in malignancy.
In the correlation analyses between each of the seven markers, the main outcome of this study is to reveal significant correlations between α-SMA and collagen I. In previous studies of fibrosis in skeletal muscle, Zhao et al. concluded that the level of α-SMA expression does not positively correlate with the level of collagen gene expression in the mdx5cv mice [43]. However, our data clearly showed that the level of α-SMA expression is closely correlated with collagen I expression.
The correlations between the expression of HLA class I, HLA-DR, PD-L1, or PD-1 and the pattern of tumor infiltrating immune cells were determined to assess the patients' prognosis [44]. In that study, it was shown that HLA-DR is not expressed on normal pancreatic ductal cells. It also shows that infiltrating CD8+ lymphocytes are significantly higher in HLA-DR–positive than in HLA-DR–negative PDAC. Our study showed that there was high expression in lower tumor grade I and III but not tumor grade II. The antitumor efficacy is closely related with infiltration of CD8+ lymphocytes corresponding to high expression of HLA-DR. Therefore, our study suggests the HLA-DR might be a potential biomarker to anticipate the outcome to immunotherapy in PDAC.
With single-cell technologies, it has already been demonstrated that there is heterogeneity across PDAC with regard to primary tumor cells and circulating tumor cells [45]. Our study confirmed again the heterogeneity in the same tumor grades. To differentiate tumor grades depending on differently expressed EMT markers, our data, for the first time, revealed that the five makers—α-SMA, vimentin, collagen I, Ki-67, and HLA-DR—had low expression on tumor grade III but high expression on tumor grade I.
Previously, we identified pancreatic cancer specific RNA aptamers (P19 and P1) using blind SELEX [30]. The P19 and P1 showed pancreatic cancer specificity. In that study, Yoon et al. showed that the staining intensity on archival human tissues is closely related with patient's survival rates [30]. However, the binding ligand of aptamer has not been identified. In this study, we successfully identified that binding ligand of P19 and P1 was mitochondrial HSP70 or mortalin. Reportedly, heat shock protein 70 (Hsp70) is localized on tumor-selective cell surface [46]. The expression of Hsp70 on plasma membrane is found on freshly isolated human carcinomas such as colorectal, neuronal, and pancreatic. But normal tissues and bone marrow of healthy human individuals do not express Hsp70 on the cell surface [46]. In our previous studies, two aptamers have also showed consistent results which are nonbinding on normal pancreatic epithelial cells and cross-binding activity on colorectal cancer and glioblastoma [30], [47]. As high expression of mortalin has been reported with low survival rates on PDAC patients [30], mortalin might be a prognostic marker in PDAC.
In conclusion, we demonstrate a multiplexed and multidimensional IMC workflow with quantification that is primarily related to EMT and cancer cell proliferation. Our data showed the different expression of each marker compared to previous results using conventional IHC methods. As M-IMC uses quantitative single-cell analysis, it provides precise quantitative data to evaluate tumor grade which might be applicable to clinical specimens. We also identified for the first time that mortalin might be a biomarker on pancreatic cancer using blind SELEX. Previously, we developed RNA aptamer-based therapeutics: aptamer conjugated with C/EBPα-saRNA [30], aptamer-drug conjugates [47], and anti-vimentin RNA aptamers [31]. Because the data acquired from single images are informative, we will investigate the changes of phenotypes and levels of expression after treatment of therapeutics to understand responsiveness to these therapeutics in future studies.
Author Contributions
S. Y, B. A., and J. J. R. developed the concept. S. Y. and H. L. analyzed data. S. Y. performed aptamer pull-down assay, binding assays with SPR, and live cell imaging. L. Q. performed Hyperion Imaging Cytometer.
Conflict of Interest
The authors declare no conflict of interest regarding the publication of this article.
Acknowledgements
We would like to thank the Light Microscopy Digital Imaging Core, and Mass Spectrometry and Proteomics core at City of Hope for support. Research reported in this publication also included work performed in shared resource facilities supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 2.Dunstan RW, Wharton KA, Quigley C, Lowe A. The use of immunohistochemistry for biomarker assessment—can it compete with other technologies? Toxicol Pathol. 2011;39:988–1002. doi: 10.1177/0192623311419163. [DOI] [PubMed] [Google Scholar]
- 3.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 4.Chang Q, Ornatsky OI, Siddiqui I, Loboda A, Baranov VI, Hedley DW. Imaging mass cytometry. Cytometry A. 2017;91:160–169. doi: 10.1002/cyto.a.23053. [DOI] [PubMed] [Google Scholar]
- 5.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 6.Baumgart M, Heinmoller E, Horstmann O, Becker H, Ghadimi BM. The genetic basis of sporadic pancreatic cancer. Cell Oncol. 2005;27:3–13. doi: 10.1155/2005/632939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 8.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schussler MH, Skoudy A, Ramaekers F, Real FX. Intermediate filaments as differentiation markers of normal pancreas and pancreas cancer. Am J Pathol. 1992;140:559–568. [PMC free article] [PubMed] [Google Scholar]
- 10.Duval JV, Savas L, Banner BF. Expression of cytokeratins 7 and 20 in carcinomas of the extrahepatic biliary tract, pancreas. and gallbladder. Arch Pathol Lab Med. 2000;124:1196–1200. doi: 10.5858/2000-124-1196-EOCAIC. [DOI] [PubMed] [Google Scholar]
- 11.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scanlon CS, Van Tubergen EA, Inglehart RC. N.J. D'Silva, Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hinz B, Phan SH, Thannickal VJ, Prunotto M, Desmouliere A, Varga J, De Wever O, Mareel M, Gabbiani G. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am J Pathol. 2012;180:1340–1355. doi: 10.1016/j.ajpath.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura T, Iguchi H, Manabe T, Ohshio G, Yoshimura T, Wang ZH, Suwa H, Ishigami S, Imamura M. Quantitative analysis of collagen and collagen subtypes I, III, and V in human pancreatic cancer, tumor-associated chronic pancreatitis, and alcoholic chronic pancreatitis. Pancreas. 1995;11:357–364. doi: 10.1097/00006676-199511000-00007. [DOI] [PubMed] [Google Scholar]
- 15.Bachem MG, Schunemann M, Ramadani M, Siech M, Beger H, Buck A, Zhou S, Schmid-Kotsas A, Adler G. Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology. 2005;128:907–921. doi: 10.1053/j.gastro.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 16.Shintani Y, Hollingsworth MA, Wheelock MJ, Johnson KR. Collagen I promotes metastasis in pancreatic cancer by activating c-Jun NH(2)-terminal kinase 1 and up-regulating N-cadherin expression. Cancer Res. 2006;66:11745–11753. doi: 10.1158/0008-5472.CAN-06-2322. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes J, Stein H, Pileri S, Rivano MT, Gobbi M, Ralfkiaer E, Nielsen KM, Pallesen G, Bartels H, Palestro G. Prognostic relevance of tumour-cell growth fraction in malignant non-Hodgkin's lymphomas. Lancet. 1987;2:448–449. doi: 10.1016/s0140-6736(87)90977-9. [DOI] [PubMed] [Google Scholar]
- 19.Pezzilli R, Partelli S, Cannizzaro R, Pagano N, Crippa S, Pagnanelli M, Falconi M. Ki-67 prognostic and therapeutic decision driven marker for pancreatic neuroendocrine neoplasms (PNENs): a systematic review. Adv Med Sci. 2016;61:147–153. doi: 10.1016/j.advms.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Baecher-Allan C, Wolf E, Hafler DA. MHC class II expression identifies functionally distinct human regulatory T cells. J Immunol. 2006;176:4622–4631. doi: 10.4049/jimmunol.176.8.4622. [DOI] [PubMed] [Google Scholar]
- 21.Viallard JF, Blanco P, Andre M, Etienne G, Liferman F, Neau D, Vidal E, Moreau JF, Pellegrin JL. CD8+HLA-DR+ T lymphocytes are increased in common variable immunodeficiency patients with impaired memory B-cell differentiation. Clin Immunol. 2006;119:51–58. doi: 10.1016/j.clim.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Dunne MR, Michielsen AJ, O'Sullivan KE, Cathcart MC, Feighery R, Doyle B, Watson JA, O'Farrell NJ, Ravi N, Kay E. HLA-DR expression in tumor epithelium is an independent prognostic indicator in esophageal adenocarcinoma patients. Cancer Immunol Immunother. 2017;66:841–850. doi: 10.1007/s00262-017-1983-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sconocchia G, Eppenberger-Castori S, Zlobec I, Karamitopoulou E, Arriga R, Coppola A, Caratelli S, Spagnoli GC, Lauro D, Lugli A. HLA class II antigen expression in colorectal carcinoma tumors as a favorable prognostic marker. Neoplasia. 2014;16:31–42. doi: 10.1593/neo.131568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saraiva DP, Jacinto A, Borralho P, Braga S, Cabral MG. HLA-DR in cytotoxic T lymphocytes predicts breast cancer patients' response to neoadjuvant chemotherapy. Front Immunol. 2018;9:2605. doi: 10.3389/fimmu.2018.02605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, Giesen C, Catena R, Varga Z, Bodenmiller B. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14:873–876. doi: 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levine JH, Simonds EF, Bendall SC, Davis KL. A.D. Amir el, M.D. Tadmor, O. Litvin, H.G. Fienberg, A. Jager, E.R. Zunder, R. Finck, A.L. Gedman, I. Radtke, J.R. Downing, D. Pe'er, G.P. Nolan, Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci U S A. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 30.Yoon S, Huang KW, Reebye V, Mintz P, Tien YW, Lai HS, Saetrom P, Reccia I, Swiderski P, Armstrong B. Targeted delivery of C/EBPalpha-saRNA by pancreatic ductal adenocarcinoma–specific RNA aptamers inhibits tumor growth in vivo. Molecular therapy : the journal of the American Society of Gene Therapy. 2016;24:1106–1116. doi: 10.1038/mt.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon S, Armstrong B, Habib N, Rossi JJ. Blind SELEX approach identifies RNA aptamers that regulate EMT and inhibit metastasis. Molecular cancer research : MCR. 2017;15:811–820. doi: 10.1158/1541-7786.MCR-16-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Painter JT, Clayton NP, Herbert RA. Useful immunohistochemical markers of tumor differentiation. Toxicol Pathol. 2010;38:131–141. doi: 10.1177/0192623309356449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klymkowsky MW, Savagner P. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588–1593. doi: 10.2353/ajpath.2009.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myoteri D, Dellaportas D, Lykoudis PM, Apostolopoulos A, Marinis A, Zizi-Sermpetzoglou A. Prognostic evaluation of vimentin expression in correlation with Ki67 and CD44 in surgically resected pancreatic ductal adenocarcinoma. Gastroenterol Res Pract. 2017;2017:9207616. doi: 10.1155/2017/9207616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–3046. doi: 10.1007/s00018-011-0735-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Hu HY, Liu H, Zhang JW, Hu K, Lin Y. Clinical significance of Smac and Ki-67 expression in pancreatic cancer. Hepatogastroenterology. 2012;59:2640–2643. doi: 10.5754/hge12071. [DOI] [PubMed] [Google Scholar]
- 38.Feig C, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18:4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinn M, Denkert C, Striefler JK, Pelzer U, Stieler JM, Bahra M, Lohneis P, Dorken B, Oettle H, Riess H. alpha-Smooth muscle actin expression and desmoplastic stromal reaction in pancreatic cancer: results from the CONKO-001 study. Br J Cancer. 2014;111:1917–1923. doi: 10.1038/bjc.2014.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olivares O, Mayers JR, Gouirand V, Torrence ME, Gicquel T, Borge L, Lac S, Roques J, Lavaut MN, Berthezene P. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat Commun. 2017;8:16031. doi: 10.1038/ncomms16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armstrong T, Packham G, Murphy LB, Bateman AC, Conti JA, Fine DR, Johnson CD, Benyon RC, Iredale JP. Type I collagen promotes the malignant phenotype of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2004;10:7427–7437. doi: 10.1158/1078-0432.CCR-03-0825. [DOI] [PubMed] [Google Scholar]
- 42.Drifka CR, Loeffler AG, Mathewson K, Keikhosravi A, Eickhoff JC, Liu Y, Weber SM, Kao WJ, Eliceiri KW. Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget. 2016;7:76197–76213. doi: 10.18632/oncotarget.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.W. Zhao, X. Wang, K.H. Sun, L. Zhou, alpha-Smooth muscle actin is not a marker of fibrogenic cell activity in skeletal muscle fibrosis, PloS one, 13 (2018) e0191031. [DOI] [PMC free article] [PubMed]
- 44.Imai D, Yoshizumi T, Okano S, Uchiyama H, Ikegami T, Harimoto N, Itoh S, Soejima Y, Aishima S, Oda Y. The prognostic impact of programmed cell death ligand 1 and human leukocyte antigen class I in pancreatic cancer. Cancer Med. 2017;6:1614–1626. doi: 10.1002/cam4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ting DT, Wittner BS, Ligorio M, Vincent Jordan N, Shah AM, Miyamoto DT, Aceto N, Bersani F, Brannigan BW, Xega K. Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 2014;8:1905–1918. doi: 10.1016/j.celrep.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hantschel M, Pfister K, Jordan A, Scholz R, Andreesen R, Schmitz G, Schmetzer H, Hiddemann W, Multhoff G. Hsp70 plasma membrane expression on primary tumor biopsy material and bone marrow of leukemic patients. Cell Stress Chaperones. 2000;5:438–442. doi: 10.1379/1466-1268(2000)005<0438:hpmeop>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon S, Huang KW, Reebye V, Spalding D, Przytycka TM, Wang Y, Swiderski P, Li L, Armstrong B, Reccia I. Aptamer-drug conjugates of active metabolites of nucleoside analogs and cytotoxic agents inhibit pancreatic tumor cell growth. molecular therapy. Nucleic acids. 2017;6:80–88. doi: 10.1016/j.omtn.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]