Abstract
Purpose
The present study was intended to identify genetic causes of infertile patients with recurrent failure of in vitro fertilization (IVF)/intracytoplasmic sperm injection (ICSI) attempts.
Methods
Infertile patients with recurrent IVF/ICSI failure from Shanghai Ji Ai Genetics & IVF Institute and the Ninth Hospital affiliated with Shanghai Jiao Tong University were recruited. Genomic DNA samples were extracted from their peripheral blood. Whole-exome sequencing and Sanger validation were performed to identify candidate variants.
Results
We identified novel transducin-like enhancer of split 6 (TLE6) gene mutations in three patients with recurrent IVF/ICSI failure. One patient carried a homozygous missense mutation (c.1226G>A; p.Arg409Gln) with subsequent fertilization failure, while the other two patients carried either a homozygous missense mutation (c.1621G>A; p.Glu541Lys) or a compound heterozygous missense mutation (c.388G>A/c.1507G>A; p.Asp130Asn/p.Val503Ile) and had viable but low-quality embryos.
Conclusions
Our study expands the mutational and phenotypic spectrum of TLE6 and suggests the important role of TLE6 during embryonic development. Our findings have implications for the genetic diagnosis of female infertility with recurrent IVF/ICSI failure.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01653-0) contains supplementary material, which is available to authorized users.
Keywords: Female infertility, Embryonic arrest, Mutation, Transducin-like enhancer of split 6 (TLE6), Subcortical maternal complex (SCMC)
Introduction
Since the invention of in vitro fertilization (IVF) in 1978, a great number of infertile couples have been able to have their children through this procedure [1]. However, an increasing number of patients have been identified with recurrent failure of IVF/intracytoplasmic sperm injection (ICSI) attempts. In recent years, a few mutant genes have been found to be associated with recurrent failure of IVF/ICSI attempts, including TUBB8 (MIM: 616768) [2] and PATL2 (MIM: 614661) [3, 4], which are associated with oocyte maturation arrest; WEE2 (MIM: 614084), which is associated with fertilization failure [5]; PANX1, which is associated with oocyte death (MIM: 608420) [6]; and PADI6 (MIM: 610363), which is associated with early embryonic arrest [7].
The subcortical maternal complex (SCMC) is specifically expressed in mammalian oocytes and early embryos and is indispensable for pre-implantation embryonic development [8]. Transducin-like enhancer of split 6 (TLE6), which is encoded by TLE6, is an essential member of the SCMC along with maternal antigen that embryos require (MATER, also known as NLR family pyrin domain-containing 5 (NLRP5)), factor located in oocytes permitting embryonic development (FLOPED, also known as oocyte expressed protein (OOEP)), peptidylarginine deiminase type VI (PADI6), and KH domain-containing protein 3 (KHDC3, also known as Filia). The evidence gathered so far supports the physiological role of SCMC components in meiotic spindle positioning, mitochondria redistribution, translation regulation, and zygotic epigenetic reprogramming [9]. Recently, mutations in TLE6 (MIM: 612399) were found to be responsible for human embryonic lethality [10].
In the present study, we used whole-exome sequencing to identify genetic causes of infertile patients with recurrent IVF/ICSI failure. We discovered novel TLE6 mutations in three patients, thereby expanding the mutational and phenotypic spectrum of TLE6.
Materials and methods
Subjects
A total of 403 infertile patients with recurrent failure of IVF/ICSI attempts were recruited from the Shanghai Ji Ai Genetics & IVF Institute and the Ninth Hospital affiliated with Shanghai Jiao Tong University. A complete medical evaluation was performed, including reproductive history, physical examination, laboratory blood work, and semen analysis. Inclusion criteria were (i) married couples failing to conceive after 1 year (or longer) of regular unprotected sex, (ii) female patients with normal hormone levels (follicle-stimulating hormone (FSH) < 10 mIU/ml) and normal ovarian reserves (anti-mullerian hormone (AMH) > 1 ng/ml), and (iii) ≥ 2 failed attempts of IVF/ICSI. Patients with other known causes of infertility, including male factors, chromosome anomalies, and sexually transmitted infections, were excluded. A total of 871 fertile female controls were recruited from the Xinhua hospital affiliated to Shanghai Jiao Tong University. The study was approved by the Ethics Committee of Medical College of Fudan University.
Genomic DNA extraction
Genomic DNA samples of patients, their family members, and controls were isolated from peripheral blood using a HiPure Blood DNA Mini Kit (Magen, Guangzhou, China). The DNA concentration and purity were measured with a NanoDrop 1000 spectrophotometer (Thermo Scientific, Waltham, Massachusetts, USA).
Sequence analysis
Whole-exome capture was carried out using the Agilent SureSelect Whole Exome Enrichment Kit (Santa Clara, California, USA), and the sequencing was performed with the Illumina HiSeq 3000 platform (San Diego, California, USA). Sequences were aligned to the human genome assembly GRCh37, and variants were annotated as previously described [11]. The frequency of corresponding mutations was determined using the Exome Aggregation Consortium (ExAC) database (http://exac.broadinstitute.org/), and the functional effects of the mutations were predicted with the in silico algorithms PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and PROVEAN (http://provean.jcvi.org). The variants were prioritized based on the following filtering criteria: (i) a frequency below 0.1% for homozygous variants or below 1% for compound heterozygous variants in the ExAC database, (ii) loss of function alleles or damaging missense variants predicted by PolyPhen-2 or PROVEAN, (iii) variants with high gene expression in human oocytes and embryos according to our in-house RNA sequencing data, and (iv) variants with an embryogenesis-related function. Homozygosity mapping was performed with HomozygosityMapper for affected individuals from family 1 and family 2 to determine the existence of candidate homozygous variants. The candidate variants were then confirmed by Sanger sequencing in patients and other family members using the primers shown in Supplementary Table 1.
Results
Clinical characteristics of patients with mutations in TLE6
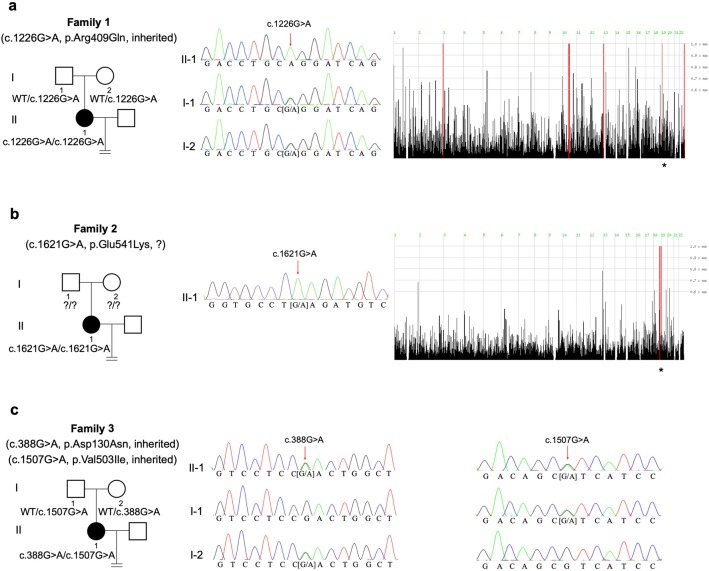
We discovered novel TLE6 mutations in three cases out of 403 infertile patients with recurrent failure of IVF/ICSI attempts. The affected individuals from three families had been diagnosed with primary infertility for several years and had undergone multiple failed IVF/ICSI cycles. Their husbands exhibited normal sperm counts, motility, and morphological features. The family pedigrees are shown in Fig. 1, and the patients’ clinical information is summarized in Table 1.
Fig. 1.
Identification of TLE6 mutations in affected individuals. Pedigrees of family 1 (a), family 2 (b), and family 3 (c) carrying TLE6 mutations that lead to embryonic arrest. Sanger sequencing confirmation and homozygosity mapping are shown next to the pedigrees. Squares denote males and circles denote females, filled symbols represent patients with primary infertility, and open symbols represent unaffected individuals. Homozygous regions harboring the strongest signal are indicated in red, and the asterisk (*) indicates the area where TLE6 is located
Table 1.
Oocyte and embryo characteristics of the IVF and ICSI attempts of the three probands
| Patient | Age (years) | Infertility duration (years) | IVF/ICSI attempts | Oocytes retrieved | Fertilized oocytes | Viable embryos on day 3 | Embryos transferred | Outcomes |
|---|---|---|---|---|---|---|---|---|
| Family 1, II-1 | 34 | 6 | 1st ICSI | 8 | 0 | 0 | 0 | Failed to fertilize |
| 2nd ICSI | 6 | 0 | 0 | 0 | Failed to fertilize | |||
| Family 2, II-1 | 26 | 4 | 1st IVF | N/A | 1 | 0 | 0 | Embryonic arrest |
| 2nd ICSI | 2 | 1 | 0 | 0 | Embryonic arrest | |||
| 3rd ICSI | 11 | 7 | 7 | 0 | Embryonic arrest | |||
| 4th ICSI | 8 | 1 | 1 | 0 | Embryonic arrest | |||
| 5th ICSI | 8 | 3 | 1 | 2 | Failed to establish pregnancy | |||
| Family 3, II-1 | 35 | 6 | 1st IVF | 8 | 4 | 0 | 0 | Embryonic arrest |
| 2nd ICSI | 8 | 7 | 0 | 0 | Embryonic arrest | |||
| 3rd ICSI | 14 | 11 | N/A | 2 | Failed to establish pregnancy |
The 34-year-old proband in family 1 had two failed ICSI attempts. Altogether, 14 oocytes were retrieved, and they all failed to be fertilized.
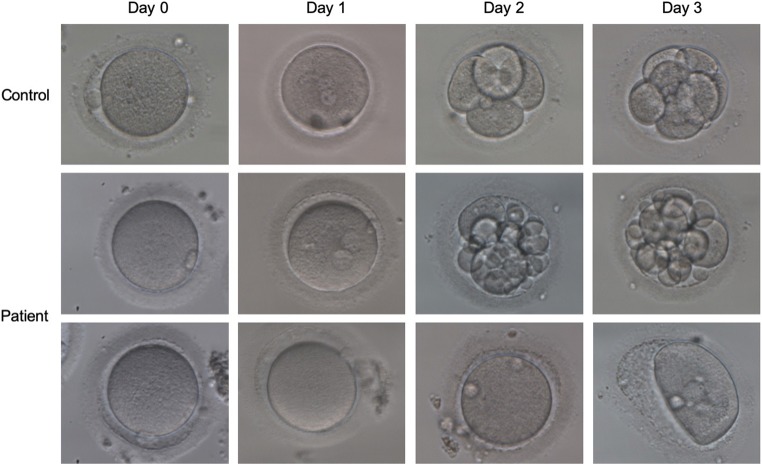
The proband in family 2 was 26 years old at examination and had undergone five failed IVF/ICSI cycles. In the first two cycles, only one oocyte was successfully fertilized but failed to develop further. In her third ICSI attempt, out of 11 oocytes, 8 appeared as normal metaphase II oocytes and were injected with single sperm, resulting in two grade III embryos and five grade IV embryos on day 3. However, all of the embryos were arrested on day 6. In her fourth ICSI attempt, 6 out of 8 oocytes were selected for sperm injection. Most of them displayed abnormal fertilization with zero-pronucleus (PN) zygotes, 3PN zygotes, or degradation, and only one oocyte was normally fertilized with 2PN, but this stopped developing on day 3. In the fifth ICSI treatment cycle, 8 oocytes were obtained. Three eggs were successfully fertilized, and they grew into one grade III and two grade IV embryos on day 2. Two embryos (one grade III and one grade IV) were selected for transplantation, but no pregnancy was established. Fig. 2 shows the phenotypic features of the patient’s oocytes and embryos.
Fig. 2.
Phenotype of oocytes/embryos from the patient from family 2 (II-1). The morphologies of the oocytes and embryos from a control and the patient from family 2 were photographed under a light microscope on day 0, day 1, day 2, and day 3 after fertilization
The proband in family 3 was 35 years old with a history of three IVF/ICSI attempts. In her first IVF attempt, 8 oocytes were obtained and 4 were fertilized successfully, but no viable embryos were available on day 3. In the second ICSI attempt, 7 out of 8 oocytes were successfully fertilized, but no high-quality embryos were obtained on day 3. In the third ICSI attempt, 11 out of 14 oocytes were fertilized and developed into poor-quality embryos. The two viable embryos (one grade IV and one grade V) were transferred but failed to establish a pregnancy.
Mutational spectrum of TLE6
A homozygous missense mutation in exon 13 of TLE6 (NM_001143986.1; c.1226G>A; p.Arg409Gln) was identified in the proband of family 1 (Fig. 1a). The mutation was verified by Sanger sequencing, and both of her parents carried a heterozygous TLE6 mutation, indicating a recessive inheritance pattern (Fig. 1a). Following the in silico analysis, the c.1226G>A (p.Arg409Gln) mutation was predicted by PolyPhen-2 to be probably damaging.
In family 2, another homozygous missense mutation c.1621G>A (p.Glu541Lys) (Fig. 1b) was identified and was predicted to be probably damaging and deleterious by PolyPhen-2 and PROVEAN, respectively.
A compound heterozygous missense mutation in TLE6 was identified in the proband of family 3 consisting of c.388G>A (p.Asp130Asn) in exon 7 inherited from her mother and c.1507G>A (p.Val503Ile) in exon 15 inherited from her father (Fig. 1c). Both missense mutations were classified as benign and neutral by PolyPhen-2 and PROVEAN, respectively. The locations, frequencies, and in silico analysis of the mutations are shown in Table 2.
Table 2.
Overview of the TLE6 mutations observed in the affected families
| Observed in families | Genomic DNA position on chr.19 (bp) | Exon | cDNA change | Protein change | PolyPhen-2 | PROVEAN | ExAC (total) |
|---|---|---|---|---|---|---|---|
| Family 1 | 2,989,765 | 13 | c.1226G>A | p.Arg409Gln | Probably damaging | Neutral | N/A |
| Family 2 | 2,994,904 | 17 | c.1621G>A | p.Glu541Lys | Probably damaging | Deleterious | N/A |
| Family 3 | 2,987,083 | 7 | c.388G>A | p.Asp130Asn | Benign | Neutral | 8.26E-06 |
| Family 3 | 2,993,550 | 15 | c.1507G>A | p.Val503Ile | Benign | Neutral | 5.99E-05 |
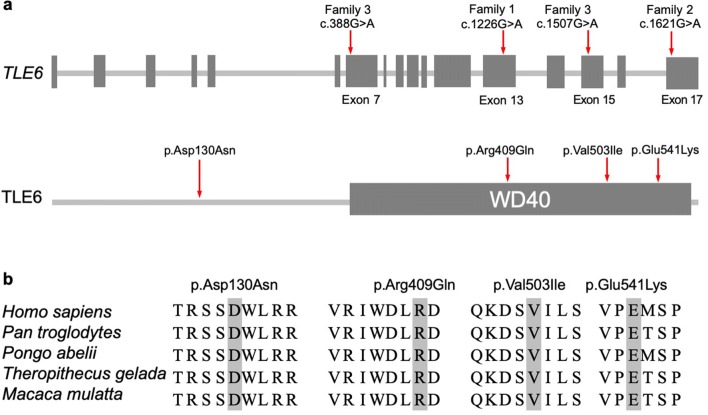
The locations of the TLE6 mutations and the conservation analysis among different species are indicated in Fig. 3. The positions of these missense mutations are highly conserved among different primates, but less so in other species (Fig. 3b).
Fig. 3.
The locations and conservation of mutated residues in TLE6. a Locations of mutations in TLE6. The positions of all mutations are indicated in the gene structure and protein structure of TLE6. b Conservation of mutations in TLE6. The affected amino acids are compared among five primate species in a conservation analysis
Discussion
In this study, we identified two homozygous missense mutations and one compound heterozygous missense mutation in TLE6 in three patients with recurrent IVF/ICSI failure. Most of the oocytes from the affected individuals failed to be fertilized. Only a few oocytes displayed normal fertilization, but these all developed into poor-quality embryos that failed to result in pregnancy.
TLE6 is a member of the Groucho/Transducin-like Enhancer of split (TLE) transcriptional co-repressor gene family in mammals [12]. In mice, Tle6 mRNA transcripts accumulate during oogenesis, peak in fully grown oocytes, degrade following meiotic maturation and ovulation, and disappear entirely during embryogenesis, while Tle6 proteins persist in preimplantation embryos up to the blastocyst stage [13]. Tle6Null female mice are completely infertile with embryos exhibiting delayed progression to the two-cell stage and abnormal development at E2.5 and E3.5, which lead to the discovery of its role in stabilizing the SCMC’s interactions with the cytoplasmic F-actin meshwork and thus ensuring the central positioning of the spindle apparatus and subsequent symmetric division in mouse zygotes [14]. TLE6 is also proposed to be a cAMP-dependent protein kinase (PKA) substrate that is phosphorylated following meiotic resumption, thus providing insights into the molecular mechanisms of this process [15].
Human TLE6 shares 44% identity with its murine homolog, and in humans, TLE6 shows an oocyte and early embryo-specific expression pattern [16]. In 2015, a single homozygous substitution (c.1529C>A) in TLE6 resulting in impaired phosphorylation by PKA and decreased binding to other SCMC proteins was found to be responsible for fertilization failure [10]. Most recently, a homozygous frameshift mutation (c.1133delC) was found to be associated with the phenotype of early embryonic arrest on day 3 [17]. In the present study, we identified novel TLE6 mutations from women with recurrent IVF/ICSI failures. Similar to previously reported cases of fertilization failure and early embryonic arrest, the oocytes from the patient of family 1 (p.Arg409Gln) failed to be fertilized, while a small fraction of oocytes from the other two patients (p.Glu541Lys; p.Asp130Asn/p.Val503Ile) could be fertilized and develop into viable but low-quality embryos for transfer, suggesting that different mutations in TLE6 cause phenotypic variability in patients.
In summary, our study identified novel mutations in TLE6 associated with fertilization failure and early embryonic arrest and thus expanded the mutational spectrum of TLE6 and the phenotypic spectrum of patients with such mutations. Our findings add new information on the genetic basis of female infertility and suggest that TLE6 might be a genetic diagnostic marker for recurrent IVF/ICSI failure.
Electronic supplementary material
(DOCX 251 kb)
Funding information
This study was supported by the National Key Research and Development Program of China (2018YFC1003800, 2017YFC1001500, and 2016YFC1000600), the National Basic Research Program of China (2015CB943300), the National Natural Science Foundation of China (81725006, 81822019, 81771581, 81571501, and 81771649), the Shanghai Rising-Star Program (17QA1400200), and the Natural Science Foundation of Shanghai (17ZR1401900).
Compliance with ethical standards
The study was approved by the Ethics Committee of Medical College of Fudan University.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Lin and Hua Xu contributed equally to this work.
Contributor Information
Xiaoxi Sun, Email: xiaoxi_sun@aliyun.com.
Qing Sang, Email: sangqing@fudan.edu.cn.
References
- 1.Harris M. Louise: the test-tube miracle. Nurs Mirror. 1978;147:xv–xvi+. [PubMed] [Google Scholar]
- 2.Feng R, Sang Q, Kuang Y, Sun X, Yan Z, Zhang S, Shi J, Tian G, Luchniak A, Fukuda Y, Li B, Yu M, Chen J, Xu Y, Guo L, Qu R, Wang X, Sun Z, Liu M, Shi H, Wang H, Feng Y, Shao R, Chai R, Li Q, Xing Q, Zhang R, Nogales E, Jin L, He L, Gupta ML Jr, Cowan NJ, Wang L. Mutations in TUBB8 and human oocyte meiotic arrest. N Engl J Med. 2016;374:223–232. doi: 10.1056/NEJMoa1510791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B, Zhang Z, Sun X, Kuang Y, Mao X, Wang X, Yan Z, Li B, Xu Y, Yu M, Fu J, Mu J, Zhou Z, Li Q, Jin L, He L, Sang Q, Wang L. Biallelic mutations in PATL2 cause female infertility characterized by oocyte maturation arrest. Am J Hum Genet. 2017;101:609–615. doi: 10.1016/j.ajhg.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddirevula S, Coskun S, Alhassan S, Elnour A, Alsaif HS, Ibrahim N, Abdulwahab F, Arold ST, Alkuraya FS. Female infertility caused by mutations in the oocyte-specific translational repressor PATL2. Am J Hum Genet. 2017;101:603–608. doi: 10.1016/j.ajhg.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sang Q, Li B, Kuang Y, Wang X, Zhang Z, Chen B, Wu L, Lyu Q, Fu Y, Yan Z, Mao X, Xu Y, Mu J, Li Q, Jin L, He L, Wang L. Homozygous mutations in WEE2 cause fertilization failure and female infertility. Am J Hum Genet. 2018;102:649–657. doi: 10.1016/j.ajhg.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sang Q, Zhang Z, Shi J, Sun X, Li B, Yan Z, et al. A pannexin 1 channelopathy causes human oocyte death. Sci Transl Med. 2019;11:eaav8731. doi: 10.1126/scitranslmed.aav8731. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Shi Y, Fu J, Yu M, Feng R, Sang Q, Liang B, Chen B, Qu R, Li B, Yan Z, Mao X, Kuang Y, Jin L, He L, Sun X, Wang L. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99:744–752. doi: 10.1016/j.ajhg.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu X, Gao Z, Qin D, Li L. A maternal functional module in the mammalian oocyte-to-mmbryo transition. Trends Mol Med. 2017;23:1014–1023. doi: 10.1016/j.molmed.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Bebbere D, Masala L, Albertini DF, Ledda S. The subcortical maternal complex: multiple functions for one biological structure? J Assist Reprod Genet. 2016;33:1431–1438. doi: 10.1007/s10815-016-0788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alazami AM, Awad SM, Coskun S, Al-Hassan S, Hijazi H, Abdulwahab FM, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240. doi: 10.1186/s13059-015-0792-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu J, Wang W, Chen B, Wu L, Li B, Mao X, Zhang Z, Fu J, Kuang Y, Sun X, Li Q, Jin L, He L, Sang Q, Wang L. Mutations in NLRP2 and NLRP5 cause female infertility characterised by early embryonic arrest. J Med Genet. 2019;56:471–480. doi: 10.1136/jmedgenet-2018-105936. [DOI] [PubMed] [Google Scholar]
- 12.Bajoghli B. Evolution of the Groucho/Tle gene family: gene organization and duplication events. Dev Genes Evol. 2007;217:613–618. doi: 10.1007/s00427-007-0167-y. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–425. doi: 10.1016/j.devcel.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, et al. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun. 2014;5:4887. doi: 10.1038/ncomms5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan FE, Padilla-Banks E, Bernhardt ML, Ord TS, Jefferson WN, Moss SB, et al. Transducin-like enhancer of split-6 (TLE6) is a substrate of protein kinase A activity during mouse oocyte maturation. Biol Reprod. 2014;90:63. doi: 10.1095/biolreprod.113.112565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu K, Yan L, Zhang X, Lu X, Wang T, Yan J, Liu X, Qiao J, Li L. Identification of a human subcortical maternal complex. Mol Hum Reprod. 2015;21:320–329. doi: 10.1093/molehr/gau116. [DOI] [PubMed] [Google Scholar]
- 17.Wang X, Song D, Mykytenko D, Kuang Y, Lv Q, Li B, Chen B, Mao X, Xu Y, Zukin V, Mazur P, Mu J, Yan Z, Zhou Z, Li Q, Liu S, Jin L, He L, Sang Q, Sun Z, Dong X, Wang L. Novel mutations in genes encoding subcortical maternal complex proteins may cause human embryonic developmental arrest. Reprod Biomed Online. 2018;36:698–704. doi: 10.1016/j.rbmo.2018.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 251 kb)