Abstract
Purpose
To identify the frequency of Y chromosome microdeletions in Indian populations and to quantitatively estimate the significance of association between these deletions and male infertility.
Methods
A total of 379 infertile males (302 azoospermic and 77 oligozoospermic infertile males) and 265 normozoospermic fertile males were evaluated for Y chromosome microdeletions (YCD) using PCR amplification and gel electrophoresis. Meta-analyses were performed on AZFa (2079 cases and 1217 controls), AZFb (2212 cases and 1267 controls), AZFc (4131 cases and 2008 controls), and AZFb+c (1573 cases and 942 controls) deletions data to quantitatively estimate the significance of association between these deletions and male infertility in Indian populations.
Results
The results revealed that out of 379 infertile azoospermic and oligozoospermic males, 38 (10.02%) had AZF deletions. No deletion was found in control samples. The highest percentage of deletions was observed in the AZFc region, followed by AZFa and AZFb. Qualitative analysis showed that AZF deletions were present in 0.59 to 32.62% (average 13.48%) of infertile cases in Indian populations. Meta-analysis revealed a significant association of AZFa (OR = 6.74, p value = 0.001), AZFb (OR = 4.694, p value = 0.004), AZFc (OR = 13.575, p value = 0.000), and AZFb+c (OR = 5.946, p value = 0.018) deletions with male infertility.
Conclusion
AZF deletions were seen in 10.02% of azoospermic and oligozoospermic cases with the highest frequency of AZFc deletions. Pooled analysis for all studies showed deletion frequency from 0.59 to 32.62% (average = 13.48%). Meta-analysis showed significant association of AZFa, AZFb, and AZFb+c deletions with male infertility. Analysis of Y chromosome microdeletions should be reckoned as an essential testing for diagnostic and therapeutic purposes.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01661-0) contains supplementary material, which is available to authorized users.
Keywords: Y-deletions, AZF deletions, Azoospermia factor, Male infertility, Oligozoospermia, Azoospermia
Introduction
Y chromosome microdeletions and their relation with azoospermia were first discovered in the year 1976 by Tiepolo and Zuffardi [1]. The Y chromosome molecular screening has prognostic value and is advised in patients with azoospermia and oligozoospermia. Y-deletion analysis in male infertility has reached a stage where its clinical adoption is increasing. Nevertheless, the availability of Y-deletion type and frequency data for various populations can pose a hindrance in its inclusion in infertility clinics workup. It is noteworthy that certain deletions are risk factors in one population, but not in others [2]. Therefore, a careful workup for the Y-deletion frequency for each population is a pre-requisite for the adoption of deletion analysis in infertility clinics.
The European Academy of Andrology (EAA) and the European Molecular Genetics Quality Network (EMQN) recommended that Y-deletion analysis should be undertaken in a phased manner, beginning with six deletion markers, followed by the analysis of further markers to define the deletion. EAA guidelines have recommended a set of six sequence-tagged sites (STS) markers covering the AZFa (sY84 and sY86), AZFb (sY127 and sY134), and AZFc (sY254 and sY255) regions, which can be analyzed using a multiplex PCR protocol. This set of markers can be used to detect almost all (over 95%) clinically important deletions in the AZF region [3].
We sought to determine the frequency of Y chromosome microdeletions in an Indian population using this set of markers. Over the past two decades, a number of studies from the Indian sub-continent have analyzed Y-deletion in Indian populations. We pooled data from all Indian studies to figure out the level of association between various Y-deletions and male infertility and to estimate the overall frequency of Y chromosome deletions in Indian populations.
Materials and methods
Subjects and sample collection
The study was approved by the Institutional Ethics Committee of CSIR-Central Drug Research Institute, Lucknow. Infertility was defined as the inability of initiating a pregnancy after 1 year or more of regular unprotected sexual intercourse. Informed written consents were obtained from all the participants of the study. The subjects were of Indo-European (Caucasians) ethnicity. Detailed medical history was obtained from each patient, and physical examination was performed before sample collection. Individuals with known history of genital tract obstruction/dysfunction (varicocele, obstructive azoospermia) and congenital defects in structure of the urogenital system (Young’s syndrome, cystic fibrosis, etc.), patients undergoing radiotherapy and those exposed to radiations as a part of their treatment, those with habitual excessive alcohol consumption and drug abuse (ecstasy, marijuana, and recreational substances), or those with known karyotype abnormalities were excluded.
Sample size was calculated using an online calculator (http://osse.bii.a-star.edu.sg/calculation1.php), setting the desired power to 80%, significance level to 95%, minor allele frequency in cases to 10%, and in controls to 0% (computed from overall data for Indian populations). This suggested a sample size of 73 each for case and control groups. Sample collection and semen analysis were performed as per the WHO criteria [4]. Semen analyses were carried out thrice, after 3 to 4 days of sexual abstinence to ascertain infertility status of the patients. The classification of patients was done as azoospermic (absence of sperm in semen) and oligozoospermic (sperm count, < 15 million/ml). After screening 391 patients, we collected 379 patient samples, consisting of 302 azoospermic and 77 oligozoospermic infertile males. Twelve patients were excluded due to reasons stated in the exclusion criteria. The diagnosis of azoospermia was confirmed after semen centrifugation followed by pellet analysis that was performed at least twice to ascertain azoospermic phenotype. For comparison purposes, 265 normozoospermic proven fertile volunteer males with age group (23–38 years) were included. The controls belonged to the same age-group and had the same ethnicity as patients. A confirmation of paternity in the last 2 years was considered as a proof of their fertility status. For genetic analysis, we collected a 3-ml blood sample from each participant in EDTA-coated tubes.
DNA isolation
Genomic DNA was isolated from peripheral blood samples using DNA/RNA extraction kits (Illumina, San Diego, CA, USA), following the manufacturer’s protocol. DNA concentration and purity were estimated using spectrophotometry (NanoDrop 2000, Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis.
Deletion analysis
Y-deletion analysis was performed according to the recommendations of the European Academy of Andrology (EAA) and the European Quality Monitoring Network Group (EMQN), which included six standard sequence-tagged site (STS) markers (sY86 and sY84 for AZFa, sY127 and sY134 for AZFb, and sY255 and sY254 for AZFc) and two control primers specific to SRY and ZFY regions [3]. The sequences of primers used in this study along with their product size and annealing temperature are given in Table 1.
Table 1.
Sequences of the sequence-tagged site (STSs) primers used for deletions analysis
| Region | Primers | Forward | Reverse | Product size (bp) | Annealing temperature (°C) |
|---|---|---|---|---|---|
| AZFa | SY84 | 5′-AGA AGG GTC CTG AAA GCA GGT-3′ | 5′-GCC TAC TAC CTG GAG GCT TC-3′ | 326 | 57 |
| SY86 | 5′-GTG ACA CAC AGA CTA TGC TTC-3′ | 5′- ACA CAC AGA GGG ACA ACC CT - 3′ | 318 | 56 | |
| AZFb | SY127 | 5′-GGC TCA CAA ACG AAA AGA AA-3′ | 5′-CTG CAG GCA GTA ATA AGG GA-3′ | 274 | 56 |
| SY134 | 5′-GTC TGC CTC ACC ATA AAA CG-3′ | 5′-ACC ACT GCC AAA ACT TTC AA-3′ | 301 | 57 | |
| AZFc | SY254 | 5′-GGG TGT TAC CAG AAG GCA AA-3′ | 5′-GAA CCG TAT CTA CCA AAG CAG C-3′ | 380 | 57 |
| SY255 | 5′-GTT ACA GGA TTC GGC GTG AT-3′ | 5′-CTC GTC ATG TGC AGC CAC-3′ | 123 | 57 | |
| Internal controls | SRY | 5′-GAA TAT TCC CGC TCT CCG GA-3′ | 5′-GCT GGT GCT CCA TTC TTG AG-3′ | 472 | 57 |
| ZFX/Y | 5′-ACC RCT GTA CTG ACT GTG ATT ACA C-3′ | 5′-GCA CYT CTT TGG TAT CYG AGA AAG T-3′ | 495 | 57 |
The total reaction volume was 10 μl, which consisted of 5 μl of master mix, 0.5 μl of both forward and reverse STS markers, and 4 μl diluted DNA samples. The mixture was centrifuged at 2000g for 10 s. The cycling conditions were as follows: initial denaturation at 95 °C for 10 min followed by 94 °C for 30 min, primer annealing at 56–57 °C for 25 s and 72 °C for 1 min for polymerization followed by a final polymerization step at 72 °C for 10 min. The analysis of the reaction products was done by electrophoresis at 100 V on 2% agarose gel (Sigma, USA). PCR bands were visualized by the ChemiDoc XRS+ system (CA, USA). In order to rule out the methodological errors, the samples with observed deletion were repeated twice to confirm the genotype as true deletions.
Statistical analysis
The frequency of Y-deletions was compared between cases and controls using an online biostatistical tool (www.vassarstats.net). p values < 0.05 were considered to be statistically significant.
Meta-analysis
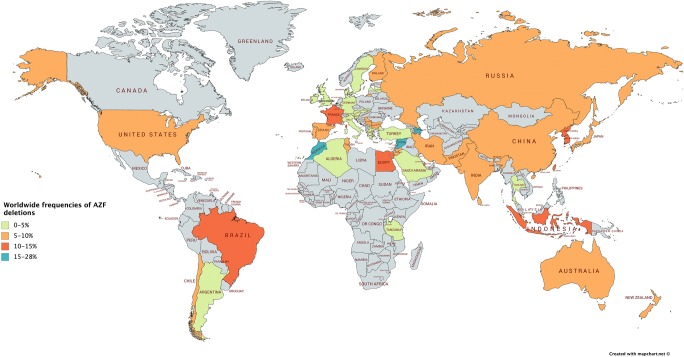
A few studies over the last two decades have analyzed Y-deletions in various populations across the world (Fig. 1) and Indian sub-continent [2, 5–14]. Most of these studies supported the association of Y-deletions with male infertility; however, the frequency of different deletions varied across the studies. Therefore, we undertook meta-analyses by pooling all the published data for Indian populations in order to figure out the overall deletion frequency and its quantitative relation with infertility.
Fig. 1.
The frequency of Y-deletions across various populations in the world
Identification of studies
Relevant studies were selected using a systematic search in public databases “PubMed (www.pubmed.com),” “GoogleScholar (scholar.google.co.in),” and “ScienceDirect (www.sciencedirect.com).” Literature search was conducted using the keywords “Y chromosome deletions,” “AZF deletions,” “AZFa deletions,” “AZFb deletions,” “AZFc deletions,” and “male infertility” in various combinations. All the articles were carefully read for the exclusion of irrelevant studies. Only the studies published in English language were included for further analysis. We did not specify a minimum sample size as the standard for inclusion of a study in the analysis. To avoid the occurrence of “double-counting” of the data and overstating of the risk estimate, we included the studies with the largest and most complete data for duplicate studies. Some of the studies had not provided data for the control group, forcing their exclusion. To prevent errors in the pooled analysis, the data extraction was performed by VS and WA, independently, and discrepancies were resolved by discussion.
Data extraction
The data against the following variables were obtained from each study: first author’s name, year of publication, source of the samples, and deletion frequency in the cases and controls.
Inclusion and exclusion criteria
The hits obtained through literature search were subjected to well-defined inclusion/exclusion criteria to select the studies for pooled analysis. Inclusion criteria were as follows: (i) the study aimed at analyzing the association of AZF deletions with male infertility risk, (ii) each of the trial was an independent case-control study, (iii) inclusion of the patients was performed according to the standard diagnostic parameters, (iv) the purpose of all the studies was similar, and (v) standard methods were used to analyze the AZF deletions. The studies that failed to provide a detailed description of the subjects, raw data, and other information required to specifically understand the study design and the data therein were excluded.
Statistical analysis
The meta-analysis was performed using the Comprehensive Meta-Analysis software (version-2). Dichotomous data in the form of the number of individuals having AZFa, AZFb, AZFc, and AZFb+c deletions in the case and control groups and the total sample size in each group were fed into the software. Odds ratio (OR) and 95% CI were chosen as the “effect size.” The heterogeneity between individual studies was tested using chi-square-based Q test, and p values > 0.10 suggested a lack of heterogeneity across the studies. The pooled OR and the corresponding 95% CIs were computed using the fixed effects model (the Mantel-Haenszel method) in the absence of heterogeneity among pooled studies, failing which, a random effects models was preferred (DerSimonian and Laird 1986). The presence of publications bias was assessed using the funnel plot of standard error (std error vs log odds ratio) and the Egger’s regression test of significance. A sensitivity analysis was performed to identify the studies that could have significantly biased the overall conclusion.
Trial sequential analysis (TSA)
The outcomes of a meta-analysis could be subject to systematic (bias) or random errors (play of chance) due to repeated significance testing, dispersed data, and potential publication bias. Bias from trials with publication bias, small trial bias, and low methodological quality may result in false p value. Therefore, we utilized trial sequential analysis (TSA) tool (Copenhagen Trial Unit, Center for Clinical Intervention Research, Denmark) to overcome these limitations, detect false-positive or false-negative errors, and calculate the required information size (RIS) for reliability of meta-analysis [15]. TSA calculates the required information size (number of samples) by adjusting the significance level for dispersed data and confirms statistical reliability of the data. Some of the earlier studies have demonstrated that the outcomes of TSA are more reliable than those of the traditional meta-analysis [16, 17]. RIS was calculated considering an overall type I error of 5% and type II error of 20%. A two-sided graph is plotted by TSA, where red straight lines are indicative of significance boundaries of the conventional meta-analysis, the blue line shows cumulative Z-score, and the red lines sloping inwards represent trial sequential monitoring boundaries with adjusted p values.
Results
Deletion analysis
The deletion analysis was performed in 644 subjects (302 azoospermic; 77 oligozoospermic, and 265 normozoospermic fertile controls). Thirty-eight (10.02%) of 379 infertile patients presented with AZF microdeletions. The frequency of microdeletions was 10.93% (33 out of 302) and 6.49% (5 out of 77) in azoospermic and oligozoospermic males, respectively. The azoospermic patients showed a higher number of deletions in comparison to oligozoospermic patients (Table 2). No deletions were detected in the control group. We computed the power of the study and found it to be 100%.
Table 2.
The frequency of AZF deletions in infertile and fertile subjects
| Group | AZFa | AZFb | AZFc |
|---|---|---|---|
| Infertile group (n = 379) | 7 (1.8%) | 8 (2.11%) | 23 (6.07%) |
| Fertile group (n = 265) | 0 (0%) | 0 (0%) | 0 (0%) |
AZFc deletions were most frequent
AZFc deletions were the most common deletions (6.07%) in comparison to the AZFa (1.8%) and AZFb (2.11%) deletions (Table 2). We found AZFc deletions in 18 azoospermic (5.9%) and five oligozoospermic men (6.49%) (Tables 3 and 4). The comparison of AZFc deletions frequency revealed a highly significant difference between cases and controls (p < 0.001) (Table 2) with an increased infertility risk in deletion carriers (Tables 3 and 4). AZFa (2.3%) and AZFb (2.6%) deletions were seen in comparable frequencies in azoospermic cases only (Table 3).
Table 3.
Statistical analysis of deletion data between azoospermic and control groups (Fisher’s exact probability test)
| Deletions | Azoospermia group | Cases N = 302 (%) | Controls N = 265 (%) | Statistical comparison/one tailed p value* |
|---|---|---|---|---|
| AZFa | With deletions | 7 (2.3) | 0 (0) | 0.0117 |
| AZFb | With deletions | 8 (2.6) | 0 (0) | 0.0061 |
| AZFc | With deletions | 18 (5.9) | 0 (0) | 0.000009 |
Table 4.
Statistical analysis of deletion data between oligozoospermic and control groups (Fisher’s exact probability test)
| Deletions | Oligozoospermia group | Cases N = 77 (%) | Controls N = 265 (%) | Statistical comparison/one-tailed p value* |
|---|---|---|---|---|
| AZFa | With deletions | 0 (0.00) | 0 (0) | - |
| AZFb | With deletions | 0 (0.00) | 0 (0) | - |
| AZFc | With deletions | 5 (6.49) | 0 (0) | 0.0005 |
Meta-analysis
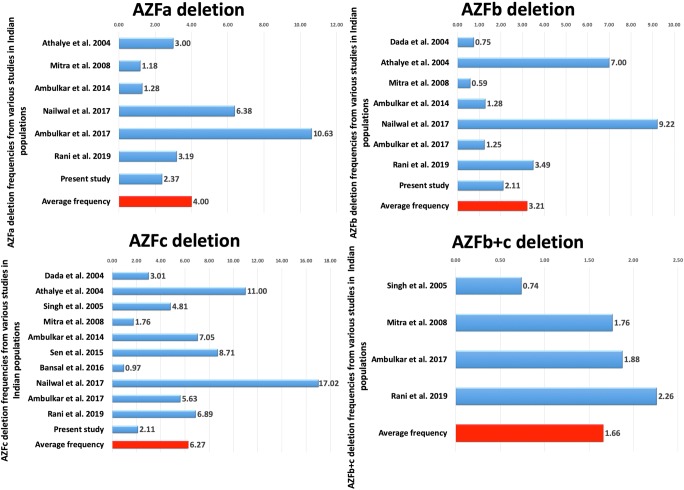
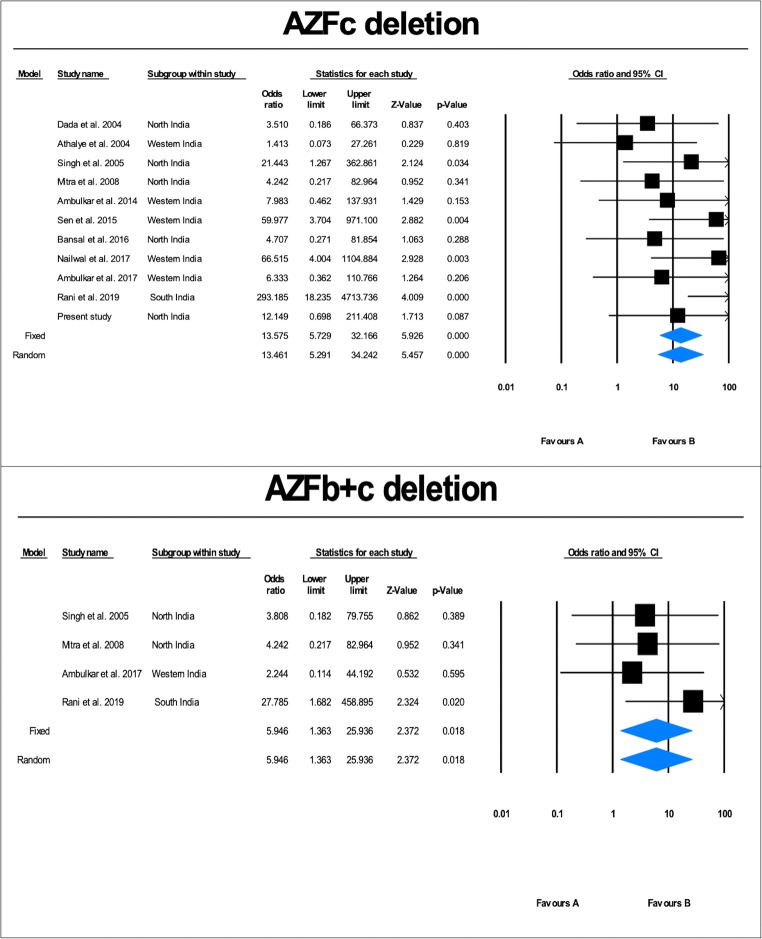
A comparison across the published studies showed the frequency of AZF deletions from 0.59 to 32.62% (average = 13.48%) in infertile patients [2, 5–14] (Fig. 2). All the cases which were included in these studies were pathologically confirmed with age-matched controls recruited from healthy populations. The main characteristics of these studies are depicted in the Supplementary data 1. As the studies pooled in the present analysis were performed across different laboratories in India, a heterogeneity test was run, which showed a lack of heterogeneity (p > 0.1). Therefore, the conclusions could be drawn on the basis of fixed effects model; however, we have presented results of both fixed and random effects models of analysis (Table 5).
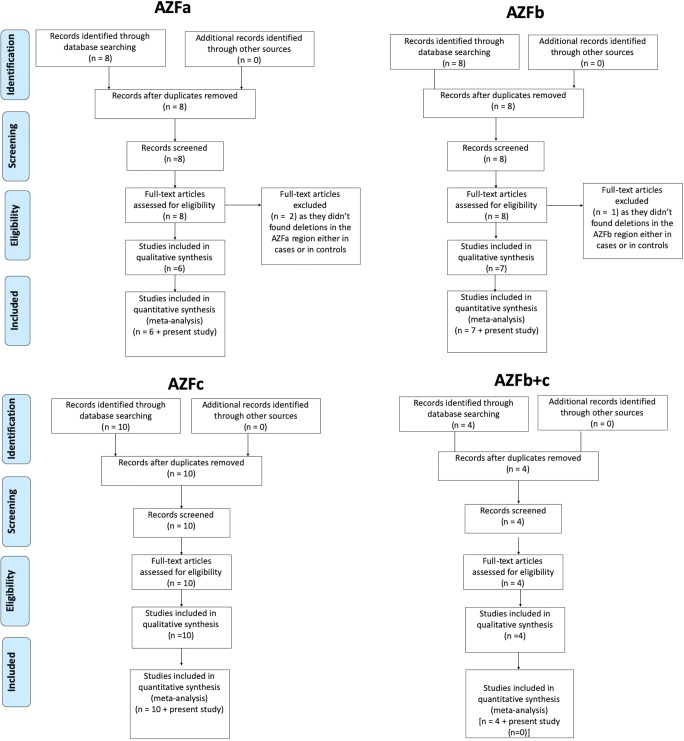
Fig. 2.
PRISMA flow diagram for screening of literature and selection of studies for meta-analysis
Table 5.
Summary of the results of meta-analyses
| Type of deletion | No. of studies | Test model | Type of association | Test of heterogeneity | Publication bias | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | p | Q | p | I2 | Egger’s p | |||
| AZFa | 7 | Fixed | 6.231 | 2.062–18.832 | 0.001 | 6.728 | 0.346 | 10.82 | 0.00 |
| Random | 6.162 | 6.162–1.909 | 0.002 | ||||||
| AZFb | 8 | Fixed | 4.694 | 1.631–13.503 | 0.004 | 7.923 | 0.339 | 11.65 | 0.10 |
| Random | 4.621 | 1.500–14.233 | 0.008 | ||||||
| AZFc | 11 | Fixed | 13.575 | 5.729–32.166 | 0.000 | 11.710 | 0.304 | 14.603 | 0.00 |
| Random | 13.461 | 5.291–34.242 | 0.000 | ||||||
| AZFb+c | 4 | Fixed | 5.946 | 1.363–25.936 | 0.018 | 1.703 | 0.636 | 82.17 | 0.30 |
| Random | 5.946 | 1.363–25.936 | 0.018 | ||||||
AZFa deletion
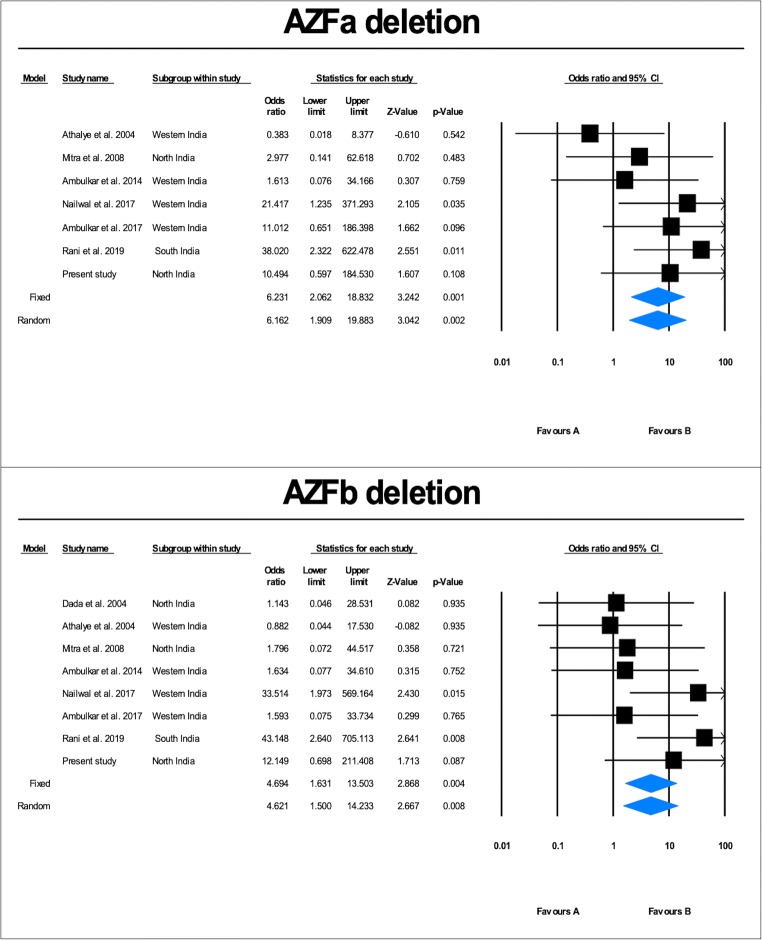
Literature search identified nine studies on AZFa deletions. Two studies, viz., Dada et al. and Singh et al., were excluded as they did not find any deletion in the AZFa region [10, 13]. Hence, seven case-control studies (2079 cases and 1217 controls), including the present study, were included in the meta-analysis (Fig. 3). Qualitative analysis of the pooled data showed that the AZFa deletion frequency ranged from 1.18 to 10.63% (average = 4.00%) (Fig. 2). Heterogeneity test showed no significant heterogeneity across the studies (Q value = 6.72, df (Q) = 6, I2 value = 10.82, p value = 0.34). Quantitative meta-analysis suggested a significant association of AZFa deletions with male infertility risk (fixed effects model: OR = 6.74, p value = 0.001; random effects model: OR = 6.64, p value = 0.02) (Fig. 4). AZFa deletions displayed the presence of publication bias (Supplementary Fig. 1). Sensitivity analysis was performed by removing studies based on sample size (less than a hundred) in either of the comparison groups. However, no notable difference in the effect size or p value was observed, ruling out the presence of sensitive studies in the pool.
Fig. 3.
The frequencies of AZF deletions in different populations in India. Blue bars and the red bars indicate the respective frequency for each study and the average thereof, respectively
Fig. 4.
Forest plot showing the odds ratio, p value, and direction of association between AZFa, and AZFb deletions and male infertility in Indian populations
AZFb deletion
Literature search identified nine studies on AZFb deletions. One of the studies by Singh and Raman [10] was excluded as they did not find any deletion in the AZFb region. Hence, a total of eight case-control studies (2212 cases and 1267 controls), including the present study, were included in the meta-analysis (Fig. 3). Qualitative analysis of the pooled data showed that the AZFb deletion frequency ranged from 0.59 to 9.22% (average = 3.21%) (Fig. 2). Heterogeneity test showed no significant heterogeneity across the studies (Q value = 7.92, df (Q) = 7, I2 value = 11.66, p value = 0.34). Quantitative meta-analysis revealed a significant association of AZFb deletions with male infertility risk (fixed effects model: OR = 4.694, p value = 0.004; random effects model: OR = 4.621, p value = 0.008) (Fig. 4). No evidence of publication bias was obtained in the case of AZFb deletion (two-tailed p value = 0.10) (Supplementary Fig. 1).
AZFc deletion
Eleven case-control studies (4131 cases and 2008 controls), including the present study, were included in the meta-analysis (Fig. 3). Qualitative analysis of the pooled data showed that the AZFc deletion frequency ranged from 0.97 to 17.02% (average = 6.27%) (Fig. 2). Heterogeneity test showed no significant heterogeneity across the studies (Q value = 11.71, df (Q) = 10, I2 value = 14.60, p value = 0.30). Quantitative meta-analysis revealed a significant association of AZFc deletions with male infertility risk (fixed effects model: OR = 13.575, p value = 0.000; random effects model: OR = 13.461, p value = 0.000) (Fig. 5). AZFc deletion displayed the presence of publication bias (Supplementary Fig. 1). Sensitivity analysis was performed based on a sample size (less than a hundred) in either of the comparison groups. However, no notable difference in the odds ratio or p value was observed, ruling out the presence of sensitive studies in the pool.
Fig. 5.
The frequencies of AZF deletions in different populations in India. Blue bars and the red bars indicate the respective frequency for each study and the average thereof, respectively
AZFb+c deletion
Four case-control studies (1573 cases and 942 controls) following a strict exclusion-inclusion criteria were included in the meta-analysis (Fig. 3). We did not observe AZFb+c deletion in the present study. Qualitative analysis of the pooled data showed that the AZFb+c deletion frequency ranged from 0.74 to 2.26% (average = 1.66%) (Fig. 2). Heterogeneity test showed no significant heterogeneity across the studies (Q value = 1.0, df (Q) = 3, I2 value = 14.60, p value = 0.63). Quantitative meta-analysis revealed a significant association of AZFb+c deletions with male infertility risk (fixed effects model: OR = 5.946, p value = 0.018; random effects model: OR = 5.946, p value = 0.018) (Fig. 5). No evidence of publication bias was seen in the case of AZFb+c (two-tailed p value = 0.30) deletion from both tests (Supplementary Fig. 1).
Trial sequential analysis (TSA)
TSA also suggested the requirement of additional trials to reach 80% study power (Supplementary Fig. 2). This simply means that further studies are required to strengthen the association observed in this meta-analysis. For AZFa, the required sample size is 3296; for AZFb, the required sample size is 3479; for AZFc, the required sample size is 6139; and for AZFb+c, the required sample size is 2515.
Discussion
Deletions in the AZF region of the Y chromosome have been recognized as the most frequent cause of spermatogenic failure [18–21]. Y-deletion analysis in azoospermic and oligozoospermic patients is strongly recommended by the European Academy of Andrology (EAA) [3]. The global frequency of Y chromosome microdeletions is reported to range from 1 to 55.5% [22]. We found microdeletions in Indian population from a minimum of 0.59% to a maximum of 32.62% with an average of 13.48% [2, 5–14]. The phenotype associated with AZF deletions varies from azoospermia to normozoospermia depending upon the size of deletion and the genetic background of the population. It has been suggested that the variations in the frequency of Y-microdeletions across studies are principally due to the variations in the ethnicity of the study participants or the stringency in the selection criteria of the patients [23].
EAA recommends that Y-deletion analysis should be undertaken in a phased manner, beginning with six primary markers, followed by the analysis of further markers to define the deletion. Six markers to initiate the deletion analysis are SY84 and SY86 for AZFa, SY127 and SY134 for AZFb, and SY254 and SY255 for AZFc regions. Failure of PCR amplification in any of these regions suggests the presence of deletion in that region, but it should be confirmed by repeating PCR. This strategy of deletion analysis using six markers has the capability of detecting more than 95% of deletion events causing male infertility and avoids the use of several primers. We found that AZFc deletion is most common and significantly affects infertility risk in the study population. In a study on AZFc partial deletions, we recently reported that gr/gr deletions significantly correlate with the loss of sperm count and fertility [2]. However, another partial deletion in this region (b2/b3) was found to be unrelated to infertility in Indian populations [24]. Therefore, Y-deletion analysis using six primers for AZFa, AZFb, and AZFc, followed by gr/gr deletion analysis may suffice for Indian infertility patients.
In the present study, we aimed to analyze the Y-deletion data for Indian populations so as to make a way for the interpretation of their clinical significance. We found the average frequency of AZFa deletions to be 4.0%, AZFb deletions to be 3.21%, AZFc deletions to be 6.27%, and AZFb+c to be 1.66% in Indian populations. The combined average frequency of AZF deletions in various Indian populations including the present study was 13.48%. To assess their frequency and correlation with infertility, we undertook meta-analysis on all Indian studies that have analyzed these deletions. Meta-analysis showed that AZFa, AZFb, AZFc, and AZFb+c deletions are significantly associated with infertility risk. However, TSA suggested that further studies on Indian populations are required to confidently establish the observed association. While azoospermic patients displayed deletions in all three regions, oligozoospermic patients had deletions only in the AZFc region. This suggests that in oligozoospermic cases, only AZFc deletion analysis could suffice, but azoospermic patients need to be subjected to analysis of all three regions. The limitations of this meta-analysis include its limited application to Indian populations only. Further, the subject recruitment criteria and quality of analysis adopted in each study may have marginally affected the overall deletion frequency. Though we adopted the most stringent meta-analysis protocol using random effects model, the manuscript is still limited by the quality of data presented in the preceding articles.
Besides its significance in clinical diagnosis of male infertility, Y-deletion analysis has recently achieved considerable importance owing to its utility in various assisted reproductive techniques (ART). Studies have shown that patients with Yq microdeletions display poor embryo quality and decreased overall success rate of ART. Furthermore, patients with Yq microdeletions show low chances of sperm retrieval in techniques such as testicular sperm extraction (TESE) or testicular sperm aspiration (TESA) [25, 26]. As Y-deletions are vertically transmitted to all of the male offspring born through assisted reproductive techniques, its clinical testing can offer genetic counseling to the couples and help them make informed choices and advance planning if they still opt for ART. All these reports highlight that Y-deletion analysis must be recommended as a routine practice in clinical diagnosis in male infertility as well as for couples opting for various ART methods. The ignorance of Y-deletion analysis by clinics hampers decision making and infertility treatment.
Conclusion
The significance of Y-deletion analysis in male infertility is proven beyond doubt. However, the clinical utility in most of the populations is restricted by the lack of advanced facilities and the availability of population frequency data. In India, Y-deletions range in frequency from 0.59 to 32.62% (average = 13.48%). We recommend a simplistic approach to Y-deletion analysis by utilizing six markers recommended by the EAA. While azoospermic samples should be subjected to deletion analysis using six markers, oligozoospermic cases can be subjected to AZFc partial deletion analysis only. Y chromosome microdeletions have prognostic value in treatment of infertility. In the case of males with microdeletion, proper genetic counseling should be provided to couple if they consider assisted reproductive techniques for infertility management. In case of males undergoing TESE, Y chromosome microdeletion screening can be useful in predicting the chances of sperm retrieval.
Electronic supplementary material
(XLSX 41 kb).
(XLSX 10 kb).
Begg’s funnel plot for analysis of publication bias. (JPG 1271 kb).
Trial sequential analysis of the association between AZFa, AZFb, AZFc and AZFb+c deletions and male infertility. (JPG 1933 kb).
Acknowledgments
Vertika Singh is thankful to the Council of Scientific and Industrial Research (CSIR), Govt. of India for SRF fellowship (09/013/(0765)/2018-EMR-I).
Funding information
Singh Rajender would like to thank the CSIR for funding (FTT-MLP0113-DeMID). Kiran Singh would like to thank the Indian Council of Medical Research (ICMR), Govt. of India, for financial support (5/10/FR/6/2018-RBMCH dt.19/060/2018).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Andrabi Syed Waseem and Vertika Singh contributed equally to this work.
Contributor Information
Kiran Singh, Email: skiran@bhu.ac.in.
Singh Rajender, Email: rajender_singh@cdri.res.in.
References
- 1.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 2.Bansal SK, Jaiswal D, Gupta N, Singh K, Dada R, Sankhwar SN, Gupta G, Rajender S. Gr/gr deletions on Y-chromosome correlate with male infertility: an original study, meta-analyses, and trial sequential analyses. Sci Rep. 2016;6:19798. doi: 10.1038/srep19798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krausz C, Hoefsloot L, Simoni M, Tüttelmann F. EAA/EMQN best practice guidelines for molecular diagnosis of Y-chromosomal microdeletions: state-of-the-art 2013. Andrology. 2014;2:5–19. doi: 10.1111/j.2047-2927.2013.00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper TG, Noonan E, von Eckardstein S, et al. World Health Organization reference values for human semen characteristics. Hum Reprod Update. 2010;16:231–245. doi: 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- 5.Dada R, Gupta NP, Kucheria K. Molecular screening for Yq microdeletion in men with idiopathic oligozoospermia and azoospermia. J Biosci. 2003;28:163–168. doi: 10.1007/BF02706215. [DOI] [PubMed] [Google Scholar]
- 6.Rani DS, Rajender S, Pavani K, Chaubey G, Rasalkar AA, Gupta NJ, Deendayal M, Chakravarty B, Thangaraj K. High frequencies of non allelic homologous recombination (NAHR) events at the AZF loci and male infertility risk in Indian men. Sci Rep. 2019;9:6276. doi: 10.1038/s41598-019-42690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen S, Ambulkar P, Hinduja I, Zaveri K, Gokral J, Pal A, Modi D. Susceptibility of gr/gr rearrangements to azoospermia or oligozoospermia is dependent on DAZ and CDY1 gene copy deletions. J Assist Reprod Genet. 2015;32:1333–1341. doi: 10.1007/s10815-015-0520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambulkar PS, Pande SS. Male infertility: screening of azoospermia factor (AZF) microdeletion in idiopathic infertile men. J Exp Biol Agric Sci. 2017;5:7–13. [Google Scholar]
- 9.Nailwal M, Chauhan JB. Gene scanning for microdeletions in the azoospermia factor region of Y-chromosome in infertile men of Gujarat, India. J Clin diagn Res. 2017;11:GC01. doi: 10.7860/JCDR/2017/26750.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh K, Raman R. Male infertility: Y-chromosome deletion and testicular aetiology in cases of azoo-/oligospermia. Indian J Exp Biol. 2005;43:1088–1092. [PubMed] [Google Scholar]
- 11.Ambulkar PS, Sigh R, Reddy MVR, Varma PS, Gupta DO, Shende MR, Pal AK. Genetic risk of Azoospermia Factor (AZF) microdeletions in idiopathic cases of azoospermia and oligozoospermia in Central Indian population. J Clin diagn Res. 2014;8:88. doi: 10.7860/JCDR/2014/7680.4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitra A, Dada R, Kumar R, Gupta NP, Kucheria K, Gupta SK. Screening for Y-chromosome microdeletions in infertile Indian males: utility of simplified multiplex PCR. Indian J Med Res. 2008;127:124–132. [PubMed] [Google Scholar]
- 13.Dada R, Gupta NP, Kucheria K. Yq microdeletions—azoospermia factor candidate genes and spermatogenic arrest. J Biomol Tech. 2004;15:176. [PMC free article] [PubMed] [Google Scholar]
- 14.Athalye AS, Madon PF, Naik NJ, Naik DJ, Gavas SS, Dhumal SB, Bandkar VM, Kawle MT, Parikh FR. A study of Y chromosome microdeletions in infertile Indian males. Int J Hum Genet. 2004;4:179–185. doi: 10.1080/09723757.2004.11885889. [DOI] [Google Scholar]
- 15.Higgins JPT, Altman DG, Gøtzsche PC, Jüni P, Moher D, Oxman AD, Savović J, Schulz KF, Weeks L, Sterne JAC. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. Bmj. 2011;343:d5928. doi: 10.1136/bmj.d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorlund K, Devereaux PJ, Wetterslev J, Guyatt G, Ioannidis JPA, Thabane L, Gluud L-L, Als-Nielsen B, Gluud C. Can trial sequential monitoring boundaries reduce spurious inferences from meta-analyses? Int J Epidemiol. 2008;38:276–286. doi: 10.1093/ije/dyn179. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Tang Q, Wu W, Yuan B, Lu C, Xia Y, Ding H, Hu L, Chen D, Sha J. Association between DAZL polymorphisms and susceptibility to male infertility: systematic review with meta-analysis and trial sequential analysis. Sci Rep. 2014;4:4642. doi: 10.1038/srep04642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reijo R, Lee T-Y, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Hum Reprod. 1996;11:27–54. doi: 10.1093/humrep/11.suppl_4.27. [DOI] [PubMed] [Google Scholar]
- 19.Repping S, van Daalen SKM, Brown LG, Korver CM, Lange J, Marszalek JD, Pyntikova T, van der Veen F, Skaletsky H, Page DC. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 20.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22:226–239. doi: 10.1210/edrv.22.2.0425. [DOI] [PubMed] [Google Scholar]
- 21.Vogt PH, Bender U, Zimmer J, Strowitzki T. Human Y chromosome and male infertility: forward and back from azoospermia factor chromatin structure to azoospermia factor gene function. Basel: Genet. Hum. Infertil. Karger Publishers; 2017. pp. 57–73. [Google Scholar]
- 22.Vijesh VV, Nambiar V, Mohammed SIK, Sukumaran S, Suganthi R. Screening for AZFc partial deletions in Dravidian men with nonobstructive azoospermia and oligozoospermia. Genet Test Mol Biomarkers. 2015;19:150–155. doi: 10.1089/gtmb.2014.0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Ma M, Li L, Zhang W, Chen P, Ma Y, Liu Y, Tao D, Lin L, Zhang S. Y chromosome haplogroups may confer susceptibility to partial AZFc deletions and deletion effect on spermatogenesis impairment. Hum Reprod. 2008;23:2167–2172. doi: 10.1093/humrep/den229. [DOI] [PubMed] [Google Scholar]
- 24.Bansal SK, Gupta G, Rajender S. Y chromosome b2/b3 deletions and male infertility: a comprehensive meta-analysis, trial sequential analysis and systematic review. Mutat Res Rev Mutat Res. 2016;768:78–90. doi: 10.1016/j.mrrev.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Brandell RA, Mielnik A, Liotta D, Ye Z, Veeck LL, Palermo GD, Schlegel PN. AZFb deletions predict the absence of spermatozoa with testicular sperm extraction: preliminary report of a prognostic genetic test. Hum Reprod. 1998;13:2812–2815. doi: 10.1093/humrep/13.10.2812. [DOI] [PubMed] [Google Scholar]
- 26.Hopps CV, Mielnik A, Goldstein M, Palermo GD, Rosenwaks Z, Schlegel PN. Detection of sperm in men with Y chromosome microdeletions of the AZFa, AZFb and AZFc regions. Hum Reprod. 2003;18:1660–16654. doi: 10.1093/humrep/deg348. [DOI] [PubMed] [Google Scholar]
Further Reading
- 27.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 28.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 41 kb).
(XLSX 10 kb).
Begg’s funnel plot for analysis of publication bias. (JPG 1271 kb).
Trial sequential analysis of the association between AZFa, AZFb, AZFc and AZFb+c deletions and male infertility. (JPG 1933 kb).