Elucidation of the mechanism of the wide dissemination of colistin-resistant bacteria in communities of developing countries is an urgent public health issue. In this study, we investigated the genetic background of the colistin resistance gene mcr in E. coli isolates from the fecal microbiota of healthy human residents living in a community in Vietnam with a high prevalence of colistin-resistant E. coli. Our study revealed for the first time, a surprisingly high percentage (36.8%) of colistin-resistant E. coli carrying chromosomal mcr-1, the emergence of which may have occurred recently, in the fecal microbiota of the community residents. The mcr-1 transposon on the chromosome may develop into a more stable genotype by the loss of insertion sequences (ISs). Our results are valuable in understanding the mechanism underlying the increasing prevalence of colistin-resistant bacteria within a community.
KEYWORDS: Escherichia coli, Vietnam, chromosomal mcr, colistin resistance, residents
ABSTRACT
The wide distribution of colistin-resistant bacteria in developing countries has become a common phenomenon. To understand the mechanisms underlying their distribution, we studied the mcr genetic background of colistin-resistant Escherichia coli isolates from the fecal microbiota of healthy human residents from a community in Vietnam with a high prevalence of colistin-resistant E. coli with mcr. Fifty-seven colistin-resistant isolates were obtained from 98 residents; one isolate was collected from each individual and analyzed for mcr. We found that 36.8% of the isolates carried chromosomal mcr-1. Further, 63.2% and 1.8% of the isolates carried mcr-1 on the plasmid and the plasmid/chromosome, respectively. Whole-genome sequencing of genetically unrelated isolates showed that the majority (6 of 7) of the isolates had the chromosomal mcr-1 in a complete ancestral mcr-1 transposon Tn6330, ISApl1-mcr-1-PAP2-ISApl1, which was inserted at various positions on the chromosomes. In addition, the majority (87.5%) of Tn6330 of mcr-1-carrying plasmids (n = 8) lacked both upstream and downstream ISApl1 transposons. The results obtained in this study indicate that plasmid-to-chromosomal transfer of mcr-1 may have occurred recently in the fecal microbiota of the residents. Additionally, Tn6330 on the chromosome may lose ISApl1 from the transposon during multiplication to gain a more stable mcr-1 state on the chromosome. Stabilization of resistance by the chromosomal incorporation of mcr-1 would be an additional challenge in combating the dissemination of resistant bacteria.
IMPORTANCE Elucidation of the mechanism of the wide dissemination of colistin-resistant bacteria in communities of developing countries is an urgent public health issue. In this study, we investigated the genetic background of the colistin resistance gene mcr in E. coli isolates from the fecal microbiota of healthy human residents living in a community in Vietnam with a high prevalence of colistin-resistant E. coli. Our study revealed for the first time, a surprisingly high percentage (36.8%) of colistin-resistant E. coli carrying chromosomal mcr-1, the emergence of which may have occurred recently, in the fecal microbiota of the community residents. The mcr-1 transposon on the chromosome may develop into a more stable genotype by the loss of insertion sequences (ISs). Our results are valuable in understanding the mechanism underlying the increasing prevalence of colistin-resistant bacteria within a community.
INTRODUCTION
Colistin is recognized as the last-resort antibiotic for the treatment of infectious diseases caused by multidrug-resistant (MDR) Gram-negative bacteria, including carbapenem-resistant bacteria. However, the wide distribution of colistin-resistant (COR) bacteria threatens the effectiveness of treatments with colistin (1).
Current reports of a wide distribution of COR Escherichia coli with a mobile resistance gene, mcr, in a community highlight the importance of the stability and possible transfer of the resistance gene to the pathogen (2). In contrast to outbreaks in nosocomial settings, the wide distribution of COR bacteria that may be occurring over a long period in the community seems to be attributable to exposure to a persistent, low-concentration antibiotic pressure. This hypothesis is supported by the frequent use of colistin-containing feed in livestock (3) and the retention of COR E. coli in the fecal microbiota of livestock (4). However, in a previous study, we showed that the wide distribution of COR E. coli in residents was not due to the clonal distribution of a certain lineage of COR bacteria (2). Therefore, the transfer of the mcr-carrying plasmid is the most likely explanation for the wide distribution of the gene. In this regard, recent reports revealed that mcr-1 could be mobilized as an ISApl1-flanked composite transposon, Tn6330 (5, 6). Therefore, the structure of the ISApl1 transposon with mcr potentially reflects the transposition of mcr in COR bacterial isolates in the community. However, further investigations are needed to confirm this hypothesis.
The long-term stability of COR bacteria in the community is another factor that affects their distribution. Chromosomal mcr may play an important role in their stability, but this has not been definitively established. To further understand the distribution mechanisms, in the present study, we investigated the mcr location, as well as the mcr transposon structure, of COR E. coli isolates obtained from healthy subjects in Vietnam residing in a community where COR bacteria are frequently detected.
RESULTS AND DISCUSSION
Location of mcr-1 in COR E. coli isolates.
The location of mcr-1 in 57 COR E. coli isolates that were originally obtained from 98 asymptomatic healthy residents of a rural community in Vietnam during a previous study (2) was assessed using S1/I-CeuI pulsed-field gel electrophoresis (PFGE) and Southern blot hybridization with mcr-1 probe analysis. All isolates used in this study were phylogenetically diverse, as determined by PFGE and multilocus sequence typing (MLST) analyses (Table 1). In addition, no clonal expansion in the community was observed, as determined in a previous study (2).
TABLE 1.
Location of mcr-1 in colistin-resistant Escherichia coli isolates testeda
Gray boxes indicate the locations where mcr was detected. ■, genome analysis was conducted; ND, not done; UN, unknown; *, data from the work of Yamamoto et al. (2).
Southern blot analysis of the isolates after S1-PFGE (see Fig. S1 in the supplemental material) revealed that 36.8% (21/57) of the isolates carried mcr-1 on their chromosomes, whereas 63.2% (36/57) carried mcr-1 on only the plasmid. (Three of the original 60 mcr-1+ E. coli isolates, from 98 residents, were excluded because of the difficulty in analysis.) In one isolate, mcr-1 was located on both the chromosome and the plasmid.
Representative results of Southern blot hybridization after S1 pulsed-field gel electrophoresis with mcr-1 and 16S rRNA probes. Row numbers indicate isolates as follows: 1, 2017.01-1CC; 2, 2018-16-2CC; 3, 2018-12-1CC; 4*, 2018-11-3CC; 5, 2018-11-2CC; 6, 2018-11-1BCC; 7, 2018-10-3BCC; 8, 2018-10-2CC; 9*, 2018-10-1CC; 10, 2018-09-5CC; 11, 2018-09-2CC; 12, 2018-08-1CC; M, marker. The asterisk indicates the isolate for which genome analysis was performed. The arrow indicates the specific probe binding position. Download FIG S1, PDF file, 0.03 MB (29.7KB, pdf) .
Copyright © 2020 Yamaguchi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
mcr-1 was originally discovered as a mobile resistance gene on the plasmid of COR bacteria (7), and it is recognized that chromosomally carried mcr-1 is very rare compared to that on the plasmids (8–13). For instance, Li et al. found that only 4% of 200 mcr-1-positive E. coli isolates from animals, food, the environment, and human fecal samples collected in China carried chromosomal mcr-1 (11). In contrast to previous reports, the results from this study showed that 36.8% of the human fecal sample isolates had chromosomal mcr-1. All isolates examined in this study were obtained from different individuals, except two isolates that were obtained from the same person at a 1-year interval. The prevalence of chromosomal mcr-1 observed in this study is extremely high compared to that reported in previous studies. Although the reasons for the high prevalence of chromosomal mcr-1-carrying isolates in human fecal microbiota are not clear, it seems likely that the high prevalence in the area of sampling could be due to the frequent use of colistin as a livestock antibiotic and feed additive (3). Additionally, most domestic livestock in that community possessed COR E. coli carrying mcr-1 (4). Under the prevailing conditions, it is conceivable that the community from which the isolates were obtained could have experienced a constant colistin pressure on the microbes over a long period, which resulted in the microbes becoming intrinsically resistant due to chromosomally encoded resistance (11). This could have resulted in chromosomal mcr-1 becoming more prevalent and stable in the community.
The chromosomal mcr-1-carrying bacteria may contribute to the emergence of untreatable MDR bacteria when MDR genes, including carbapenem resistance genes on the plasmid (14), are transferred to bacteria possessing chromosomal mcr. In fact, this likelihood is supported by our current study, wherein several extended-spectrum β-lactamase (ESBL)-producing E. coli isolates that had MDR with mcr-1 were obtained from fecal microbiota of healthy human residents of the community (15).
Genetic structure of chromosomally and/or plasmid-carried mcr-1 transposon.
Recently, it has been proposed that mcr-1 can be mobilized as an ISApl1-flanked composite transposon (Tn6330) (5, 16). The study also reported that transmission is possible both upstream and downstream of ISApl1. The structure of this composite transposon is considered to be stabilized by the structure in which insertion sequences (ISs) have dropped out (6, 17). Therefore, the structure of mcr-1 transposon Tn6330 in the bacterium is important not only for the transmission among microbes but also for the stability of mcr-1.
To elucidate the mcr-1 transposon structure of the isolates in this study, 14 representative strains (6 of 18 chromosomal mcr-1 isolates, one chromosomal/plasmid mcr-1 isolates, and 7 of 30 plasmid mcr-1 isolates) were subjected to genome analysis. Several strains were obtained from the same household (Table 2). In the case of household 2, two mcr-1 isolates were obtained from the same member at a 1-year interval. All these isolates were phylogenetically different (2). Table 2 shows the bacterial characteristics of chromosomally and/or plasmid-carried mcr-1 transposons for these 14 isolates.
TABLE 2.
Characterization of chromosomally and/or plasmid-carried mcr-1 transposon of colistin-resistant Escherichia coli
| Household | Household member |
Yr of isolation |
Isolate | MLST typea |
No. of plasmid |
mcr
carriage |
Size (kbp)a |
mcr location (kbp) |
mcr transposon | Transposon typeb |
Plasmid Inc type |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A | 2017 | 2017.01.04CC | ST206 | 2 | Chromosome | 4,615 | 3215 | ISApl1-mcr1- PAP2-ISApl1 |
A | |
| B | 2018 | 2018-01-1CC | ST206 | 3 | Chromosome | 4,711 | 1754 | ISApl1-mcr1- PAP2-ISApl1 |
A | ||
| 2 | A | 2017 | 2017.02.01CC | ST48 | 4 | Plasmide | 231 | mcr1-PAP2 | C | IncHI2 | |
| B | 2017 | 2017.02.02CC | UNc | 4 | Plasmid | 34 | mcr1-PAP2 | C | IncX4 | ||
| 2018 | 2018-02-2CC | ST10 | 3 | Chromosome | 4,574 | 1271 | ISApl1-mcr1-PAP2 | B | |||
| Plasmid | 47 | mcr1-PAP2 | C | IncP1 | |||||||
| 3 | A | 2017 | 2017.03.03CC | ST46 | 4 | Plasmid | 294 | mcr1-PAP2 | C | IncHI2 | |
| 6 | A | 2018 | 2018-06-4CC | ST189 | 4 | Chromosome | 4,753 | 3569 | ISApl1-mcr1- PAP2-ISApl1 |
A | |
| B | 2018 | 2018-06-1CC | ST590 | 4 | Plasmid | 33 | mcr1-PAP2 | C | IncX4 | ||
| 10 | A | 2018 | 2018-10-1CC | ST165 | 4 | Chromosome | 4,701 | 923 | ISApl1-mcr1- PAP2-ISApl1 |
A | |
| B | 2018 | 2018-10-2CC | ST155 | 2 | Plasmid | 60 | mcr1-PAP2 | C | IncI2 | ||
| 11 | A | 2018 | 2018-11-3CC | ST206-liked | 1 | Chromosome | 4,557 | 1449 | ISApl1-mcr1- PAP2-ISApl1 |
A | |
| 15 | A | 2017 | 2017.15.01CC | ST201 | 4 | Chromosome | 4,869 | 2670 | ISApl1-mcr1- PAP2-ISApl1 |
A | |
| B | 2017 | 2017.15.03CC | ST189 | 3 | Plasmid | 104 | ISApl1-mcr1-PAP2 | B | IncY | ||
| 19 | A | 2017 | 2017.19.01CC | ST542 | 10 | Plasmid | 33 | mcr1-PAP2 | C | IncX4 | |
Size of chromosome or plasmid.
Data from work of Snesrud et al. (6).
UN, unknown.
Only adk was different, compared to ST206.
Plasmids with similar levels of shading are similar to one another.
Plasmid-carried mcr-1 transposon.
Two isolates obtained from different residents showed different host bacterium MLST types but possessed very similar IncHI2 plasmids with the same mcr-1 transposon, mcr-1-PAP2. Similarly, three other isolates showed different E. coli MLST types but had the same IncX4 plasmid with the mcr-1-PAP2 transposon (Table 2). These results indicate that the transfer of plasmids among bacteria may occur frequently in the human bacterial flora.
Other plasmids showed different mcr-1 transposons, including ISApl1-mcr-1-PAP2, which was carried by different Inc types, such as IncP1, IncI2, and IncY. These results confirmed that mcr-1 was retained in various Inc-type plasmids in bacteria from fecal microbiota of residents in the community, as previously shown by the diversity of mcr-1-carrying plasmids (9). In addition, the majority (7 of 8) of plasmids that were assessed had the simplest mcr-1 transposon structure, mcr-1-PAP2. One remaining plasmid had the ISApl1-mcr-1-PAP2 transposon. Our results indicate that mcr-1 on the plasmid of COR E. coli strains in the fecal microbiota of humans may be stabilized by the loss of ISApl1 in many cases, as described by Wang et al. (17).
Chromosomally carried mcr-1 transposon Tn6330.
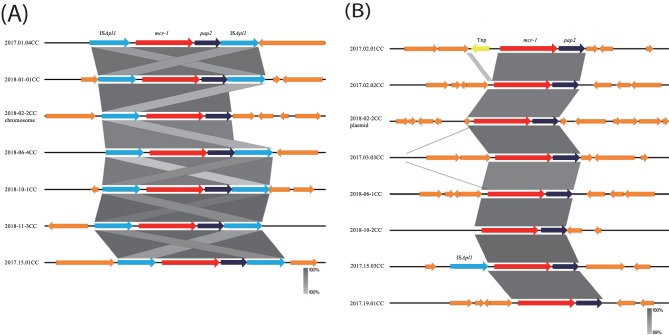
The mcr-1 transposon structure in the chromosome of the seven COR E. coli isolates assessed in this study showed that the majority (6 of 7) of the isolates possessed a complete ancestral mcr-1 transposon, Tn6330 (6), without the loss of ISApl1 in their chromosomes (Fig. 1). The Tn6330 insertion sites on the chromosomes of these isolates were random. Besides, the AT- and CG-rich sequences at either end of Tn6330 were not found on the chromosomes of the isolates (data not shown). Because the insertion sites of Tn6330 are notable for AT- and CG-rich regions (6), it is presumable that the absence of such 2-bp target site duplications in these isolates may be caused by mutations after the transposition of Tn6330, even though there was no evidence to support it. The one remaining isolate lost the ISApl1 present downstream of Tn6330, which may contribute to stabilizing the mcr-1 gene (16).
FIG 1.
Comparison of the genetic structures of the mcr-1 transposon in Escherichia coli isolates. (A) Isolates possessing chromosomal mcr-1. (B) Isolates possessing plasmid mcr-1.
The genome analysis of these isolates revealed that there was only one Tn6330 in the chromosome of the isolates tested. In addition, there was no independent mcr-1 without an IS in the chromosomes. Thus, the results from the genome assessment of COR E. coli isolates of fecal microbiota from the community residents support a previous finding that ISApl1 facilitates mcr-1 transmission (11).
The dominant fully intact Tn6330, ISApl1-mcr-1-PAP2-ISApl1, on the chromosome among COR E. coli isolates from fecal microbiota of community residents may indicate that this transposon insertion into the chromosome occurred recently. If the high prevalence of chromosomally carried mcr-1 has occurred recently, it may indicate that the wide distribution of COR bacteria in the community has progressed to a stable state. Furthermore, although it was found in one isolate, the loss of ISApl1 from the mcr-1 transposon Tn6330 indicates that the resistance gene on the chromosome has shifted to a more stable state.
One isolate was found to harbor mcr-1 on both the chromosome and plasmid. Because the origin of the mcr-1 transposon is unclear from the transposon elements, the relationship between mcr-1 on the chromosome and the plasmid in this isolate is unknown. In this regard, the following can be speculated. Because truncated Tn6330 was found in all plasmids assessed in this study, it is likely that COR E. coli isolates with plasmids harboring Tn6330 may have prevailed in the community for a long time. Moreover, it can be presumed that the transposition of Tn6330 from the plasmid to the chromosome occurred, followed by the loss of the IS during the process of stabilizing mcr-1. Therefore, this isolate may be a transitional intermediate-type strain. The finding of the intermediate-type isolate suggests that the process for stabilizing mcr-1 is in progress.
The assessment of mcr-1 location in COR E. coli isolates in this study showed that only one isolate carried it both on the chromosome and on the plasmid, whereas the other isolates carried the mcr-1 either on the chromosome or on the plasmid. It is not clear why the Tn6330 was carried on either the plasmid or the chromosome and only rarely on both. Because large antibiotic-resistant plasmids may be lost during their multiplication in an antibiotic-free environment due to their significant metabolic burden on the host strain, it can be speculated that after the transposition of Tn6330 from the plasmid to the chromosome, the plasmid may no longer need to carry Tn6330 or the plasmid itself may not be needed (18).
MATERIALS AND METHODS
Sample collection for COR E. coli isolates.
A total of 57 COR E. coli isolates with mcr-1 were assessed in this study. All the COR E. coli isolates were initially obtained from healthy residents in a rural community in Vietnam between November 2017 and February 2018. One isolate was obtained from each resident. It was found that a high percentage (70.4%) of the residents were carrying COR E. coli with mcr-1 in their stool, as reported in a previous study (2).
Assessment of mcr-1.
Digoxigenin (DIG)-labeled DNA probes used for the detection of mcr-1 and 16S rRNA genes in Southern blot hybridization were prepared using the PCR DIG probe synthesis kit (Sigma-Aldrich, St. Louis, MO) with the primers shown in Table S1 in the supplemental material. The location of mcr-1 in the examined isolates was determined by S1 nuclease PFGE and Southern blot hybridization, as previously described (19). For some bacterial isolates, I-CeuI PFGE and Southern blot hybridization were also performed to determine mcr-1 locations according to the methods described in previous studies (20, 21).
Primers used for amplifying the probes for Southern blot hybridization. Download Table S1, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2020 Yamaguchi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genome sequencing.
Of the 57 COR E. coli isolates with mcr, a total of 14 isolates were chosen for genome sequencing (Table 1). Whole-genome sequencing and assembly of isolates, along with a plasmid harboring mcr-1, were performed on the Illumina MiSeq (Illumina Inc., CA) and MinION (Oxford Nanopore Technologies, London, United Kingdom) sequencers, as described previously (22). The genomes were annotated using DDBJ Fast Annotation and Submission Tool pipeline (https://dfast.nig.ac.jp). Genome analysis was performed using the Geneious R11 software (Biomatters, Ltd., Auckland, New Zealand), Easyfig (23), and BRIG (24).
Accession number(s).
The draft genome sequences of the colistin-resistant E. coli strains 2017.01.04CC (chromosome), 2018-01-1CC (chromosome), 2017.02.01CC (plasmid), 2017.02.02CC (plasmid), 2018.02.2CC (chromosome and plasmid), 2017.03.03CC (plasmid), 2018-06-4CC (chromosome), 2018-06-1CC (plasmid), 2018-10-1CC (chromosome), 2018-10-2CC (plasmid), 2018-11-3CC (chromosome), 2017.15.01CC (chromosome), 2017.15.03CC (plasmid), and 2017.19.01CC (plasmid) were deposited in DDBJ/GenBank under the accession numbers AP021891, AP021892, LC511657, LC511656, AP021896 (chromosome), AP021897 (chromosome), LC511658, AP021893, LC511661, AP021894, LC511662, AP021895, AP021890, LC511659, and LC511660, respectively.
ACKNOWLEDGMENT
This study was supported by the Japan Society for the Promotion of Science KAKENHI (grant 17H01687).
REFERENCES
- 1.Kaye KS, Pogue JM, Tran TB, Nation RL, Li J. 2016. Agents of last resort: polymyxin resistance. Infect Dis Clin North Am 30:391–414. doi: 10.1016/j.idc.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto Y, Kawahara R, Fujiya Y, Sasaki T, Hirai I, Khong DT, Nguyen TN, Nguyen BX. 2019. Wide dissemination of colistin-resistant Escherichia coli with the mobile resistance gene mcr in healthy residents in Vietnam. J Antimicrob Chemother 74:523–524. doi: 10.1093/jac/dky435. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama T, Jinnai M, Kawahara R, Diep KT, Thang NN, Hoa TT, Hanh LK, Khai PN, Sumimura Y, Yamamoto Y. 2017. Frequent use of colistin-based drug treatment to eliminate extended-spectrum beta-lactamase-producing Escherichia coli in backyard chicken farms in Thai Binh Province, Vietnam. Trop Anim Health Prod 49:31–37. doi: 10.1007/s11250-016-1154-y. [DOI] [PubMed] [Google Scholar]
- 4.Kawahara R, Fujiya Y, Yamaguchi T, Khong DT, Nguyen TN, Tran HT, Yamamoto Y. 2019. Most domestic livestock possess colistin-resistant commensal Escherichia coli harboring mcr in a rural community in Vietnam. Antimicrob Agents Chemother 63:e00594-19. doi: 10.1128/AAC.00594-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, Zheng Z, Chan EW, Chen S. 2017. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother 72:393–401. doi: 10.1093/jac/dkw411. [DOI] [PubMed] [Google Scholar]
- 6.Snesrud E, McGann P, Chandler M. 2018. The birth and demise of the ISApl1-mcr-1-ISApl1 composite transposon: the vehicle for transferable colistin resistance. mBio 9:e02381-17. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Liu JY, Liao TL, Huang WC, Liu YM, Wu KM, Lauderdale TL, Tsai SF, Kuo SC, Kuo HC. 2018. Increased mcr-1 in pathogenic Escherichia coli from diseased swine, Taiwan. J Microbiol Immunol Infect doi: 10.1016/j.jmii.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Sun J, Li XP, Fang LX, Sun RY, He YZ, Lin J, Liao XP, Feng Y, Liu YH. 2018. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents 51:842–847. doi: 10.1016/j.ijantimicag.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Zurfluh K, Tasara T, Poirel L, Nordmann P, Stephan R. 2016. Draft genome sequence of Escherichia coli S51, a chicken isolate harboring a chromosomally encoded mcr-1 gene. Genome Announc 4:e00796-16. doi: 10.1128/genomeA.00796-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Yu H, Xie M, Chen K, Dong N, Lin D, Chan EW, Chen S. 2018. Genetic basis of chromosomally-encoded mcr-1 gene. Int J Antimicrob Agents 51:578–585. doi: 10.1016/j.ijantimicag.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HW, Zhang T, Ma JH, Fang Y, Wang HY, Huang ZX, Wang Y, Wu C, Chen GX. 2017. Occurrence of plasmid- and chromosome-carried mcr-1 in waterborne Enterobacteriaceae in China. Antimicrob Agents Chemother 61:e00017-17. doi: 10.1128/AAC.00017-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dona V, Bernasconi OJ, Pires J, Collaud A, Overesch G, Ramette A, Perreten V, Endimiani A. 2017. Heterogeneous genetic location of mcr-1 in colistin-resistant Escherichia coli isolates from humans and retail chicken meat in Switzerland: emergence of mcr-1-carrying IncK2 plasmids. Antimicrob Agents Chemother 61:e01245-17. doi: 10.1128/AAC.01245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolejska M, Papagiannitsis CC. 2018. Plasmid-mediated resistance is going wild. Plasmid 99:99–111. doi: 10.1016/j.plasmid.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara R, Khong DT, Le HV, Phan QN, Nguyen TN, Yamaguchi T, Kumeda Y, Yamamoto Y. 2019. Prevalence of mcr-1 among cefotaxime-resistant commensal Escherichia coli in residents of Vietnam. Infect Drug Resist 12:3317–3325. doi: 10.2147/IDR.S224545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snesrud E, He S, Chandler M, Dekker JP, Hickman AB, McGann P, Dyda F. 2016. A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob Agents Chemother 60:6973–6976. doi: 10.1128/AAC.01457-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun 9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nang SC, Morris FC, McDonald MJ, Han ML, Wang J, Strugnell RA, Velkov T, Li J. 2018. Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J Antimicrob Chemother 73:1604–1610. doi: 10.1093/jac/dky061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirai I, Fukui N, Taguchi M, Yamauchi K, Nakamura T, Okano S, Yamamoto Y. 2013. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents 42:500–506. doi: 10.1016/j.ijantimicag.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Liu SL, Hessel A, Sanderson KE. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci U S A 90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harada S, Ishii Y, Saga T, Kouyama Y, Tateda K, Yamaguchi K. 2012. Chromosomal integration and location on IncT plasmids of the blaCTX-M-2 gene in Proteus mirabilis clinical isolates. Antimicrob Agents Chemother 56:1093–1096. doi: 10.1128/AAC.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsumoto Y, Kinjo T, Motooka D, Nabeya D, Jung N, Uechi K, Horii T, Iida T, Fujita J, Nakamura S. 2019. Comprehensive subspecies identification of 175 nontuberculous mycobacteria species based on 7547 genomic profiles. Emerg Microbes Infect 8:1043–1053. doi: 10.1080/22221751.2019.1637702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative results of Southern blot hybridization after S1 pulsed-field gel electrophoresis with mcr-1 and 16S rRNA probes. Row numbers indicate isolates as follows: 1, 2017.01-1CC; 2, 2018-16-2CC; 3, 2018-12-1CC; 4*, 2018-11-3CC; 5, 2018-11-2CC; 6, 2018-11-1BCC; 7, 2018-10-3BCC; 8, 2018-10-2CC; 9*, 2018-10-1CC; 10, 2018-09-5CC; 11, 2018-09-2CC; 12, 2018-08-1CC; M, marker. The asterisk indicates the isolate for which genome analysis was performed. The arrow indicates the specific probe binding position. Download FIG S1, PDF file, 0.03 MB (29.7KB, pdf) .
Copyright © 2020 Yamaguchi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used for amplifying the probes for Southern blot hybridization. Download Table S1, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2020 Yamaguchi et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.