Abstract
The phylogeny and biogeography of the genus Paphiopedilum were evaluated by using phylogenetic trees derived from analysis of nuclear ribosomal internal transcribed spacer (ITS) sequences, the plastid trnL intron, the trnL-F spacer, and the atpB-rbcL spacer. This genus was divided into three subgenera: Parvisepalum, Brachypetalum, and Paphiopedilum. Each of them is monophyletic with high bootstrap supports according to the highly resolved phylogenetic tree reconstructed by combined sequences. There are five sections within the subgenus Paphiopedilum, including Coryopedilum, Pardalopetalum, Cochlopetalum, Paphiopedilum, and Barbata. The subgenus Parvisepalum is phylogenetic basal, which suggesting that Parvisepalum is comprising more ancestral characters than other subgenera. The evolutionary trend of genus Paphiopedilum was deduced based on the maximum likelihood (ML) tree and Bayesian Evolutionary Analysis Sampling Trees (BEAST). Reconstruct Ancestral State in Phylogenies (RASP) analyses based on the combined sequence data. The biogeographic analysis indicates that Paphiopedilum species were firstly derived in Southern China and Southeast Asia, subsequently dispersed into the Southeast Asian archipelagoes. The subgenera Paphiopedilum was likely derived after these historical dispersals and vicariance events. Our research reveals the relevance of the differentiation of Paphiopedilum in Southeast Asia and geological history. Moreover, the biogeographic analysis explains that the significant evolutionary hotspots of these orchids in the Sundaland and Wallacea might be attributed to repeated migration and isolation events between the south-eastern Asia mainland and the Sunda Super Islands.
Keywords: Paphiopedilum, molecular phylogeny, biogeography, evolutionary trend, dispersal events
Introduction
The orchid genus Paphiopedilum Pfitzer belongs to the subfamily Cypripedioideae Lindley. This subfamily has been considered a distinct lineage since Lindley (1840) separated them from other orchids based on the characteristic of having two separated fertile anthers [see (Cribb, 1998)]. This subfamily includes only five genera: Cypripedium, Mexipedium, Paphiopedilum, Phragmipedium, and Selenipedium. Mexipedium and Selenipedium are monotypic genera (Albert and Chase, 1992), which was a finding supported by ITS sequence analysis (Cox et al., 1997). These five genera are distributed in separate and restricted geographical ranges (Cribb, 1998). Paphiopedilum is distinguished from genera Cypripedium and Selenipedium by its conduplicate coriaceous leaves, as opposed to the plicate persistent leaves of the latter two genera. Furthermore, Paphiopedilum differs from Phragmipedium and Mexipedium, as they display imbricate sepal vernation, different chromosome base numbers and a unilocular ovary (Albert and Chase, 1992; Albert, 1994).
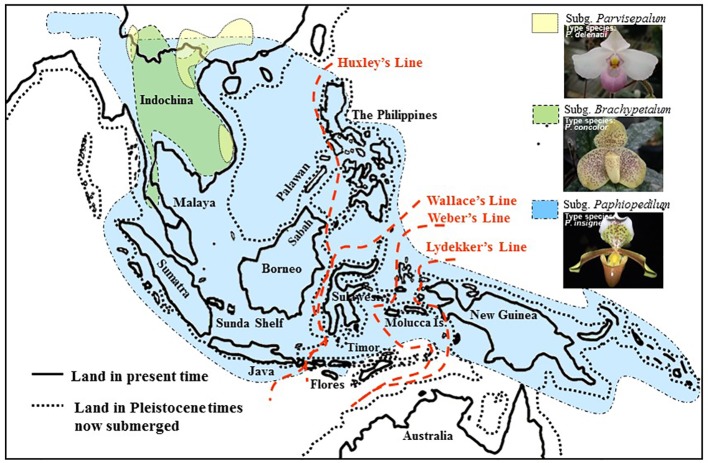
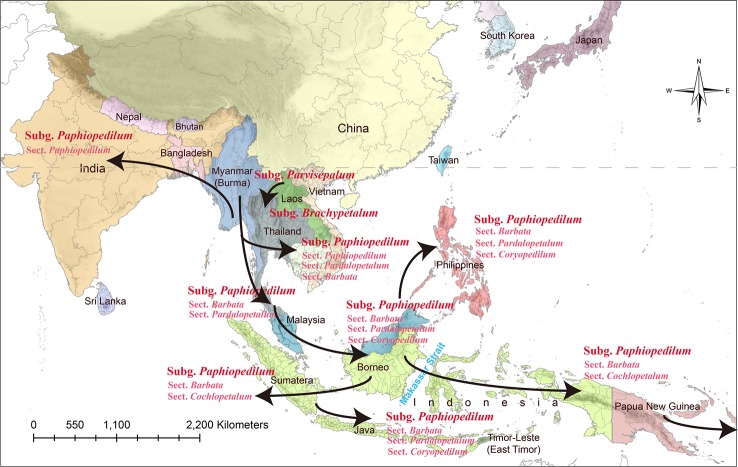
The systematics of the genus Paphiopedilum proposed by Cribb (1997b) are largely consistent with Atwood (1984), except that Cribb placed the Parvisepalum group within subgenus Brachypetalum. Cribb (1997b) accepted the suggestion of Karasawa (1982) and Karasawa and Saito (1982) to promote the Parvisepalum group (e.g., Parvisepalum delenatii, Parvisepalum armeniacum, Parvisepalum micranthum, Parvisepalum malipoense, and Parvisepalum emersonii) to the subgeneric rank, since the two relatively new species (i.e., P. malipoense and P. emersonii) found in this group have been described. According to the classification of Cribb (1997b), the genus Paphiopedilum comprised of approximately 69 species worldwide. Cribb divided this genus into three subgenera, Parvisepalum, Brachypetalum, and Paphiopedilum, which are mainly based on the morphological characteristics of flower inflorescence, leaf type, floral morphology, and molecular data on ITS sequences (Cox et al., 1997). Recently, several new species and treatment have been described for this genus. The genus Paphiopedilum was described as containing approximately 98 species worldwide by the year 2000 (Koopowitz, 2000). In this genus, Paphiopedilum was divided into five sections: Coryopedilum, Pardalopetalum, Cochlopetalum, Paphiopedilum, and Barbata. Subgenera of the genus Paphiopedilum distribute in distinct geographic regions (Cribb, 1997b). The subgenera Parvisepalum and Brachypetalum, as well as the section Paphiopedilum of the subgenus Paphiopedilum, are found only in mainland Asia. The Parvisepalum subgenus is concentrated in southern China and Vietnam, the subgenus Brachypetalum is mostly found in Thailand ( Figure 1 ). Among the subgenus Paphiopedilum, Paphiopedilum ranges from India to southern China, Thailand and Indo-China, and the species diversity found in southern China was the most concentrated. The section Cochlopetalum is restricted to the islands of Sumatra and Java. The section Pardalopetalum is widespread in Southeast Asia, the Malay Archipelago, as far east as Sulawesi, and Luzon in the Philippines. The enormous species diversity of the section Coryopedilum locates in Borneo, and this section range from the Philippines to Sulawesi in New Guinea. The section Barbata is widespread from eastern Nepal, across to Hong Kong and the Philippines, south to the Malay Archipelago, New Guinea, and the Solomon Islands (Cribb, 1998).
Figure 1.
Map of the geographical distribution of Paphiopedilum based on the phylogeny of Cribb (1998). Comparison of Southeast Asian landmasses between the Pleistocene era and the present. During the Pleistocene, Indochina, Malaya, Sumatra, Java, Borneo, and the Philippines were interconnected and were separated from Sulawesi by the Makassar Strait.
Paphiopedilum is a genus of tropical Asiatic origin, and its range extends eastward, reaching the Philippines, Southeast Asia, Borneo, and the Malay Archipelago, crossing Wallace's Line into Sulawesi, the Moluccas, New Guinea, and the Solomon Islands (Cribb, 1998). Tracking back to the geological history of Southeast Asian, the Palawan, Mindoro, Zamboanga, and the adjacent small islands are the older islands of the Southern Philippines. These regions are located on the border of the Eurasian Plate and have been shifting away from the mainland mass by tectonic collision since the early Miocene (~30 Mya) and the shell of the older plate was merged to Borneo until 5~10 Mya (Karig et al., 1986; Stephan et al., 1986; Hall, 1996). In contrast, most of the Philippine islands formed less than 5 Mya (Aurelio et al., 1991; Quebral et al., 1994). In addition, the Sundaland was comprised of the Malay Peninsula, Sumatra, Java, Sulawesi, and Borneo and merged with Bali, the Philippines, and even New Guinea/Australia into Sunda Superland interconnecting by land bridge during the last glacial period (0.01~1.8 Mya) (van Oosterzee, 1997). Since the last glacial period, species migrated forward and backwards between these regions and isolated after the last glacial maximum (LGM), causing the broken of Sunda Superland (Tsai et al., 2015).
The chloroplast primers for the atpB-rbcL, trnL-trnF spacer, and trnL intron are useful for phylogenetic studies at the intrageneric level. The primers for the trnL-trnF spacer and trnL intron developed by Taberlet et al. (1991) have been applied for inferring phylogenies at the intrageneric level (Goldblatt et al., 2002; Hodkinson et al., 2002; Mogensen, 1996; Van Raamsdonk et al., 2003), and have also been used successfully on Orchidaceae (Tsai et al., 2012). The atpB-rbcL regions are high length differences due to frequent occurrence of indels and are often used in combination with other primers to provide more information (Yoshinaga et al., 1992; Chiang et al., 1998; Von Konrat et al., 2010). Therefore, this study aims to further elucidate the phylogeny of Paphiopedilum through analysis ITS (internal transcribed spacer) sequences and three non-coding plastid DNA sequences (trnL intron, trnL-F, and atpB-rbcL spacers). In addition, the biogeography of this genus is clarified based on the phylogenetic tree derived from the molecular evidence.
Materials and Methods
Plant Materials
Seventy-eight taxa of Paphiopedilum and two outgroups from genus Phragmipedium were used in this study ( Table 1 ). All leaf materials were taken from living plants in the greenhouse of the Kaohsiung District Agricultural Improvement Station (KDAIS) in Taiwan.
Table 1.
Names of specimens, geographical distribution, source, and GenBank accession numbers for sequences of the internal transcribed spacer (ITS) of ribosomal DNA (rDNA), the plastid trnL intron, the trnL-F spacer, and the atpB-rbcL spacer.
| Taxa and systematic classification a | Geographical distribution | Voucher b | GenBank accession no. | |||
|---|---|---|---|---|---|---|
| ITS | trnL intron | trnL-F spacer | atpB-rbcL spacer | |||
| Genus Paphiopedilum | ||||||
| Subgenus Parvisepalum | ||||||
| Paphiopedilum armeniacum S.C. Chen & F.Y. Liu | Southwest China | C. C. Tsai 2021 | EF156086 | EF156001 | EF156171 | GQ850803 |
| Paphiopedilum delenatii Guill. | Vietnam | C. C. Tsai 2073 | EF156096 | EF156011 | EF156181 | GQ850813 |
| Paphiopedilum emersonii Koop. & P.J. Cribb | China | C. C. Tsai 2351 | EF156099 | EF156014 | EF156184 | GQ850816 |
| Paphiopedilum hangianum Perner & Gruss | China | C. C. Tsai 2201 | EF156109 | EF156024 | EF156194 | GQ850826 |
| Paphiopedilum jackii H.S. Hua | China, Vietnam | C. C. Tsai 2330 | EF156118 | EF156033 | EF156203 | GQ850832 |
| Paphiopedilum malipoense S.C. Chen & Z.H. Tsi | China, Vietnam | C. C. Tsai 2024 | EF156125 | EF156040 | EF156210 | GQ850839 |
| Paphiopedilum micranthum T. Tang & F. T. Wang | China, Vietnam | C. C. Tsai 2020 | EF156128 | EF156043 | EF156213 | GQ850842 |
| Paphiopedilum micranthum var. eburneum Fowlie | China | No voucher | EF156127 | EF156042 | EF156212 | GQ850841 |
| Paphiopedilum vietnamense Perner & Gruss | Vietnam | C. C. Tsai 2110 | EF156158 | EF156073 | EF156243 | GQ850871 |
| Subgenus Brachypetalum | ||||||
| Paphiopedilum concolor (Bateman) Pfitzer | China, Burma, Thailand, Laos, Vietnam | C. C. Tsai 2307 | EF156093 | EF156008 | EF156178 | GQ850810 |
| Paphiopedilum godefroyae (God.-Leb.) Stein | Thailand | C. C. Tsai 2321 | EF156107 | EF156022 | EF156192 | GQ850824 |
| Paphiopedilum godefroyae var. leucochilum (Masters) Hallier | Thailand | C. C. Tsai 2031 | EF156106 | EF156021 | EF156191 | GQ850823 |
| Paphiopedilum niveum (Rchb.f.) Stein | Southern Thailand, Malay peninsula | C. C. Tsai 2039 | EF156130 | EF156045 | EF156215 | GQ850844 |
| Subgenus Paphiopedilum | ||||||
| Section Coryopedilum | ||||||
| Paphiopedilum adductum Asher | Philippines | C. C. Tsai 2025 | EF156082 | EF155997 | EF156167 | GQ850799 |
| Paphiopedilum anitum Golamco | Philippines | C. C. Tsai 2295 | EF156083 | EF155998 | EF156168 | GQ850800 |
| Paphiopedilum gigantifolium Braem, M.L. Baker & C.O. Baker | Sulawesi | No voucher | EF156103 | EF156018 | EF156188 | GQ850821 |
| Paphiopedilum kolopakingii Fowlie | Borneo | C. C. Tsai 2057 | EF156121 | EF156036 | EF156206 | GQ850835 |
| Paphiopedilum ooii Koopowitz | Borneo | no voucher | EF156138 | EF156046 | EF156216 | GQ850845 |
| Paphiopedilum philippinense (Rchb.f.) Stein | Northeast Borneo, Philippines | C. C. Tsai 2007 | EF156142 | EF156050 | EF156220 | GQ850848 |
| Paphiopedilum glanduliferum (Blume) Stein | New Guinea | C. C. Tsai 2040 | EF156104 | EF156019 | EF156189 | GQ850822 |
| Paphiopedilum randsii fowlie | Mindanao, Philippines | C. C. Tsai 2297 | EF156132 | EF156053 | EF156223 | GQ850851 |
| Paphiopedilum rothschildianum (Rchb.f.) Stein | Borneo | C. C. Tsai 2249 | EF156135 | EF156056 | EF156226 | GQ850853 |
| Paphiopedilum sanderianum (Rchb.f.) Stein | Borneo | C. C. Tsai 2309 | EF156136 | EF156057 | EF156227 | GQ850854 |
| Paphiopedilum stonei (Hook.) Stein | Borneo | C. C. Tsai 2310 | EF156146 | EF156061 | EF156231 | GQ850858 |
| Paphiopedilum supardii Braem & Loeb | Borneo | C. C. Tsai 2189 | GQ505309 | GQ505312 | GQ505315 | GQ850860 |
| Paphiopedilum wilhelminae L.O. Williams | New Guinea | C. C. Tsai 2205 | GQ505310 | GQ505313 | GQ505316 | GQ850875 |
| Section Pardalopetalum | ||||||
| Paphiopedilum dianthum T. Tang & F.T. Wang | China | C. C. Tsai 2085 | EF156097 | EF156012 | EF156182 | GQ850814 |
| Paphiopedilum haynaldianum (Rchb.f.) Stein | Philippines | No voucher | EF156110 | EF156025 | EF156195 | GQ850827 |
| Paphiopedilum lowii (Lindl.) Stein | Peninsular Malaysia, Sumatra, Borneo, Sulawesi | C. C. Tsai 2285 | EF156124 | EF156039 | EF156209 | GQ850838 |
| Paphiopedilum parishii (Rchb.f.) Stein | Southwest China, Burma, Thailand | C. C. Tsai 2276 | EF156140 | EF156048 | EF156218 | GQ850847 |
| Paphiopedilum richardianum Asher & Beaman | Sulawesi | C. C. Tsai 2068 | EF156133 | EF156054 | EF156224 | GQ850852 |
| Section Cochlopetalum | ||||||
| Paphiopedilum victoria-regina (Sander) M.W. Wood | Sumatra | C. C. Tsai 2045 | EF156157 | EF156072 | EF156242 | GQ850870 |
| Paphiopedilum liemianum (Fowlie) Karas. & Saito | Northern Sumatra | C. C. Tsai 2133 | EF156123 | EF156038 | EF156208 | GQ850837 |
| Paphiopedilum moquetteanum (J.J. Smith) Fowlie | Southwest Java | C. C. Tsai 2314 | EF156129 | EF156044 | EF156214 | GQ850843 |
| Paphiopedilum primulinum M.W. Wood & P. Taylor | Northern Sumatra | C. C. Tsai 2359 | EF156143 | EF156051 | EF156221 | GQ850849 |
| Paphiopedilum victoria-mariae (Rolfe) Rolfe | Sumatra | No voucher | EF156156 | EF156071 | EF156241 | GQ850869 |
| Section Paphiopedilum | ||||||
| Paphiopedilum barbigerum Tang & Wang | China, northern Vietnam | C. C. Tsai 2023 | EF156088 | EF156003 | EF156173 | GQ850805 |
| Paphiopedilum charlesworthii (Rolfe) Pfitzer | Burma, northern Thailand, southwest China | C. C. Tsai 2192 | EF156091 | EF156006 | EF156176 | GQ850808 |
| Paphiopedilum druryi (Bedd.) Stein | Southern India | C. C. Tsai 2093 | EF156098 | EF156013 | EF156183 | GQ850815 |
| Paphiopedilum exul (Ridl.) Rolfe | Peninsular Thailand | C. C. Tsai 2083 | EF156101 | EF156016 | EF156186 | GQ850818 |
| Paphiopedilum esquirolei Schltr. | China, India, Bhutan (Southeast Asia) | C. C. Tsai 2335 | EF156100 | EF156015 | EF156185 | GQ850817 |
| Paphiopedilum fairrieanum (Lindl.) Stein | India, Bhutan | C. C. Tsai 2079 | EF156102 | EF156017 | EF156187 | GQ850819 |
| Paphiopedilum gratrixianum (Masters) Guillaumin | Laos, Vietnam | C. C. Tsai 2155 | EF156108 | EF156023 | EF156193 | GQ850825 |
| Paphiopedilum helenae Aver. | Northern Vietnam | C. C. Tsai 2053 | EF156111 | EF156026 | EF156196 | GQ850828 |
| Paphiopedilum henryanum Braem | China, northern Vietnam | C. C. Tsai 2277 | EF156112 | EF156027 | EF156197 | GQ850829 |
| Paphiopedilum herrmannii Fuchs & Reisinger | Northeast India | C. C. Tsai 2109 | EF156113 | EF156028 | EF156198 | GQ850880 |
| Paphiopedilum hirsutissimum (Lindl. ex Hook.) Stein | China, India, Bhutan (Southeast Asia) | C. C. Tsai 2240 | EF156114 | EF156029 | EF156199 | GQ850830 |
| Paphiopedilum spicerianum (Rchb.f.) Pfitzer | Northeast India | C. C. Tsai 2229 | EF156145 | EF156060 | EF156230 | GQ850857 |
| Paphiopedilum tigrinum Koop. & N. Haseg. | China | C. C. Tsai 2218 | EF156149 | EF156064 | EF156234 | GQ850862 |
| Paphiopedilum tranlienianum Gruss & Perner | Unknown | C. C. Tsai 2042 | EF156151 | EF156066 | EF156236 | GQ850864 |
| Paphiopedilum villosum (Lindl.) Stein | India, Burma, Thailand (Southeast Asia) | C. C. Tsai 2216 | EF156159 | EF156074 | EF156244 | GQ850872 |
| Section Barbata | ||||||
| Paphiopedilum acmodontum Schoser ex M.W. Wood | Philippines | C. C. Tsai 2094 | EF156081 | EF155996 | EF156166 | GQ850879 |
| Paphiopedilum appletonianum (Gower) Rolfe | China, Thailand, Cambodia, Laos, Vietnam (Southeast Asia) | C. C. Tsai 2153 | EF156084 | EF155999 | EF156169 | GQ850801 |
| Paphiopedilum argus (Rchb.f.) Stein | Philippines | C. C. Tsai 2282 | EF156085 | EF156000 | EF156170 | GQ850802 |
| Paphiopedilum braemii Mohr | Northern Sumatra, Indonesia | C. C. Tsai 2151 | EF156089 | EF156004 | EF156174 | GQ850806 |
| Paphiopedilum barbatum (Lindl.) Pfitzer | Southern Thailand, peninsular Malaysia, Sumatra | C. C. Tsai 2227 | EF156087 | EF156002 | EF156172 | GQ850804 |
| Paphiopedilum callosum (Rchb.f.) Stein | Thailand, Cambodia, Laos, Vietnam (south-east Asia) | C. C. Tsai 2267 | EF156090 | EF156005 | EF156175 | GQ850807 |
| Paphiopedilum ciliolare (Rchb.f.) Stein | Philippines | C. C. Tsai 2078 | EF156092 | EF156007 | EF156177 | GQ850809 |
| Paphiopedilum curtisii (Rchb. f.) Stein | Sumatra | C. C. Tsai 2107 | EF156094 | EF156009 | EF156179 | GQ850811 |
| Paphiopedilum dayanum (Lindl.) Stein | Borneo | C. C. Tsai 2280 | EF156095 | EF156010 | EF156180 | GQ850812 |
| Paphiopedilum fowliei Birk | Philippines | No voucher | GQ505311 | GQ505314 | GQ505317 | GQ850820 |
| Paphiopedilum hookerae (Rchb.f.) Stein | Borneo | C. C. Tsai 2089 | EF156116 | EF156031 | EF156201 | GQ850831 |
| Paphiopedilum volonteanum (Sander) Stein | Borneo | No voucher | EF156115 | EF156030 | EF156200 | GQ850873 |
| Paphiopedilum javanicum (Reinw. ex Lindl.) Pfitzer | Borneo, southeast Sumatra, Java | C. C. Tsai 2326 | EF156120 | EF156035 | EF156205 | GQ850834 |
| Paphiopedilum javanicum var. virens (Rchb. f.) Stein | North Borneo | No voucher | EF156119 | EF156034 | EF156204 | GQ850833 |
| Paphiopedilum lawrenceanum (Rchb.f.) Stein | Borneo | C. C. Tsai 2013 | EF156122 | EF156037 | EF156207 | GQ850836 |
| Paphiopedilum mastersianum (Rchb.f.) Stein | Moluccas | C. C. Tsai 2341 | EF156126 | EF156041 | EF156211 | GQ850840 |
| Paphiopedilum papuanum (Ridl.) Ridl. | New Guinea | No voucher | EF156139 | EF156047 | EF156217 | GQ850846 |
| Paphiopedilum purpuratum (Lindl.) Stein | China, Vietnam | C. C. Tsai 2049 | EF156131 | EF156052 | EF156222 | GQ850850 |
| Paphiopedilum sangii Braem | Sulawesi | C. C. Tsai 2088 | EF156137 | EF156058 | EF156228 | GQ850855 |
| Paphiopedilum schoseri Braem | Moluccas | No voucher | EF156144 | EF156059 | EF156229 | GQ850856 |
| Paphiopedilum sukhakulii Schoser & Senghas | Northern Thailand | C. C. Tsai 2226 | EF156147 | EF156062 | EF156232 | GQ850859 |
| Paphiopedilum superbiens (Rchb.f.) Stein | Sumatra | C. C. Tsai 2082 | EF156148 | EF156063 | EF156233 | GQ850861 |
| Paphiopedilum tonsum (Rchb.f.) Stein | Northern Sumatra, Indonesia | C. C. Tsai 2087 | EF156150 | EF156065 | EF156235 | GQ850863 |
| Paphiopedilum urbanianum Fowlie | Philippines | C. C. Tsai 2161 | EF156152 | EF156067 | EF156237 | GQ850865 |
| Paphiopedilum veniferum Koop. & Haseg | Unknown | C. C. Tsai 2253 | EF156153 | EF156068 | EF156238 | GQ850866 |
| Paphiopedilum venustum (Wall. ex Sims) Pfitzer ex Stein | Bhutan, India, Nepal | C. C. Tsai 2032 | EF156154 | EF156069 | EF156239 | GQ850867 |
| Paphiopedilum wardii Summerh | Burma, southwest China | C. C. Tsai 2139 | EF156161 | EF156076 | EF156246 | GQ850874 |
| Genus Phragmipedium | ||||||
| Phragmipedium pearcei Garay | Ecuador, Peru | C. C. Tsai 2009 | EF156163 | EF156078 | EF156248 | GQ850877 |
| Phragmipedium longifolium Rchb. f. & Warsc | Costa Rica, Panama, Colombia, Ecuador | C. C. Tsai 2043 | EF156165 | EF156080 | EF156250 | GQ850876 |
The systematics of Phalaenopsis are based on Christenson (2001).
Voucher specimens were deposited at the herbarium of the National Museum of Natural Science, Taiwan (TNM).
DNA Extraction, PCR Amplification, and Sequencing
Total DNA was extracted from fresh etiolated leaves by using the cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). Approximate DNA yields were determined by using the spectrophotometer (model U-2001, Hitachi).
The PCR reaction was used to amplify nuclear ribosomal ITS sequence and chloroplast (cp) DNA fragments trnL intron and the trnL-trnF spacer, atpB-rbcL spacer. ITS primers were designed from conserved regions of the 3' end of the 18S rRNA gene and the 5'end of the 26S rRNA gene using sequences from different species present in GenBank. Universal primers for trnL intron and the trnL-trnF spacer were referenced from Taberlet et al. (1991). Primer sequences for amplifying of the atpB-rbcL spacer were designed from the conserved regions of the 3' end of the atpB gene and the 5'end of the rbcL gene of chloroplast DNA using sequences of different species obtained from GenBank. Detailed amplification conditions and primer sequences are given in Supplementary Table S1 . All PCR products were separated by agarose gel electrophoresis (1.0%, w/v in TBE) and were recovered using glassmilk (BIO 101, California).
PCR products were directly sequenced using the dideoxy chain-termination method on an ABI377 automated sequencer with the Ready Reaction Kit (PE Biosystems, California) of the BigDye™ Terminator Cycle Sequencing. The PCR reaction primer sequences were used as sequencing primers. Each sample was sequenced two or three times to confirm the sequences. Reactions were performed as recommended by the product manufacturers.
Sequence Alignment and Phylogenetic Reconstruction
The sequence alignment was determined using the ClustalW multiple alignment program in BioEdit (Hall, 1999), and four regions were combined for the following analysis. The alignment was checked, and apparent alignment errors were corrected by hand. Indels (insertion/deletions) were treated as missing data. For phylogenetic reconstruction, two Phragmipedium taxa treating as outgroups were sequenced to resolve whether all in-group taxa formed a monophyletic lineage. The best-fitting substitution model was selected ( Supplementary Table S2 ) by a model test using MEGA 6.0 (Tamura et al., 2013). Tamura 3-parameter model (T92) using a discrete Gamma distribution (+G) was selected for following neighbor-joining (NJ) phylogenetic reconstruction. The general time reversible (GTR) using a discrete gamma distribution (+G) and considering the proportion of invariable sites (+I) were chosen for following divergence time estimation using the Yule model methods in BEAST 1.8.0 (Drummond and Rambaut, 2007; Drummond et al., 2012). The phylogenetic tree for the combined multiple sequence datasets used equally weighted characters. Moreover, because the sequence data of the four genera (Mexipedium, Selenipedium, Cypripedium, and Goodyera) in NCBI is limited, only two sets of fragment data (ITS: Mexipedium xerophyticum-MK161260.1; Selenipedium aequinoctiale-JF825977.1; Cypripedium_macranthos-KT338684.1; Goodyera_procera-MK451741.1and trnL-trnF spacer: Mexipedium xerophyticum-FR851215.1; Selenipedium aequinoctiale-JF825975.1; Cypripedium_macranthos-JF797026.1; Goodyera_procera-MK451782.1) are used as an additional analysis and compared with the data using only genus Phragmipedium as outgroup. The results of six outgroups are showed in Supplementary Data . Genetic relationships were determined using NJ in the MEGA 6.0 (Tamura et al., 2013), maximum parsimony (MP) in PHYLIP 3.68 (Felsenstein, 2004), and maximum likelihood (ML) in MEGA 6.0 (Tamura et al., 2013). Bootstrapping (1,000 replicates) was carried out to estimate the support for NJ, MP, and ML topologies (Felsenstein, 1985; Hillis and Bull, 1993). The strict consensus parsimonious tree was then constructed by using the MEGA 6.0 (Tamura et al., 2013).
Divergence Time Estimation
The combined chloroplast DNA (cpDNA) dataset was used to estimate the divergence times using the Bayesian Yule model methods (BEAST version 1.7.5). The characteristic of uniparental inheritance in cpDNA prevents the interference of recombination introgression on phylogenetic reconstruction (Drummond et al., 2012). The general-time reversible (GTR) model with estimates of invariant sites (+I) and gamma-distributed among site rate variation (+G) in all matrices without partitions model was determined by the nucleotide substitution model test, conducted in MEGA 6.0 (Tamura et al., 2013).
To estimate the divergence time, two strategies, the strict and relaxed clock models, were adopted. For the relaxed clock, the calibration point at the most recent common ancestor (MRCA) of Paphiopedilum and Phragmipedium for 22 Ma (Gustafsson et al., 2010) were used to calculate the divergence times of each node. However, since there is no suitable fossil record to correct the calibration of divergence times for the ingroup, we recalculate the divergence time by strict clock model for consistency. For the strict clock, the mean substitution rate of 1.82 × 10−9 subs/site/year with the lower and upper limits 1.11 × 10−9 subs/site/year and 2.53 × 10−9 subs/site/year, respectively, were used for the cpDNA spacers in Phalaenopsis amabilis complex (Tsai et al., 2015).
We conducted four independent runs of a Yule prior and four Markov Chain Monte Carlo (MCMC) chains with a different starting seed. The first 10% simulations were discarded (burn-in) in a total of 108 generations. For thinning, one tree was reserved every 10,000 trees, and finally, 10,000 trees were left to calculate the posterior probability of each node. The effective sample size (ESS) > 200 was used as a criterion to check whether the sampling (simulations) is proper and is reaching a stationary distribution by Tracer v1.6 (Rambaut et al., 2018). Four independent-runs results, including the log file and tree file, were combined with the assistance of LogCombiner 1.6.1 (Drummond and Rambaut, 2007). TreeAnnotator 1.6.1 (Drummond and Rambaut, 2007) was used to summarize a consensus tree with a criterion of the maximum clade reliability using the mean heights option. The final consensus tree was drawn by FigTree 1.3.1 (Rambaut, 2009).
Biogeographic Inference Using Reconstruct Ancestral State in Phylogenies
The Statistical dispersal–vicariance analysis was used to assess the biogeographic patterns of Paphiopedilum species [Statistical Dispersal-Vicariance Analysis (S-DIVA), (Yu et al., 2010)]. Bayes–Lagrange Statistical dispersal–extinction–cladogenesis (S-DEC) model (Ree and Smith, 2008) was performed in Reconstruct Ancestral State in Phylogenies (RASP) 3.2 (Yu et al., 2015) to distinguish the events of vicariance, dispersal, and extinction. Five geographic areas were determined mainly according to Myers et al. (2000) with a little modification to illuminate the vicariance, dispersal and extinction events of Paphiopedilum species. The hotspot areas in South-Central China and Indo-Burma with the Malay Peninsula were combined as area A, consisting of China, Nepal, India, Bhutan, Burma, Thailand, Malaysia, Cambodia, Vietnam, and Laos. The hotspot “Sundaland” including Borneo, Java, and Sumatra were set as the area B. We move the Malay Peninsula from the area “Sundaland” to area A due to the integrity of the current landmass. The hotspots “Wallacea” (include Sulawesi and Moluccas) and “Philippines” were set as area C and area E. Respectively, islands of New Guinea eastern from the Wallacea are defined as area D. Species of outgroup were all defined as the area I. These two outgroup species are distributed in Ecuador, Peru, Costa Rica, Panama, and Colombia. The real distribution of outgroups is too far from the areas of the species in this study. Therefore, the ranges of outgroups are assigned to a new area in which none of the ingroup species occurs (Yu et al., 2014). The ML tree topologies were used in S-DIVA analysis.
Results
Sequence Alignment and Characteristics
The lengths of the ITS sequences obtained from the Paphiopedilum and outgroup samples were similar to those reported for a broad example of angiosperms (Baldwin, 1992; Baldwin et al., 1995). The alignment length of the ITS sequence is 735 nucleotides, of which 343 were identified as variable sites with 235 potentially parsimony informative sites. The average genetic distance between the 78 Paphiopedilum samples was 0.039 in ITS, and the average genetic distance between the 78 Paphiopedilum species was 0.01 in cpDNA. The alignment of combined plastid DNA fragments contained a total of 2,409 characters, of which 872 were identified as variable sites with 588 potentially parsimony informative sites. Since the sequences of three samples of every species are the same within species, only one sequence per species was used for the analyses and deposited in NCBI GenBank. The accession numbers of the nuclear ribosomal ITS sequences and the three fragments of plastid DNA from the 78 Paphiopedilum taxa and the two outgroup samples (from the genus Phragmipedium) are shown in Table 1 .
Phylogeny Reconstruction
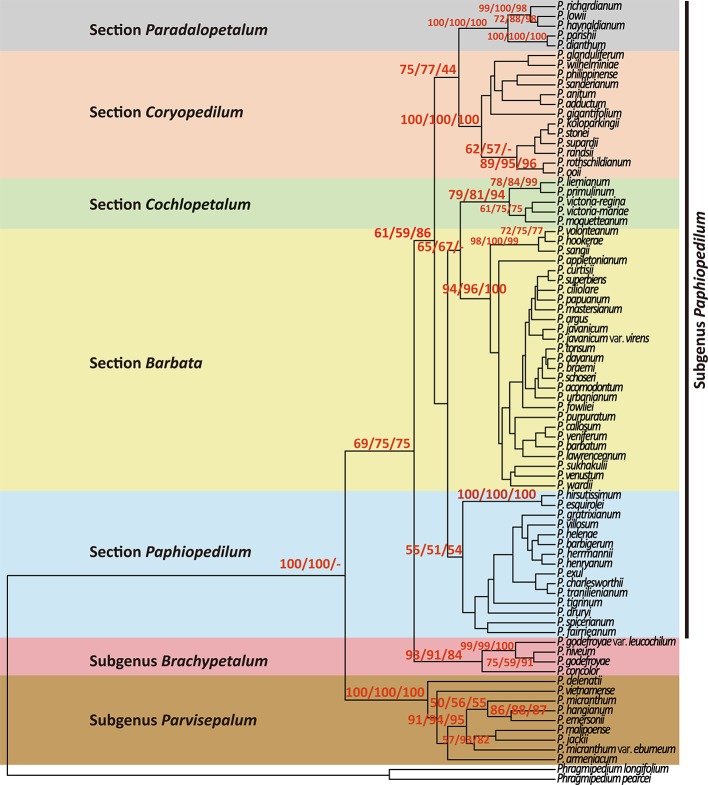
Both NJ and MP trees revealed a monophyletic relationship of 78 Paphiopedilum taxa with high bootstrap supports ( Figure 2 ). Moreover, the use of six outgroups for phylogeny reconstruction showed similar bootstrap supports ( Supplementary Figure S1 ). In the Paphiopedilum monophyletic clade, three subgenera Parvisepalum, Brachypetalum, and Paphiopedilum formed independent monophyletic clades with 100/100/100, 98/91/84, and 61/59/86% bootstrap supporting values in NJ/MP/ML trees, in which subgenus Parvisepalum was diverged firstly from other lineages ( Figure 2 , Supplementary Figure S1 ). In subgenus Paphiopedilum, sections Barbata, Cochlopetalum, Pardalopetalum, and Coryopedilum are monophyletic with 94/96/100, 79/81/94, 100/100/100, and 100/100/100% bootstrap supporting values in NJ, MP, and ML trees. Additionally, the use of six outgroups for phylogeny reconstruction showed similar patterns ( Supplementary Figure S1 ). However, section Paphiopedilum showed a low support [56/51/54% bootstrap values ( Supplementary Figure S2 ); 59% bootstrap values ( Supplementary Figure S1 )] for the monophyly.
Figure 2.
Phylogenetic relationships using neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) resulting from analysis of the combined data matrix (nuclear ribosomal ITS, plastid trnL intron, trnL-F spacer, and atpB-rbcL spacer) from 78 Paphiopedilum and 2 outgroup species. Only the strict consensus of all most parsimonious trees (MP trees) are showed in this figure, and the bootstrap values > 50% are shown on each branch for NJ/MP/ML between major lineage.
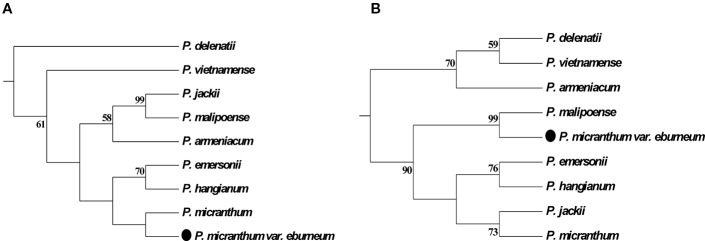
In addition, the molecular data demonstrates that a newly described variety, P. micranthum var. eburneum, is closely related to P. malipoense based on the plastid DNA within subgenus Parvisepalum, which is inconsistent with the inference by nuclear ITS and combined data. In ITS tree, P. micranthum var. eburneum is sister with P. micranthum var. micranthum ( Figures 2 and 3 ). Therefore, we infer a hybridization event between the ancestor of P. micranthum and P. malipoense that lead to a plastid capture in P. micranthum var. eburneum.
Figure 3.
Parsimonious phylogenetic tree of the Paphiopedilum subgenus Parvisepalum derived from ITS sequences of nuclear ribosomal DNA (nrDNA) (A) and plastid DNA (B). The solid circle (․) represents a putative natural hybrid (based on molecular evidence).
Divergence Time Estimates
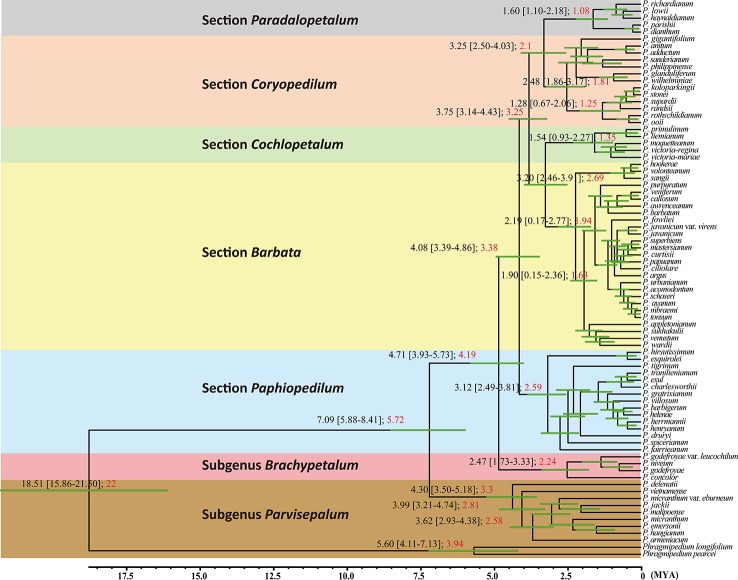
The coalescence time of the genus Paphiopedilum was estimated to be 7.09 Mya with 95% confidence intervals (95% CI) of 5.88–8.41 Mya ( Figure 4 ) according to the substitution rate referenced from Tsai et al. (2015). If calibrating by relaxed clock referring to Gustafsson et al. (2010), the estimated coalescence time of the genus Paphiopedilum was 5.72 Mya ( Figure 4 ). In the genus Paphiopedilum, the coalescence times estimated by strict clock were 4.30 Mya (95% CI: 3.50–5.18 Mya), 2.47 Mya (95% CI: 1.73–3.33 Mya), and 4.08 Mya (95% CI: 3.39–4.86 Mya) for subgenera Parvisepalum, Brachypetalum, and Paphiopedilum, respectively ( Figure 4 ). After re-calibrating by a relaxed clock, the coalescence time was estimated to be 3.3 Mya, 2.24, and 3.38 Mya Mya for subgenera Parvisepalum, Brachypetalum, and Paphiopedilum, respectively ( Figure 4 ). In addition, the strict clock suggested that the coalescence times were tracked back to 2.19 Mya (95% CI: 0.17–2.77 Mya), 1.54 Mya (95% CI: 0.93–2.27 Mya), 3.12 Mya (95% CI: 2.49–3.81 Mya), 2.48 (95% CI: 1.86–3.17 Mya), and 1.60 Mya (95% CI: 1.10–2.18 Mya) for clades of subgenera Barbata, Cochlopetalum, Paphiopedilum, Coryopedilum, and Pardalopetalum of subgenus Paphiopedilum, respectively. By relaxed clock, the coalescence time was estimated to be 1.94, 1.3, 2.59, 1.81, and 1.08 Mya for clades of subgenera Barbata, Cochlopetalum, Paphiopedilum, Coryopedilum, and Pardalopetalum of subgenus Paphiopedilum, respectively. Additionally, the use of six outgroups for divergence time estimation also showed similar supports ( Supplementary Figure S2 ). In short, the estimates of the coalescence times by strict and relaxed clocks are similar, but the time calculated is slightly shorter by relaxed clocks. Regardless, the coalescence time of the genus Paphiopedilum will not be earlier than Upper Miocene.
Figure 4.
Results of calescence time estimations performed with BEAST 1.8.0 for the from 78 Paphiopedilum taxa based on the combined data matrix (nuclear ribosomal ITS, plastid trnL intron, trnL-F spacer, and atpB-rbcL spacer). The black numbers in each node are using the mean rate of 1.82 × 10−9 subs/site/year, with the lower and upper limits 1.11 × 10−9 subs/site/year and 2.53 × 10−9 subs/site/year (Tsai et al., 2015). The red numbers in each node are using the Gustafsson et al. (2010) fossil calibration time data to calibrate the divergence time.
Demographic History and Historical Biogeography Inference
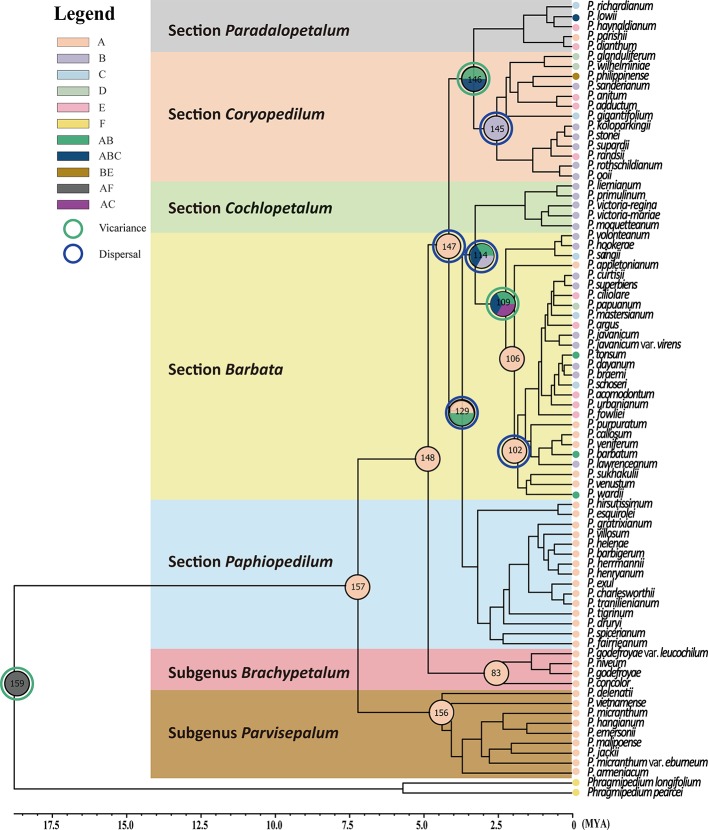
Complicated evolutionary processes of continuous and episodic dispersal, vicariance, and extinctions determined the current geographic distribution of genus Paphiopedilum. Since the most probable ancestral areas located on continental Asia (area A in Figure 5 ), dispersal events seem to determine the extant distributions of subgenera largely. The results supported vicariance events on nodes 159, 146, and 109 shown in Table 2 and Figure 5 , and on nodes 163, and 132 ( Supplementary Table S3 and Figure S3 ). The node 147 revealed dispersal events among section Coryopedilum/Pardalopetalum of Paphiopedilum and other sections of Paphiopedilum causing by migration route from area A (China, Nepal, India, Bhutan, Burma, Thailand, Malaysia, Cambodia, Vietnam, and Laos) to B area (Sumatra, Borneo, and Java). Meanwhile, the nodes 145, 129, 114, and 102 also revealed dispersal events from north to south, according to Sundaland and Sunda Super Islands.
Figure 5.
Ancestral distributions reconstructed by the Statistical dispersal–vicariance analysis [S-DIVA, (Yu et al., 2010)] and Bayes–Lagrange Statistical dispersal–extinction–cladogenesis (S-DEC) model (Ree and Smith, 2008) performed in Reconstruct Ancestral State in Phylogenies (RASP) 3.2 (Yu et al., 2015). Phylogenetic relationships of the 78 Paphiopedilum species, plus the two outgroups Phragmipedium pearcei and Ph. longifolium, obtained from sequence judgments of the combined sequence and generated by BEAST. The distribution areas of extant the 78 Paphiopedilum species are marked in capitals A–E and I [(A) China, Nepal, India, Bhutan, Burma, Thailand, Malaysia, Cambodia, Vietnam, and Laos; (B) Sumatra, Borneo and Java; (C) Sulawesi and Moluccas; (D) New Guinea; (E) Philippines; and (I) outgroup], respectively. The green and blue circles indicate the vicariance and dispersal events obtained from the RASP analysis, respectively.
Table 2.
The ancestral areas and dispersal–vicariance analysis inferred through Reconstruct Ancestral State in Phylogenies (RASP). Ancestral areas for the node and the number of dispersal (Dis), vicariance (Vic), and extinction (Ext) events are shown.
| Node | Ancestral areas | RASP ROUTE | Dis | Vic | Ext | Prob |
|---|---|---|---|---|---|---|
| 159 | [AF] | AF- > A|F | 0 | 1 | 0 | 1.00 |
| 157 | [A] | A- > A^A- > A|A | 0 | 0 | 0 | 1.00 |
| 156 | [A] | A- > A^A- > A|A | 0 | 0 | 0 | 1.00 |
| 148 | [A] | A- > A^A- > A|A | 0 | 0 | 0 | 1.00 |
| 147 | [A] | A- > A^A^B- > ABC^A^B- > AB|ABC | 3 | 0 | 0 | 0.99 |
| 146 | [ABC|AB] | ABC- > AC|B | 0 | 1 | 0 | 1.00 |
| 145 | [B] | B- > B^B- > BDE^B- > BDE|B | 2 | 0 | 0 | 0.91 |
| 129 | [AB|A] | AB- > AB^A- > ABC^A- > A|ABC | 2 | 0 | 0 | 0.74 |
| 114 | [ABC|AB|B] | ABC- > ABC^B- > B|ABC | 1 | 0 | 0 | 0.60 |
| 109 | [ABC|AC|AB] | ABC- > BC|A | 0 | 1 | 0 | 1.00 |
| 106 | [A] | A- > A^A- > A|A | 0 | 0 | 0 | 0.89 |
| 102 | [A] | A- > A^A- > ABE^A- > A|ABE | 2 | 0 | 0 | 0.99 |
| 83 | [A] | A- > A^A- > A|A | 0 | 0 | 0 | 1.00 |
Furthermore, the use of six outgroups for dynamic historical inference showed similar patterns ( Supplementary Table S3 and Figure S3 ). Only the nodes 146 and 109 were detected vicariance event causing by the geological separation between Indochina and Sumatra/Borneo/Java. In addition, in subgenus Paphiopedilum, 2 vicariance and 10 dispersal events were detected, which suggesting a significant dispersal process affected biogeographical patterns in shaping the current distribution in the subgenus Paphiopedilum. Areas A and B might be the two possible ancestral areas and likely shaped by several complicated dispersal events in the subgenus Paphiopedilum.
Discussion
Systematics Revision of Genus Paphiopedilum
In general, our phylogenetic inference is mostly congruent with that of Cox et al. (1997), Cribb (1997b), and Guo et al. (2015). In the genus Paphiopedilum, tessellated leaves, single flowers with broad elliptic to subcircular petals, and a sizeable thin-textured lip characterize subgenus Parvisepalum in southwest China and Vietnam (Cribb, 1998). Within this subgenus, the phylogenetic topography and the divergence time of at least 4.30 Mya rejected the previous hypothesis of the sister-species relationship between P. armeniacum and P. delenatii (Cribb, 1983) ( Figure 4 ). The geographical distribution of these two species is also separated (Yunnan, China for P. armeniacum, and Vietnam for P. delenatii) (Cribb, 1998).
Furthermore, a newly described variety, P. micranthum var. eburneum, is phylogenetically close to P. malipoense in maternal-inherited plastid DNA but close to P. micranthum in biparental-inherited nuclear ITS sequences ( Figure 5 ), suggesting that P. micranthum var. eburneum is a natural hybrid between the maternal parent P. malipoense and the paternal parent P. micranthum and experienced the event of chloroplast capture. The overlap of the geographical distribution of these three taxa also supports this hypothesis (Cribb, 1998). In addition, ITS sequences are usually concertedly evolved via unequal crossing-over (Schlotterer and Tautz, 1994) and biased gene conversion (Hillis et al., 1991), which results in sequence homogeneity between paralogs (Maynard, 1989).
The monophyly of subgenus Brachypetalum inferred in this study is congruent with the inference by Cox et al. (1997). The subgenus Brachypetalum is geographically confined to Southeast Asia (Cribb, 1998). Albeit overlapping distribution with subgenus Parvisepalum (Cribb, 1998), subgenus Brachypetalum is phylogenetically separated, consistent with the distinguishable leaf anatomy between these two subgenera (Cribb, 1998). Both molecular and morphological evidences support the independent taxonomic treatment between subgenera Brachypetalum and Parvisepalum (Karasawa, 1982; Karasawa and Saito, 1982; Cribb, 1997b), but object with Atwood's (1984) opinion of taking the subgenus Parvisepalum as a synonym of Brachypetalum.
The monophyletic subgenus Paphiopedilum can be morphologically and phylogenetically subdivided into five sections: Coryopedilum, Pardalopetalum, Cochlopetalum, Paphiopedilum, and Barbata (Cox et al., 1997; Cribb, 1997b). Section Coryopedilum is characterized by its plain green, strap-like leaves, and multi-flowered inflorescences, which flowers are blooming simultaneously (Cribb, 1998). This section distributes throughout Borneo, Sulawesi, New Guinea, and the Philippines (Cribb, 1998). Except placing Paphiopedilum parishii and Paphiopedilum dianthum into section Pardalopetalum from section Coryopedilum, Cribb (1997b) agreed with Atwood (1984) that section Pardalopetalum is independent from section Coryopedilum taxonomically, according to the ITS analysis (Cox et al., 1997) and similar green strap-like leaves and staminodes (Cribb, 1997b), which is consistent with our phylogenetic inference. However, the only character that separates sections of Pardalopetalum and Coryopedilum is the morphology of staminode. Whether this single character is sufficient to characterize them as separating sections should be re-evaluated with more evidence.
Unlike the simultaneous bloom of section Coryopedilum, section Cochlopetalum flower in succession, and their flowers bear elliptic bracts, linear, spirally twisted, spreading, ciliate petals, and a pot-shaped spotted lip (Cribb, 1998). Section Cochlopetalum distributes in Sumatra and Java only (Cribb, 1998). The extensive section Barbata is the sister of section Cochlopetalum, also characterized by a solitary flower with a lip and prominent incurved side-lobes, but the leaf tessellated (Cribb, 1998). The morphological dissimilarity and reciprocally monophyletic relationship indicate that, despite recently diverged, these two sections should be independent taxonomically.
Biogeography and Evolutionary Trends
The clade of genus Paphiopedilum is coalesced to 7.09 or 5.72 Mya, similar to the estimate of 7.62 Mya by Guo et al. (2015). The flower morphology of subgenus Parvisepalum is intermediate between other subgenera of Paphiopedilum and Cypripedium (Chen and Tsi, 1984), which could be explained by the earlier divergence of subgenus Parvisepalum in genus Paphiopedilum (Guo et al., 2012). Presently, the genus Cypripedium is distributed throughout worldwide temperate zones (Cox et al., 1997), with China as a center for species diversity (Cribb, 1997a). Therefore, genera Paphiopedilum and Cypripedium have most likely diverged in mainland Asia (Chen and Tsi, 1984).
However, genus Paphiopedilum was suggested as the sister with two American genera Phragmipedium and Mexipedium according to morphology, plastid rbcL (Albert, 1994), ITS (Cox et al., 1997), and both nuclear and plastid genes (Guo et al., 2012). These inferences are conflict to the hypothesis of the divergence between Paphiopedilum and Cypripedium in China, but implied the divergence of Paphiopedilum from the group of Phragmipedium + Mexipedium, by which the slipper orchids (Cypripedioideae) were hypothesized widespread throughout North America and Asia in the past (Atwood, 1984; Albert, 1994; Cox et al., 1997).
Subgenus Parvisepalum in southwest China and Vietnam diverged earlier from the other subgenera of genus Paphiopedilum. The coalescence time of subgenera Parvisepalum, Brachypetalum, and Paphiopedilum were tracking back to the Upper Miocene (Guo et al., 2015). Subgenus Brachypetalum in mainland Southeast Asia was descended from the subgenus Parvisepalum inferred by S-DIVA ( Figure 5 ), which agrees with other disjunctions at the Southern China and Indochina (Guo et al., 2015) or Sunda Shelf and New Guinea/Australia (Tougard, 2001; Lohman et al., 2011; Tsai et al., 2015).
The subgenus Paphiopedilum is further descended and evolved quickly in the Sunda Shelf. A land bridge might connect Mindoro, Palawan, Borneo, the Malay Peninsula, Borneo, Sumatra, Java, Bali, and various parts of the Philippines when sea levels falling during Pleistocene (about 0.01~1.8 Mya) (van Oosterzee, 1997) ( Figure 1 ). Surfaced land bridge connected these regions and was beneficial to the interisland and continent-island dispersal in Southeast and South Asia (van Oosterzee, 1997). The clade of Coryopedilum + Pardalopetalum was the first derived in subgenus Paphiopedilum based on the phylogenetic tree, which reflects in the sympatric distribution of subgenus Brachypetalum and section Pardalopetalum ( Figure 6 ). Following this clade formation, sections Paphiopedilum and Barbata were subdivided and dispersed throughout Southeast Asian archipelagoes across the land bridge during the glacials. The southward expansion from continental Asia into the Greater Sunda Islands through the Indochina and Malay Peninsulas were also reported in other taxa, e.g., Lithocarpus (Fagaceae) (Yang et al., 2018). Such colonization events between continental Asia and the Greater Sunda Islands mostly occurred during Miocene and Plio-Pleistocene (de Bruyn et al., 2014). As a “corridor,” Indochina reveals high flora diversity and the high species richness, which facilitates the in situ speciation (de Bruyn et al., 2014).
Figure 6.
The possible evolutionary routes of the genus Paphiopedilum.
Another flora diversity hotspot is Borneo (de Bruyn et al., 2014), which is also important for the genus Paphiopedilum. The section Cochlopetalum, which is found only in Sumatra and Java, might represent a group derived from Borneo. The S-DIVA inferred multiple times dispersal events sourced from Borneo with two vicariances to illustrate the current distribution of the five sections within subgenus Paphiopedilum ( Figure 5 and Table 2 ). The Borneo is the second original center of Paphiopedilum. The tropical forests and rugged topography harbor diversified niches opened to the speciation of organisms. The repeated submergence and emergence of land bridges could promote repeated genetic isolation and gene flow between closed related taxa. During the Plio-Pleistocene glacial oscillations. This process accelerates the diversification rates in the Sunda Super Islands.
Dispersal and vicariance events that exposed geographic isolation among taxa might be due to the land bridge submergence (Chiang et al., 2009; Chiang et al., 2013; Ge et al., 2012; Ge et al., 2015; Hsu et al., 2013) in Sunda Shelf and Sunda Super Island during the Pleistocene ( Figure 5 and Table 2 ) (Guo et al., 2015; Tsai et al., 2015). Cenozoic collision accompanied by a cyclical climate (glacial oscillations) caused by the fragmentation of the Sunda Super Islands (de Bruyn et al., 2014). The Borneo, Java, Sumatra, and the southern Philippines belong to the Sunda plate, Sulawesi is composed of broken plates, and the Moluccas and New Guinea belong to the Australian plate (Hall, 1998). The deep trenches between these plates cause segregation of species between Sundaland and the islands in the east. Because of the seed germination relies on symbiotic fungi, the geographical isolation maybe not only influences orchid itself but also in symbiotic fungi. However, these inferences still need further verification.
Conclusions
In summary, the origin and coalescence time of genus Paphiopedilum tracked back to Southern China/Eastern Indochina since late Miocene and early Pliocene, while the range expansion and species divergence were related to sea-level fluctuations during the Plio-Pleistocene glacial cycles. Historical geological barriers shaped a pattern of vicariance among disjunct distributed subgenera after isolated ancestral populations. The ancestral taxa of subgenus Paphiopedilum migrated from Southern China/Eastern Indochina to south which developed quickly in the Sunda Shelf. Due to the submergence of the Sunda Shelf and Sunda Super Island, species of subgenus Paphiopedilum dispersed with isolation between islands as well as subsequent in situ speciation within islands from other taxa within section or subgenus, which accelerated species divergence in subgenus Paphiopedilum. Paphiopedilum distributes in four of 25 biodiversity hotspots (Myers et al., 2000), the Indo-Burma, Sundaland, Wallacea, and Philippines, where are also the “major evolutionary hotspots” (de Bruyn et al., 2014). It is suggests that rich and fascinating historical biogeographic events have created rich species diversity there, such as the case of Paphiopedilum. However, deforestation has caused the so-called “empty forest syndrome” (de Bruyn et al., 2014). We hope that these areas will not become extinction hotspots, even though they are almost now.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
Conceived and designed the experiments: C-CT and Y-CC. Performed the experiments: C-CT, P-CL, Y-ZK, C-HC, and Y-CC. Analyzed the data: C-CT, P-CL, Y-ZK, C-HC, and Y-CC. Contributed reagents/materials/analysis tools: C-CT, P-CL, Y-ZK, C-HC, and Y-CC. Wrote the paper: C-CT, P-CL, and Y-CC. Conceived of the study, edited the manuscript, and approved the final manuscript: C-CT, P-CL, Y-ZK, C-HC, and Y-CC.
Funding
This research was supported by funding from the Ministry of Science and Technology, Taiwan (MOST 105-2621-B-110-003-MY3 and MOST 105-2621-B-110-001) to Y-CC and by partial financing (the Higher Education Sprout Project) of NSYSU.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The achievement of this study is dedicated to the memory of the scientific contributions of the first author C-CT, who died of a stroke on November 05, 2015. We are grateful to Dr. F. H. Lin for his valuable comments and helpful discussion in the course of the study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00126/full#supplementary-material
Phylogenetic relationships using Maximum Likelihood resulting from analysis of the combined data matrix (nuclear ribosomal ITS, and trnL-F spacer) from 78 Paphiopedilum and 6 outgroup species.
Results of calescence time estimations performed with BEAST 1.8.0 for the from 78 Paphiopedilum taxa and 6 outgroup species based on the combined data matrix (nuclear ribosomal ITS, and trnL-F spacer).
Ancestral distributions reconstructed by the Statistical dispersal–vicariance analysis (S-DIVA, Yu et al., 2010) and Bayes–Lagrange Statistical dispersal–extinction–cladogenesis (S-DEC) model (Ree and Smith, 2008) performed in RASP 3.2 (Yu et al., 2015). Phylogenetic relationships of the 78 Paphiopedilum species, plus the six outgroups, obtained from sequence judgments of the combined sequence and generated by BEAST. The distribution areas of extant the 78 Paphiopedilum species species are marked in capitals A–E and I ((A) China, Nepal, India, Bhutan, Burma, Thailand, Malaysia, Cambodia, Vietnam, and Laos; (B) Sumatra, Borneo and Java; (C) Sulawesi and Moluccas; (D) New Guinea; (E) Philippines; and (I) outgroup), respectively. The green and blue circles indicate the vicariance and dispersal events obtained from the RASP analysis, respectively.
References
- Albert V. A., Chase M. A. (1992). Mexipedium: a new genus of slipper orchid (Cypripedioideae: Orchidaceae). Lindleyana 7, 172–176. [Google Scholar]
- Albert V. A. (1994). Cladistic relationships of the slipper orchids (Cypripedioideae: Orchidaceae) from congruent morphological and molecular data. Lindleyana 9, 115–132. [Google Scholar]
- Atwood J. T. (1984). The relationships of the slipper orchids (subfamily Cypripediodeae), Orchidaceae. Selbyana 7, 129–247. [Google Scholar]
- Aurelio M. A., Barrier E., Rangin C., Muller C. (1991). The philippine fault in the late cenozoic tectonic evolution of the bondoc-masbate-N. leyte area, central philippines. J. Southeast Asian Earth Sci. 6, 221–238. 10.1016/0743-9547(91)90069-A [DOI] [Google Scholar]
- Baldwin B. G., Sanderson M. J., Wojciechowski M. F., Campbell C. S., Donoghue M. J. (1995). The ITS region of nuclear ribosomal DNA-a valuable source of evidence on angiosperm phylogeny. Ann. Missouri Bot. Garden 82, 247–277. 10.2307/2399880 [DOI] [Google Scholar]
- Baldwin B. G. (1992). Phylogenetic utility of the internal transcribed sequences of nuclear ribosomal DNA in plants: an example from the Compositae. Mol. Phylogenet. Evol. 1, 3–16. 10.1016/1055-7903(92)90030-K [DOI] [PubMed] [Google Scholar]
- Chen S. C., Tsi Z. H. (1984). On Paphiopedilum malipoense sp. nov.-an intermediate from between and Cypripedium. Acta Phytotaxonomica Sin. 22, 119–124. [Google Scholar]
- Chiang T. Y., Schaal B. A., Peng C. I. (1998). Universal primers for amplification and sequencing a noncoding spacer between the atpB and rbcL genes of chloroplast DNA. Bot. Bull. Academia Sin. 39, 245–250. [Google Scholar]
- Chiang Y. C., Hung K. H., Moore S. J., Ge X. J., Huang S., Hsu T. W., et al. (2009). Paraphyly of organelle DNAs in Cycas Sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol. Biol. 9, 161. 10.1186/1471-2148-9-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y. C., Huang B. H., Chang C. W., Wan Y. T., Lai S. J., Huang S., et al. (2013). Asymmetric introgression in the horticultural living fossil Cycas Sect, Asiorientales using a genome-wide scanning approach. Int. J. Mol. Sci. 14, 8228–8251. 10.3390/ijms14048228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A. V., Pridgeon A. M., Albert V. A., Chase M. W. (1997). Phylogenetics of the slipper orchids (Cypripediodeae: Orchidaceae); Nuclear rDNA sequences. Plant Syst. Evol. 208, 197–223. 10.1007/BF00985442 [DOI] [Google Scholar]
- Cribb P. (1983). A synopsis of the genus Paphiopedilum . Plantsman 4, 193–212. [Google Scholar]
- Cribb P. (1997. a). The Genus Cypripedium (Portland, Oregon: Timber Press; ). [Google Scholar]
- Cribb P. (1997. b). The Genus Paphiopedilum (Portland, Oregon: Timber Press; ). [Google Scholar]
- Cribb P. (1998). The Genus Paphiopedilum. 2nd ed. (Kota Kinabalu, Sabah, Malaysia: Natural History Publications (Borneo) in association with Royal Botanic Gardens, Kew; ), 427. [Google Scholar]
- de Bruyn M., Stelbrink B., Morley R. J., Hall R., Carvalho G. R., Cannon C. H., et al. (2014). Borneo and Indochina are major evolutionary hotspots for Southeast Asian biodiversity. Syst. Biol. 63, 879–901. 10.1093/sysbio/syu047 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Drummond A. J., Rambaut A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214–221. 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A. J., Suchard M. A., Xie D., Rambaut A. (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. BMC Evol. Biol. 29, 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (2004). PHYLIP (Phylogeny Inference Package) Version 3.6 (Seattle: Department of Genome Sciences, University of Washington; ). [Google Scholar]
- Ge X. J., Hsu T. W., Hung K. H., Lin C. J., Huang C. C., Huang C. C., et al. (2012). Inferring multiple refugia and phylogeographical patterns in Pinus massoniana based on nucleotide sequence variation and fingerprinting. PLoS One 7, e43717. 10.1371/journal.pone.0043717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X. J., Hung K. H., Ko Y. Z., Hsu T. W., Gong X., Chiang T. Y., et al. (2015). Genetic divergence and biogeographical patterns in Amentotaxus argotaenia species complex. Plant Mol. Biol. Rep. 33 (2), 264–280. 10.1007/s11105-014-0742-0 [DOI] [Google Scholar]
- Goldblatt P., Savolainen V., Porteous O., Sostaric I., Powell M., Reeves G., et al. (2002). Radiation in the Cape flora and the phylogeny of Racock irises Moraea (Iridaceae) based on four plastid DNA regions. Mol. Phylogenet. Evol. 25, 341–360. 10.1016/S1055-7903(02)00235-X [DOI] [PubMed] [Google Scholar]
- Guo Y. Y., Luo Y. B., Liu Z. J., Wang X. Q. (2012). Evolution and biogeography of the slipper orchids: eocene vicariance of the conduplicate genera in the old and new world tropics. PLoS One 7 (6), e38788. 10.1371/journal.pone.0038788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y. Y., Luo Y. B., Liu Z. J., Wang X. Q. (2015). Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids (Paphiopedilum) in South-East Asia. Mol. Ecol. 24, 2838–2855. 10.1111/mec.13189 [DOI] [PubMed] [Google Scholar]
- Gustafsson A. L. S., Verola C. F., Antonelli A. (2010). Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evol. Biol. 10, 177. 10.1186/1471-2148-10-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. (1996). “Reconstructing Cenozoic SE Asia,” in Tectonic Evolution of SE Asia. Eds. Hall R., Blundell D. J. (London, UK: Geological Society of London Special Publication No. 106; ), 153–184. [Google Scholar]
- Hall R. (1998). “The plate tectonics of Cenozoic SE Asia and the distribution of land and sea,” in Biogeography and Geological Evolution of SE Asia. Eds. Hall R., Holloway J. D. (Leiden, The Netherlands: Backhuys Publishers; ), 99–131. [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hillis D. M., Bull J. J. (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42, 182–192. 10.1093/sysbio/42.2.182 [DOI] [Google Scholar]
- Hillis D. M., Moritz C., Porter C. A., Baker R. J. (1991). Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science 251, 308–310. 10.1126/science.1987647 [DOI] [PubMed] [Google Scholar]
- Hodkinson T. R., Chase M. W., Lledo M. D., Salamin N., Renvoize S. A. (2002). Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinate, Andropogoneae, Poaceae) based on the DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacer. J. Plant Res. 115, 381–392. 10.1007/s10265-002-0049-3 [DOI] [PubMed] [Google Scholar]
- Hsu T. W., Shih H. C., Kuo C. C., Chiang T. Y., Chiang Y. C. (2013). Characterization of 42 microsatellite markers from poison ivy, Toxicodendron radicans (Anacardiaceae). Int. J. Mol. Sci. 214, 20414–20426. 10.3390/ijms141020414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa K., Saito K. (1982). A revision of the genus Paphiopedilum (Orchidaceae). Bull. Hiroshima Bot. Garden 5, 1–69. [Google Scholar]
- Karasawa K. (1982). The Genus Paphiopedilum (Karasawa, Hiroshima, Japan: Seibundo Shinkosha Publisher; ). [Google Scholar]
- Karig D. E., Sarewitz D. R., Iiaeck G. D. (1986). Role of strike-slip faulting in the evolution of allochthonous terraces in the Philippines. Geology 14, 852–855. [DOI] [Google Scholar]
- Koopowitz H. (2000). A revised checklist of the genus Paphiopedilum . Orchid Digest 64, 155–179. [Google Scholar]
- Lindley J. (1840). The genera and species of orchidaceous plants (Pp554. Ridgways, Piccadilly, London: printed by W. Nicoi., 60, Pali Mall). [Google Scholar]
- Lohman D. J., de Bruyn M., Page T., von Rintelen K., Hall R., Ng P. K. L., et al. (2011). Biogeography of the Indo-Australian Archipelago. Annu. Rev. Ecol. Evol. Syst. 42, 205–226. 10.1146/annurev-ecolsys-102710-145001 [DOI] [Google Scholar]
- Maynard S. (1989). Evolutionary Genetics (Oxford, UK: Oxford University Press; ), 325. [Google Scholar]
- Mogensen H. L. (1996). The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 83, 383–404. 10.1002/j.1537-2197.1996.tb12718.x [DOI] [Google Scholar]
- Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A. B., Kent J. (2000). Biodiversity hotspots for conservation priorities. Nature 403, 853–858. 10.1038/35002501 [DOI] [PubMed] [Google Scholar]
- Quebral R. D., Pubellier M., Rangin C. (1994). The Mindanao: a transition from collision to strike-slip environment. Tectonics 15, 713–726. 10.1029/95TC00480 [DOI] [Google Scholar]
- Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. (2009). FigTree version 1.3.1 (University of Edinburgh: Institute of Evolutionary Biology; ). [http://tree.bio.ed.ac.uk/software/FigTree/]. [Google Scholar]
- Ree R. H., Smith S. A. (2008). Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. 10.1080/10635150701883881 [DOI] [PubMed] [Google Scholar]
- Schlotterer C., Tautz D. (1994). Chromosomal homogeneity and Drosophila ribosomal DNA arrays suggests intrachromosomal exchanges drive concerted evolution. Curr. Biol. 4, 777–783. 10.1016/S0960-9822(00)00175-5 [DOI] [PubMed] [Google Scholar]
- Stephan J. F., Blanchet R., Rangin C., Pelletir B., Letouzey J., Muller C. (1986). Geodynamic evolution of the Taiwan-Luzon-Mindoro belt since the Late Eocene. Tectonophysics 125, 245–268. 10.1016/0040-1951(86)90017-X [DOI] [Google Scholar]
- Taberlet P., Gielly L., Pautou G., Bouvet J. (1991). Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 17, 1105– 1109. 10.1007/BF00037152 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular evolutionary genetics analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tougard C. (2001). Biogeography and migration routes of large mammal faunas in south-east asia during the late middle pleistocene: focus on the fossil and extant faunas from thailand. Palaeogeogr Palaeoclimatol. Palaeoecol. 168, 337–358. 10.1016/S0031-0182(00)00243-1 [DOI] [Google Scholar]
- Tsai C. C., Chiang Y. C., Lin Y. S., Liu W. L., Chou C. H. (2012). Plastid trnL intron polymorphisms among Phalaenopsis species used for identifying the plastid genome type of Phalaenopsis hybrids. Sci. Hortic. 142, 84–91. 10.1016/j.scienta.2012.05.004 [DOI] [Google Scholar]
- Tsai C. C., Chou C. H., Wang H. V., Ko Y. Z., Chiang T. Y., Chiang Y. C. (2015). Biogeography of the Phalaenopsis amabilis species complex inferred from nuclear and plastid DNAs. BMC Plant Biol. 15, 202. 10.1186/s12870-015-0560-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterzee P. (1997). Where Worlds Collide: The Wallace Line (Ithaca, NY: Cornell Univ. Press; ). [Google Scholar]
- Van Raamsdonk L. M., Ensink W., Van Heusden A. W., Vrielink-Van Ginkel M., Kik C. (2003). Biodiversity assessment based on cpDNA and crossability analysis in selected species of Allium subgenus Rhizirideum . Theor. Appl. Genet. 107, 1048–1058. 10.1007/s00122-003-1335-8 [DOI] [PubMed] [Google Scholar]
- Von Konrat M., Shaw A. J., Renzaglia K. S. (2010). Bryophytes: The closest living relatives of early land plants. Phytotaxa 9, 1–278. 10.11646/phytotaxa.9.1.1 [DOI] [Google Scholar]
- Yang C. K., Chiang Y. C., Huang B. H., Ju L. P., Liao P. C. (2018). Nuclear and chloroplast DNA phylogeography suggests an early miocene southward expansion of Lithocarpus (Fagaceae) on the Asian continent and islands. Bot. Stud. 59, 27. 10.1186/s40529-018-0244-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K., Kubota Y., Ishii T., Wada K. (1992). Nucleotide sequence of atpB, rbcL, trnR, dedB and psaL chloroplast genes from a fern Angiopteris lygodiifolia: a possible emergence of Spermatophyta lineage before the separation of Bryophyta and Pteridophyta. Plant Mol. Biol. 18, 79–82. 10.1007/BF00018458 [DOI] [PubMed] [Google Scholar]
- Yu Y., Harris A. J., He X. J. (2010). S-DIVA (statistical dispersal–vicariance analysis): a tool for inferring biogeographic histories. Mol. Phylogenet. Evol. 56, 848–850. 10.1016/j.ympev.2010.04.011 [DOI] [PubMed] [Google Scholar]
- Yu Y., Harris A. J., He X. J. (2014). A rough guide to RASP 2.1 (Beta) (Brazil: University of Sao Paulo; ). [Google Scholar]
- Yu Y., Harris A. J., Blair C., He X. (2015). RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49. 10.1016/j.ympev.2015.03.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic relationships using Maximum Likelihood resulting from analysis of the combined data matrix (nuclear ribosomal ITS, and trnL-F spacer) from 78 Paphiopedilum and 6 outgroup species.
Results of calescence time estimations performed with BEAST 1.8.0 for the from 78 Paphiopedilum taxa and 6 outgroup species based on the combined data matrix (nuclear ribosomal ITS, and trnL-F spacer).
Ancestral distributions reconstructed by the Statistical dispersal–vicariance analysis (S-DIVA, Yu et al., 2010) and Bayes–Lagrange Statistical dispersal–extinction–cladogenesis (S-DEC) model (Ree and Smith, 2008) performed in RASP 3.2 (Yu et al., 2015). Phylogenetic relationships of the 78 Paphiopedilum species, plus the six outgroups, obtained from sequence judgments of the combined sequence and generated by BEAST. The distribution areas of extant the 78 Paphiopedilum species species are marked in capitals A–E and I ((A) China, Nepal, India, Bhutan, Burma, Thailand, Malaysia, Cambodia, Vietnam, and Laos; (B) Sumatra, Borneo and Java; (C) Sulawesi and Moluccas; (D) New Guinea; (E) Philippines; and (I) outgroup), respectively. The green and blue circles indicate the vicariance and dispersal events obtained from the RASP analysis, respectively.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.