Abstract
Novel genomes are today often annotated by small consortia or individuals whose background is not from bioinformatics. This audience requires tools that are easy to use. Such need has been addressed by several genome annotation tools and pipelines. Visualizing resulting annotation is a crucial step of quality control. The UCSC Genome Browser is a powerful and popular genome visualization tool. Assembly Hubs, which can be hosted on any publicly available web server, allow browsing genomes via UCSC Genome Browser servers. The steps for creating custom Assembly Hubs are well documented and the required tools are publicly available. However, the number of steps for creating a novel Assembly Hub is large. In some cases, the format of input files needs to be adapted, which is a difficult task for scientists without programming background. Here, we describe MakeHub, a novel command line tool that generates Assembly Hubs for the UCSC Genome Browser in a fully automated fashion. The pipeline also allows extending previously created Hubs by additional tracks. MakeHub is freely available for downloading at https://github.com/Gaius-Augustus/MakeHub.
Keywords: Genome annotation, Annotation visualization, RNA-seq, Genome browser
Introduction
With decreasing sequencing costs, sequencing the genomes of non-model organisms that are of interest to individuals or small research consortia has become affordable. Pipelines and tools that enable scientists with diverse backgrounds to easily annotate protein-coding genes in novel genomes have been developed and are frequently used, for example, the tools AUGUSTUS [1], [2], [3], [4], [5], GeneMark-ES/ET [6], [7], [8], GlimmerHMM [9], SNAP [10], and GeMoMa [11], [12], [13], as well as the pipelines BRAKER [14], [15], WebAUGUSTUS [16], and MAKER [17], [18]. The output of such gene prediction tools and pipelines is in a table-like text file in gene transfer format (GTF) or general feature format 3 (GFF3). Visualization of predicted gene structures in context with available extrinsic evidence is a crucial step of quality control in any genome annotation project [19]. A number of genome browsers are available for this task, for example, the UCSC Genome Browser [20], JBrowse [21], and GBrowse2 [22]. While JBrowse and GBrowse2 require installation of the browser software on a server or on a local computer, the UCSC Genome Browser bypasses the requirement for software installation. Instead, it offers the opportunity of visualizing any genome through so-called locally hosted ‘Assembly Hubs’ combined with existing UCSC Genome Browser servers (e.g., at https://genome.ucsc.edu) [23].
An Assembly Hub is simply a directory that contains configuration files required by the UCSC Genome Browser as well as track data files with the data to be visualized. The steps for creating custom Assembly Hubs are well documented (http://genomewiki.ucsc.edu/index.php/Assembly_Hubs) and the required tools are publicly available. An experienced bioinformatician is able to create Assembly Hubs with ease. However, a scientist with limited programming background may find it troublesome to manually create the required configuration files, to adapt the output of gene prediction pipelines to the demands of UCSC tools for creating data tracks, and to run all required tools in the correct order.
Recently, the workflow G-OnRamp for fully automated generation of UCSC Assembly Hubs (and JBrowse instances) through the usage of Galaxy web forms became available [24]. The tool that generates UCSC Assembly Hubs within G-OnRamp is called Hub Archive Creator and works seamlessly with genome annotation output files in the G-OnRamp Galaxy framework. However, Hub Archive Creator is difficult to use as a stand-alone command line tool outside of G-OnRamp, because it relies on strict input file format consistency, which is ensured when bioinformatics tools are called inside of G-OnRamp but is not guaranteed when using the same tools in their original release form on the command line.
Therefore, we here describe the novel command line tool MakeHub for the fully automated generation of UCSC Assembly Hubs on the command line from the output of BRAKER, MAKER, GlimmerHMM, SNAP, and GeMoMa for genomes of single species.
Implementation
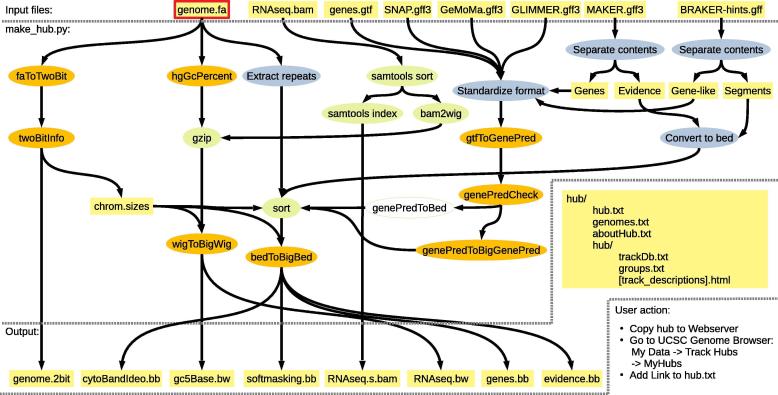
The MakeHub pipeline is implemented in Python and is compatible with Linux and Mac OS X x86_64 computers. The pipeline is illustrated in Figure 1. It provides a command line interface for creating fully functional Assembly Hubs from a genome file (and optionally files with gene and evidence information) and for adding tracks to existing hubs.
Figure 1.
Illustration of the MakeHub pipeline
Input, intermediate, and output files and directories are shown in yellow rectangles. The only compulsory input file for creating a new hub is highlighted with borders in red, whereas all other input files are optional. UCSC tools are indicated with ovals in orange, whereas other external tools are indicated with ovals in green. MakeHub components are indicated with ovals in blue.
Genome files in FASTA format are converted to twoBit format using UCSC’s faToTwoBit. Chromosome (or contig) sizes that serve as input to tools for the later creation of bigWig and bigBed files are extracted using UCSC’s twoBitInfo. GC-content information is collected from the genome with UCSC’s hgGcPercent and written into wiggle (WIG) format. Subsequently, the WIG file is converted to bigWig format using UCSC’s wigToBigWig [25]. A cytoband track that shows the location of the browser window in a target sequence is generated using the same twoBitInfo output and bedToBigBed with an automatically obtained cytoBand AutoSQL file. The genome is screened for softmasked repeat information by MakeHub and repeats are written to browser extensible data (BED) format, sorted, and then converted to bigBed format with UCSC’s bedToBigBed.
Hub configuration files, most prominently the hub/hub.txt file, are created and initialized upon completion of genome processing.
Transcriptome alignment files in binary alignment map (BAM) format can be visualized in two ways. By default, BAM files are sorted with SAMtools [26] and converted to WIG format. This conversion step can be performed either with the AUGUSTUS tool bam2wig [27] or, in its absence, with SAMtools and built-in MakeHub functionality. WIG files are subsequently converted to bigWig format as described above. This generates tracks that allow for an intuitive interpretation of gene structures in context with RNA-seq coverage information.
Optionally, BAM files can be displayed from native BAM format. The required SAM index is automatically generated with SAMtools. Viewing native BAM files gives immediate access to alignment quality information of single reads.
MakeHub seamlessly integrates with output files of the popular genome annotation tools and pipelines AUGUSTUS, GeneMark-ES/ET, GlimmerHMM, SNAP, BRAKER, MAKER, and GeMoMa. GTF and GFF3 files of these tools and pipelines are standardized to a UCSC-compatible GTF format by MakeHub. Subsequently, UCSC's gtfToGenePred is used to convert the GTF file to GenePred data, which is checked for consistency by UCSC's genePredCheck and passed to genePredToBigGenePred and bedToBigBed, generating tracks that allow browsing predicted proteins at the amino acid level. If not available, genePredToBigGenePred is automatically replaced by genePredToBed, generating a track that does not allow browsing amino acids in predicted genes but still visualizes gene structures. In both cases, the final output is in bigBed format.
MakeHub accepts the output directory of a BRAKER run as an input argument and automatically identifies the gene prediction files for visualization in that directory (alternatively, AUGUSTUS and GeneMark-ES/ET predictions can be passed as arguments to separate options). MakeHub automatically extracts gene models from the MAKER GFF3 output file (it usually contains evidence for gene models as well). MakeHub accepts the native GFF3 output file of GeMoMa and GlimmerHMM, and the output of SNAP’s zff2gff3.pl script.
Visualization of the evidence that goes into gene model inference is crucial. This evidence often exceeds the information that can be seen in a RNA-seq WIG or BAM track. For example, annotators are often interested in viewing splice junctions from RNA-seq and/or protein alignments with coverage and strand information in a concise overview. On the other hand, alignments from cDNA, assembled transcriptomes, and proteins need to be visualized in a gene-structure-like fashion. MakeHub automatically generates suitable tracks with evidence from MAKER output and from BRAKER hint files in GFF format. Gene-structure-like evidence (e.g., full length protein alignments) is visualized similarly as gene models, while other evidence, such as splice junctions, is visualized as segments. All resulting evidence track data files are in the indexed bigBed format.
MakeHub automatically generates HTML template files for describing a hub and its tracks. These files are required for public hubs (http://genomewiki.ucsc.edu/index.php/Public_Hub_Guidelines, February 10th 2019). These pages should be edited to appropriately describe genome projects and individual tracks before adding a hub to the list of public hubs at UCSC.
Automatically generated Assembly Hubs must be copied to a publicly available web server for deployment. The hyperlink to hub.txt can be provided to the UCSC Genome Browser for data visualization (see instructions in Figure 1).
Conclusion
In summary, MakeHub is a command line tool that enables scientists with little experience in bioinformatics to generate Assembly Hubs of their genome data and annotations of interest with ease.
Availability
MakeHub is freely available for downloading at https://github.com/Gaius-Augustus/MakeHub.
Author’s contributions
KJH developed the idea, implemented the software, wrote the manuscript, and approved the final version.
Competing interests
The author has declared no competing interests.
Acknowledgments
The international collaboration between the groups of Mark Borodovsky and Mario Stanke, supported by US National Institutes of Health (Grant No. HG000783), gave rise to the development of MakeHub. This project was funded by Universität Greifswald, Germany. I thank Mario Stanke, Matthis Ebel, and Malte Wellnitz for proofreading.
Handled by Ting Wang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanke M., Schöffmann O., Dahms S., Morgenstern B., Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanke M., Diekhans M., Baertsch R., Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 5.König S., Romoth L.W., Gerischer L., Stanke M. Simultaneous gene finding in multiple genomes. Bioinformatics. 2016;32:3388–3395. doi: 10.1093/bioinformatics/btw494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M. Gene prediction novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lomsadze A., Burns P.D., Borodovsky M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab inito eukaryotic gene-finders. Bioinformatics. 2004;10:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 10.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keilwagen J., Wenk M., Erickson J.L., Schattat M.H., Grau J., Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keilwagen J., Hartung F., Paulini M., Twardziok S.O., Grau J. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics. 2018;19:189. doi: 10.1186/s12859-018-2203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keilwagen J., Hartung F., Grau J. GeMoMa: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol. 2019;1962:161–177. doi: 10.1007/978-1-4939-9173-0_9. [DOI] [PubMed] [Google Scholar]
- 14.Hoff K.J., Lange S., Lomsadze A., Borodovsky M., Stanke M. BRAKER1: unsupervised RNA-Seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics. 2015;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoff K.J., Lomsadze A., Borodovsky M., Stanke M. Whole-genome annotation with BRAKER. Methods Mol Biol. 2019;1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoff K.J., Stanke M. WebAUGUSTUS-a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013;41:W123–W128. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B. MAKER: an easy-to-use annotation pipeline for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt C., Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff K.J., Stanke M. Current methods for automated annotation of protein-coding genes. Curr Opin Insect Sci. 2015;7:8–14. doi: 10.1016/j.cois.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 20.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–1638. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein L.D. Using GBrowse 2.0 to visualize and share next-generation sequence data. Brief Bioinform. 2013;14:162–171. doi: 10.1093/bib/bbt001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2013;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Sargent L., Leung W., Elgin S.C.R., Goecks J. G-OnRamp: a Galaxy-based platform for collaborative annotation of eukaryotic genomes. Bioinformatics. 2019;35:4422–4423. doi: 10.1093/bioinformatics/btz309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed data sets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoff K.J., Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics. 2019;65 doi: 10.1002/cpbi.57. [DOI] [PubMed] [Google Scholar]