Abstract
Background
Cancer cell metabolism can be characterised by adaptive metabolic alterations, which support abnormal proliferative cell growth with high energetic demand. De novo nucleotide biosynthesis is essential for providing nucleotides for RNA and DNA synthesis, and drugs targeting this biosynthetic pathway have proven to be effective anticancer therapeutics. Nevertheless, cancers are often able to circumvent chemotherapeutic interventions and become therapy resistant. Our understanding of the changing metabolic profile of the cancer cell and the mode of action of therapeutics is dependent on technological advances in biochemical analysis.
Scope of review
This review begins with information about carbon- and nitrogen-donating pathways to build purine and pyrimidine moieties in the course of nucleotide biosynthesis. We discuss the application of stable isotope resolved metabolomics to investigate the dynamics of cancer cell metabolism and outline the benefits of high-resolution accurate mass spectrometry, which enables multiple tracer studies.
Conclusion
With the technological advances in mass spectrometry that allow for the analysis of the metabolome in high resolution, the application of stable isotope resolved metabolomics has become an important technique in the investigation of biological processes. The literature in the area of isotope labelling is dominated by 13C tracer studies. Metabolic pathways have to be considered as complex interconnected networks and should be investigated as such. Moving forward to simultaneous tracing of different stable isotopes will help elucidate the interplay between carbon and nitrogen flow and the dynamics of de novo nucleotide biosynthesis within the cell.
Keywords: Cancer metabolism, Isotope resolved metabolomics, Flux analysis
1. Metabolic alterations in the context of nucleotide biosynthesis
Cancer cells reprogram their metabolism to enable cell growth, biomass synthesis, and proliferation due to the induction of oncogenes or the loss of tumour suppressors. Otto Warburg conducted one of the first quantitative metabolic studies in the 1920s [1]. Warburg observed that tumours consume excessive amounts of glucose and ferment the majority to lactic acid even in the presence of oxygen. Warburg assumed that the shift from mitochondrial respiration to aerobic glycolysis in cancer cells may be caused by impaired mitochondria [2]. Indeed, this was disproved by Weinhouse through one of the early examples of isotopic labelling in respect to tumour metabolism. Weinhouse observed that the quantity of labelled CO2 produced by oxidative phosphorylation, performed by functioning mitochondria, occurs at the same rate in cancer cells as in normal cells [3]. Through decades of research on the “Warburg Effect,” it has become clear that the enhanced rerouting of glucose into lactic acid is accompanied by a decelerated influx of carbons into the tricarboxylic acid (TCA) cycle, rather than being a result of impaired mitochondria [4]. Regardless, the hallmark of a cancer cell with respect to the Warburg Effect is an increased glucose uptake to fuel adenosine triphosphate (ATP) production via lactate. The preference for the production of ATP via lactate rather than oxidative phosphorylation seems counterintuitive; 2 mol of ATP are generated per mole of glucose via lactate, whereas oxidative phosphorylation provides 36 mol of ATP. The advantages that this metabolic alteration confers have been highly disputed.
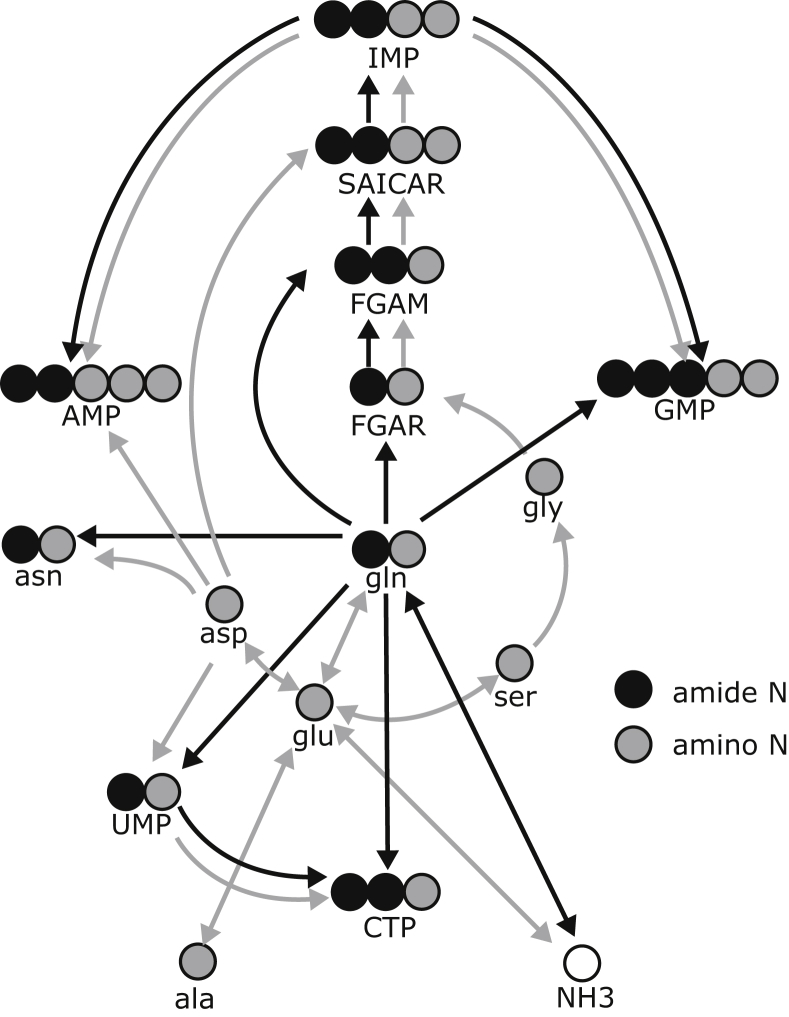
Metabolic alterations in the cancer cell are not limited to increased glucose metabolism. To accommodate the demand of nitrogen and carbon for nucleotide synthesis, cancer cells enhance the consumption of both glucose and glutamine. While glucose is one of the most important carbon sources, glutamine provides both carbon and nitrogen. Usually, metabolic networks are constructed from the carbon flow perspective through 13C isotope labelling experiments. The inclusion of further isotope labels (e.g., 15N nitrogen) promises to obtain a more comprehensive metabolic picture (Figure 1). Simultaneous labelling experiments may provide new insights into the interaction of metabolic pathways. A better understanding of alterations in cancer cell metabolism could facilitate the development of novel therapeutics and may aid treatment strategies, especially to prevent drug resistance. To analyse the entire interactive and dynamic network of a cancer cell, an approach providing an overall systemic perspective is required. Traditional linear thinking has been proven to be too limited to understand the multifaceted network of these molecular interactions [5]. In this framework, systems biological studies are needed to improve our understanding of the biological and physiological complexity of the disease. Mathematical and computational techniques are indispensable to process comprehensive data sets, obtained by high-throughput methods in proteomics, metabolomics, and genomics [6,7]. The basic knowledge on metabolic pathways often fails to explain therapeutic resistances, because metabolic reprogramming is difficult to predict, and the metabolic crosstalk between several cell types within a tumour enhances the metabolic repertoire of cancer.
Figure 1.
Glutamine-centric nitrogen metabolism with focus on amino acids and nucleotides. Abbreviations: inosine-monophosphate (IMP), guanosine-monophosphate (GMP), adenosine monophosphate (AMP), cytidin triphosphate (CTP), uridine monophosphate (UMP), glutamine (gln), asparagine (asn), aspartate (asp), glutamate (glu), serine (ser), glycine (gly), alanine (ala), formyl-glycine-amide-ribonucleotide (FGAR), formyl-glycine-amidine-ribonucleotide (FGAM), 5amino-imidazol-4-N-succino-carboxamid-ribonucleotide (SAICAR).
1.1. Glycolysis
An enhanced glucose uptake and metabolism fuels cancer cells with the necessary ATP and carbons for growth and proliferation. The measurement of this dynamic system is paramount in understanding the metabolic differences between cell types and their pathologies. The metabolism of glucose branches into multiple pathways and requires the analysis of the flux of carbon through central carbon metabolism: glycolysis, pentose phosphate pathway (PPP), TCA cycle, and amino acid metabolism. Because of the rapidity of the flow of carbon through these pathways, complex in vitro strategies are required to produce reliable models [8].
In many cancer cells, enzymatic elements of these pathways are overexpressed to compensate for their metabolic demands. As an example, in the last step of glycolysis, pyruvate kinase (PKM) converts phosphoenol pyruvate into pyruvate. Cancer cells commonly increase the expression of the PKM splice variant PKM2. The lower activity of PKM2 contributes to an accumulation of upstream glycolytic intermediates, leading to an increased flux into adjacent pathways, for example, the PPP and the one-carbon metabolism [9]. Through isotopic tracing, it has been observed that the PKM2 variant supports the synthesis of nucleotides, and the deletion of PKM2 causes a nucleotide crisis via thymidine [10].
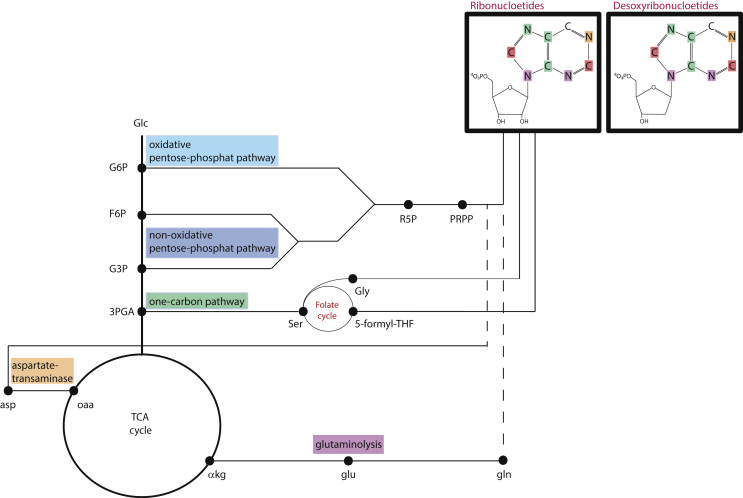
The PPP branches at two distinct points in the preparatory phase of glycolysis, using different intermediates for the production of ribose-5-phosphate (R5P). The oxidative pathway branches from glucose-6-phosphate (G6P), and the nonoxidative pathway branches from fructose-6-phosphate (F6P), combining with glyceraldehyde-3-phosphate (G3P) to cycle carbons to R5P (Figure 2). Both these pathways have been found to be elevated in cancers, to support NADH and nucleotide intermediates synthesis [11]. However, through [1,2–13C] glucose tracing, it has been observed that the non-oxidative pathway takes precedence in, at least, leukaemic and colon cancer cells [12,13]. Clearly, there is a wealth of research addressing the carbon flow through the PPP to R5P from glucose. Yet, there is scope to describe the role of nitrogen sources for nucleotide synthesis, particularly in the context of the mode of action of therapeutics.
Figure 2.
Pathways carrying the nucelotide biosynthesis. The ribonucleotides show carbon and nitrogen derived from independent pathways illustrated by the corresponding colours.
Cancer cells with elevated PKM2 harbour increased 3-phosphoglyceric acid (3-PGA). 3-PGA is an intermediate of glycolysis and fuels serine and glycine production. Serine is synthesised by three steps emerging from 3-PGA. As a subsequent step, glycine is synthesised via the enzyme serine hydroxymethyltransferase (SHMT). Serine and glycine provide precursors for nucleic acids via refuelling the one-carbon metabolism (Figure 2). The conversion of serine to glycine via SHMT is part of the one-carbon metabolism, which turns into the folate cycle. Simultaneously to glycine production, the enzyme SHMT converts tetrahydrofolate (THF) to 5,10-methylene-THF (5-formyl-THF). Subsequently, 5-formyl-THF is converted to 10-formyltetrahydrofolate (F-THF) (Figure 2). In addition, glycine is incorporated into 5-phosphoribosyl-1-amine to produce formyl-glycine-amide-ribonucleotide an early intermediate in purine biosynthesis. In summary, glycine and the folate cycle together provide one nitrogen and two carbon atoms for the building of the purine ring.
Recent research addressing the role of phosphoglycerate dehydrogenase, which catalyses the first step of 3-PGA to serine metabolism, showed that its inhibition results in a depletion of nucleotides. Furthermore, by tracing both the carbon and nitrogen flow, it was found that serine, not glycine, is essential for cancer cell proliferation [14,15]. Functional tracing of carbon and nitrogen flow through the cancer metabolic profile requires the understanding of the roles of glutamine and asparagine, in combination with glucose studies.
1.2. Glutamine and asparagine metabolism
In addition to glucose, glutamine is a major nutrient used by cancer cells [16]. Glutamine fuels anaplerotic reactions to maintain the TCA cycle function and mitochondrial integrity. It is known that the depletion of glutamine causes cell death [16]. This phenomenon, termed glutamine addiction, has been observed in a variety of cancer types in vitro and in vivo [17]. Glutamine provides the carbon skeleton for the TCA cycle and contributes to protein synthesis (Figure 2). Glutamine is essential as a nitrogen donor within de novo nucleotide biosynthesis, for both purines and pyrimidines. In addition, it donates nitrogen via aminotransferases to produce, amongst others, aspartate. Oxaloacetate- and glutamine-derived glutamate is converted via the enzyme aspartate transaminase to alpha-ketoglutarate and aspartate (Figure 2). The latter is an important nitrogen donor to purine and pyrimidine synthesis. This metabolic dependency has been extensively studied to elucidate novel therapeutic approaches that are based on the concept of glutamine addiction. The present knowledge seems not to be sufficient to identify novel targets and to explain therapeutic resistances. Investigating the flow of glutamine's nitrogen, in addition to glucose's carbon, could help to gain further insight in this regard.
Several cancer cells have the ability to produce glutamine via glutamine synthase (GS). It has been found that high GS-expressing glioblastoma cells were less sensitive to glutamine deprivation compared with low-GS-expressing cells [18]. High GS expression led to more stable purine levels under glutamine starvation, while pyrimidine levels were barely affected in the studied cell types. 15N-ammonia labelling revealed a direct incorporation of ammonia into nucleotides. While glutamine and aspartate are involved in both purine and pyrimidine synthesis, glycine contributes only to purine synthesis. This raises the question as to whether the impact of glutamine deprivation on glycine synthesis impedes the formation of purines. Furthermore, the suppression of glutamine synthesis in high-glutamine synthetase-expressing cancer types may improve the outcome of chemotherapy. This requires a detailed understanding of how glutamine production is facilitated within the cells described, in particular the origin of glutamines amido group. To investigate the role of nitrogen donors in de novo purine synthesis, labelling with 15N-glutamine will allow tracing of nitrogen into purine intermediates and elucidate mechanisms of regulation [19].
Zhang et al. identified asparagine-producing pathways in glioblastoma to be advantageous for survival under glutamine starvation [20]. The enzyme citrate synthase catalyses the condensation of acetyl-CoA and oxaloacetate to citrate in the first step of the TCA cycle. Silencing of this enzyme induced the production of aspartate and asparagine from oxaloacetate and rescued for glutamine deprivation. The role of the aspartate nitrogen has to be clarified in this context. Aspartate supports the formation of glutamate, the precursor for glutamine synthesis, on the one hand, and the formation of purine and pyrimidine rings for nucleotide synthesis on the other hand. The enzyme pyruvate dehydroxygenase is involved in the formation of acetyl-CoA from pyruvate, and its inhibition prevents apoptosis under glutamine starvation. Asparagine deprivation sensitised cells to glutamine withdrawal, while asparagine addition prevented apoptosis. This leads to the hypothesis that the ability to redirect oxaloacetate consumption to aspartate and asparagine production could circumvent the detrimental effects of glutamine deprivation. This assumption was confirmed by Cheng et al., who identified pyruvate carboxylase (PC) activity to be compensating for glutaminase silencing [21]. PC provides another pathway for pyruvate to enter the TCA cycle. This step forms oxaloacetate, the precursor for aspartate and asparagine. Pavlova et al. further investigated the role of asparagine in cancer metabolism [22]. They observed an increased GS expression after asparagine addition only when it was not catabolised to aspartate. Krall et al. also emphasised that the beneficial role of asparagine cannot be explained by catabolism and is more likely to be a result of its ability to facilitate amino acid uptake [23]. Interestingly, a 15N2-asparagine isotope tracing experiment resulted in approximately 10% nitrogen incorporation into purines, while no other incorporation events have been observed. Asparagine nitrogen is not directly involved in purine biosynthesis, and the amino acids that normally contribute their nitrogen to purine synthesis remained unlabelled [23]. This raises the question regarding which mechanisms introduced the observed labels. Dynamic isotope labelling strategies that enable the analysis of in-stationary isotope incorporation will be beneficial to investigate such phenomena.
1.3. Fuelling purine and pyrimidine biosynthesis
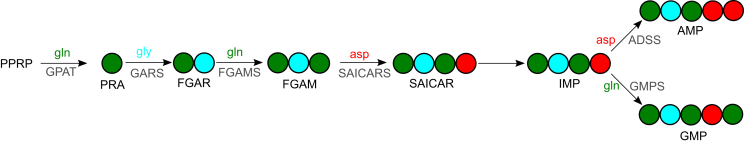
Glycolysis, PPP, glutaminolysis, and amino acid metabolism are connected and converge to provide precursors for nucleotide biosynthesis. The amino acids glutamine, glycine, and asparagine play an important role as nitrogen donors in inosine-monophosphate (IMP) biosynthesis. IMP is the precursor for purine nucleotides: adenosine monophosphate (AMP) and guanosine monophosphate (GMP). IMP synthesis requires two amide nitrogen from glutamine, one amino nitrogen from glycine, and one further amino nitrogen from aspartate. The conversion to AMP uses a further aspartate nitrogen, while glutamine provides an additional nitrogen to GMP production (Figure 3). The synthesis of pyrimidine nucleotides involves up to three amino acid–dependent steps. First, glutamine donates its nitrogen in a rate-limiting step to the formation of carbamoylphosphate (CBMP). CBMP subsequently condenses with aspartate, and further reactions provide uridine monophosphate (UMP). UMP-derived uridine triphosphate (UTP) can be converted to cytidin triphosphate (CTP) in a glutamine-dependent reaction (Figure 4). In a concerted manner, these central metabolic pathways fuel de novo nucleotide biosynthesis. A quantitative and time-resolved analysis of these metabolic branching points may provide new insights into metabolic reprogramming and drug resistance.
Figure 3.
Amino acid–dependent steps of purine nucleotide biosynthesis. Nitrogens are depicted as circles. Colours of nitrogens correspond to the colours of amino acids from which they were derived. Abbreviations: 5-phosphoribosyl-1-pyrophosphate (PRPP), 5-phosphoribosyl-1 -amine (PRA), formyl-glycine-amide-ribonucleotide (FGAR), formyl-glycine-amidine-ribonucleotide (FGAM), 5amino-imidazol-4-N-succino-carboxamid-ribonucleotide (SAICAR), inosine monophosphate (IMP), adenosine monophosphate (AMP), guanosine monophosphate (GMP), glutamine (gln), glycine (gly), aspartate (asp), glutamine-PRPP-amidotransferase (GPAT), GAR-synthetase (GARS), FGAM-synthase (FGAMS), SAICAR-synthase (SAICARS), adenylosuccinate synthetase (ADSS), GMP-synthase (GMPS).
Figure 4.
Amino acid–dependent steps of pyrimidine nucleotide biosynthesis. Nitrogens are depicted as circles. Colours of nitrogen correspond to the colours of the amino acids from which they were derived. Abbreviations: carbamoylphosphate (CBMP), uridine monophosphate (UMP), uridine triphosphate (UTP), cytidin-triphosphate (CTP), carbamoylphosphate synthetase-2 (CPS2), aspartate transcarbamylase (ATCase), CTP-synthase-1 (CTPS1), glutamine (gln), aspartate (asp).
2. Targeting nucleotide biosynthesis for cancer therapy
Dividing cells require nucleotides to synthesise RNA and DNA. Central metabolic pathways such as glycolysis, one-carbon metabolism, glutamine, and amino acid metabolism contribute carbon and nitrogen atoms in a concerted manner to fuel de novo nucleotide biosynthesis.
2.1. Targeting de novo nucleotide biosynthesis
A number of chemotherapeutics are antimetabolites that target de novo nucleotide biosynthesis. They either mimic a metabolite and inhibit the reaction or are modified metabolites that cannot be further metabolised and subsequently stop the respective pathway. Transformed cells optimise nucleotide biosynthesis for rapid cell proliferation. To disturb this process, the well-investigated compound 5-fluoruracil (5-FU) is used for the treatment of different cancer types. By mimicking uracil, 5-FU inhibits the enzyme thymidylate synthase (TYMS), which methylates deoxyuridin phosphate to deoxythymidin phosphate using me-THF [24]. Thus, 5-FU blocks DNA synthesis and subsequently cell division; however, the efficacy of 5-FU in the clinics is limited. This is surprising, as no other reactions are known to contribute to thymidine-nucleotide biosynthesis. The inhibition of 5-formyl–THF synthesis, an important cofactor of TYMS, seems to be more effective. For example, the drug methotrexate (which inhibits dihydrofolate reductase [DHFR]), and the antimetabolite pemetrexed (which inhibits TYMS, DHFR, and glycinamide ribonucleotide formyltransferase [GARFT]) target the synthesis of folate cofactors and thymidinenucleotides [25,26]. These drugs are mostly used in combination with cis-platin [27]. Cisplatin can affect DNA replication and induces DNA damage. Another DNA synthesis inhibitor is hydroxyurea (HU), which targets ribonucleotide reductase (RNR), first established in 1963 as new antitumor agent by Stearns et al. [28]. RNR is the only enzyme converting ribonucleotides to deoxyribonucleotides and presents a promising target; as a result, it has been the focus of many studies. RNR provides dNTP pools for DNA replication and repair [29]. However, the application of HU as a therapeutic induces severe side effects in patients. Although nucleotide biosynthesis is an essential metabolic process, therapeutic targeting of this metabolic network did not yield the final results that a biochemist would predict. A subsection of tumours are refractory after initial therapy, and relapsing tumours can establish a resistance against drugs targeting de novo nucleotide biosynthesis.
In addition to antimetabolites and drugs that target nucleotide biosynthesis directly, the precursor phase of nucleotide biosynthesis can be targeted with small molecules. Carbon and nitrogen atoms for nucleotide biosynthesis are provided by central metabolic pathways and converge in a multistep process to from purine and pyrimidine nucleotides. Thus, targeting central metabolic pathways also affects nucleotide synthesis. However, the plasticity of the central metabolic network can often circumvent blockages induced by drug treatments.
2.2. Targeting glutamine metabolism
Glutamine is a nonessential amino acid and is required for de novo nucleotide biosynthesis. Several enzymes that directly participate in nucleotide synthesis use glutamine as nitrogen donor. The enzyme glutaminase was found induced in cancer [30,31] and was anticipated as an effective metabolic target. Glutaminase is a deaminating enzyme that converts glutamine to glutamate. Glutamate is further metabolised to alpha-ketoglutarate and channelled into the TCA cycle. However, targeting glutaminase using small-molecule inhibitors was not effective to significantly reduce carbon flow into the TCA cycle [32]. It may be postulated that additional reactions that convert glutamine to glutamate could bypass glutaminase inhibition. Furthermore, Leone et al. stated that the therapeutic response in vivo to targeted blockade of glutaminase is limited [33]. In this study, the authors focused on 6-diazo5-oxo-l-norleucine (DON). DON is a bacterial compound and structural analogue of glutamine that is expected to inhibit a wide range of glutamine-metabolising enzymes. In vitro application of DON led to a reduced glutamine-derived 15N incorporation in formyl glycinamide ribonucleotide (FGAR); an early intermediate of purine biosynthesis (Figure 3). The authors could also show that the effect of DON on protein level of the MYC oncogene [32] exceeds the shRNA knock down of single nucleotide biosynthetic enzymes by far. However, the target(s) of DON remained elusive. To better understand the dynamics of this network and the stoichiometric contribution of nitrogen and carbon providing pathways at their points of convergence, a dynamic analysis of carbon and nitrogen flow will be indeed beneficial. The method pulsed stable isotope resolved metabolomics (pSIRM), an approach to create time series of isotope incorporation before the isotopic steady state has been reached [8], may be the foundation for further developments that allow the analysis of multiple tracers at the same time. pSIRM may also enable the support of the identification of drug actions (e.g., DON) at complex metabolic networks.
3. Analysis of central metabolic networks using stable isotope resolved metabolomics
Central carbon metabolism and nucleotide biosynthesis are complex metabolic pathways. A single measurement “snapshot” can offer only a static picture of an actually dynamic system. The principles that lead to the metabolic homeostasis remain rather elusive. For example, an increased concentration of a certain metabolite may be caused by accelerated production, reduced degradation, or both. To understand these underlying mechanisms, it is important to decipher the cellular response to perturbations (e.g., drug treatments). To resolve such dynamics of intracellular metabolic pathways, isotope labelling can be applied. Different technologies enable an isotope-assisted analysis of such networks. Nuclear magnetic resonance (NMR) spectroscopy is a nondestructive approach that allows for absolute quantification and in vivo analysis [34,35]. However, mass spectrometry (MS) is currently favoured because of its higher sensitivity and lower cost. NMR spectroscopy allows for the analysis of positional isotope enrichment. In contrast, MS does not provide positional information without additional efforts (e.g., fragmentation). Each peak in a mass spectrum is related to a set of isotopologues that can differ in their positional composition of isotopes. Fragmentation of molecules, induced through electron impact ionisation, allows for the identification of positional isotope enrichment [36].
3.1. Isotope labelling experiments
Isotope labelling experiments are suitable to observe relative pathway contributions as well as the metabolic fate of small molecules within a cell. The concept is based on the fact that molecules of the same species differ naturally in their molecular mass. Molecules that differ in their isotope composition are termed isotopologues. Isotopologues that carry the same number of isotopes but on different positions are defined as positional isotopologues or isotopomers. Thus, nonpositional isotopologues can be easily distinguished by their masses, while separation of isotopomers requires at least the application of a MS technique that allows for fragmentation.
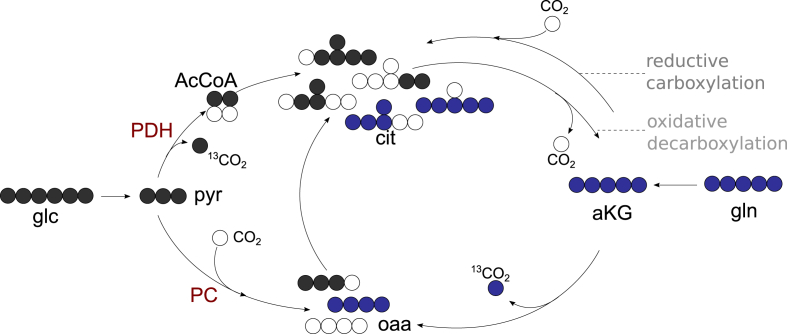
13C labelling experiments are a frequent choice to investigate metabolism [37,38]. The naturally high abundant carbon isotope is 12C, and the heavier stable 13C accounts for approximately 1.1%. Thus, under normal conditions, the vast majority of carbon compounds will be measurable as 12C isotopologues. The consumption of isotopic labelled substrates induces 13C enrichment in downstream intermediates. The enrichment can be observed by MS analysis as a mass shift (i.e., an increase in the intensities of heavier isotopologue peaks). Comparing such observations under different conditions gives a first impression on how nutrient availability or drug treatment affects metabolic processes. Single metabolites can be produced by multiple redundant pathways. Such pathways can differ in how they use carbons to form a certain metabolite. Figure 5 shows different ways to form citrate or oxaloacetate and how labelling patterns differ, depending on the carbon flow, when tracing with 13C-u glucose or 13C-u glutamine. The appropriate choice of number and position of isotopic labelled carbons within a substrate may elucidate the activity of different pathways by producing distinct isotope patterns. In this regard, the choice of tracer can depend on the pathway of interest [39,40]. Jang et al. outlined how to interpret the results of several pathway-specific labelling experiments [41].
Figure 5.
13C-u glucose or glutamine routing. The flux of 13C-u glucose–derived pyruvate through PDH results in m+2 citrate, the flux through PC results in m+3 citrate, and the combination of both results in m+5 citrate. 13C-u glutamine tracing leads to m+4 or m+5 citrate. Solid circles depict 13C carbons; empty circles depict 12C carbons. Abbreviations: glucose (glc), pyruvate (pyr), acetyl-CoA (AcCoA), cytidin triphosphate (CTP), oxaloacetate (oaa), α-ketoglutarate (αKG), glutamine (gln), pyruvate carboxylase (PC), pyruvate dehydroxygenase (PDH). Adapted from [59].
3.2. Flux analysis
The determination of the metabolic flux promises a more comprehensive understanding of the dynamics within a metabolic network. Metabolic fluxes describe the turnover of chemical reactions in terms of reaction rates. Extracellular fluxes can be measured by secretion and consumption, yet the determination of intracellular fluxes is not as trivial. Simple balanced networks can be described by linear equations, given their stoichiometry, and fluxes can be calculated by flux balance analysis [42]. This approach fails for more complex systems that exhibit cycles, reversible reactions, or parallel fluxes. Measurements from labelling experiments are used to simulate fluxes for those networks by metabolic flux analysis (MFA). Simulations are based on the assumption that the investigated organism meets the metabolic steady-state condition, in which fluxes and metabolite concentrations are constant. Fluxes are determined by an optimisation method that fits initially estimated values to the measured data [43]. Once a set of fluxes has been determined, confidence intervals describe the probability that a variant set of fluxes would produce similar measurements [44].
3.3. Metabolic and isotopic steady state and in-stationary MFA
Most isotope labelling approaches assume the system fulfils the metabolic steady-state condition. In this state, metabolite concentrations do not change over time. While overall metabolite concentrations are assumed to be constant in the metabolic steady state, the introduction of isotopic labelled substrates to the system results in a dynamic enrichment of isotopes. This is a finite process, and at some point, metabolite pools are constantly enriched with isotopes. This is the onset of the isotopic steady state. Nondynamic tracing studies choose labelling times to reach the isotopic steady state. The duration until this state is reached depends on the pathways under consideration and the experimental setups. Glycolysis, for example, can equilibrate within 10 min after glucose labelling, while TCA cycle intermediates are normally not balanced before 2 h. Labelling times for nucleotides should be at least 24 h [41]. More reliable results may be obtained by in-stationary MFA [[45], [46], [47]]. However, the experimental requirements are high with regard to measured time points and repetitions. While classical metabolic flux analyses are based on measurements taken at the isotopic steady state, the in-stationary MFA is based on multiple measurements taken before the isotopic steady state has been reached. Such time series may better represent the dynamics of the metabolic system. The experimental design needs to be adapted to perform multiple measurements before the system enters the isotopic steady state. This increases the effort of sample collection and preparation as well as computational demands. For mathematical modelling, the metabolic steady state is still assumed. Multiple measurements of the metabolic profile in the presence of a drug, through time, presents uncertainties on designing the correct experimental conditions. The primary metabolic response to drug treatment is dependent on the target itself and the metabolic activity of the cell. After the initial metabolic disturbance, the system will adapt at all regulatory layers, and it cannot be ensured that metabolism fulfils the steady state condition at the time point of measurement, or if the obtained results just reflect the metabolic target.
3.4. Pulsed stable isotope resolved metabolomics for analysis of pathway activity and drug response
Stable isotope-assisted methods such as pSIRM [8] or kinetic flux profiling [48] apply stable isotope labelled substrates for short time periods and analyse isotope incorporation into metabolism, even if the isotopic steady state is not reached. The information of isotope enrichment and quantity of a certain metabolite together allow a real estimate of isotope incorporation over time. Specifically, if the action of a metabolic inhibitor should be analysed, such analysis is informative. The action of metabolic inhibitors may target the biochemical network first but will provoke adaptation of the metabolic network itself. Ultimately, this leads to adaptations of cellular signalling and subsequently transcription and translation. To resolve time-dependent processes, it is necessary to obtain dynamic metabolic measurements at early, intermediate, and late time points. For example, Pietzke and Zasada et al. analysed the action of 2-deoxyglucose (2-DG), an often used metabolic inhibitor, within 15 min [49]. Using the pSIRM technology after a short application of 2-DG revealed that 2-DG is converted by hexokinase to 2-DG phosphate. 2-DG phosphate is produced at a millimolar concentration and thereby sequesters the cellular ATP pool. However, 2-DG is not an inhibitor of hexokinase per se, as usually stated in the literature. In addition, an activation of citrate production, probably through activation of AMPK and pyruvate dehydrogenase, could be observed. Thus, the combination of short time label incorporation and measurement of absolute metabolite concentrations allow quantitative and dynamic insights into metabolism and to uncover actions of drugs or oncogenes [8,50].
3.5. Towards multiple tracer studies
Current tracer applications are based on 13C studies to investigate the central carbon metabolism and its branching pathways. The implementation of further stable isotopes such as 2H, 18O, 34S, or 15N predominantly allows for additional insights into the metabolic behaviour of cells. To understand the role of glucose and glutamine as both carbon and nitrogen donors, it may be beneficial to simultaneously trace 13C and 15N. Common MS techniques are coupled to separation methods such as gas chromatography (GC) or liquid chromatography (LC). GC is most often used in combination with low- or medium-resolution mass analysers such as time of flight (ToF) or quadrupole. However, it is not possible to distinguish carbon- and nitrogen-induced mass shifts using GC–MS technologies, which produce only nominal mass spectra (Figure 6). The same issue emerges using instruments with high mass accuracy but only low or medium resolution. LC-MS Orbitrap systems or high-resolution multi-reflectron ToFs can provide enough accuracy, resolution, and scan speed to distinguish isotopologues that contribute to the same isotopic peak in nominal spectra (Figure 7). For some years, commercial GC instruments have been available that also enable the analysis of simultaneous tracer studies [51,52].
Figure 6.
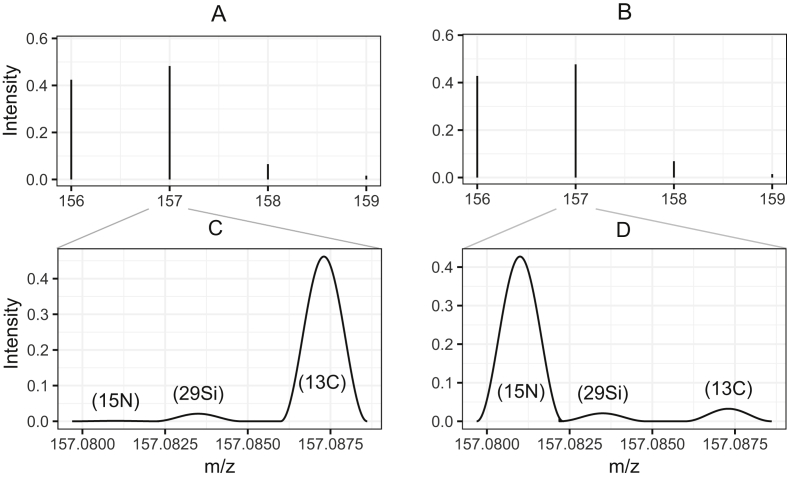
Comparison of carbon and nitrogen induced mass shifts in either nominal spectra (integer mass values) or exact spectra (masses with a precision of about 5 decimal places). A) The simulated nominal spectrum of a glutamine fragment with a 50% incorporation of one carbon. B) The simulated nominal spectrum of a glutamine fragment with a 50% incorporation of one nitrogen. There is no difference from A). C) The m/z = 157 peak of A) resolved to exact masses. The 13C peak is dominant and shows that the mass shift in A) was mainly induced by the 13C incorporation. D) The m/z = 157 peak of B) resolved to exact masses. The 15N peak is dominant and shows that the mass shift in B) was mainly induced by 15N incorporation.
Figure 7.
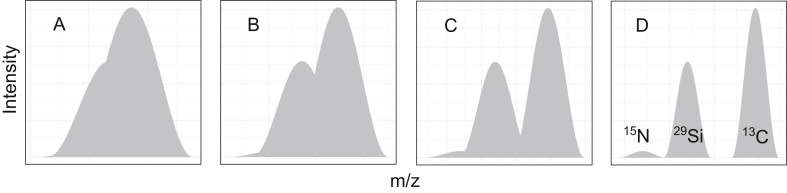
Mass spectra of identical samples under different resolutions: A) 15,000, B) 30,000, C) 60,000, D) 120,000. Ultra-high resolution allows for a reliable distinction of 13C, 15N, and 29Si isotopologues.
An important step in the evaluation of labelling experiments is the correction for natural isotope abundances. To determine the experimental isotope enrichment, data points first have to be adjusted according to the natural abundances of the used tracer. Several approaches to accomplish data correction have already been published [53,54]. These approaches predominantly focus on data containing nominal spectra, derived from single-tracer experiments, and mostly using reference measurements. Carreer et al. presented a computational framework to correct highly resolved accurate mass spectra, derived from multiple tracers for natural abundances [55]. The correction of an 13C, 15N enriched analyte/fragment spectrum requires the following steps:
-
1.
Identify in each isotope peak the number of heavy carbons and nitrogens.
-
2.
Generate a data set D = (Ik,l), so that Ik,l equals the sum of all intensities of peaks that contain exactly k 13C and l 15N atoms.
-
3.
Correct the data set D to obtain a data set D' = (I'k,l)). After the correction, the values I'k,l correspond to the actual experimental enrichment of isotopologues that carry exactly k 13C and l 15N atoms.
The correction is based on the following principles: A certain amount of Ik,l corresponds to isotopologues with an experimental 13C and 15N incorporation smaller that k and/or l, respectively. The measured incorporation sums up to k and l only by natural abundance. This amount has to be subtracted from Ik,l. There may also exist a certain amount of isotopologues that exhibit an experimental incorporation of k and l 13C and 15N isotopes plus some additional heavy carbons and nitrogens by natural abundance. This amount is not tracked by Ik,l and has to be added (Figure 8). For completeness, it has to be mentioned that derivatisation methods for GC introduce additional atoms to the molecule, for example, through trimethylsilyl or tert-butyldimethylsilyl groups. As silicon has two high abundant isotopes, this has to be considered within the natural abundance correction when only nominal spectra are available. Regarding the correction of high-resolution accurate mass spectra according to the mentioned algorithm, GC–MS derivates require no additional attention.
Figure 8.
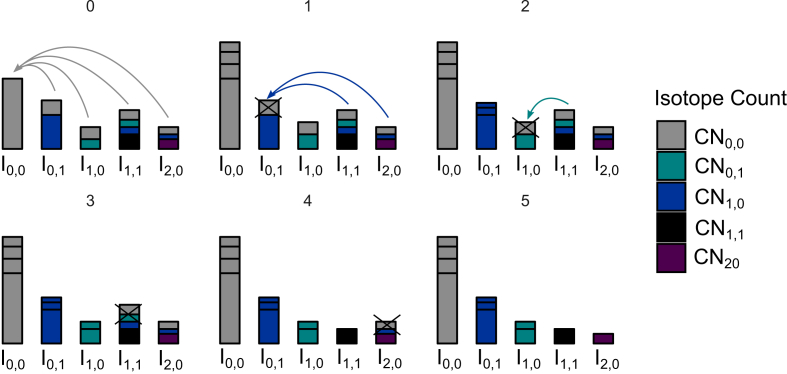
Visualization of the correction for natural abundancies of isotopes. The correction of a 13C, 15N enriched analyte/fragment spectrum requires the following steps: i) Identify in each isotope peak the number of heavy carbons and nitrogen. ii) Generate a data set D = (Ik,l), so that Ik,l equals the sum of all intensities of peaks that contain exactly k 13C and l 15N atoms. iii) Correct the data set D to obtain a data set D' = (I'k,l). After the correction, the values I'k,l correspond to the actual experimental enrichment of isotopologues that carry exactly k 13C and l 15N atoms. The correction is based on the following principles: A certain amount of Ik,l corresponds to isotopologues with an experimental 13C and 15N incorporation smaller than k and/or l, respectively. The measured incorporation sums to k and l only by natural abundance. This amount has to be subtracted from Ik,l. There may also exist a certain amount of isotopologues that exhibit an experimental incorporation of k and l13C and 15N isotopes plus some additional heavy carbon and nitrogen atoms by natural abundance. This amount is not tracked by Ik,l and has to be added. Notations CNk,l indicate an incorporation of k13C and l15N atoms.
4. Concurrent tracing: elucidating the crossroads of carbon and nitrogen metabolism
Nucleotide biosynthesis requires an appropriate supply of carbons (e.g., glucose) that feed the PPP as well as nitrogen from amino acids that can be synthesised using glutamine among other sources. The simultaneous labelling of glucose carbons and glutamine nitrogen will provide detailed insights into the nucleotide metabolism of cancer cells. To date, only a few studies focused on the concurrent tracing of carbon and nitrogen [[56], [57], [58]]. They applied GC/combustion/isotope ratio (GC-C-IR) or Fourier transform ion cyclotron resonance (FT-ICR)MS. An appropriate separation and efficient identification could also be provided by a GC ultra-high-resolution MS. Computational methods have to be established to process the highly complex data gained from such labelling experiments. Using this advanced methodology, it is possible to investigate the dynamics of carbon and nitrogen incorporation at their convergence within the metabolic network. Such studies will help to understand the flexibility of the cancer metabolic network and hopefully will highlight metabolic bottlenecks that can be explored for novel therapeutic interventions. The structures of metabolic pathways were discovered decades ago; today's challenge is to understand the regulatory principles that underlie these pathways. Many different cancers have been studied with regard to their pathway usage and metabolic phenotype. It became evident that inhibition of cell growth is not sufficient to finally kill the cancer cells in vitro and in vivo [32]. Besides the metabolic interaction of cancer cells with the tumour microenvironment, cancer cells can reprogram their metabolism to circumvent metabolic shortcomings introduced by therapeutic drugs. However, our understanding of the metabolic flexibility of the cancer metabolic network is sparse. Cancers may employ a variety of mechanisms that confer drug resistance, for example, exclusion of the drug from the cells, metabolising of the drug through cytochrome oxidases, glutathione conjugation, or induction of alternative signalling pathways. All these processes are energy demanding and may provoke detectable metabolic adaptations. Although small-molecule inhibitors of the de novo nucleotide biosynthesis are among the first chemotherapeutics, their clinical efficacy cannot be predicted with certainty. An improved understanding of the metabolic mode of individual tumours and their capacity to rewire the metabolic network to induce therapy resistance is necessary. In this regard, the analysis of carbon and nitrogen metabolism and their convergence in the de novo nucleotide biosynthesis, and at best simultaneously, will be instrumental in future cancer research.
Acknowledgements
The work was supported by the Sander Foundation (SB), DKTK (German consortium for translational cancer research) (SF), and the Enable project funded by the Deutsche Krebshilfe (MF). It was made possible by the generous support of the Helmholtz foundation through the AMPRO program and BIMSB (Berlin Institute for medical systems biology). We thank Prof. Markus Landthaler for providing the GC-Orbitrap MS.
Conflict of interest
None declared.
Abbreviations
- PRPP
phosphoribosy pyrophosphate
- gln
glutamine
- glu
glutamate
- ala
alanine
- asp
aspartate
- asn
asparagine
- ser
serine
- gly
glycine
- αKG
ketoglutarate
- oaa
oxaloacetate
- pyr
pyruvate
- glc
glucose
- GS
glutamine-synthetase
- CPS2
carbamoylphosphate synthetase-2
- GPAT
glutamine-PRPP-amidotransferase
- FGAMS
FGAM-synthase
- GMPS
GMP-synthase
- CTPS1
CTP-synthase-1
- GARS
GAR-synthetase
- SAICARS
SAICAR-synthase
- ADSS
adenylosuccinate synthetase
- ATCase
aspartate transcarbamylase
- PDH
pyruvate dehydroxygenase
- PC
pyruvate carboxylase
- PRA
5-phosphoribosyl-1 -amine
- FGAR
formyl-glycine-amide-ribonucleotide
- FGAM
formyl-glycine-amidine-ribonucleotide
- SAICAR
5-amino-imidazol-4-N-succino-carboxamid-ribonucleotide
- IMP
inosine monophosphate
- UMP
uridine monophosphate
- GMP
guanosine monophosphate
- AMP
adenosine monophosphate
- UTP
uridine triphosphate
- CTP
cytidin triphosphate
- CBMP
carbamoylphosphate
- AcCoA
acetyl-CoA
- MS
mass spectrometry
- GC
gas chromatography
- LC
liquid chromatography
- FT-ICR
Fourier transform ion cyclotron resonance
- GC-C-IR
gas chromatography/combustion/isotope ratio
- MFA
metabolic flux analysis
References
- 1.Warburg O., Wind F., Negelein N. The metabolism of tumors in the body. The Journal of General Physiology. 1926;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warburg O. On the origin of cancer cells. Science. 1956;123 doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 3.Weinhouse S. Studies on the fate of isotopically labeled metabolites in the oxidative metabolism of tumors. Cancer Research. 1951;11:585–591. [PubMed] [Google Scholar]
- 4.Heiden M.G.V., Cantley L.C., Thompson C.B., Mammalian P., Exhibit C., Metabolism A. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1034. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller O., Stocking C., Wagener C. Wiley-Blackwell; 2017. Cancer Signaling: from molecular biology to targeted therapy. [Google Scholar]
- 6.Wang E. Cancer systems biology. 2010. A roadmap of cancer systems biology; pp. 3–22. [Google Scholar]
- 7.Zasada C., Kempa S. Metabolism in cancer. Springer; 2016. Quantitative analysis of cancer metabolism: from pSIRM to MFA; pp. 207–220. [DOI] [PubMed] [Google Scholar]
- 8.Pietzke M., Zasada C., Mudrich S., Kempa S. Decoding the dynamics of cellular metabolism and the action of 3-bromopyruvate and 2-deoxyglucose using pulsed stable isotope-resolved metabolomics. Cancer and Metabolism. 2014;2:9. doi: 10.1186/2049-3002-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheong H., Lu C., Lindsten T., Thompson C.B. Therapeutic targets in cancer cell metabolism and autophagy. Nature Biotechnology. 2012;30:671–678. doi: 10.1038/nbt.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunt S.Y., Muralidhar V., Hosios A.M., Israelsen W.J., Gui D.Y., Newhouse L. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Molecular Cell. 2015;57:95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Langbein S., Frederiks W.M., Zur Hausen A., Popa J., Lehmann J., Weiss C. Metastasis is promoted by a bioenergetic switch: new targets for progressive renal cell cancer. International Journal of Cancer. 2008;122:2422–2428. doi: 10.1002/ijc.23403. [DOI] [PubMed] [Google Scholar]
- 12.Boros L.G., Puigjaner J., Cascante M., Lee W.N.P., Brandes J.L., Bassilian S. Oxythiamine and dehydroepiandrosterone inhibit the nonoxidative synthesis of ribose and tumor cell proliferation. Cancer Research. 1997;57:4242–4248. [PubMed] [Google Scholar]
- 13.Diaz-Moralli S., Aguilar E., Marin S., Coy J.F., Dewerchin M., Antoniewicz M.R. A key role for transketolase-like 1 in tumor metabolic reprogramming. Oncotarget. 2016;7:51875–51897. doi: 10.18632/oncotarget.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid M.A., Allen A.E., Liu S., Liberti M.V., Liu P., Liu X. Serine synthesis through PHGDH coordinates nucleotide levels by maintaining central carbon metabolism. Nature Communications. 2018;9 doi: 10.1038/s41467-018-07868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Labuschagne C.F., van den Broek N.J., Mackay G.M., Vousden K.H., Maddocks O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Reports. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 16.Eagle H. Nutrition needs of mammalian cells in tissue culture. Science (New York, N.Y.) 1955;122:501–514. doi: 10.1126/science.122.3168.501. [DOI] [PubMed] [Google Scholar]
- 17.Wise D.R., Thompson C.B. Glutamine addiction: a new therapeutic target in cancer. Trends in Biochemical Sciences. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tardito S., Oudin A., Ahmed S.U., Fack F., Keunen O., Zheng L. Glutamine synthetase activity fuels nucleotide biosynthesis and supports growth of glutamine-restricted glioblastoma. Nature Cell Biology. 2015;17:1556–1568. doi: 10.1038/ncb3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Sahra I., Hoxhaj G., Ricoult S.J., Asara J.M., Manning B.D. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J., Fan J., Venneti S., Cross J., Takagi T., Bhinder B. Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Molecular Cell. 2014;56:205–218. doi: 10.1016/j.molcel.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng T., Sudderth J., Yang C., Mullen A.R., Jin E.S., Mat´es J.M. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlova N.N., Hui S., Ghergurovich J.M., Fan J., Intlekofer A.M., White R.M. As extracellular glutamine levels decline, asparagine becomes an essential amino acid. Cell Metabolism. 2018;27:428–438.e5. doi: 10.1016/j.cmet.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.KrCall A.S., Xu S., Graeber T.G., Braas D., Christofk H.R. Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor. Nature Communications. 2016;7:11457. doi: 10.1038/ncomms11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shahinian A.H., Spears C.P., Moran R.G. In vivo kinetics of thymidylate synthetase inhibition in 5-FU sensitive and resistance murine colon adneocarcinomas. Cancer Research. 1982;42:450–456. [PubMed] [Google Scholar]
- 25.Daidone F., Florio R., Rinaldo S., Contestabile R., di Salvo M.L., Cutruzzol`a F. In silico and in vitro validation of serine hydroxymethyltransferase as a chemotherapeutic target of the antifolate drug pemetrexed. European Journal of Medicinal Chemistry. 2011;46:1616–1621. doi: 10.1016/j.ejmech.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Ducker G.S., Ghergurovich J.M., Mainolfi N., Suri V., Jeong S.K., HsinJung Li S. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of di↵use large B-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:11404–11409. doi: 10.1073/pnas.1706617114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg V.C.L., Trosko J.L. Others, Platinum compounds: a new class of potent anti tumors agent. Nature. 1969;222:385–386. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 28.Stearns B., Losee K.A., Bernstein J. Hydroxyurea. A new type of potential antitumor agent. Journal of Medicinal Chemistry. 1963;6:201. doi: 10.1021/jm00338a026. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad M.F., Alam I., Hu↵ S.E., Pink J., Flanagan S.A., Shewach D. Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:8241–8246. doi: 10.1073/pnas.1620220114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medina M.A., Sanchez-Jimenez F., Marquez F.J., Perez-Rodriguez J., Quesada A.R., de Castro Nu´ñez I. Glutamine and glucose as energy substrates for Ehrlich ascites tumour cells. Biochemistry International. 1988;16:339–347. [PubMed] [Google Scholar]
- 31.Baggetto L.G. Deviant energetic metabolism of glycolytic cancer cells. Biochimie. 1992;74:959–974. doi: 10.1016/0300-9084(92)90016-8. [DOI] [PubMed] [Google Scholar]
- 32.Dejure F.R., Royla N., Herold S., Kalb J., Walz S., Ade C.P. The MYC mRNA 3’-UTR couples RNA polymerase II function to glutamine and ribonucleotide levels. The EMBO Journal. 2017;36:1854–1868. doi: 10.15252/embj.201796662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leone R.D., Zhao L., Englert J.M., Sun I.-M., Oh M.-H., Sun I.-H. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019 doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markley J.L., Brüschweiler R., Edison A.S., Eghbalnia H.R., Powers R., Raftery D. The future of NMR-based metabolomics. Current Opinion in Biotechnology. 2017;43:34–40. doi: 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emwas A.-H.M. Metabonomics: methods and protocols, methods in molecular biology. Humana Press; New York, NY: 2015. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research; pp. 161–193. [DOI] [PubMed] [Google Scholar]
- 36.Choi J., Grossbach M.T., Antoniewicz M.R. Measuring complete isotopomer distribution of aspartate using gas chromatography/tandem mass spectrometry. Analytical Chemistry. 2012;84:4628–4632. doi: 10.1021/ac300611n. [DOI] [PubMed] [Google Scholar]
- 37.Antoniewicz M.R. 2018. A guide to 13C metabolic flux analysis for the cancer biologist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buescher J.M., Maddocks O.D.K., Lunt S.Y., Metallo C.M., Kibbey R.G., Vousden K. A roadmap for interpreting 13 C metabolite labeling patterns from cells. Current Opinion in Biotechnology. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antoniewicz M.R. 2013. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. [DOI] [PubMed] [Google Scholar]
- 40.Metallo C.M., Walther J.L., Stephanopoulos G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. Journal of Biotechnology. 2009;144:167–174. doi: 10.1016/j.jbiotec.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jang C., Chen L., Rabinowitz J.D. 2018. Metabolomics and isotope tracing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curran K.A., Crook N.C., Alper H.S. Using flux balance analysis to guide microbial metabolic engineering. Methods in Molecular Biology. 2012;834:197–216. doi: 10.1007/978-1-61779-483-4_13. [DOI] [PubMed] [Google Scholar]
- 43.Wiechert W., de Graaf A.A. Bidirectional reaction steps in metabolic networks: ii. Flux estimation and statistical analysis. Biotechnology and Bioengineering. 1997;55:101–135. doi: 10.1002/(SICI)1097-0290(19970705)55:1<118::AID-BIT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 44.Antoniewicz M.R., Kelleher J.K., Stephanopoulos G. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metabolic Engineering. 2006;8:324–337. doi: 10.1016/j.ymben.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Nöh K., Grönke K., Luo B., Takors R., Oldiges M., Wiechert W. Metabolic flux analysis at ultra short time scale: isotopically non-stationary 13C labeling experiments. Journal of Biotechnology. 2006;129:249–267. doi: 10.1016/j.jbiotec.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 46.Wiechert W., N¨oh K. 2013. Isotopically non-stationary metabolic flux analysis: complex yet highly informative. [DOI] [PubMed] [Google Scholar]
- 47.Noack S., N¨oh K., Moch M., Oldiges M., Wiechert W. Stationary versus non-stationary 13C-MFA: a comparison using a consistent dataset. Journal of Biotechnology. 2011;154:179–190. doi: 10.1016/j.jbiotec.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Yuan J., Bennett B.D., Rabinowitz J.D. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nature Protocols. 2008;3:1328. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pietzke M., Kempa S. Pulsed stable isotope-resolved metabolomic studies of cancer cells. Methods in Enzymology. 2014;543:179–198. doi: 10.1016/B978-0-12-801329-8.00009-X. [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Ulbrich J., Müller J., Wüstefeld T., Aeberhard L., Kress T.R. Others, Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 51.Griep-Raming J., Peterson A.C., McAlister G.C., Coon J.J., Quarmby S.T. Development and characterization of a GC-enabled QLT-orbitrap for HighResolution and high-mass accuracy GC/MS. Analytical Chemistry. 2010;82:8618–8628. doi: 10.1021/ac101757m. [DOI] [PubMed] [Google Scholar]
- 52.Weidt S., Haggarty J., Kean R., Cojocariu C.I., Silcock P.J., Rajendran R. A novel targeted/untargeted GC-Orbitrap metabolomics methodology applied to Candida albicans and Staphylococcus aureus biofilms. Metabolomics. 2016;12 doi: 10.1007/s11306-016-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niedenführ S., ten Pierick A., van Dam P.T.N., Suarez-Mendez C.A., Nöh K., Wahl S.A. Natural isotope correction of MS/MS measurements for metabolomics and 13C fluxomics. Biotechnology and Bioengineering. 2016;113:1137–1147. doi: 10.1002/bit.25859. [DOI] [PubMed] [Google Scholar]
- 54.Midani F.S., Wynn M.L., Schnell S. The importance of accurately correcting for the natural abundance of stable isotopes. Analytical Biochemistry. 2017;520:27–43. doi: 10.1016/j.ab.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carreer W.J., Flight R.M., Moseley H.N.B. A computational framework for high-throughput isotopic natural abundance correction of omics-level ultra-high resolution FT-MS datasets. Metabolites. 2013;3:853. doi: 10.3390/metabo3040853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blank L.M., Desphande R.R., Schmid A., Hayen H. Analysis of carbon and nitrogen co-metabolism in yeast by ultrahigh-resolution mass spectrometry applying 13C- and 15N-labeled substrates simultaneously. Analytical and Bioanalytical Chemistry. 2012;403:2291–2305. doi: 10.1007/s00216-012-6009-4. [DOI] [PubMed] [Google Scholar]
- 57.Molero G., Aranjuelo I., Teixidor P., Araus J.L., Nogu´es S. Measurement of 13C and 15N isotope labeling by gas chromatography/combustion/isotope ratio mass spectrometry to study amino acid fluxes in a plant-microbe symbiotic association. Rapid Communications in Mass Spectrometry. 2011;25:599–607. doi: 10.1002/rcm.4895. [DOI] [PubMed] [Google Scholar]
- 58.Nilsson R., Jain M. Simultaneous tracing of carbon and nitrogen isotopes in human cells. Molecular BioSystems. 2016;12:1929–1937. doi: 10.1039/c6mb00009f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Badur M.G., Metallo C.M. Reverse engineering the cancer metabolic network using flux analysis to understand drivers of human disease. Metabolic Engineering. 2018;45:95–108. doi: 10.1016/j.ymben.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]