Significance
As nucleosomes prevent binding of transcription factors (TFs) to DNA, their position needs to be actively modulated. Nucleosomes can also reposition spontaneously, but this process and its effect on TF binding have not been extensively studied. Here, we developed a method based on single-molecule optical tweezers to simultaneously measure nucleosome diffusion and TF binding to the same DNA molecule. We show that nucleosomes undergo confined diffusion on the DNA, and that the confinement is relieved upon incorporation of the histone variant H2A.Z leading to an increase in TF binding, which then further biases nucleosome diffusion. Our results shed light on a previously uncharacterized mechanism of transcriptional regulation.
Keywords: nucleosomes, transcription factors, chromatin, optical tweezers
Abstract
The structure of promoter chromatin determines the ability of transcription factors (TFs) to bind to DNA and therefore has a profound effect on the expression levels of genes. However, the role of spontaneous nucleosome movements in this process is not fully understood. Here, we developed a single-molecule optical tweezers assay capable of simultaneously characterizing the base pair-scale diffusion of a nucleosome on DNA and the binding of a TF, using the luteinizing hormone β subunit gene (Lhb) promoter and Egr-1 as a model system. Our results demonstrate that nucleosomes undergo confined diffusion, and that the incorporation of the histone variant H2A.Z serves to partially relieve this confinement, inducing a different type of nucleosome repositioning. The increase in diffusion leads to exposure of a TF’s binding site and facilitates its association with the DNA, which, in turn, biases the subsequent movement of the nucleosome. Our findings suggest the use of mobile nucleosomes as a general transcriptional regulatory mechanism.
Packaging of the DNA into chromatin reduces its accessibility to regulatory proteins, such as transcription factors (TFs) and RNA polymerase (RNAP), making the structure and dynamics of nucleosomes an essential part of gene expression regulation in higher organisms (1, 2). Although atomic-resolution structures of the nucleosome reveal strong contacts between the histone core and the DNA (3), evidence has accumulated indicating that these interactions are often disrupted spontaneously, and that the crystal structure represents only a snapshot of a complex conformational dynamics. Atomic force microscopy (AFM) studies observed nucleosomal DNA looping (4), and intermediates composed of tetramers and hexamers (5), while conformational changes of the histone octamer inside the nucleosome were recently detected using cryo-electon microscopy (6). Moreover, nucleosomes have been shown to spontaneously unwrap DNA from its ends in a process termed “thermal breathing” (5, 7–12). The momentary exposure of the DNA was shown to facilitate the invasion of regulatory proteins to nucleosomal DNA (7, 8, 10) and the elongation by RNAP (4, 13), highlighting the potential effect of these dynamics on the process of transcription.
Previous studies have indicated the existence of an additional type of thermally driven dynamics: the spontaneous “mobility” or “thermal sliding” of nucleosomes by which their center of mass repositions on the DNA in an unprompted longitudinal-like movement. Initial studies using 2D gel electrophoresis (14, 15) reported that nucleosomes are able to reposition over timescales of hours when incubated at 37 °C, but not at 4 °C. Later experiments used chemically modified H4 histones, capable of inducing a cleavage at the nucleosome’s dyad (16), and found that the repositioning rates depend on the positioning sequence and the length of the DNA fragment. Others showed that sin mutations, which weaken the binding of the histone octamer close to the dyad (17), and also deletion of the histone tails (18), alter the inherent mobility of nucleosomes. Altogether, these experiments established the existence of spontaneous thermal sliding by the nucleosome, and motivated the development of theoretical models (19–21) and computational studies (22). However, these experiments suffered from important limitations: First, most were performed using nucleosome positioning sequences such as 5S rDNA or the “601” sequence (23), which provide an initial homogeneous population of reconstituted nucleosomes, but introduce specific features that may hinder the dynamics that normally occur on natural, biologically relevant sequences. Second, they cannot resolve the potential base pair-scale dynamics of nucleosome movement. Third, since diffusion is inherently stochastic, its fine details can be lost by averaging over an ensemble of molecules. Recent studies using high-speed AFM were successful in visualizing spontaneous sliding events of individual nucleosomes at high temporal resolution (5, 24), but lack the longitudinal resolution required to systematically study the mechanisms of sliding.
The inclusion of histone variants in nucleosomes is a critical mechanism for regulating gene expression (25). In particular, the evolutionarily conserved and essential H2A.Z is localized to the regulatory regions of both heterochromatin and open chromatin associated with TF binding (26–30), with contrasting results reported on its effect on nucleosome stability (31) and transcription (32, 33). In our previous study (34), we showed that H2A.Z is incorporated into the nucleosome positioned at the TSS of the Lhb gene, which covers binding sites for important TFs involved in transcriptional activation. Using single-molecule DNA unzipping with optical tweezers, we also showed that H2A.Z-containing nucleosomes display a higher positional dispersion, and initial experiments revealed that this increased dispersion is a reflection of their higher mobility.
Clearly, the mobility is expected to modulate the association rate of proteins to binding sites inside the nucleosome. In addition, it was recently shown that nucleosomes also accelerate the dissociation of TFs (35), suggesting a delicate interplay between the movement of nucleosomes and the binding of proteins. However, no experimental method has been demonstrated that can monitor simultaneously these two dynamic processes. In this work, we follow the spontaneous movement of individual nucleosomes, at base pair-scale resolution and on biologically relevant DNA sequences, to elucidate the role of thermal sliding in the regulation of TF binding. By repetitively and partially unzipping the nucleosomal DNA, thus revealing the position of the Lhb TSS nucleosome without perturbing the major DNA-histone interactions, we are able to follow the base pair-scale movements of individual nucleosomes over extended periods of time, while simultaneously monitoring the binding of the TF Egr-1 to its site in the nucleosomal DNA. Our results provide a detailed characterization of the interplay between H2A.Z-dependent nucleosomal mobility and binding of a TF, and suggest a potential and general regulatory role for mobile nucleosomes.
Results
Real-Time Measurements of Nucleosomal Diffusion.
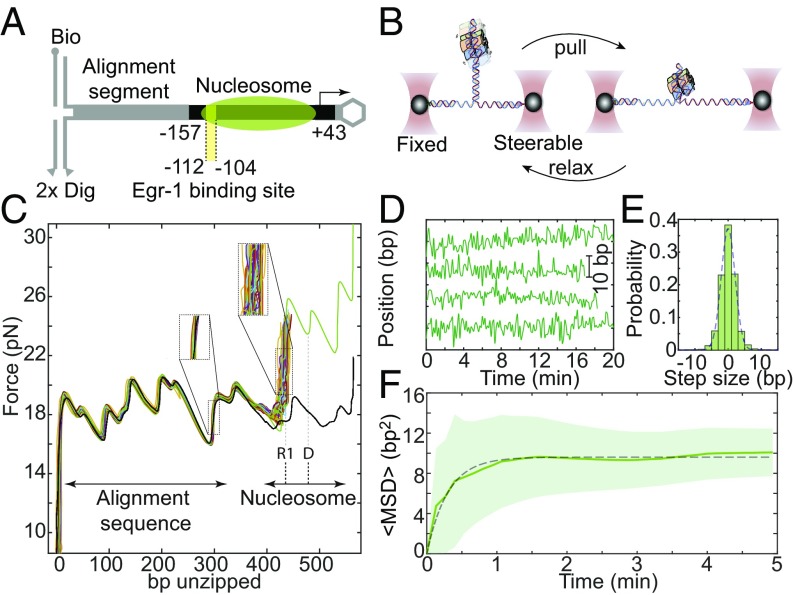
To track the positional dynamics of nucleosomes on a biologically relevant DNA sequence, we assembled nucleosomes using canonical mouse histones expressed in bacteria and a ∼200-bp DNA fragment corresponding to the −157/+43 region of the mouse Lhb gene promoter, previously shown to be packaged into a nucleosome (34). We ligated the reconstituted nucleosomes to a naked DNA segment that functions as an alignment sequence (34, 36) and is connected to two ∼2,000-bp dsDNA handles harboring a single biotin and two digoxigenin terminal modifications, respectively (Fig. 1A). Next, we connected the handles to two ∼1-μm beads with matching anti-digoxigenin or streptavidin modifications, and trapped the beads by the two focused laser beams of a dual-trap optical tweezers setup (34, 37). Moving one trap away from the other leads to an increase in the tension between the two strands of the dsDNA. Applying forces higher than ∼16–17 pN is sufficient to overcome the interactions between them, leading to “unzipping” of the DNA (Fig. 1B).
Fig. 1.
Real-time measurements of the base pair-scale diffusion of nucleosomes. (A) Nucleosomes reconstituted on the –157/+43 Lhb sequence are ligated to an alignment segment and connected to dsDNA handles. (B) The construct is attached to polystyrene beads trapped in two separate optical traps. One of the traps is moved to unzip the tethered construct until a force of ∼23–24 pN is reached, indicating the presence of the nucleosome, and then relaxed. This process is repeated with a total cycle time of 8 s. (C) Repetitive partial unzipping cycles of a nucleosome reconstituted on the Lhb TSS sequence (colored). The last cycle is used to dissociate the nucleosome irreversibly (green) and is followed by an additional cycle of unzipping of the resulting naked DNA (black). Note the broader distribution of nucleosome’s position (Right Inset) relative to the distribution of the position of an alignment segment (Left Inset). The position of region 1 (R1) and the dyad (D) are indicated. (D) Individual traces of the position of nucleosomes as a function of time, sampled every 8 s. (E) Probability distribution function of the step size, i.e., the relative position of the nucleosome between times separated by five unzipping cycles. Data filtered with a five cycles running average window. A Gaussian fit to the histogram is shown (dashed line). The skewness of the distribution is <0.1. (F) Ensemble-averaged mean squared displacement (MSD) for nucleosomes reconstituted on Lhb TSS, as a function of time. Data shown as mean ± SEM, low-pass filtered with a five-points window running average. The number of experiments is shown in SI Appendix, Table S3. The dashed line is a fit to the expression , with a diffusion constant D = 1.3 ± 0.14 bp2⋅s−1 and confining potential spring constant k = 1.2 ± 0.2 pN⋅bp−1.
It has been shown previously that the disruption of protein–DNA interactions by unzipping the DNA requires forces that are higher than those needed to unzip DNA alone (38–42). In particular, when an unzipping fork encounters a nucleosome, the latter is disrupted with a characteristic signature that highlights the position and strength of two regions of strong histone–DNA interactions (36, 43). The first interaction (region 1) is attributed to contacts of the DNA with the H2A/H2B dimer, and the second (region 2) with the H3/H4 tetramer. We and others have shown (34, 36) that disruption of region 1, as opposed to that of region 2, is a reversible process, as the interactions between the H2A/H2B dimer and the DNA reform when the force is relaxed. This fact was exploited in our previous work (34) in which we probed the position of a single nucleosome, every 30 s, for a total of ∼5–35 times, and showed that nucleosomes are able to reposition spontaneously. However, the relatively small number of times nucleosomes were probed before they disassembled prevented us from further characterizing the properties of this spontaneous movement. Here, by applying forces of ∼23–24 pN, which are a few piconewtons lower than those required to disrupt region 1, we were able to probe the position of this region without breaking it, thus minimizing any potential perturbation of the nucleosome structure (Fig. 1C). The high stability and resolution of our instrument, combined with the use of an alignment sequence as a set of “fiducial marks,” allowed as to determine the position of a nucleosome in every single probing cycle with ∼2-bp accuracy (SI Appendix, Fig. S1A). Complemented with a double (instead of single) digoxigenin tag in the DNA handle, the lifetime of the tethers was substantially increased. Hence, we were able to measure precisely and repetitively the position of a single nucleosome many times (∼40–240 cycles, every 8 s) over a long period (5–30 min; SI Appendix, Fig. S2), allowing us to follow the trajectories of individual nucleosomes as they reposition spontaneously on the DNA (Fig. 1D). Importantly, our measurements do not perturb the spontaneous dynamics of nucleosomes and are not affected by the breathing dynamics (SI Appendix).
To study the mechanism of sliding, we calculated the mean square displacement (, where is the instantaneous position of the nucleosome) as a function of time, for each nucleosome relative to its initial position, and averaged over the whole ensemble of nucleosomes. For “normal” 1D diffusion, the MSD is expected to depend linearly on time, according to , where D is the diffusion constant. However, we observed a nonlinear dependence on time (Fig. 1F). The MSD reaches a steady state, and thus a subdiffusion model, described by a power-law , with α < 1, cannot fit the data either. The MSD is well fitted by a model that assumes diffusion in a confining harmonic potential (44). Hence, we conclude that the spontaneous movement of nucleosomes is spatially confined. Notably, the mean position of the nucleosomes in the measured ensemble is distributed over a much larger region (between approximately −150 and −90) than the region sampled by an individual nucleosome (SI Appendix, Fig. S3). Moreover, the energy surfaces on which the nucleosomes diffuse (SI Appendix, Fig. S4) do not overlap. Hence, the observed confinement is not the result of a specific sequence effect, which defines a common potential energy surface for the movement of all of the nucleosomes, but rather represents the inability of each individual nucleosome to explore regions beyond the close vicinity of its initial position at the time of reconstitution. This may suggest that certain structural elements in the nucleosome do not participate in the sliding process and function as “anchors” attached to the DNA. These results are in line with previous results showing that sliding is a “local” effect (15) and that the absence of the N-tail of H2B enhances repositioning (18). Interestingly, the observed confinement region is larger than the one measured for nucleosomes reconstituted on 601 DNA (SI Appendix, Fig. S1B). From the fit of the MSD curve we can estimate a diffusion constant of ∼1.3 bp2⋅s−1. This result is consistent with previous estimations from gel electrophoresis studies and with the predictions of models based on the propagation of twist defects (20).
Notably, the observed confinement in the repositioning of the nucleosome limits the effect that this movement can have on modulating the accessibility of the transcription machinery to DNA. However, in transcriptionally active regions, the canonical histones are often replaced by histone variants (25), as we previously showed specifically for the Lhb TSS nucleosome, which is enriched with H2A.Z (34). Hence, we wondered whether the incorporation of this histone variant can modulate the diffusional properties of the nucleosome.
H2A.Z Increases Nucleosome Diffusion.
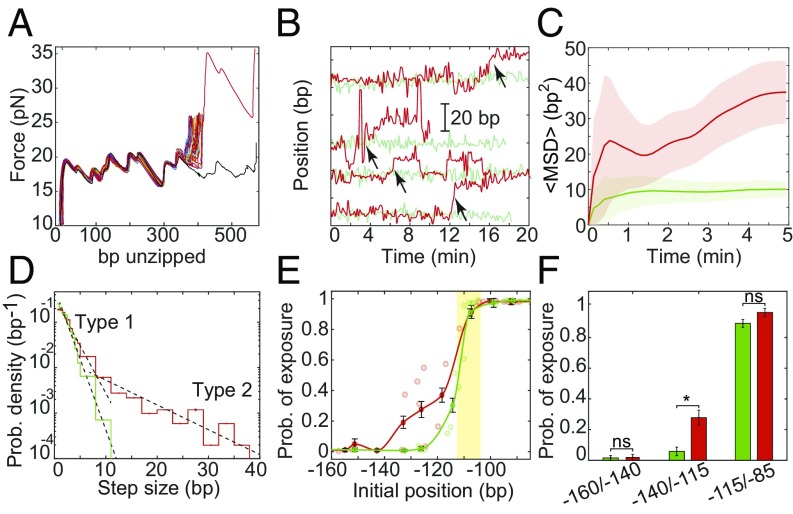
We reconstituted nucleosomes using H2A.Z-contatining octamers and subjected them to multiple unzipping analysis to probe their positional dynamics (Fig. 2A). By looking at the individual trajectories of single particles (Fig. 2B), it was clear that the H2A.Z nucleosomes exhibit enhanced mobility compared with their H2A counterparts. Remarkably, the movement of the H2A.Z-nucleosomes was characterized by sporadic and relatively large translocation events (Fig. 2B and SI Appendix, Fig. S3). Moreover, their MSD is significantly higher than that of H2A nucleosomes and did not reach a plateau in the timescale of our experiments (Fig. 2C and SI Appendix, Fig. S6), suggesting that they are able to escape, at least partially, the confinement. In addition, the short-time part of the MSD curve reveals that the diffusion constant of these nucleosomes is about a factor of 2 higher than that of H2A nucleosomes.
Fig. 2.
H2A.Z increases the diffusion of nucleosomes. (A) Repetitive partial unzipping curves of a nucleosome reconstituted with H2A.Z. Data are presented as in Fig. 1C. (B) Position of individual H2A (green) and H2A.Z (red) nucleosomes over time, presented as in Fig. 1D. The H2A traces (green) are identical to those in Fig. 1D. Examples of repositioning events larger than 5 bp (“type 2”) are marked with arrows. (C) Ensemble-averaged MSD of nucleosomes as a function of time, presented as in Fig. 1F. The number of experiments is shown in SI Appendix, Table S3. (D) Probability distribution function of the step size. Single-exponential fits (dashed black lines) are shown for steps sizes <5 bp (“type 1”) and >5 bp (“type 2”). nsteps, H2A = 2,274 and nsteps, H2A.Z = 1,308. (E) Probability of exposure of the Egr-1 binding site (yellow strip), calculated for experiments longer than 7 min (i.e., the number of nucleosome’s positions whose 5′ edge is positioned downstream the Egr-1 binding site, out of the total number of positions in the same experiment); nH2A = 75 and nH2A.Z = 88; mean ± SEM. (F) The average probability of exposure for nucleosomes whose initial 5′ edge is located between −160/−140 (nH2A = 3; nH2A.Z = 3), −140/−115 (nH2A = 5; nH2A.Z = 6), and −115/−85 (nH2A = 12; nH2A.Z = 4) is shown; mean ± SEM; *P < 0.05, Student’s t test.
As opposed to H2A nucleosomes, the probability density function (PDF) of the step size for H2A.Z nucleosomes cannot be fitted to a single exponential. A double exponential fits the data reasonably well (Fig. 2D). This may indicate that H2A.Z repositioning is associated with two types of movement: The first type (“type 1”), similar to the motion of H2A nucleosomes, occurs on faster timescales, comparable to the sampling rate of one per 8 s, and is characterized by movements smaller than ∼5 bp. The second type of motion (“type 2”), which is unique to H2A.Z nucleosomes, is characterized by movements larger than ∼5 bp and occurs in a timescale of minutes. Clearly, these rare events dominate the overall movement of the H2A.Z nucleosomes on long timescales.
The observed increase in nucleosome diffusion imposed by the presence of H2A.Z can affect the time-averaged accessibility of regulatory elements on the Lhb TSS. In gonadotrope cells, the 5′ edge of the Lhb TSS nucleosome is positioned at approximately −110/−130 (34), indicating that packaging covers most of the TF binding sites important for the gene’s transcriptional activation. One of these sites is located at −104/−112, where the TF Egr-1 was shown to bind (42, 45). Hence, we expect the accessibility of this site to increase greatly if the H2A nucleosome is exchanged by H2A.Z, with its increased and distinct diffusion. To address this point, we used the data from the diffusion experiments to estimate the instantaneous position of the nucleosomes 5′ edge (SI Appendix, Fig. S10), and thus calculate the probability of exposure, i.e., the number of probing events where the 5′ end of the nucleosome moved downstream and exposed the Egr-1 site, as a function of nucleosome initial position relative to the TSS. For both types of nucleosome, we found, as expected, that those initially positioned upstream of −140 cover the binding site, and those positioned downstream of −100 expose it (Fig. 2E). Interestingly, differences in the probability of exposure were observed in the biologically relevant region −110/−130, where we observed a significantly higher probability of exposure for H2A.Z nucleosomes (Fig. 2F). This observation made us wonder whether Egr-1 is indeed able to exploit this “window of opportunity” created by the momentary exposure of its binding site to bind to the Lhb promoter.
Nucleosome Diffusion Facilitates TF Binding.
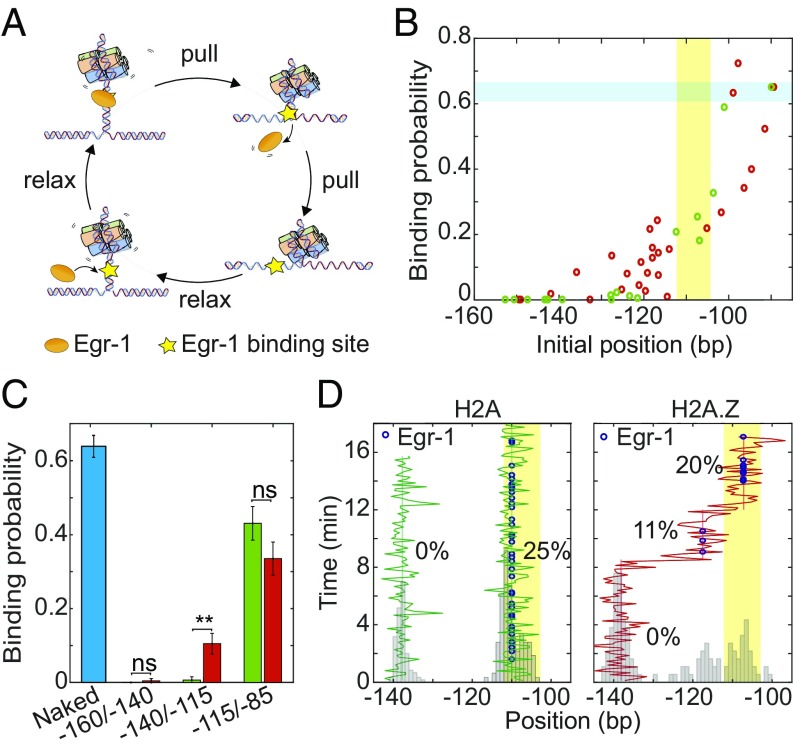
To elucidate the effect of nucleosome movement on TF binding, we exploited the laminar flow cell in our optical tweezers and exposed a nucleosome construct to the DNA binding domain of Egr-1 (referred as Egr-1 for simplicity), at typical concentrations of 500 nM. In a previous study using an unzipping assay, we were able to characterize the mean breaking force and the binding probability of Egr-1 to its binding site in the naked Lhb TSS (42). Notably, the characteristic breaking force required to dissociate Egr-1 from the −104/−112 site (site “−2” in ref. 42) is significantly lower than the typical force required for breaking the nucleosome’s region 1. Hence, our current ∼23- to 24-pN force threshold allowed us to displace Egr-1 from the DNA, while leaving the downstream nucleosome intact (Fig. 3A). Binding of Egr-1 to its site was readily distinguishable by a force “rip” upstream of the nucleosome's region 1 (SI Appendix, Fig. S11). Accordingly, in each unzipping cycle, we were able to detect not only the position of the nucleosome but also the presence of a bound Egr-1 on the same DNA molecule.
Fig. 3.
Nucleosome diffusion facilitates TF binding. (A) Experimental scheme for the repetitive probing of nucleosome positioning in the presence of Egr-1. (B) Calculated binding probability for Egr-1 as a function of nucleosome initial position, for experiments longer than 7 min. Data are shown as the result of individual experiments; nH2A = 25 and nH2A.Z = 20. The Egr-1 binding site is highlighted in yellow. The binding probability of Egr-1 to naked DNA (highlighted in blue) is shown for comparison. (C) Average binding probability for H2A (green) and H2A.Z (red) nucleosomes whose initial position is located between −160/−140 (nH2A = 3; nH2A.Z = 4), −140/−115 (nH2A = 5; nH2A.Z = 15), and −115/−85 (nH2A = 12; nH2A.Z = 6). Shown as mean ± SEM. The binding probability of Egr-1 to naked DNA (blue) is shown for comparison (n = 262; eight experiments). (D) Repositioning trajectory of two H2A (Left, green) and a single H2A.Z (red, Right) nucleosomes. Histograms of the positions are shown in gray. The location of the Egr-1 binding site is marked in yellow. The blue dots represent detection of a bound Egr-1 in the corresponding cycle, and the numbers, the percent of the probing cycles where a protein was bound, for different mean positions of the nucleosome.
Multiple unzipping cycles, performed on naked or nucleosomal Lhb TSS DNA in the presence of Egr-1, allowed us to calculate the effect of the nucleosome on the dynamics of Egr-1 binding (Fig. 3 B and C). The average binding probability on naked DNA for this binding site was ∼0.6, which is consistent with our previously published results (42). However, for nucleosomal DNA, this probability depends on the nucleosome position: For both types of nucleosome, when the nucleosome’s initial position was upstream of −140 no binding could be observed, and for nucleosomes positioned downstream of −100 the binding probability was similar to that on naked DNA. Notably, significant differences between the nucleosomes were observed in the intermediate region: H2A.Z nucleosomes positioned initially between −140/−110 showed an increased time-averaged binding probability of Egr-1 compared with H2A nucleosomes located initially at the same positions (Fig. 3C). This trend was similar to the one observed for the calculated probability of exposure based on the diffusion of nucleosomes without Egr-1, suggesting that the increased diffusion of H2A.Z nucleosomes is responsible for the increased binding. Interestingly, when we examined the trajectories of individual nucleosomes initially positioned between −140/−110 (Fig. 3D), it was evident that the increase in Egr-1 binding probability is a consequence of the large, type 2 movements of H2A.Z nucleosomes, during which they partially uncover the Egr-1 site, creating distinct degrees of accessibility. In contrast, H2A nucleosomes positioned in this region did not show significant repositioning or Egr-1 binding. These results suggest that the increase in nucleosomal diffusion mediated by the type 2 events, which is characteristic of nucleosomes containing the H2A.Z variant, leads to increased site exposure and TF binding.
TF Binding Biases the Spontaneous Movement of the Nucleosome.
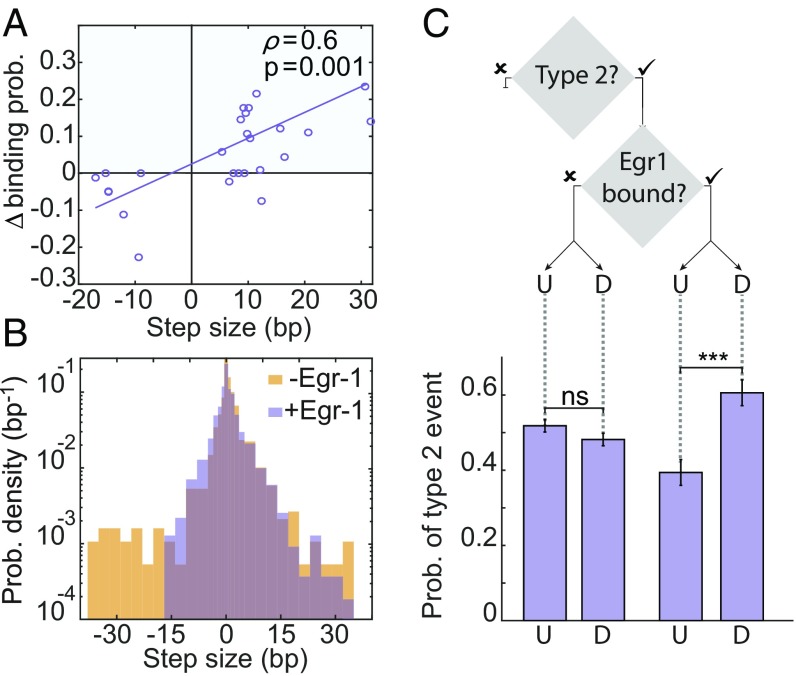
In principle, the large repositioning events of H2A.Z nucleosomes can also have a negative effect on TF binding if access to the site is blocked, and we indeed observed such events (SI Appendix, Fig. S5). However, events involving a decrease in binding probability were approximately three times less frequent compared with those of probability increase (Fig. 4A), suggesting a possible role for Egr-1 binding in breaking the symmetry of the nucleosome’s movement. Interestingly, when we compared the MSD as a function of time in the presence or absence of Egr-1, a decrease in nucleosome diffusion due to the presence of Egr-1 was evident (SI Appendix, Fig. S6A). This suggests that Egr-1 affects nucleosome repositioning by suppressing long repositioning events, via one of two possible scenarios. One possibility is that Egr-1 molecules, by their mere presence in the solution or nonspecific binding to the DNA, repress type 2 movements (e.g., by affecting intermolecular crowding). Alternatively, the specific association of Egr-1 to its binding motif, located at −104/−112, may create a barrier for nucleosome movement. To clarify this question, we compared the PDF for the step size for H2A.Z when Egr-1 was present or absent in the solution. Notably, in the presence of Egr-1, there is a clear suppression in the long repositioning events toward −104/−112 (Fig. 4B). Importantly, this asymmetry is position dependent, as it was not observed for nucleosomes whose 5′ edge is located in the region between −140/−160, which completely covers the binding site (SI Appendix, Fig. S6B). If specific binding of Egr-1 is indeed responsible for the bias in nucleosome repositioning downstream, we expect that, once Egr-1 is bound, the probability of type 2 events in the downstream direction will increase over those occurring upstream. To explore this possibility, we calculated the conditional probability for type 2 events, upstream or downstream, given that the TF is specifically bound, or not bound but present in the solution (Fig. 4C). The probability of type 2 events shows no clear preference in direction when the TF is not specifically bound. In contrast, when Egr-1 is bound, the probability of downstream type 2 events is significantly higher than upstream events. Taken together, the data suggest that, once Egr-1 binds to its site, the nucleosome movement is biased toward downstream repositioning.
Fig. 4.
Binding of a TF biases the diffusion of the nucleosome. (A) Two-dimensional scattering plot for the change in binding probability vs. the size of a type 2 repositioning step, for H2A.Z-containing nucleosomes. Positive changes (blue area) correspond to an increase in the binding probability, while negative changes (white area) to a decrease (nH2A = 6; nH2A.Z = 27). A linear fit to the scatter plot data are shown; Pearson correlation ρ = 0.6, P = 0.01. (B) Probability distribution function of the step size for the experiments with H2A.Z-containing nucleosomes, with initial position between −140/−115, in the absence (yellow; nsteps = 1,308) or the presence (purple; nsteps = 2,916) of Egr-1. (C) Conditional probability for upstream (U) or downstream (D) type 2 events, given that Egr-1 is bound (Right) or not (Left) to its binding site. ntype 2 = 974, nupstream,bound = 56, ndownstream,bound = 84, nupstream, not bound = 429, and ndownstream, not bound = 405. ***P < 0.001, χ2 test.
Notably, given that Egr-1 binds to this site with koff ∼ 1 s−1 and kon ∼ 3–10 × 106 M−1⋅s−1 (SI Appendix, Fig. S7, ref. 46), at typical cellular concentrations the binding dynamics of Egr-1 are much faster than the typical type 2 repositioning times, implying that binding and dissociation are in rapid equilibrium with respect to these movements. Hence, it is likely that, in vivo, nucleosome movements are limited by the mean occupancy of the binding site by Egr-1.
Discussion
Our experimental assay allows us to probe the instantaneous position of a nucleosome repetitively, with nearly base pair resolution and over long times, thus enabling measurement of the thermal mobility of reconstituted nucleosomes. Notably, our measurements are based on probing the position of the nucleosome’s first interaction region, and therefore cannot rule out completely that conformational changes in the nucleosome structure play a role in the observed repositioning of this region. In fact, previous works suggesting the importance of octamer plasticity for nucleosome dynamics suggest that some conformational changes are required for the nucleosome to move (3). However, our data do suggest that the observed dynamics are, at least essentially, a center-of-mass motion (SI Appendix). Our results indicate that, once initially localized (during in vitro reconstitution in our experiments or by the action of remodeling machinery in the cell), canonical H2A-containing nucleosomes undergo confined diffusion, exploring a small region of the DNA. Deposition of H2A.Z into the nucleosome dramatically changes its diffusional properties. In addition to the short and frequent repositioning steps similar to those observed for H2A nucleosomes, H2A.Z nucleosomes exhibit an additional type of step, less frequent but longer, which dominates the repositioning of the nucleosomes and confer them the ability to overcome the confinement and translocate over longer distances. As the concurrent breaking of all octamer–DNA interactions would require an energy of at least ∼75 kBT (47), theoretical models suggest that the spontaneous mobility of nucleosomes is driven by structural defects, either loops (19) or twists (21), created as the DNA spontaneously unwraps and rewraps. Although the prevalence of each type of defect is still under debate, it is clear that twist defects, the most likely being of 1 bp, should form faster than loops, which are energetically advantageous at the length of 10 bp. Moreover, recent computational models predict that the formation of loops is restricted to specific regions, such as SHL ± 2 (48) located between the nucleosome's region 1 and 2, but DNA overtwisting enables accommodation of an additional extra base pair that can from in any position along the DNA (49). Hence, it is possible that the two types of repositioning events we report for H2A.Z-containing nucleosomes represent the dynamics induced by the two types of structural defects proposed: the small-scale movements may correspond to the energetically lower and more abundant twist defects, generated at second and perhaps subsecond timescales, while the long-range repositioning events that occur at minute timescales might correspond to the dynamics facilitated by the formation of loop defects.
What makes H2A.Z nucleosomes distinct? Although the crystal structures of both canonical and H2A.Z-containing nucleosomes are similar, a number of key differences exist. In particular, the interactions of the H3/H4 tetramer with the docking domain of H2A.Z are destabilized due to substitution of a glutamine by the smaller glycine at position 104 in H2A.Z (50). In light of recent studies indicating that nucleosome thermal repositioning requires octamer plasticity (51), we propose that such plasticity can be achieved by the destabilization of the H2A.Z/H2B docking domain, which leads to conformational changes in the histone octamer. Such changes will affect not only the local interactions between the H3/H4 tetramer, but can also propagate allosterically to the octamer interactions with DNA. In support of this hypothesis, our previous work revealed a decrease in strength of the H3/H4 interaction with DNA upon H2A.Z incorporation (34).
By simultaneously measuring the nucleosome’s position and the presence of a bound TF at the binding site for Egr-1, we have shown that the spontaneous movements of the nucleosome, and in particular the long repositioning events typical for H2A.Z nucleosomes, modulate the exposure of the Egr-1 binding site and thus modulate TF binding to the DNA (Fig. 5). In the context of transcriptional regulation, this may serve as a means to fine-tune the basal accessibility, as opposed to the more drastic effect that results from the creation of a nucleosome-depleted region. Facilitated by the nucleosome’s intrinsic properties and independent of other factors, this mechanism may control the initial binding of a pioneer TF to recruit remodeling machinery to a specific DNA locus. Interestingly, this possibility is in line with previous studies that demonstrated that H2A.Z nucleosomes are necessary for the recruitment of pioneer TFs (26, 52, 53). Specifically for the Lhb gene, the binding site of Sf-1, located at −119/−127 and responsible for the gonadotrope differentiation and basal activation of Lhb (54), also resides inside the nucleosome. It is possible that the pioneer binding of Sf-1 is facilitated by the exposure of this binding site, induced by the mobility of the nucleosome.
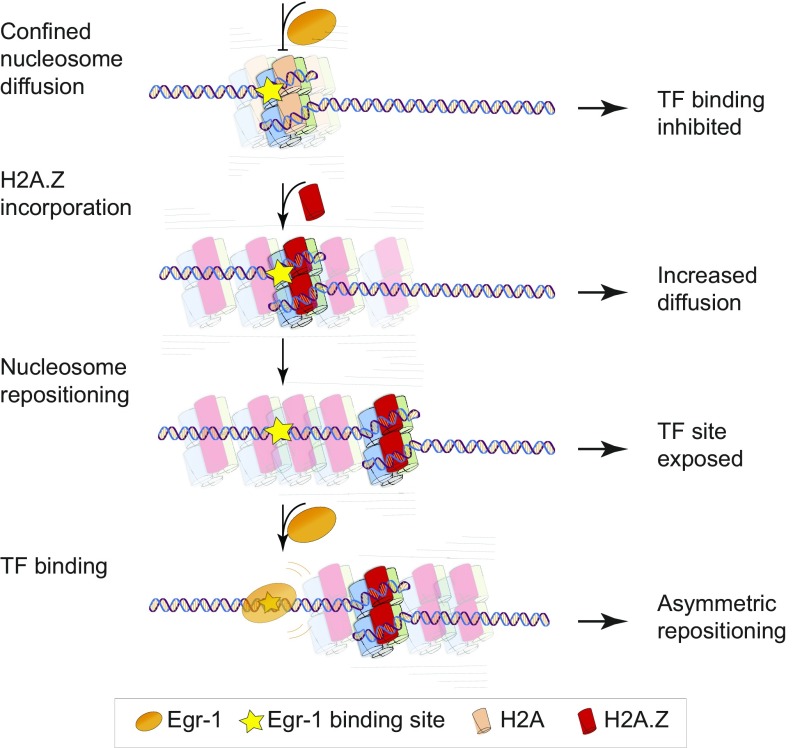
Fig. 5.
A model for the interplay between H2A.Z-induced diffusion and TF binding. Nucleosomes undergo confined diffusion on the DNA and limit binding of a TF. The incorporation of the histone variant H2A.Z serves to partially relieve this confinement, via a different type of thermal diffusion. The increased diffusion leads to exposure of the TF’s binding site and facilitates its association with the DNA. The bound TF then restricts nucleosomal movements upstream, thus asymmetrically biasing the repositioning downstream.
We observed that TF binding biases the direction of the mobility away from the binding site. Although it is possible that this is the result of steric interactions between the nucleosome and the bound TF, it is also possible that binding of the TF suppresses the formation of loops or twists by locally modifying the mechanical properties of DNA. In any case, the effect of TF binding on the mobility suggests that binding can serve as an autoregulatory mechanism to further increase TF accessibility. Moreover, this can provide the basis for nucleosome-mediated cooperativity in the binding of different TFs, as binding of one TF may modify the accessibility of another TF binding site. Together, this makes the use of a mobile nucleosome a more versatile and potent regulation mechanism than simply shifting the nucleosome’s position.
In a wider perspective, mobile nucleosomes may be exploited to facilitate distinct outcomes when incorporated into specific genomic regions during different biological contexts. H2A.Z was shown to localize at regions other than gene promoters, among them enhancer regions (54), regions of heterochromatin (29), and DNA damage sites (55). As these processes require extensive DNA accessibility to maintain important cellular activities, it is likely that H2A.Z incorporation plays a role in establishing the long-term changes that self-sustain the basal accessibility of regions of DNA for prolonged periods. Finally, the incorporation of an essential H2A.Z in the +1 position in virtually all eukaryotes (30) suggests that its specific physical properties may fulfill a role also in transcriptional elongation, and that the usage of nucleosome mobility for regulation is a general and evolutionarily conserved mechanism.
Experimental Procedures
The measurements were performed using high-resolution optical tweezers (37). Detailed experimental procedures are described in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge support from the Israel Science Foundation (Grants 1782/17 and 1902/12 [to A.K.] and 1850/17 [to P.M.]) and the J. S. Frankford Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815424116/-/DCSupplemental.
References
- 1.Lorch Y., LaPointe J. W., Kornberg R. D., Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Wray G. A., et al. , The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Bintu L., et al. , The elongation rate of RNA polymerase determines the fate of transcribed nucleosomes. Nat. Struct. Mol. Biol. 18, 1394–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyagi A., Ando T., Lyubchenko Y. L., Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry 50, 7901–7908 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Bilokapic S., Strauss M., Halic M., Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 25, 101–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polach K. J., Widom J., Mechanism of protein access to specific DNA sequences in chromatin: A dynamic equilibrium model for gene regulation. J. Mol. Biol. 254, 130–149 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Li G., Levitus M., Bustamante C., Widom J., Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Ngo T. T. M., et al. , Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 7, 10813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G., Widom J., Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11, 763–769 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Wei S., Falk S. J., Black B. E., Lee T.-H., A novel hybrid single molecule approach reveals spontaneous DNA motion in the nucleosome. Nucleic Acids Res. 43, e111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., et al. , Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 42, 8767–8776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges C., Bintu L., Lubkowska L., Kashlev M., Bustamante C., Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pennings S., Meersseman G., Bradbury E. M. M., Mobility of positioned nucleosomes on 5 S rDNA. J. Mol. Biol. 220, 101–110 (1991). [DOI] [PubMed] [Google Scholar]
- 15.Meersseman G., Pennings S., Bradbury E. M., Mobile nucleosomes—a general behavior. EMBO J. 11, 2951–2959 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaus A., Luger K., Tan S., Richmond T. J., Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. U.S.A. 93, 1370–1375 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaus A., Rencurel C., Ferreira H., Wiechens N., Owen-Hughes T., Sin mutations alter inherent nucleosome mobility. EMBO J. 23, 343–353 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamiche A., Kang J. G., Dennis C., Xiao H., Wu C., Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. U.S.A. 98, 14316–14321 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiessel H., Widom J., Bruinsma R. F., Gelbart W. M., Polymer reptation and nucleosome repositioning. Phys. Rev. Lett. 86, 4414–4417 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Kulić I. M., Schiessel H., Nucleosome repositioning via loop formation. Biophys. J. 84, 3197–3211 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulić I. M., Schiessel H., Chromatin dynamics: Nucleosomes go mobile through twist defects. Phys. Rev. Lett. 91, 148103 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Fathizadeh A., Berdy Besya A., Reza Ejtehadi M., Schiessel H., Rigid-body molecular dynamics of DNA inside a nucleosome. Eur. Phys. J. E Soft Matter 36, 21 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Anderson J. D., Widom J., Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 296, 979–987 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Stumme-Diers M. P., Banerjee S., Hashemi M., Sun Z., Lyubchenko Y. L., Nanoscale dynamics of centromere nucleosomes and the critical roles of CENP-A. Nucleic Acids Res. 46, 94–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber C. M., Henikoff S., Histone variants: Dynamic punctuation in transcription. Genes Dev. 28, 672–682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gévry N., Chan H. M., Laflamme L., Livingston D. M., Gaudreau L., p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21, 1869–1881 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu G., et al. , H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12, 180–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Illingworth R. S., Botting C. H., Grimes G. R., Bickmore W. A., Eskeland R., PRC1 and PRC2 are not required for targeting of H2A.Z to developmental genes in embryonic stem cells. PLoS One 7, e34848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarcinella E., Zuzarte P. C., Lau P. N. I., Draker R., Cheung P., Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell. Biol. 27, 6457–6468 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlatanova J., Thakar A., H2A.Z: View from the top. Structure 16, 166–179 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Marques M., Laflamme L., Gervais A. L., Gaudreau L., Reconciling the positive and negative roles of histone H2A.Z in gene transcription. Epigenetics 5, 267–272 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Bönisch C., Hake S. B., Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res. 40, 10719–10741 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian V., Fields P. A., Boyer L. A., H2A.Z: A molecular rheostat for transcriptional control. F1000Prime Rep 7, 01 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudnizky S., et al. , H2A.Z controls the stability and mobility of nucleosomes to regulate expression of the LH genes. Nat. Commun. 7, 12958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Y., North J. A., Rose S. D., Poirier M. G., Nucleosomes accelerate transcription factor dissociation. Nucleic Acids Res. 42, 3017–3027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall M. A., et al. , High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16, 124–129 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik O., Khamis H., Rudnizky S., Marx A., Kaplan A., Pausing kinetics dominates strand-displacement polymerization by reverse transcriptase. Nucleic Acids Res. 45, 10190–10205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch S. J., Shundrovsky A., Jantzen B. C., Wang M. D., Probing protein-DNA interactions by unzipping a single DNA double helix. Biophys. J. 83, 1098–1105 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koch S. J., Wang M. D., Dynamic force spectroscopy of protein-DNA interactions by unzipping DNA. Phys. Rev. Lett. 91, 028103 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Jiang J., et al. , Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by single-molecule unzipping force analysis. Mol. Cell 20, 771–781 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Jin J., et al. , Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat. Struct. Mol. Biol. 17, 745–752 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudnizky S., et al. , Single-molecule DNA unzipping reveals asymmetric modulation of a transcription factor by its binding site sequence and context. Nucleic Acids Res. 46, 1513–1524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shundrovsky A., Smith C. L., Lis J. T., Peterson C. L., Wang M. D., Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat. Struct. Mol. Biol. 13, 549–554 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Jeon J.-H., Metzler R., Inequivalence of time and ensemble averages in ergodic systems: Exponential versus power-law relaxation in confinement. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 85, 021147 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Tremblay J. J., Drouin J., Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone beta gene transcription. Mol. Cell. Biol. 19, 2567–2576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geertz M., Shore D., Maerkl S. J., Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc. Natl. Acad. Sci. U.S.A. 109, 16540–16545 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blossey R., Schiessel H., The dynamics of the nucleosome: Thermal effects, external forces and ATP. FEBS J. 278, 3619–3632 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Lequieu J., Schwartz D. C., de Pablo J. J., In silico evidence for sequence-dependent nucleosome sliding. Proc. Natl. Acad. Sci. U.S.A. 114, E9197–E9205 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandani G. B., Niina T., Tan C., Takada S., DNA sliding in nucleosomes via twist defect propagation revealed by molecular simulations. Nucleic Acids Res. 46, 2788–2801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suto R. K., Clarkson M. J., Tremethick D. J., Luger K., Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7, 1121–1124 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Bilokapic S., Strauss M., Halic M., Structural rearrangements of the histone octamer translocate DNA. Nat. Commun. 9, 1330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallant-Behm C. L., et al. , ΔNp63α represses anti-proliferative genes via H2A.Z deposition. Genes Dev. 26, 2325–2336 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Z., et al. , Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151, 1608–1616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melamed P., et al. , Multifaceted targeting of the chromatin mediates gonadotropin-releasing hormone effects on gene expression in the gonadotrope. Front. Endocrinol. (Lausanne) 9, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y., et al. , Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 48, 723–733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.