Significance
Despite advances in treatment options, breast cancer remains the second leading cause of cancer-related death in women. Extensive efforts are underway to develop targeted therapies for specific subsets of breast cancer patients, including those with triple-negative breast cancer. Because the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway is known to be oncogenic in tumor cells, inhibitors of this pathway are being extensively examined as candidates for targeted therapy in breast and other cancers. We demonstrate here that despite the antitumor effect of JAK/STAT inhibition on tumor cells, JAK/STAT inhibition acts on the tumor microenvironment to increase production of protumorigenic inflammatory factors, which promote therapeutic resistance. Furthermore, targeting inflammatory mediators improves responsiveness of tumors to JAK/STAT inhibition.
Keywords: breast cancer, mammary tumor, JAK, STAT, inflammation
Abstract
Tumor-associated macrophages contribute to tumor progression and therapeutic resistance in breast cancer. Within the tumor microenvironment, tumor-derived factors activate pathways that modulate macrophage function. Using in vitro and in vivo models, we find that tumor-derived factors induce activation of the Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) pathway in macrophages. We also demonstrate that loss of STAT3 in myeloid cells leads to enhanced mammary tumorigenesis. Further studies show that macrophages contribute to resistance of mammary tumors to the JAK/STAT inhibitor ruxolitinib in vivo and that ruxolitinib-treated macrophages produce soluble factors that promote resistance of tumor cells to JAK inhibition in vitro. Finally, we demonstrate that STAT3 deletion and JAK/STAT inhibition in macrophages increases expression of the protumorigenic factor cyclooxygenase-2 (COX-2), and that COX-2 inhibition enhances responsiveness of tumors to ruxolitinib. These findings define a mechanism through which macrophages promote therapeutic resistance and highlight the importance of understanding the impact of targeted therapies on the tumor microenvironment.
Tumor-associated macrophages (TAMs) contribute to tumor progression through a variety of mechanisms that include promotion of angiogenesis, tumor cell invasion, matrix remodeling, and immunosuppression (1). Macrophages have also been linked to resistance of tumors to chemotherapy (2, 3). Recent efforts have focused on developing therapeutic strategies that reduce macrophage recruitment and inhibit protumorigenic macrophage function within the tumor microenvironment (4). However, less is known regarding the impact of macrophages on therapeutic responsiveness to agents designed to target oncogenic signaling pathways in tumor cells. Because tumor cells and tumor infiltrating macrophages are exposed to similar factors, such as cytokines and growth factors, it is likely that signaling pathways that are active in tumor cells are also activated in macrophages. Given that macrophages are key components of the inflammatory environment in tumors, understanding the effects of targeted therapies on the function of these cells is important for developing strategies that enhance therapeutic responsiveness and ultimately reduce recurrence.
Macrophage polarization within the tumor microenvironment is thought to play a key role in tumor progression (5). Conventionally, macrophages are polarized toward the classical phenotype in response to IFN-γ and the alternatively activated phenotype by IL-4 (6–8). The two phenotypes, referred to as M1 and M2 (9), or M(IFN-γ) and M(IL-4) to more specifically define macrophage phenotypes based on stimulus (7), are critical in response to infection. Classically activated macrophages [M1, M(IFN-γ)] drive the proinflammatory protective cascade, while alternatively activated macrophages [M2, M(IL-4)] develop to antagonize inflammation and orchestrate the wound-healing response. While TAM polarization is often associated with an alternatively activated-like phenotype, recent studies have suggested that TAMs can express markers associated with both polarization states depending on tumor type and stage, suggesting that macrophage phenotype and function within the tumor microenvironment are complex (10–12). Signal transducer and activator of transcription (STAT) factors are important regulators of macrophage polarization. IFN-γ–activated STAT1 has been linked to M1 polarization, while STAT6 and STAT3, activated by IL-4/IL-13 and IL-10, respectively, have been linked to M2 polarization (13). Activated STAT3 has been observed in up to 30% of myeloid cells in human breast cancers (14) and has been implicated in regulating myeloid cell function in a number of tumor models. While the majority of studies support a protumor role for myeloid STAT3 (15–20), other studies have suggested that loss of myeloid STAT3 may contribute to tumorigenesis (21). These studies highlight the complexities of STAT-mediated regulation of myeloid cells during tumor initiation and growth.
Activation of the JAK/STAT pathway in tumor cells is known to contribute to tumor growth and progression. Both STAT3 and STAT5 have been shown to promote breast cancer growth and progression and the JAK/STAT pathway is being explored as a potential therapeutic target for breast cancer patients (22–29). However, the impact of inhibiting JAK/STAT signaling in cells of the tumor microenvironment has not been extensively investigated. Based on the knowledge that myeloid cells, such as macrophages, are critical components of the tumor microenvironment and that these cells are functionally modulated by the JAK/STAT pathway, there is a clear need to understand the role of JAK/STAT signaling in immune cell populations to effectively target this pathway in cancer patients.
In these studies, we demonstrate that soluble factors from triple-negative breast cancer (TNBC) cells induce STAT3 activation in macrophages. Using genetic approaches, we demonstrate that STAT3 deletion using the c-fms-iCre model leads to enhanced formation of mammary tumors. Using pharmacological approaches, we demonstrate that the presence of macrophages within mammary tumors contributes to resistance of tumors to the clinically relevant JAK inhibitor, ruxolitinib. Additionally, we find that either genetic deletion of STAT3 or treatment of macrophages with ruxolitinib results in induction of the protumorigenic factor prostaglandin synthase 2 (PTGS2)/cyclooxygenase-2 (COX-2), which contributes to resistance of tumor cells to JAK inhibition. Furthermore, we provide evidence that the COX-2 inhibitor, celecoxib, enhances the efficacy of ruxolitinib in mammary tumors. Taken together, these studies suggest that inhibition of JAK/STAT signaling in myeloid cells leads to altered expression of factors that contribute to therapeutic resistance, and suggest that combination therapies that target these factors may enhance the efficacy of JAK inhibitors in breast cancer.
Results
Tumor Cell-Derived Factors Activate STAT3 in Macrophages.
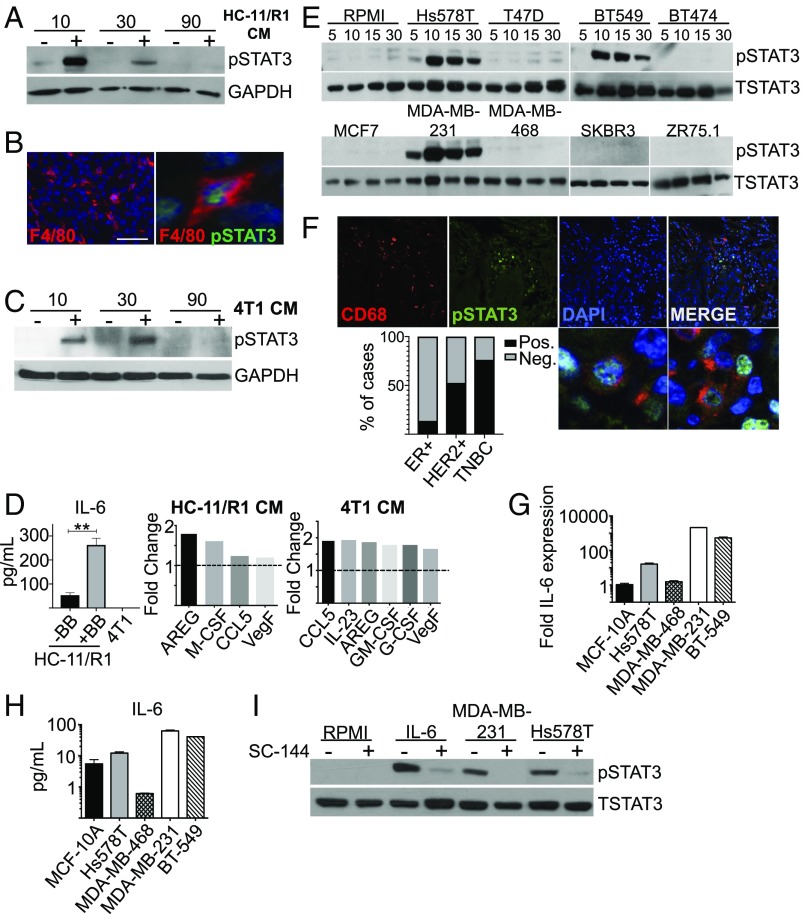
We have previously demonstrated that activation of fibroblast growth factor receptor 1 (FGFR1) in breast cancer cells leads to production of IL-6 family cytokines, which promotes tumor cell proliferation, survival, and migration in a STAT3-dependent manner (30). Furthermore, activation of FGFR1 in mammary epithelial cells leads to macrophage recruitment and activation (31, 32). Therefore, we hypothesized that activation of FGFR1 would lead to the production of soluble factors that activate STAT3 in macrophages. Initial studies were performed using the previously characterized HC-11/R1 mammary epithelial cell line (33, 34). HC-11/R1 cells express a chemically inducible FGFR1 (iFGFR1) that is activated by dimerization following treatment with a B/B homodimerizer (34). Treatment of bone marrow-derived macrophages (BMDMs) with conditioned medium (CM) from B/B-treated HC-11/R1 cells led to rapid STAT3 activation (Fig. 1A), demonstrating that iFGFR1 activation in these cells leads to production of soluble factors that activate STAT3 in macrophages. To verify that STAT3 is also activated in macrophages in vivo, HC-11/R1–derived mammary tumors were analyzed by immunofluorescence. Analysis of the macrophage marker F4/80 demonstrated extensive infiltration of tumors by F4/80+ cells (Fig. 1B). Quantification of pSTAT3 staining found that ∼23% of F4/80+ cells are also pSTAT3+, suggesting that this pathway is activated in a subset of macrophages in the tumor microenvironment. To confirm these findings in another model, similar studies were performed using 4T1 cells, a well-established mammary tumor cell line that has also been shown to depend upon both FGFR and STAT3 activity for growth in vivo (35, 36). Similar to the HC-11/R1 cells, treatment of BMDMs with 4T1 cell CM led to rapid activation of STAT3 (Fig. 1C).
Fig. 1.
Tumor cell-derived factors activate STAT3 in macrophages. (A) CM from B/B or control stimulated HC-11/R1 cells was used to stimulate BMDMs for the indicated times in minutes. Immunoblot analysis of pSTAT3 and GAPDH (loading control). (B) HC-11/R1 tumor sections were stained for F4/80 (red) and pSTAT3 (green). Higher-magnification panel indicates costained cells. (Scale bar: 50 μm.) (Magnification: Right, 112 ×.) (C) 4T1 CM was used to stimulate BMDMs for the indicated times. Immunoblot analysis of pSTAT3 and GAPDH protein. (D) IL-6 ELISA of HC-11/R1 cells treated with control (−B/B) or B/B and 4T1 cells. Select cytokines from HC-11/R1 or 4T1 CM fold-change expression relative to control media condition as baseline (dotted line). **P < 0.01. (E) Immunoblot for pSTAT3 and total STAT3 (TSTAT3) protein in THP-1 cells treated with the CM from the indicated human breast cancer cells. (F) TN human breast cancer samples stained for CD68 (red) and pSTAT3 (green). Higher-magnification images show colabeled cells and bar graph depicting the percentage of ER+, HER2+, and percent of TNBC cases in which costained cells were identified. (Magnification: 40×; higher power images, 106×.) (G) qRT-PCR analysis for IL-6 mRNA in TNBC cells. (H) ELISA analysis for IL-6 protein in TNBC CM. (I) Immunoblot analysis of pSTAT3 and total STAT3 protein in THP-1 macrophages pretreated with SC-144 inhibitor or control before exposure to breast cancer cell line CM, no CM (RPMI), or 20 ng/mL recombinant IL-6 (positive control) for 15 min.
In previous studies, we have demonstrated that iFGFR1-activated HC-11/R1 cells produce IL-6 family cytokines, which contribute to STAT3 activation in epithelial cells in a gp130-dependent manner (30). Inhibition of gp130 in BMDMs led to a partial reduction in pSTAT3 following exposure to HC-11/R1 media (SI Appendix, Fig. S1A), suggesting that HC-11/R1–derived IL-6 family cytokines contribute to STAT3 activation in BMDMs, although additional factors may also contribute. The 4T1 cells produced comparatively less IL-6 as shown by ELISA (Fig. 1D). To identify additional potential STAT3-activating cytokines in HC-11/R1 and 4T1 CM, we performed a proteome profiler mouse XL cytokine array (SI Appendix, Fig. S1B). Several cytokines and growth factors were found in HC-11/R1 and 4T1 CM compared with control CM that are known to activate STAT3 in various cell types and models—such as Amphiregulin, G-CSF, GM-CSF, and VEGF (Fig. 1D)—several of which correlate with poor prognosis in patients (37–40). These data suggest that there are several cytokines and growth factors present in mouse tumor cell CM that may contribute to activation of the STAT3 signaling pathway in macrophages.
To determine whether human breast cancer cells also activate STAT3 in macrophages, differentiated THP-1 cells were incubated with CM collected from a panel of human breast cancer cell lines consisting of estrogen receptor (ER)-positive cells (MCF7, T47D, ZR75.1), HER2+ cells (BT474, SKBR3), and triple-negative cell lines (Hs578T, MDA-MB-231, MDA-MB-468, and BT549). We found that MDA-MB-231, Hs578T, and BT-549 breast cancer cell lines, which represent the TNBC subtype, rapidly and robustly activated STAT3 signaling in macrophages (Fig. 1E). In contrast, ER+ (T47D, MCF7, ZR75.1), HER2+ (BT474, SKBR3), and triple-negative MDA-MB-468 cell lines did not induce activation of STAT3 in macrophages. These findings were confirmed using human peripheral blood mononuclear cell (PBMC)-derived macrophages (SI Appendix, Fig. S1C). To determine whether STAT3 is activated in macrophages in human breast cancer samples, patient samples were costained for CD68 and pSTAT3. CD68+pSTAT3+ cells were identified in 3 of 21 (13.6%) of ER+ samples, 11 of 21 (52.5%) of HER2+, and 16 of 21 of (76.2%) of TNBC samples (Fig. 1F). These findings confirm the presence of pSTAT3+ macrophages in human breast tumors and also confirm the findings that these cells are present in the majority of TNBCs.
Further studies using a Bio-Plex cytokine assay were performed to identify the specific TNBC-derived factors that activate STAT3 in macrophages (SI Appendix, Fig. S1D). STAT3 can be activated by various cytokines in human macrophages, including IL-6 and IL-10 (41). Analysis of cytokine levels revealed increased levels of IL-6, but not IL-10, in CM obtained from TNBC cell lines compared with MCF-10A cells, which represent a normal-like breast epithelial cell line (SI Appendix, Fig. S1D). Further validation of IL-6 levels revealed the highest expression levels at both the mRNA and protein levels in the TNBC cells capable of activating STAT3 in macrophages (Fig. 1 G and H). Treatment of macrophages with a gp130 inhibitor led to reduction in pSTAT3 activation following treatment with CM from MDA-MB-231 and Hs578T cells (Fig. 1I), suggesting that human breast cancer cell-derived IL-6 family cytokines contribute to STAT3 activation in macrophages. Taken together, our findings demonstrate that both mouse and human tumor cells produce factors that activate STAT3 in macrophages.
Deletion of STAT3 in Myeloid Cells Enhances Tumor Onset and Growth.
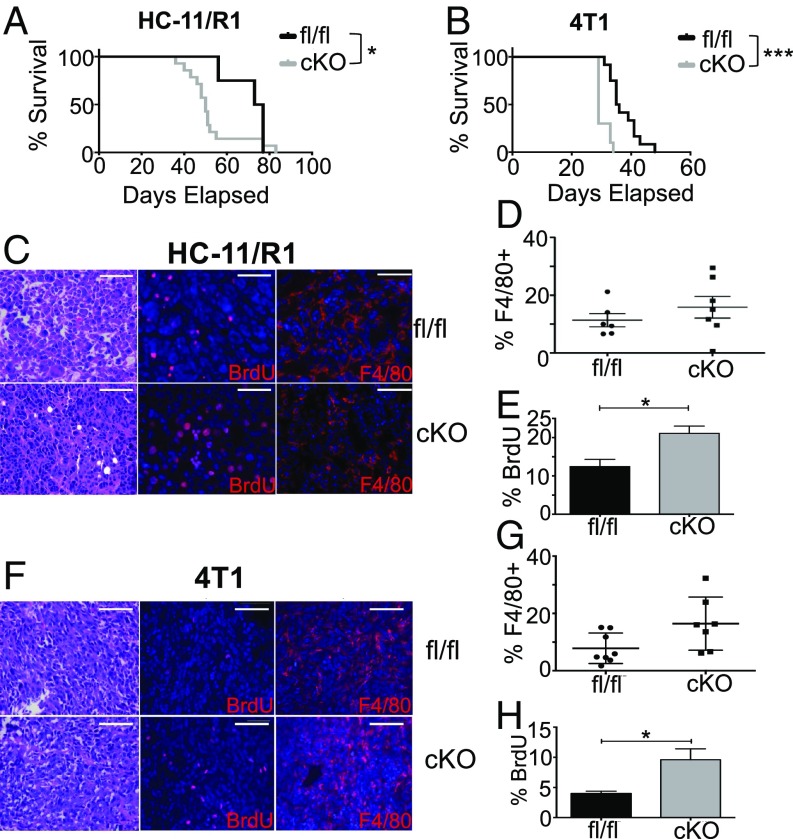
Based on our findings that tumor cell-derived factors activate STAT3 in macrophages, further studies were performed to determine the impact of myeloid STAT3 deletion on mammary tumor formation and growth. For these studies, STAT3-floxed mice were crossed with c-fms-iCre mice to generate STAT3cKO mice. Previous characterization of c-fms-iCre mice demonstrated Cre expression primarily in monocytes, macrophages, and granulocytes with lower levels of expression found in splenic T cells (21). Analysis of spleens of nontumor bearing STAT3cKO mice suggested that there are no overt alterations in the number of immune populations in these mice (SI Appendix, Fig. S2A). Analysis of total STAT3 expression in immune cell populations in HC-11/R1–derived mammary tumors by flow cytometry revealed reduced STAT3 expression in myeloid cells, including macrophages and granulocytes (SI Appendix, Fig. S2B), demonstrating selective deletion in these populations in cells within the tumor microenvironment. HC-11/R1 cells were injected into mammary fat pads of recipient STAT3fl/fl and STAT3cKO to assess the impact of myeloid STAT3 deletion on mammary tumor growth. There was an increase in the number of recipient mice that developed tumors in the STAT3cKO mice (87%) compared with littermate STAT3fl/fl controls (33%). Furthermore, overall survival in the STAT3cKO mice that developed tumors was poorer than in control mice (P = 0.046) (Fig. 2A). To confirm these findings using an additional model, orthotopic transplants were performed using 4T1 cells. These cells represent a highly aggressive tumor cell line, and tumors formed in 100% of mice of both genotypes. However, we observed more rapid tumor growth and reduced survival in the STAT3cKO mice injected with 4T1 cells compared with STAT3fl/fl littermate controls (P ≤ 0.0001) (Fig. 2B). To ensure that these phenotypes are not due to nonspecific effects of Cre recombinase expression, additional control experiments were performed in which tumor cells were injected into fat pads of c-fms-iCre mice. No differences in survival or tumor growth rate were observed when comparing mice that received tumors with or without Cre recombinase present in their myeloid lineage (SI Appendix, Fig. S2 C and D).
Fig. 2.
Deletion of STAT3 in myeloid cells enhances tumor onset and growth. (A) Kaplan–Meier curves of HC-11/R1 cells transplanted into STAT3fl/fl (n = 4) or STAT3cKO (n = 14) mice. (B) Kaplan–Meier curves of 4T1 cells transplanted into STAT3fl/fl (n = 12) or STAT3cKO (n = 10) mice. (C) HC-11/R1 or 4T1 (F) tumor sections stained for H&E, BrdU, and F4/80 from STAT3fl/fl and STAT3cKO mice. (Scale bars: 50 μm.) (D) Percentage of F4/80+ cells in HC-11/R1 or 4T1 tumors (G) among all single live cells of STAT3fl/fl and STAT3cKO mice. Means not statistically different. (E) Percentage of BrdU+ cells in HC-11/R1 or 4T1 tumors (H) among total DAPI-stained nuclei of STAT3fl/fl and STAT3cKO mice. Numbers were quantified from counts across at least five images in three mice per genotype. *P < 0.05, ***P < 0.001.
Tumors from STAT3cKO and STAT3fl/fl mice were histologically similar. Both demonstrated epithelioid tumor cells growing in a solid, vaguely alveolar pattern with pushing tumor borders. The cells had high nuclear:cytoplasmic ratios, irregular nuclear contours, and coarse chromatin with moderately sized nucleoli. Most tumors demonstrated areas of geographic necrosis with moderate to marked neutrophilic infiltrates; the extent of necrosis tended to be larger in tumors from STAT3fl/fl animals in comparison with STAT3cKO mice. Areas of myxoid degeneration were often present adjacent to the necrosis. Analysis of BrdU incorporation revealed increased rates of cell proliferation in tumors generated in the STAT3cKO hosts compared with their respective controls [Fig. 2 C and E (P = 0.0258), Fig. 2 F and H (P = 0.0103)]. Further analysis by immunofluorescence and flow cytometry demonstrated that F4/80+ cells represented a similar percentage of total cells between STAT3fl/fl and STAT3cKO tumors, suggesting that macrophage recruitment is not impacted by loss of STAT3 function (Fig. 2 C, D, F, and G). These findings suggest that STAT3 deletion in myeloid populations leads to enhanced mammary tumor formation associated with increased rates of proliferation within the tumors.
Macrophage Depletion Enhances the Efficacy of Ruxolitinib in Mammary Tumor Models.
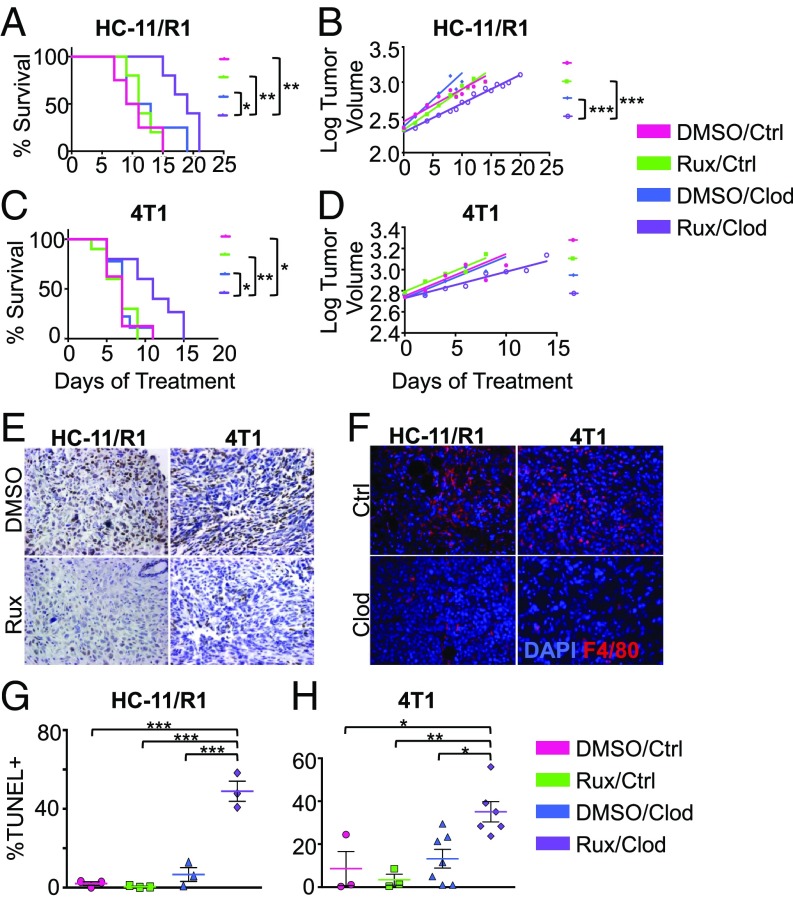
Based on the results that genetic deletion of STAT3 in myeloid cells enhances mammary tumorigenesis, we hypothesized that loss of STAT3 activity in myeloid cells would result in reduced responsiveness of tumors to STAT inhibition. To inhibit the STAT pathway pharmacologically, further studies focused on the JAK inhibitor, ruxolitinib, which is being examined in clinical trials for breast and other cancers (22, 24, 26, 27). HC-11/R1 or 4T1 cells were injected into the mammary fat pads of wild-type BALB/c mice and once tumors were established, mice were treated with ruxolitinib. We found that treatment of mammary tumor-bearing mice with ruxolitinib did not impact survival (Fig. 3 A and C), which is consistent with other studies (42). Reduced pSTAT3 staining was found in tumor sections from mice that received ruxolitinib treatment (Fig. 3E), demonstrating drug efficacy within the tumors. To determine whether macrophages impact responsiveness of tumors to ruxolitinib, mice were treated with either clodronate liposomes to deplete phagocytic cells or empty liposomes as a control. Analysis of tumor sections for F4/80+ cells demonstrated effective macrophage depletion in clodronate liposome-treated mice (Fig. 3F). While treatment with clodronate liposomes alone did not impact survival, administration of clodronate liposomes in conjunction with ruxolitinib led to a significant increase in overall survival in both tumor models (Fig. 3 A and C). When comparing the log-transformed tumor growth rates among treatment groups, there was a significant difference between the Rux/Clodronate group with both DMSO/Clodronate and Rux/Control groups in HC-11/R1–bearing mice (Fig. 3B). The growth rates were not different between the DMSO/Control and Rux/Clodronate groups; however, the tumor volumes at any given point were significantly different. These results suggest that macrophages may be more important at the very early stages of tumor development and that once tumors reach a certain size, the growth rates progress similarly. The same interpretation applies to 4T1-bearing mice. While the growth rates were not statistically significantly different, there was a slowing of tumor growth in the Rux/Clodronate group compared with the other groups (Fig. 3D). Although analysis of BrdU incorporation revealed no statistically significant differences in proliferation among the treatment groups (SI Appendix, Fig. S3), apoptosis as determined using a TUNEL assay was significantly enhanced in the mice randomized to Rux/Clodronate compared with the control or single-treatment groups (Fig. 3 G and H). Taken together, these data suggest that macrophages promote resistance of mammary tumors to ruxolitinib.
Fig. 3.
Macrophage depletion enhances the efficacy of ruxolitinib in mammary tumor models. (A) Kaplan–Meier curves of HC-11/R1 or 4T1-transplanted (C) WT BALB/c mice treated with DMSO or Rux with Control (n = 4–5 mice per group) or Clodronate liposomes (n = 8–10 mice per group). (B) Tumor volume (log-transformed) and linear regression trendlines for each treatment group of HC-11/R1 or 4T1 (D) tumor-bearing mice. (E) Immunohistochemistry for pSTAT3 (brown) on HC-11/R1 or 4T1 tumor-bearing mice treated with DMSO/Control liposomes (vehicle) and Rux/Control liposomes. (F) Immunofluorescence of F4/80+ cells (red) in HC-11/R1 (Left) or 4T1 (Right) tumor-bearing mice treated with DMSO/Control liposomes and DMSO/Clodronate liposomes. (G) TUNEL staining from each treatment group in the HC-11/R1 or 4T1 (H) tumor-bearing mice. *P < 0.05, **P < 0.01, ***P < 0.001. Images were taken at 40× magnification.
Inhibition of JAK Activity in Tumor-Associated Macrophages Leads to Increased Resistance of Tumor Cells to Ruxolitinib Treatment.
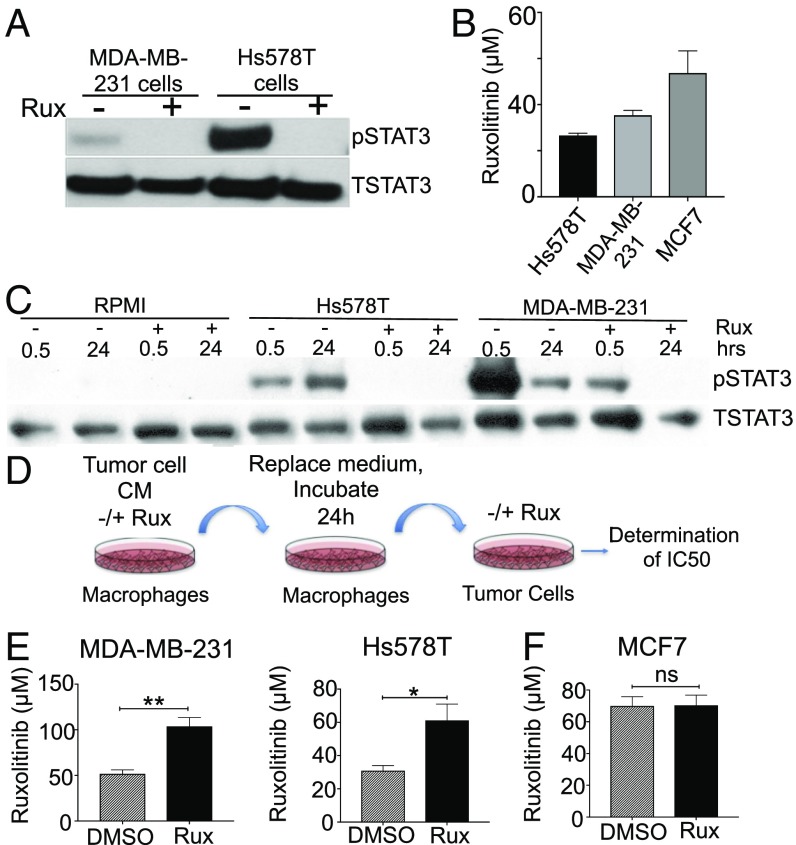
As shown in Fig. 2, genetic deletion of STAT3 signaling in myeloid cells enhances mammary tumor onset and growth. Furthermore, pharmacological inhibition of JAK/STAT signaling in mammary tumors, where both tumor cells and cells within the stroma are exposed to inhibitor, results in no impact on overall survival when macrophages are present (Fig. 3). Taken together, these findings suggest that the efficacy of ruxolitinib on the tumor cells is dampened by the presence of macrophages within the tumor microenvironment, possibly due to the production of protumorigenic factors by macrophages following loss of STAT3 activity. To assess this possibility, an in vitro assay was developed to determine whether inhibition of the JAK/STAT pathway in macrophages induces the production of soluble factors that act in a paracrine manner on tumor cells to reduce their responsiveness to ruxolitinib. MDA-MB-231 and Hs578T cells both exhibit basal levels of JAK-dependent STAT3 activation (Fig. 4A). Treatment of these cells with ruxolitinib resulted in reduced viability at concentrations comparable to those observed in TNBC cell lines in other studies (Fig. 4B) (43). Because STAT3 can be activated in a JAK-independent manner (44), studies were performed to confirm that ruxolitinib effectively inhibits STAT3 activation in macrophages following treatment with TNBC CM in THP-1 cells and human primary macrophages (Fig. 4A and SI Appendix, Fig. S4A). Further in vitro studies were performed to determine whether treatment of macrophages with ruxolitinib induces the production of factors that interfere with the ability of ruxolitinib to inhibit tumor cell growth. For these studies, THP-1 cells were treated with tumor cell CM in the presence of either ruxolitinib or solvent control. After 24 h, we replaced the medium with serum-free RPMI to collect the factors secreted by these TAMs alone and to eliminate potential confounding effects of soluble factors present in the original tumor cell CM. This collected supernatant is referred to as either DMSO TAM CM or Rux TAM CM. We then determined whether the DMSO TAM CM and Rux TAM CM impacted the ability of ruxolitinib to reduce tumor cell viability. Tumor cells required approximately twofold more ruxolitinib to achieve 50% cell death following treatment with Rux TAM CM compared with cells treated with DMSO TAM CM (P = 0.0081 and P = 0.0427 for MDA-MB-231 and Hs578T, respectively) (Fig. 4E). These findings were also observed in human primary macrophages (see, for example, Fig. 7 A and B). When using CM from MCF7 cells, which does not strongly stimulate STAT3 activity in macrophages (Fig. 1E), we observed no differences between DMSO TAM CM and Rux TAM CM conditions in response to ruxolitinib treatment (Fig. 4F), suggesting that the effects of ruxolitinib are specific for JAK/STAT-activated macrophages. These findings demonstrate that treatment of macrophages with ruxolitinib results in increased production of soluble factors that act on tumor cells in a paracrine manner to promote resistance to ruxolitinib.
Fig. 4.
Inhibition of JAK activity in tumor-associated macrophages leads to increased resistance of tumor cells to ruxolitinib treatment. (A) Immunoblot analysis of pSTAT3 and total STAT3 protein in MDA-MB-231 and Hs578T TNBC cells. (B) Rux IC50 (μM) assay at 48 h in Hs578T, MDA-MB-231, and MCF7 (ER+) breast cancer cells. (C) Immunoblot analysis of pSTAT3 and total STAT3 of THP-1 macrophages treated with RPMI, Hs578T, or MDA-MB-231 CM with or without Rux for 30 min and 24 h. (D) Schematic of tumor cells treated with TAM CM (Rux or DMSO) to assess the IC50 of the inhibitor. (E) IC50 concentration (μM) of ruxolitinib required for MDA-MB-231 (Left), Hs578T (Right), and MCF7 (F) cells following exposure to the indicated CM. *P < 0.05, **P < 0.01, ns, not significant.
Fig. 7.
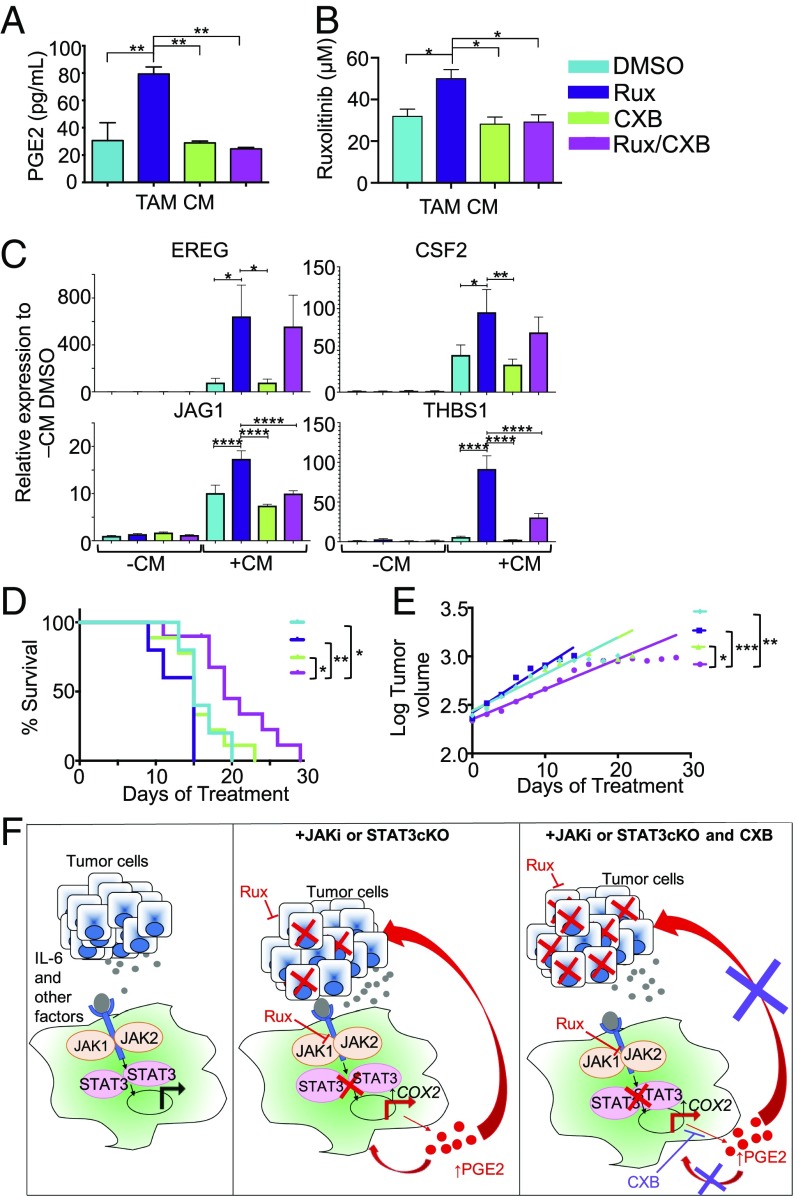
Celecoxib in combination with ruxolitinib leads to a decrease in tumor growth rate and an increase in overall survival. (A) PGE2 ELISA in DMSO, celecoxib (CXB), Rux, and ruxolitinib plus celecoxib (Rux/CXB) TAM CM (TAMs from human primary macrophages originally treated with Hs578T CM). (B) Ruxolitinib concentration required for 50% cell death of Hs578T cells in the presence of DMSO TAM CM, CXB TAM CM, Rux TAM CM, or Rux/CXB TAM CM. (C) qRT-PCR analysis for EREG, CSF2, THBS1, and JAG1 in human primary macrophages in either control or tumor CM with DMSO, Rux, CXB, or Rux/CXB. (D) Kaplan–Meier curves of 4T1-transplanted wild-type BALB/c mice treated with DMSO, CXB, Rux, or Rux/CXB (n = 5–10 mice per group). (E) Tumor volume (log-transformed) and linear regression trendlines of 4T1 tumor-bearing mice. (F) Graphical abstract illustrating the key concepts that tumor cell-derived factors, including IL-6 family cytokines, contribute to activation of STAT3 signaling in macrophages (Left). Inhibition of STAT3, either genetically or pharmacologically, leads to increased production of PGE2, which potentially acts via autocrine and/or paracrine mechanisms on tumor cells and macrophages to promote therapeutic resistance (Center). Targeting PGE2 production with celecoxib enhances responsiveness of tumor cells to ruxolitinib (Right). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ****P < 0.0001.
JAK Inhibition in Macrophages Induces Expression of a Subset of Protumorigenic Factors.
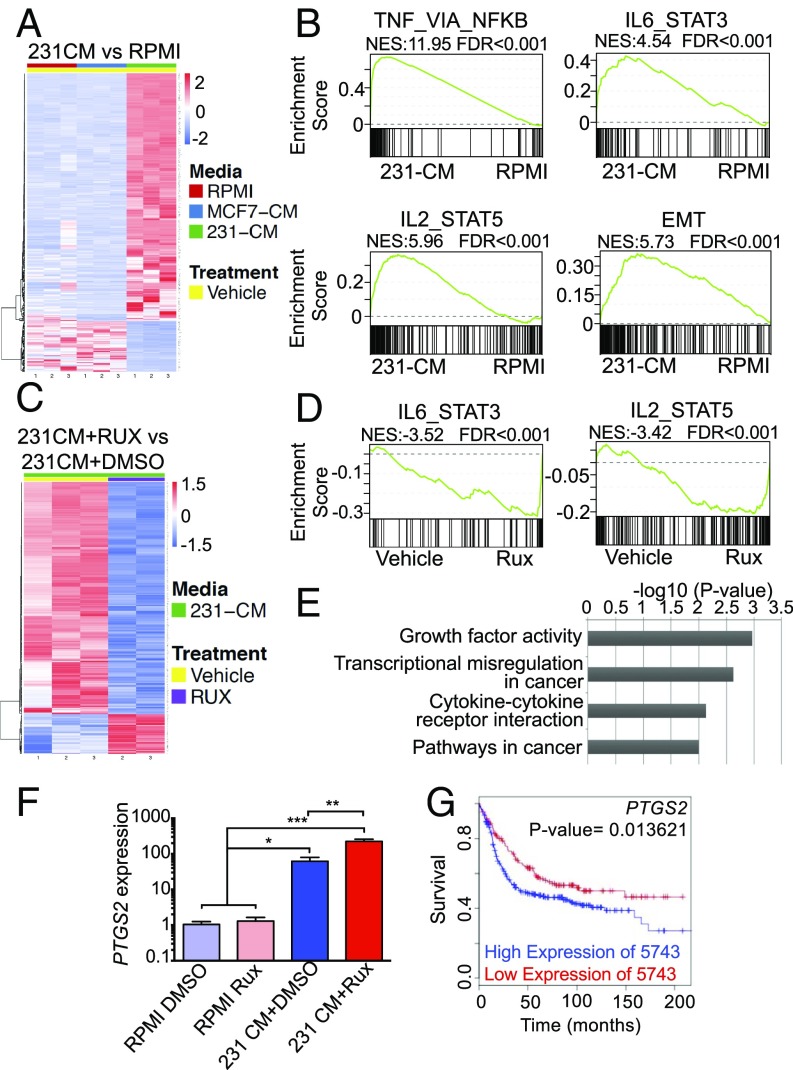
RNA-sequencing (RNA-seq) studies were performed to identify specific genes that are regulated following ruxolitinib treatment in macrophages (45). Human primary macrophages were stimulated for 2 h with CM from MDA-MB-231 or MCF7 cells containing either ruxolitinib or solvent and total RNA was harvested for RNA-seq. Analysis of differentially expressed genes revealed that treatment of macrophages with CM obtained from MDA-MB-231 cells led to regulation of numerous genes that were not similarly regulated in macrophages treated with CM from MCF7 cells, as observed in both the heat map and principal components analysis (Fig. 5A and SI Appendix, Fig. S5A). Further gene set enrichment analyses (GSEA) comparing macrophages treated with MDA-MB-231 CM vs. control medium demonstrated enriched expression of genes associated with the IL-6/STAT3 pathway (Fig. 5B), as expected based on our findings that CM from these cells activates this pathway (Fig. 1E). GSEA analysis also revealed genes associated with the IL-2/STAT5 pathway in macrophages (Fig. 5B), and immunoblot analysis confirmed activation of STAT5 following treatment of macrophages with tumor cell CM (SI Appendix, Fig. S4), providing additional validation for the RNA-seq analysis. Additional pathways that are established regulators of macrophage function, such as TNFA/NFκB, Myc, and IFN (9, 46), were also increased in macrophages treated with MDA-MB-231 CM (Fig. 5B and SI Appendix, Fig. S5B). Interestingly, genes associated with epithelial-mesenchymal transition (EMT) were enriched after treatment with MDA-MB-231 CM (Fig. 5B), many of which are soluble factors that promote EMT and invasion in a paracrine manner, such as IL6, CXCL1, MMP1, and VEGFA (47–50).
Fig. 5.
Ruxolitinib enhances the expression of a subset of protumorigenic factors in macrophages. (A and B) Heatmap of differentially regulated genes and significant GSEA pathways comparing human primary macrophages treated with RPMI or MDA-MB-231 CM. FDR < 0.05, log2FC > 3.5. (C and D) Heatmap of differentially regulated genes (FDR < 0.05, log2FC ≥ 0.5) and significant GSEA pathways in macrophages treated with MDA-MB-231 CM in the presence of either Rux or DMSO. (E) Gene ontology analysis showing pathways altered based on genes that are induced in 231CM+Rux samples. (F) qRT-PCR analysis for PTGS2 expression in macrophages treated with control or tumor CM in the presence of absence of ruxolitinib. *P < 0.05, **P < 0.01, ***P < 0.001. (G) Kaplan–Meier curve of basal subtype patient samples stratified by high or low expression of PTGS2 as determined by the MTCI Breast Cancer Survival Analysis Tool.
These findings are consistent with other studies demonstrating that MDA-MB-231 cells induce tumor-promoting phenotypes in macrophages (51). While some genes in this pathway were down-regulated following ruxolitinib treatment, the EMT gene set was not uniformly altered by ruxolitinib treatment (SI Appendix, Fig. S5D), suggesting that tumor cell-induced signaling pathways other than the JAK/STAT pathway likely contribute to regulation of this gene set. Numerous studies have suggested that breast cancer cells drive alternative activation/M2 polarization of macrophages. However, analysis of established human M1/M2 markers revealed induction of genes associated with both polarization states (SI Appendix, Fig. S5A), suggesting that soluble factors from MDA-MB-231 cells induce a mixed polarization phenotype in macrophages. These findings are consistent with recent studies demonstrating that tumor-associated macrophages exhibit a mixed phenotype in vivo (12, 52). Overall, these results demonstrate that multiple signaling pathways are activated in macrophages following exposure to tumor cell-derived factors, which may have both pro- and antitumor effects on macrophage function, highlighting the complex nature by which tumor cells regulate macrophage phenotype.
To identify genes that are specifically regulated by the JAK pathway, a comparison of macrophages treated with MDA-MB-231 CM in the presence or absence of ruxolitinib was performed. These studies found that ruxolitinib treatment led to a reduction in the expression levels of numerous genes (Fig. 5C). GSEA demonstrated that ruxolitinib specifically reduced expression of pathways induced by MDA-MB-231 CM, including the IL-6/STAT3 and IL-2/STAT5 pathways demonstrating efficacy of ruxolitinib as expected (Fig. 5D and SI Appendix, Fig. S5C). Interestingly, ruxolitinib treatment also induced expression of a subset of genes (Fig. 5C and SI Appendix, Fig. S6B). Gene ontology analysis revealed that the genes induced by ruxolitinib are enriched in cancer-associated pathways (Fig. 5E). Using the MTCI Breast Cancer Analysis Survival Tool (53), we determined whether the genes induced by ruxolitinib are associated with reduced disease-free survival specifically in breast cancer patients with PAM50-defined basal breast cancer. We recognize that the gene lists used in these analyses are from breast cancer samples that contain both parenchyma and stroma, rather than tumor-associated macrophages specifically, but we reasoned that if high levels of a gene are associated with poor outcome, this would be consistent with potential tumor-promoting function. Of the genes found to be significantly induced by ruxolitinib [false-discovery rate (FDR) < 0.05, log2 fold-change (FC) ≥ 0.5], genes were found to be significantly associated with reduced survival included THBS1, EREG, JAG1, PID1, and GPR157 (SI Appendix, Fig. S6B). These findings demonstrate that treatment of tumor cell-conditioned macrophages with ruxolitinib results in increased expression of a subset of genes with potential tumor-promoting function.
Reduced JAK/STAT Activity Correlates with Increased PTGS2 Expression.
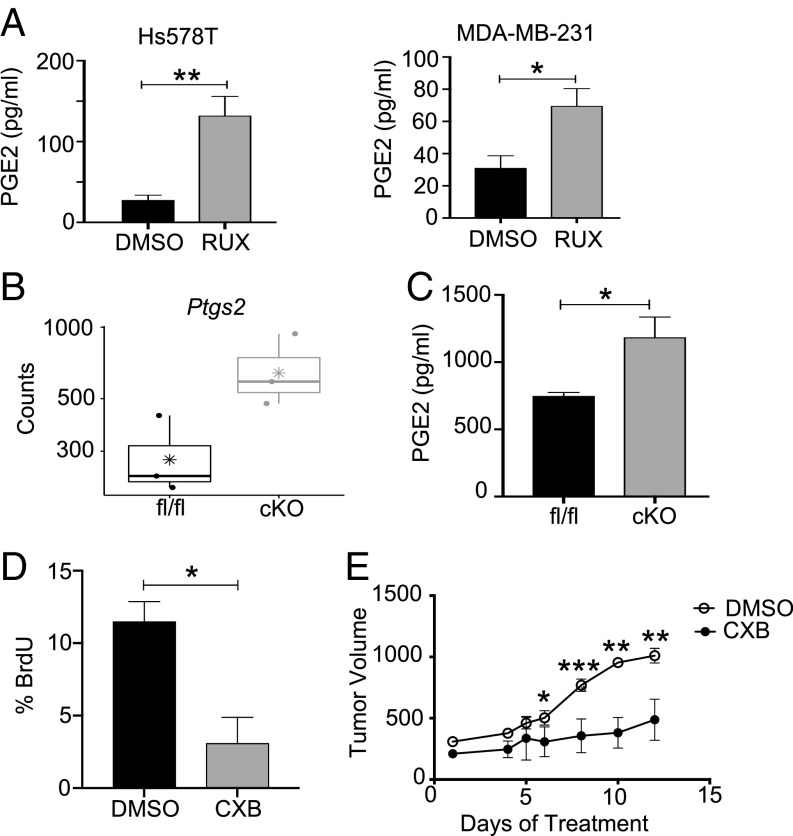
To identify potentially targetable pathways, we further analyzed the dataset for genes associated with pathways for which clinically relevant inhibitors are available. Analysis of our gene list revealed increased expression of PTGER2, the receptor for prostaglandin E2 (PGE2), in response to ruxolitinib. Further analysis of the data also demonstrated increased expression of PTGS2, which encodes for COX-2, the key driver of PGE2 production (SI Appendix, Fig. S6 B and C). This was confirmed by qRT-PCR analysis in an independent validation experiment (Fig. 5F). Analysis of PTGS2 expression demonstrated reduced disease-free survival in patients with basal breast cancer (Fig. 5G), demonstrating its potential as a tumor-promoting factor in this breast cancer subtype. Consistent with the increase in PTGS2 expression, increased PGE2 levels were found in the CM obtained from PBMC-derived macrophages treated with tumor cell CM and ruxolitinib compared with macrophages treated with tumor cell CM and solvent (P = 0.0019, P = 0.0439, respectively) (Fig. 6A).
Fig. 6.
Reduced JAK/STAT activity correlates with increased PTGS2 expression. (A) PGE2 ELISA of DMSO or Rux TAM CM from human primary macrophages treated with Hs578T CM (Left) or MDA-MB-231 CM (Right). (B) Basal Ptgs2 gene expression transcripts in STAT3fl/fl and STAT3cKO BMDMs. (C) Basal PGE2 ELISA levels in STAT3fl/fl and STAT3cKO BMDMs. (D) Percentage of BrdU+ cells of total DAPI stained nuclei from 4T1-transplanted STAT3fl/fl and STAT3cKO mice treated with DMSO or Celecoxib. Cells were counted across at least five images in three to four mice per treatment group. (E) Tumor volume (mm3) of 4T1 tumor-bearing STAT3cKO mice given treatments of DMSO (n = 4) or Celecoxib (n = 7) via daily oral gavage. *P < 0.05, **P < 0.01, ***P < 0.001.
Based on these results, we hypothesized that genetic deletion of STAT3 would also lead to increased Ptgs2 expression. Analysis of data generated by RNA-seq (45) confirmed increased Ptgs2 expression in STAT3cKO BMDMs compared with STAT3fl/fl BMDMs (Fig. 6B). Furthermore, there was a significant increase in PGE2 production by STAT3cKO BMDMs compared with STAT3fl/fl BMDMs (Fig. 6C). Based on these findings, we sought to determine whether targeting COX-2 using the COX-2 selective inhibitor celecoxib reduces tumor growth in the STAT3cKO mice. Analysis of 4T1 tumors from celecoxib-treated STAT3cKO mice revealed a significant decrease in proliferation and tumor growth compared with solvent control (Fig. 6 D and E). Notably, celecoxib does not impact 4T1 tumor growth in wild-type mice (Fig. 7E). These findings demonstrate that COX-2 activity is increased in macrophages with either pharmacologic or genetic loss of STAT3 signaling and that COX-2 activity contributes to proliferation and tumor growth observed in the STAT3cKO mice.
Celecoxib in Combination with Ruxolitinib Leads to Enhanced Therapeutic Efficacy.
Because COX-2 is associated with tumor promotion and therapeutic resistance (54, 55), we hypothesized that targeting COX-2 activity would inhibit the ability of ruxolitinib-treated macrophages to promote resistance of TNBC cells to ruxolitinib. Celecoxib effectively reduced the PGE2 production by ruxolitinib-treated human primary macrophages (Fig. 7A). Further studies using the in vitro model described in Fig. 4 demonstrated that treatment of human primary macrophages celecoxib restored responsiveness of tumor cells to ruxolitinib (Fig. 7B). These findings demonstrate that treatment of tumor cell-conditioned macrophages with ruxolitinib leads to increased COX-2 activity, which contributes to therapeutic resistance of tumor cells.
Based on the observation that the receptor for PGE2, PTGER2, is increased in ruxolitinib-treated macrophages (SI Appendix, Fig. S5B), additional studies were performed to determine whether celecoxib acts in an autocrine manner to modulate the expression levels of ruxolitinib-induced protumorigenic genes in macrophages identified by RNA-seq analysis. While celecoxib treatment did not impact ruxolitinib-induced expression of CSF2 or EREG, there was a significant reduction in expression of THBS1 and JAG1 (Fig. 7C). These data also provide additional validation of key target genes identified by RNA-seq analysis and also demonstrate that induction of these genes by ruxolitinib only occurs in the context of tumor cell-derived factors. Taken together, these results suggest that macrophage-derived PGE2 can potentially act through autocrine or paracrine mechanisms to promote therapeutic resistance following treatment with ruxolitinib.
Further studies were performed to evaluate the effect of ruxolitinib and celecoxib combination therapy on tumor growth. The 4T1 tumor-bearing mice were treated with ruxolitinib and celecoxib either alone or in combination. Survival in mice from solvent control, ruxolitinib-only, and celecoxib-only treatment groups were not statistically significant from each other (Fig. 7D). However, overall survival in the combination group (Rux/CXB) was statistically significantly longer than the control groups (Fig. 7D) and tumor growth rates were slower (Fig. 7E). These data suggest that inhibition of JAK/STAT signaling in myeloid cells increases their expression of protumor factors, such as COX-2 and its enzymatic product, PGE2. Targeting these factors can suppress macrophage-driven tumor cell therapeutic resistance and improve overall survival and outcome (Fig. 7F).
Discussion
The JAK/STAT pathway is activated within tumor cells and exhibits oncogenic activity in breast and other cancers. Given that this pathway is often activated by soluble factors within the tumor microenvironment, we hypothesized that this pathway is also activated in tumor-infiltrating myeloid cells. Therefore, we investigated the ability of mammary tumor cells to activate STAT signaling in macrophages. Our results demonstrate that tumor cell-derived soluble factors, including IL-6 family cytokines, induce robust activation of STAT3. Analysis of human breast cancer samples demonstrated the presence of CD68+pSTAT3+ cells in both HER2+ and TNBC populations. While these studies focused on TNBC based on in vitro findings, it would be interesting to further assess STAT3 activation in macrophages using additional HER2+ models to determine whether this activation is due to tumor cell-derived factors or other mechanisms. For example, fibroblasts produce cytokines such as IL-6 (56), which may also contribute to STAT3 activation in vivo. While the identification of CD68+pSTAT3+ cells is confirmatory of the findings in the mouse models, it is important to note that while CD68 is primarily considered a macrophage marker, it has been found on other cell types in breast cancers (57).
The JAK/STAT pathway regulates gene-expression changes in both tumor cells and immune cells. STAT3 activation in tumor cells contributes to tumor cell growth and inhibition of STAT3 in mammary tumor models leads to reduced tumor cell proliferation and a decrease in tumor burden (30, 58). Activated STAT3 has been observed in up to 30% of myeloid cells in human breast cancers and has been implicated in regulating myeloid cell function in tumors (14). Our results, in which STAT3 deletion is driven in myeloid lineages using the c-fms-iCre model, demonstrate that STAT3 deletion in immune populations leads to enhanced tumor onset and growth. Due to limitations with specificity of conditional myeloid deletion models (59), we are unable to distinguish between the effects of STAT3 deletion in macrophages and granulocytes in vivo. However, the identification of increased COX-2 activity in STAT3 knockout macrophages supports the conclusion that STAT3-mediated alterations in macrophages contribute to the COX-2–dependent increase in tumor growth in this model. Further studies are required to define potential contributions of STAT3 regulation of granulocytes in the tumor microenvironment. Overall, the results from these studies suggest that STAT3 deletion in the cell types that are affected by c-fms–mediated Cre expression results in a protumorigenic environment.
STAT3 has been implicated in suppressing M1/proinflammatory and enhancing M2/antiinflammatory polarization of macrophages (60). This suggests that loss of STAT3 in the myeloid cell population would induce an M1/proinflammatory environment that promotes antitumor immune responses. Indeed, inducible Mx1-cre–mediated STAT3 deletion enhances antigen presentation by dendritic cells and promotes antitumor T cell activity, resulting in reduced tumor growth in mouse models of melanoma and urothelial carcinoma (15). STAT3 in hematopoietic cells has also been found to enhance accumulation of myeloid-derived suppressor cells, promote angiogenesis, and suppress T cell activation (16, 17). Additional studies of LysM-Cre–mediated deletion of STAT3 have also demonstrated reduced tumor growth by modulating mechanisms associated with tumor cell invasion in pancreatic neuroendocrine tumors (19) and promoting T cell-mediated antitumor immune responses in models of lung and colorectal tumors (18, 20). Together, these studies have led to the general consensus in the literature that STAT3 in myeloid cells enhances immunosuppression and other protumor functions (61). In contrast, other studies of conditional STAT3 deletion have suggested that STAT3 regulation of myeloid cells may be more complex than expected. LysM-Cre–mediated deletion of STAT3 was not found to impact tumor growth in a medulloblastoma model (61). Furthermore, Deng et al. (21) reported the development of spontaneous colon tumors in the Csf1r-iCre model, suggesting that STAT3 deletion in myeloid cells enhances tumorigenesis, which is consistent with our findings in the mammary tumor models. Additionally, LysM-Cre–mediated deletion of SOCS3, a negative regulator of STAT3 activity, led to increased STAT3 activation in macrophages, reduced tumor growth, and increased survival in a mouse model of glioma (62). Finally, studies of myeloid STAT3 in the context of viral infection have demonstrated impaired activation of T cells and NK cells (63), suggesting that STAT3 regulation of myeloid cells is highly context-dependent. Loss of STAT3 in macrophages has been linked to increased production of proinflammatory cytokines, including IL-1β, IL-6, and TNF-α (64). Thus, it is possible that STAT3 deletion in myeloid cells enhances the formation of tumors that are sensitive to the tumor-promoting effects of inflammatory cytokines (21, 65) and in less immunogenic models that not as susceptible to T cell-mediated antitumor responses. Taken together, these findings suggest that STAT3 regulation of myeloid cells is complex and conflicting results in experimental models may be a result of factors such as cell-type specificity and timing of deletion, tumor type, stage of tumor development, background strain, and immunogenicity.
The JAK/STAT pathway has emerged as an attractive therapeutic target due to its high levels of activity in breast cancer cells (66). As a result, several clinical trials have started testing STAT3-specific, as well as broader JAK-targeting, inhibitors in cancer patients (67–69). However, given the importance of JAK/STAT signaling in regulating immune function, it is important to consider the potential effects of JAK/STAT inhibitors on immune populations. JAK/STAT inhibition in mammary tumor-bearing mice has been shown to inhibit NK cell proliferation, activation, and degranulation (42), which was associated with increased metastasis. Inhibition of the JAK/STAT pathway has also been shown to promote expression of VEGF from NK cells and enhance angiogenesis in hematopoietic tumors (70). However, the impact of JAK inhibition on myeloid cell populations within the mammary tumor microenvironment has not been previously investigated. Our findings demonstrate that macrophages not only contribute to therapeutic resistance of tumor cells to JAK inhibition, but that treatment of macrophages with ruxolitinib leads to increased expression of factors with known protumorigenic properties that contribute to resistance. Interestingly, genetic deletion of STAT3 in myeloid cells led to increased tumor growth, while pharmacological inhibition of JAK/STAT signaling led to no change in tumor growth or survival. Because pharmacological inhibition targets both tumor and stromal cells, these findings are consistent with the conclusion that the presence of macrophages in the tumor microenvironment blunts the ability of ruxolitinib to effectively kill tumor cells, resulting in no net change in tumor growth in ruxolitinib-treated mice. Taken together, these studies highlight the importance of understanding the impact of targeted therapies on distinct nontumor cell types within the tumor microenvironment, which provides important insights into mechanisms of therapeutic resistance.
Ruxolitinib is currently being evaluated in clinical trials for efficacy in multiple solid tumor types. Eight of these trials have already been terminated due to lack of effect and evidence of disease progression in patients receiving the inhibitor either as a single agent or in combination with chemotherapeutic agents (ClinicalTrials.gov). Two of these trials were using the inhibitor to treat patients with advanced, metastatic HER2− and TNBCs. One of these studies recruited patient cohorts based on pSTAT3 expression within triple-negative tumors and found no objective responses throughout the trial when using ruxolitinib as a single agent, despite evidence of drug efficacy within the tumor (69). Our data, in combination with other studies (42, 70), provide insights into potential mechanisms that contribute to low levels of patient response in the clinical trials and provide opportunities for combinatorial approaches to effectively enhance the efficacy of JAK-targeted therapies. Of the many factors produced from JAK-inhibited TAMs that may contribute to therapeutic resistance, we chose to further explore PGE2 as elevated levels of the COX-2/PGE2 pathway indicate poor prognosis in breast cancer patients, especially in those with TNBC (71). Furthermore, there are multiple clinically relevant and approved pharmacological inhibitors of COX-2 available in addition to celecoxib. We demonstrate in vitro that targeting COX-2 activity with celecoxib decreased PGE2 production in TAMs and that this also resulted in increased tumor cell susceptibility to ruxolitinib treatment. The mechanisms by which PGE2 can have an impact on increasing tumor cell therapeutic resistance, such as chemoresistance, have been recently studied (72). PGE2 has been shown to lead to activation of AKT via the PI3K pathway, which is associated with tumor cell survival and proliferation (73, 74). The in vivo studies described here reveal that combination therapy with celecoxib and ruxolitinib significantly extended survival compared with either drug alone, suggesting that combining these drugs may represent a therapeutically relevant approach for alleviating the protumorigenic impact of JAK inhibition on macrophages.
While JAK/STAT inhibitors are entering clinical trials and being evaluated for clinical benefit, there are knowledge gaps regarding the functional properties of STATs in nontumor cells that may impact therapeutic efficacy. Ruxolitinib treatment alone did not impact tumor growth in either wild-type or STAT3cKO mice (Fig. 7E and SI Appendix, Fig. S7), suggesting that the presence of pSTAT3+ macrophages in the tumor microenvironment may not effectively predict whether patients will respond to ruxolitinib. Instead, the extent of macrophage infiltration may be a critical determinant of responsiveness to JAK/STAT inhibitors. Determining whether lack of pSTAT3+ macrophages correlates with higher levels of COX-2 in human breast cancer samples would provide rationale for combining ruxolitinib and celecoxib in patients with high levels of macrophage infiltration regardless of the presence of pSTAT3+ macrophages. Previous studies (42, 70), together with our current findings, suggest that targeting JAK/STAT activation alone can lead to unanticipated effects on the tumor microenvironment. Careful analysis of the effects of these inhibitors on nontumor cells within the tumor microenvironment will provide rationale for combination therapy that reverse the negative impacts of JAK/STAT inhibition on immune cell function.
Materials and Methods
Mice.
FVB c-fms-iCre Tg (Csf1r-icre)1Jwp/J mice and B6.129S1-STAT3flox/flox (STAT3tm1Xyfu) mice were purchased from The Jackson Laboratory and backcrossed to the BALB/c background via the speed congenic technology provided by IDEXX RADIL. STAT3fl/fl and c-fms-iCre mice were crossed to generate conditional knockout mice (STAT3cKO). Controls include STAT3fl/fl littermates and c-fms-iCre mice. All experiments were performed with 6- to 8-wk-old female mice in specific pathogen-free environment facilities. All animal care and procedures were approved by the Institutional Animal Care and Use Committee of the University of Minnesota and were in accordance with the procedures detailed in the Guide for the Care and Use of Laboratory Animals (75).
Human Breast Cancer Samples.
Specimens and associated clinical data were obtained after approval from the University of Minnisota Institutional Review Board, as described in ref. 30. All materials were de-identified before use in this study. Sample analysis if described in the SI Appendix.
Cell Culture.
HC-11/R1 cells were obtained from Jeffrey Rosen, Baylor College of Medicine, Houston, TX, and maintained as described previously (33). BMDMs were obtained as described previously (31). The 4T1 cells were obtained from Thomas Griffith, University of Minnesota, Minneapolis, MN. MDA-MB-231, Hs578T, MDA-MB-468, BT-474, MCF7, ZR-75-1, T47D, SKBR3, BT-549, MCF-10A, and THP-1 cells were obtained from ATCC. Human primary macrophages were derived from PBMCs, as described in SI Appendix, along with all other cell culture conditions and stimulations.
Immunoblot Analysis.
Cells were lysed in RIPA buffer containing protease inhibitors and protein lysates were subjected to SDS/PAGE and immunoblot analysis as previously described (30). Antibodies are listed in SI Appendix.
In Vivo Studies.
For BALB/c mice tumor induction, 1 × 106 HC-11/R1 cells (30) or 1 × 104 4T1 cells in 50% Matrigel (BD Biosciences) were injected into the inguinal mammary fat pads of 6-wk-old mice. All mice receiving HC-11/R1 tumors received 1 mg/kg B/B Homodimerizer, intraperitoneally, twice weekly. All mice were examined for tumor development by palpation and considered tumor-bearing once tumor size reached ∼100 mm3. Once wild-type mouse tumors reached 200 mm3 (HC-11/R1) or 500 mm3 (4T1), mice were randomly assigned into one of four treatment groups: DMSO/Control liposome, Rux/Control liposomes, DMSO/Clodronate liposomes, Rux/Clodronate liposomes. For combination therapy experiments, when tumors reached 200 mm3 mice were randomized into treatment groups: DMSO (control for both inhibitors), CXB (celecoxib and DMSO), Rux (ruxolitinib and DMSO), and Rux/CXB (combination therapy). Treatments are described in SI Appendix. Tumor volume was calculated using the following equation: V = (L × W2)/2. Mice were killed when tumors reached 1,000 mm3 and survival was recorded as number of days treated until tumor size endpoint.
Quantitative RT-PCR.
RNA was extracted from cells using TriPure (Roche) and cDNA was prepared using the qScript cDNA synthesis kit (Quanta Biosciences) according to the manufacturers’ protocols. qRT-PCR was performed using PerfeCTa SYBR Green (Quanta Biosciences) and the Bio-Rad iQ5 system. The 2−ΔΔCt method was used to determine relative quantification of gene expression and normalized to cyclophilin B (CYBP). Primer sequences are in SI Appendix.
RNA-Seq Analysis.
Total RNA was collected using TriPure reagent (Roche) from primary human PBMC-derived macrophages treated with MDA-MB-231 CM in the presence of DMSO or 0.5 µM ruxolitinib, as described above or from STAT3fl/fl and STAT3cKO BMDMs. Samples were submitted in biological triplicate to the University of Minnesota Genomics Center for quality control, library creation, and next-generation sequencing. Details regarding quality control of raw data, identification of differentially expressed genes, survival analysis, and methods for GSEA are provided in SI Appendix.
Tissue Analysis.
For analysis of frozen sections, tumors were snap-frozen in OCT; 5-µm-thick sections were cut and fixed in acetone for 5 min at room temperature. Tissues sections were stained for F4/80, BrdU, and TUNEL, as described in SI Appendix. For staining of human samples, sections from a previously described tissue microarray (30) were stained for CD68 and pSTAT3, as described in the SI Appendix.
Statistical Analysis.
Experiments were performed at least three times. Box-and-whisker plots shows median (long cross) and mean (short cross) with minimum (25%) and maximum quartile (75%). Continuous outcomes were compared by treatment group using two-sided two-sample Student’s t tests, Mann–Whitney tests, or ANOVA, as appropriate. Patterns of tumor growth, on the log scale, were compared by treatment group using linear regression models; P values represent comparisons of slopes. Means ± SEs are presented unless otherwise stated. Overall survival data were summarized using Kaplan–Meier curves and compared by treatment groups using log-rank tests (GraphPad PRISM v6). P values < 0.05 were considered statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Online Supplemental Material.
Supplemental figures associated with this study include: cytokine measurement of various TNBC CM (SI Appendix, Fig. S1), immune cell markers in nontumor bearing mice (SI Appendix, Fig. S2), BrdU analysis of tumor bearing mice (SI Appendix, Fig. S3), additional immunoblot analysis of primary PBMC-derived macrophages and pSTAT5 (SI Appendix, Fig. S4), PCA and GSEA analysis of tumor CM (SI Appendix, Fig. S5), M1/M2 target genes in PBMC macrophages (SI Appendix, Fig. S6), and effects of ruxolitinib on tumors in STAT3cKO mice (SI Appendix, Fig. S7).
Supplementary Material
Acknowledgments
The authors thank Dr. Jeffrey Rosen (Baylor College of Medicine) and Dr. Thomas Griffith (University of Minnesota) for providing cell lines; and Dr. Danielle Renner (University of Minnesota) for scientific discussions related to the manuscript and data analysis. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health Award UL1TR000114. Funding was also provided by NIH/National Institute of Allergy and Infectious Diseases T32AI007313 and NIH/National Cancer Institute F31CA220746 (to E.A.I.); NIH/National Cancer Institute T32CA009138 (to C.M.L.); and NIH/National Cancer Institute R01CA215052, NIH/Eunice Kennedy Shriver National Institute of Child Health and Development R01HD095858, and Department of Defense W81XWH-16-1-0034 funding (to K.L.S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131300).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816410116/-/DCSupplemental.
References
- 1.Noy R., Pollard J. W., Tumor-associated macrophages: From mechanisms to therapy. Immunity 41, 49–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeNardo D. G., et al. , Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 1, 54–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruffell B., Coussens L. M., Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P., Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399–416 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams C. B., Yeh E. S., Soloff A. C., Tumor-associated macrophages: Unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer 2, 15025 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez F. O., Gordon S., The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 6, 13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray P. J., et al. , Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stein M., Keshav S., Harris N., Gordon S., Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 176, 287–292 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A., Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Movahedi K., et al. , Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70, 5728–5739 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Van Ginderachter J. A., et al. , Classical and alternative activation of mononuclear phagocytes: Picking the best of both worlds for tumor promotion. Immunobiology 211, 487–501 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Chevrier S., et al. , An immune atlas of clear cell renal cell carcinoma. Cell 169, 736–749.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N., Liang H., Zen K., Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front. Immunol. 5, 614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Q., et al. , The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia 15, 848–862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kortylewski M., et al. , Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 11, 1314–1321 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Kortylewski M., et al. , Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell 15, 114–123 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kujawski M., et al. , Stat3 mediates myeloid cell-dependent tumor angiogenesis in mice. J. Clin. Invest. 118, 3367–3377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou J., et al. , Myeloid STAT3 promotes lung tumorigenesis by transforming tumor immunosurveillance into tumor-promoting inflammation. Cancer Immunol. Res. 5, 257–268 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan D., Wang H. W., Bowman R. L., Joyce J. A., STAT3 and STAT6 signaling pathways synergize to promote cathepsin secretion from macrophages via IRE1α activation. Cell Rep. 16, 2914–2927 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathria P., et al. , Myeloid STAT3 promotes formation of colitis-associated colorectal cancer in mice. OncoImmunology 4, e998529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng L., et al. , A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. Am. J. Pathol. 176, 952–967 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Britschgi A., et al. , JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: A rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 22, 796–811 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Creamer B. A., et al. , Stat5 promotes survival of mammary epithelial cells through transcriptional activation of a distinct promoter in Akt1. Mol. Cell. Biol. 30, 2957–2970 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosford S. R., Miller T. W., Clinical potential of novel therapeutic targets in breast cancer: CDK4/6, Src, JAK/STAT, PARP, HDAC, and PI3K/AKT/mTOR pathways. Pharm. Genomics Pers. Med. 7, 203–215 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iavnilovitch E., Cardiff R. D., Groner B., Barash I., Deregulation of Stat5 expression and activation causes mammary tumors in transgenic mice. Int. J. Cancer 112, 607–619 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Marotta L. L., et al. , The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J. Clin. Invest. 121, 2723–2735 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Shea J. J., Holland S. M., Staudt L. M., JAKs and STATs in immunity, immunodeficiency, and cancer. N. Engl. J. Med. 368, 161–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ren S., Cai H. R., Li M., Furth P. A., Loss of Stat5a delays mammary cancer progression in a mouse model. Oncogene 21, 4335–4339 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Schmidt J. W., et al. , Stat5 regulates the phosphatidylinositol 3-kinase/Akt1 pathway during mammary gland development and tumorigenesis. Mol. Cell. Biol. 34, 1363–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohrer L. R., et al. , Activation of the FGFR-STAT3 pathway in breast cancer cells induces a hyaluronan-rich microenvironment that licenses tumor formation. Cancer Res. 74, 374–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohrer L. R., Schwertfeger K. L., Macrophages promote fibroblast growth factor receptor-driven tumor cell migration and invasion in a CXCR2-dependent manner. Mol. Cancer Res. 10, 1294–1305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwertfeger K. L., et al. , A critical role for the inflammatory response in a mouse model of preneoplastic progression. Cancer Res. 66, 5676–5685 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Xian W., Schwertfeger K. L., Vargo-Gogola T., Rosen J. M., Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model. J. Cell Biol. 171, 663–673 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welm B. E., et al. , Inducible dimerization of FGFR1: Development of a mouse model to analyze progressive transformation of the mammary gland. J. Cell Biol. 157, 703–714 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Issa A., et al. , Combinatorial targeting of FGF and ErbB receptors blocks growth and metastatic spread of breast cancer models. Breast Cancer Res. 15, R8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling X., Arlinghaus R. B., Knockdown of STAT3 expression by RNA interference inhibits the induction of breast tumors in immunocompetent mice. Cancer Res. 65, 2532–2536 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Hollmén M., et al. , G-CSF regulates macrophage phenotype and associates with poor overall survival in human triple-negative breast cancer. OncoImmunology 5, e1115177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu Y. L., et al. , Lung tumor-associated dendritic cell-derived amphiregulin increased cancer progression. J. Immunol. 187, 1733–1744 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Lu W., Chen H., Yel F., Wang F., Xie X., VEGF induces phosphorylation of STAT3 through binding VEGFR2 in ovarian carcinoma cells in vitro. Eur. J. Gynaecol. Oncol. 27, 363–369 (2006). [PubMed] [Google Scholar]
- 40.Thorn M., et al. , Tumor-associated GM-CSF overexpression induces immunoinhibitory molecules via STAT3 in myeloid-suppressor cells infiltrating liver metastases. Cancer Gene Ther. 23, 188–198 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Niemand C., et al. , Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J. Immunol. 170, 3263–3272 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Bottos A., et al. , Decreased NK-cell tumour immunosurveillance consequent to JAK inhibition enhances metastasis in breast cancer models. Nat. Commun. 7, 12258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H., et al. , Dual Aurora A and JAK2 kinase blockade effectively suppresses malignant transformation. Oncotarget 5, 2947–2961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David M., et al. , STAT activation by epidermal growth factor (EGF) and amphiregulin. Requirement for the EGF receptor kinase but not for tyrosine phosphorylation sites or JAK1. J. Biol. Chem. 271, 9185–9188 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Irey E. A., et al. , JAK/STAT inhibition in macrophages promotes therapeutic resistance by inducing expression of protumorigenic factors. NCBI GEO Database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE131300. Deposited 15 May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pello O. M., et al. , Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood 119, 411–421 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Boire A., et al. , PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 120, 303–313 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Kim M., et al. , VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene 36, 5199–5211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo P. L., Shen K. H., Hung S. H., Hsu Y. L., CXCL1/GROα increases cell migration and invasion of prostate cancer by decreasing fibulin-1 expression through NF-κB/HDAC1 epigenetic regulation. Carcinogenesis 33, 2477–2487 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Sullivan N. J., et al. , Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sousa S., et al. , Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 17, 101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Azizi E., et al. , Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madden S. F., et al. , BreastMark: An integrated approach to mining publicly available transcriptomic datasets relating to breast cancer outcome. Breast Cancer Res. 15, R52 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howe L. R., Inflammation and breast cancer. Cyclooxygenase/prostaglandin signaling and breast cancer. Breast Cancer Res. 9, 210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong D., et al. , The roles of the COX2/PGE2/EP axis in therapeutic resistance. Cancer Metastasis Rev. 37, 355–368 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Studebaker A. W., et al. , Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 68, 9087–9095 (2008). [DOI] [PubMed] [Google Scholar]
- 57.Ruffell B., et al. , Leukocyte composition of human breast cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 2796–2801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thakur R., Trivedi R., Rastogi N., Singh M., Mishra D. P., Inhibition of STAT3, FAK and Src mediated signaling reduces cancer stem cell load, tumorigenic potential and metastasis in breast cancer. Sci. Rep. 5, 10194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abram C. L., Roberge G. L., Hu Y., Lowell C. A., Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sica A., Mantovani A., Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 122, 787–795 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu H., Pardoll D., Jove R., STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 9, 798–809 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McFarland B. C., et al. , Loss of SOCS3 in myeloid cells prolongs survival in a syngeneic model of glioma. Oncotarget 7, 20621–20635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsia H. C., Stopford C. M., Zhang Z., Damania B., Baldwin A. S., Signal transducer and activator of transcription 3 (Stat3) regulates host defense and protects mice against herpes simplex virus-1 (HSV-1) infection. J. Leukoc. Biol. 101, 1053–1064 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takeda K., et al. , Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Goldberg J. E., Schwertfeger K. L., Proinflammatory cytokines in breast cancer: Mechanisms of action and potential targets for therapeutics. Curr. Drug Targets 11, 1133–1146 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Yu H., Lee H., Herrmann A., Buettner R., Jove R., Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer 14, 736–746 (2014). [DOI] [PubMed] [Google Scholar]
- 67.Oh D. Y., et al. , Phase I study of OPB-31121, an oral STAT3 inhibitor, in patients with advanced solid tumors. Cancer Res. Treat. 47, 607–615 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong A. L., et al. , Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann. Oncol. 26, 998–1005 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Stover D. G., et al. , Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer 4, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gotthardt D., et al. , STAT5 is a key regulator in NK cells and acts as a molecular switch from tumor surveillance to tumor promotion. Cancer Discov. 6, 414–429 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Kochel T. J., Goloubeva O. G., Fulton A. M., Upregulation of cyclooxygenase-2/prostaglandin E2 (COX-2/PGE2) pathway member multiple drug resistance-associated protein 4 (MRP4) and downregulation of prostaglandin transporter (PGT) and 15-prostaglandin dehydrogenase (15-PGDH) in triple-negative breast cancer. Breast Cancer (Auckl.) 10, 61–70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin M. C., Chen S. Y., He P. L., Herschman H., Li H. J., PGE2/EP4 antagonism enhances tumor chemosensitivity by inducing extracellular vesicle-mediated clearance of cancer stem cells. Int. J. Cancer 143, 1440–1455 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Greenhough A., et al. , The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis 30, 377–386 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Hill R., et al. , Cell intrinsic role of COX-2 in pancreatic cancer development. Mol. Cancer Ther. 11, 2127–2137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.