Abstract
The CRISPR (clustered regularly interspaced short palindromic repeats) and Cas9 (CRISPR associated protein 9) system has been successfully adopted as a versatile genetic tool for functional manipulations, due to its convenience and effectiveness. Genetics lesions induced by single guide RNA (gRNA) are usually small indel (insertion-deletion) DNA mutations. The impact of this type of CRISPR-induced DNA mutation on the coded mRNA transcription processing and protein translation can be complex. Unexpected or unknown transcripts, generated through alternative splicing, may impede the generation of successful loss-of-function mutants. To create null or null-like loss-of-function mutant zebrafish, we employed simultaneous multiple gRNA injection into single-cell stage embryos. We demonstrated that DNA composed of multiple exons, up to 78kb in length, can be deleted in the smarca2 gene locus. Additionally, two different genes (rnf185 and rnf215) were successfully mutated in F1 fish with multiple exon deletions using this multiplex gRNA injection strategy. We expect this approach will be useful for knock-out studies in zebrafish and other vertebrate organisms, especially when the phenotype of a single gRNA-induced mutant is not clear.
Keywords: CRISPR-Cas9, large knockout, rnf185, rnf215, smarca2, zebrafish
The CRISPR (clustered regularly interspaced short palindromic repeats) and Cas9 (CRISPR-associated protein 9) system can induce double-strand breaks in a DNA sequence-specific manner, and generate indel (insertion-deletion) mutations after non-homologous end-joining repair. Thus, it was rapidly implemented into modern biological research since the first successful demonstration in human cell lines due to its simplicity and easy use (Cong et al. 2013; Mali et al. 2013). Zinc finger nuclease and TALEN (transcription activator-like effector nucleases) are currently being phased out, due to the convenience, simplicity, and efficiency of CRISPR-Cas9, which frequently induce indel mutations around PAM sequences. Originally, only model organisms such as fruit fly, nematodes, mice, and zebrafish (Ma and Liu 2015) were used for mechanistic and evolutionary research, but with CRISPR, other non-model organisms such as axolotl and lamprey have been extended for these interests (Square et al. 2015; Flowers et al. 2014; Martin et al. 2016).
Zebrafish are a powerful model system for vertebrate embryogenesis and human diseases (Dooley and Zon 2000; Lieschke and Currie 2007). They have many advantages, such as a large number of offspring, rapid external development, early transparent embryogenesis, tractable genetics, and relatively low cost compared to murine models. They have been extensively used in developmental studies, and to study a variety of human diseases including cancers. CRISPR has been successfully adopted in the zebrafish research community and has become a cornerstone in many zebrafish research laboratories (Jao et al. 2013; Liu et al. 2019; Hwang et al. 2013). However, the size of these indel genetic lesions induced by CRISPR is usually small (e.g., deletion and/or insertion of a few nucleotides). Thus, altered mRNA processing and other unexpected transcripts may impede the successful rate of CRISPR generating zebrafish null mutants (Tuladhar et al. 2019; Anderson et al. 2017).
To definitively generate null or null-like loss-of-function zebrafish mutants, multiple exons or whole coding regions ideally should be deleted, but this approach is relatively challenging technically. It has been demonstrated that TALEN can remove up to 1Mb in zebrafish (Xiao et al. 2013; Ignatius et al. 2018). In this same paper, CRISPR was able to delete 1,423bps within the mir17a-mir92a region in mixed injected fish embryos (Xiao et al. 2013). Although TALEN can be effective for the purpose of large deletions, the tedious effector assembly makes it less efficient compared to the ease of CRISPR guide RNA (gRNA) synthesis. Therefore, generating large genomic deletions with CRISPR is the more convenient choice. The feasibility of this method was also demonstrated in a mouse knockout model (Zhang et al. 2015).
Here, we report a convenient way of creating 3 zebrafish gene mutant lines with large genomic deletions using the CRISPR-Cas9 system. We successfully created deletions up to 78kbp, as well as simultaneously mutating two genes in a single injection. We expect this approach will be useful for future zebrafish knock-out studies, especially when there is no evident phenotype associated with indel mutations induced by a single gRNA.
Materials and Methods
Zebrafish strains and husbandry
Our zebrafish were raised and maintained following the procedures described in the zebrafish book (Westerfield 2000). All experiments were carried out according to the protocols approved by PACUC (Purdue Animal Care and Use Committee). The Purdue animal housing facility is an AAALAC-approved animal facility. The wild type line used in this study is of the TAB (also called Tübingen/AB, Tub/AB, or Tu/AB) background.
Bioinformatics, CRISPR design, and gRNA synthesis
Zebrafish gene coding and transcript information were based on current zebrafish genome annotation (GRCZ11) in Ensembl. CHOPCHOP was employed for gRNA design (Montague et al. 2014). For gene-specific oligonucleotides, SP6 promoter sequence was added before the CRISPR RNA sequence followed by an overlap adaptor, which is complementary to the 5′ end of an 80bp constant oligonucleotide according to the published protocol (Gagnon et al. 2014). All the oligonucleotides (Supplementary Table 1) were synthesized by IDT (Integrated DNA Technologies). Double-strand DNAs were created by T4 DNA polymerase (New England Biolabs Inc.) after annealing both gene-specific and constant oligonucleotides. In vitro transcription was employed for gRNA synthesis using HiScribe SP6 RNA Synthesis Kit (New England Biolabs Inc.). Usually, >10µg RNA can be generated per reaction within 2-4 hr at 37°. All the gRNAs were then purified using a Zymo RNA concentrator-5 kit (Zymo Research) or MEGAclear Transcription Clean-Up Kit (TheromFisher) following manufacturer’s instructions. PCR primers upstream and downstream of the gRNA targeting sites for T7E1 assays were designed in ApE program using default settings and synthesized by IDT. All the PCR primers were optimized using gradient PCR (55-70°) with wildtype genomic DNAs. ORF (open reading frame) analyses were performed in cBioPortal (Cerami et al. 2012) and SMART (Simple Modular Architecture Research Tool) (Schultz et al. 2000).
Zebrafish embryo microinjection
Zebrafish embryo microinjections were performed as we described previously (Silic and Zhang 2018). Briefly, adult fish were separated overnight in a fish mating tank by a divider, and 1-cell stage fish embryos were collected in the early morning of the second day immediately before injection. An injection solution was prepared as following: 25ng/µl for each gRNA; 20ng/µl Cas9 Protein (PNA Bio Inc); 0.01% Phenol Red (Sigma). For each embryo, 2nL solution was injected. Dead and deformed fish embryos were removed and healthy ones were raised to adult fish as F0 founders.
Genomic DNA isolation, PCR, and T7 endonuclease 1 (T7E1) assay
Genomic DNAs (gDNA) were isolated by the Hotshot method (Truett et al. 2000). For fish embryos, 20 to 30 embryos were pooled 1 day after microinjection. Briefly, each pooled fish embryo was incubated in 100µL 50 mM NaOH at 95° for 1 hr in a thermocycler and then neutralized with 10µL 1M Tris-HCl buffer pH 8.0. For adult fish, gDNAs were isolated with the same method from caudal fin clip instead of whole fish embryos.
For each gRNA, PCRs with flanking primers (Supplementary Table 1) were performed with Taq polymerase at the optimized PCR conditions from gradient PCR tests. Generally, the annealing temperature ranged from 55-70°. To detect large deletions, forward primers from earlier exons and reverse primers from the later exons were used for PCR. The annealing temperature was decided by the overlap range of annealing temperatures of each forward and reverse primers for both targeted exons. Electrophoresis was used for checking PCR results on 1.5–2% agarose gels in sodium boric acid buffer (Brody and Kern 2004).
Guide RNAs’ mutagenesis efficiency was estimated by T7E1 assays as published with some modifications (Jao et al. 2013). Briefly, each PCR product was denatured by incubating in 95° for 10 min and then renatured to facilitate heteroduplex formation in NEB buffer 2 (New England Biolabs Inc.) (95°-85°, -2°/s, 85°, 1 min; 85°-75°,-0.3°/s, 75°, 1 min; 75°-65°,-0.3°/s, 65°, 1 min; 65°-55°,-0.3°/s, 55°, 1 min; 55°-45°,-0.3°/s, 45°, 1 min; 45°-35°,-0.3°/s, 35°, 1 min; 35°-25°,-0.3°/s, 25°, 1 min; 12°, hold). We did not perform gel purification, as the PCRs yielded clear target bands. For each reaction, 0.5μL T7E1 enzyme (5units, New England Biolabs Inc.) was added and then incubated at 37° for 1 hr in a thermocycler. Once the incubation is done, the samples were immediately examined on a 2% agarose gel.
The germline transmission rate of mutations induced by each gRNA was assessed by numbers of F1 embryos carrying a mutation of interest from the cross of F0 founders with wildtype. A pool of F1 zebrafish embryos was harvested for gDNA preparation (Truett et al. 2000). After PCR and T7E1 assay for each gRNA, the transmission rate was measured by intensity of both wildtype and mutated DNA bands according to previous reports (Guschin et al. 2010). Briefly, the gel images of T7E1 were quantified with ImageJ. Fraction cleaved was calculated as the total signal associated with the cleaved peaks divided by the sum of the signal of the cleaved bands and the un-cleaved band (wildtype). Fraction cleaved was then used to calculate the germline transmission rate using the equation, .
Gene cloning and plasmid preparation for sequencing
Adult F1 fish with mutations detected by PCR were sequenced to confirm DNA sequence changes of the targeted genes. The gDNA sequences around the targeted regions were amplified using primers for T7E1 assays by high fidelity FlashPfu DNA Polymerase (Tonk Bioscience). PCR products were purified with NucleoSpin Gel and PCR Clean-Up (Takara Bio) according to its manufactory manual. The purified PCR products were ligated with the linearized pJet1.2 vector by T4 DNA ligase from CloneJET PCR Cloning Kit (Thermo Scientific). The ligation reaction was transformed into Top10 E. coli by heat shock. Single isolated colonies were chosen for plasmid mini-preparation and Bgl II endonuclease (New England Biolabs Inc.) diagnosis since the vector has two Bgl II cutting sites at both ends. Three positive plasmid clones were sequenced at the Purdue Genomics Core Facility by T7 sequencing primer.
Data availability
Reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11541960.
Results
Zebrafish smarca2 small and large gene mutations from multiple gRNAs targeting the same gene
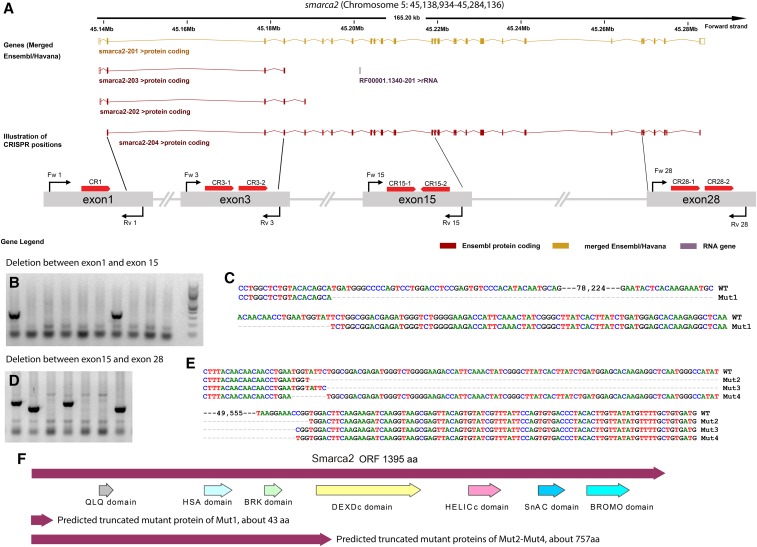
Mutations from a single gRNA are usually small indel genetic changes resulting from double-stranded DNA break repair via non-homologous end-joining mechanism. Whether these DNA mutations have an evident impact on protein translation depends on RNA splicing, RNA nonsense-mediated decay, mRNA misregulation, and other factors (Mou et al. 2017; Anderson et al. 2017; El-Brolosy et al. 2019; Ma et al. 2019; Tuladhar et al. 2019). We have found several CRISPR-induced zebrafish mutants within the early exons of a few genes that do not have any apparent phenotypes (G. Zhang, unpublished data). Thus, it is difficult to distinguish whether this phenomenon is caused by the incomplete loss-of-function of the small indel mutations, or if the gene is biologically dispensable. This is further complicated given there are only limited available commercial antibodies for zebrafish proteins. One possible solution is to delete all or most of the coding exons of the gene of interest. Especially when there are multiple transcripts coded by a gene locus such as the smarca2 gene (a.k.a. BRM, Figure 1), it could be difficult to target all the transcripts by a single gRNA. Here, we first chose smarca2 as an example, since it is a core component of SWI/SNF chromatin remodeler complex (important for embryonic development and cancer). The zebrafish smarca2 gene is located on chromosome 5. We designed 7 gRNAs to target different exons of the zebrafish smarca2 gene (Figure 1A), and simultaneously injected these 7 gRNAs into the cytoplasm of 1-cell stage zebrafish embryos: 1 gRNA targeting exon1 and 2 gRNAs targeting each of exons 3, 15, and 28. To examine the individual gRNA efficiency, we performed T7E1 assays on pooled 1-day old injected fish embryos. Indeed, we detected the expected sizes of cleaved DNA bands (Supplementary Figure 1) on agarose gels, suggesting most of these gRNAs are indeed functional although there are some noise bands likely from genetic polymorphism of the PCR amplified region.
Figure 1.
Multiple gRNAs targeting smarca2 exons induced LKO deletions in zebrafish. (A) Schematic diagram of gRNAs’ targeting smarca2 exons based on Ensembl annotation. Red bars with arrows indicate the positions of gRNAs against the smarca2 coding DNA. Red arrows match the gRNA directions. The lengths of illustrated exons are not proportional to the real size of corresponding exons. Black lines and arrows represent primers and their directions. The exon number is based on the longest transcript, smarca2-204. Note: Some of the exons may look like one exon on the graph due to their small size and close proximity. (B) Representatives of positive adult F1 fish by PCR with primers Fw 1 and Rv 15. No PCR products in wildtype due to the large size of amplicon. PCR works if there is a deletion between exon 1 and exon 15. Each lane is corresponding to an F1 adult fish. (C) Large deletion (78,288bp) between exon 1 and exon15 was confirmed by Sanger sequencing. 78,224bp is the size of the omitted region including the dashes. (D) Representatives of positive mutants identified by PCR between exon 15 and 28 with primers Fw 15 and Rv 28. Three different sized PCR bands were identified. Each lane is corresponding to an F1 adult fish. (E) Three large deletions (Mut2-Mut4) between exon 15 and exon 28 from different fish were confirmed by Sanger sequencing. Each line represents a type of mutation. 49,555bp is the size of the omitted region including the dashes. (F) Predicted zebrafish Smarca2 wildtype and mutant proteins based on the mutation positions. Protein domains were based on cBioPortal and SMART annotations. Only wildtype protein from the longest transcript (smarca2-204) is shown here. Mutant protein sizes were calculated by the ORF finder.
The remainder of F0 injected fish embryos were raised to adulthood. We randomly selected 11 adult F0 founders, and crossed them with wildtype fish for generating F1 embryos. From each founder, 20-30 fish embryos were pooled for T7E1 assays to estimate germline transmission rate of mutations induced by each gRNA (Table 1). The rest of the F1 embryos were raised to adulthood for further large knockout (LKOs) screening by PCR. As there are two different gRNAs in the targeted smarca2 exons (3, 15 and 28), small knockouts (SKOs) can occur if two gRNAs worked simultaneously on the same exon. To demonstrate this kind of SKO, individual fins were collected from F1 adult fish for genomic DNA. PCR was performed to identify small knock-out mutations between 2 nearby gRNA target sites within each exon (Figure 1A). Nine fish were identified as SKOs within exon 15, and 4 fish with SKOs were identified within exon 28. However, we could not identify any SKOs within exon 3 (Table 3).
Table 1. The F0 to F1 transmission rates of induced mutations by the smarca2 gene gRNAs.
| F0 founder | CR1 % | CR3–1 % | CR3–2 % | CR15–1 % | CR15–2 % | CR28–1 % | CR28–2 % | 1-15 LKO | 15-28 LKO | # F1 adults analyzed | # F1 adults with an LKO deletion | % F1 adults with an LKO deletion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.95 | 7.36 | 17.19 | YES | 8 | 1 | 12.50 | |||||
| 2 | 8.16 | 1.37 | 6.26 | 3.61 | YES | 8 | 4 | 50 | ||||
| 3 | 10.98 | 2.80 | 6.86 | 2.61 | 23.19 | 21 | ||||||
| 4 | 7.09 | 12.50 | 16 | |||||||||
| 5 | 6.00 | 16.63 | 14.42 | 20 | ||||||||
| 6 | 4.39 | 4.87 | 3.92 | YES | 19 | 3 | 15.80 | |||||
| 7 | 3.73 | 0.78 | 0 | |||||||||
| 8 | 3.18 | 1.08 | 1.20 | 19 | ||||||||
| 9 | 4.62 | 0.86 | 9.04 | 3.23 | 26 | |||||||
| 10 | 2.12 | 1.90 | 0.23 | 0 | ||||||||
| 11 | 2.66 | 1.05 | 1.05 | 0.16 | 17 |
Note: Empty space indicates that no or uncertain activities detected. LKO: large knockout. The percentage of mutant alleles at each gRNA target site were measured by analyzing T7E1 results from pools of F1 embryos, and the percentage of LKO deletion rate was calculated based on F1 adult fish.
Table 3. Identification of deletion mutations in F1 adults.
| Gene | Forward primer | Reverse primer | Expected PCR size bp | Mutation type | Distance bp | # F1 adults with an LKO deletion | # F1 adults analyzed | % F1 adults with an LKO deletion |
|---|---|---|---|---|---|---|---|---|
| smarca2 | Fw 1 | Rv 3 | 340-430 | — | — | 0 | 154 | 0 |
| Fw 1 | Rv 15 | ∼310 | LKO | 78, 224 | 3 | 154 | 1.95 | |
| Fw 1 | Rv 28 | 250-320 | — | – | 0 | 154 | 0 | |
| Fw 15 | Rv 28 | 260-410 | LKO | 49,644 | 5 | 154 | 3.25 | |
| Fw 3 | Rv 3 | ∼340 | — | — | 0 | 154 | 0 | |
| Fw 15 | Rv 15 | ∼240 | SKO | 77 | 11 | 154 | 7.14 | |
| Fw 28 | Rv 28 | ∼280 | SKO | 66 | 4 | 154 | 0.26 | |
| rnf185 | Fw 2 | Rv 4 | ∼240 | LKO | 3,366 | 2 | 48 | 4.2 |
| rnf215 | Fw 6 | Rv 7 | ∼320 | LKO | 1,037 | 3 | 48 | 6.25 |
Note: SKO: small knockout; LKO, large knockout.
Our above data indicated that it is likely that multiple gRNAs can work simultaneously on the same chromosome. Thus, large chromosomal deletions between smarca2 exons may also be present in adult F1 fish. We reasoned that LKOs between exon 15 and exon 28 are most likely since these two exons contain SKO mutations. Besides, we have omitted exon 3 because there were no SKO mutations detected. Therefore, we have tested all the possible combinations between, exon 1 and exon 15, exon 15 and exon 28, and exon 1 and exon 28. The forward primers of earlier exons and the reverse primers of later exons were used for PCR screening to identify LKO mutants. As expected, we were able to identify three different fish with LKO deletion (Figure 1B) between exon 1 and 15 out of 154 adult F1 fish from the 9 different F0 founders (Tables 1 and 3). In addition, 5 out of 154 fish were also identified to have LKO mutations between exon 15 and 28 (Figure 1D) from different F0 founder fish (Tables 1 and 3). Unfortunately, we could not find any fish that had LKO mutations from exon 1 to exon 28. Overall, about 5% of the total number of F1 fish contained LKO mutations from the injection of multiple CRIPSRs (Table 3). To further confirm these LKO mutations, we cloned and sequenced the PCR products. Sequencing results confirmed deletions occurred between two gRNA target sites: Mut1 (78,288bp), Mut2 (49,660bp), Mut3 (49,644bp), and Mut4 (49,528bp). (Figure 1 C & E). ORF analysis suggests truncated proteins may result from these LKOs (Figure 1F).
Simultaneous large-deletion mutations of two genes on the same chromosome from a single injection
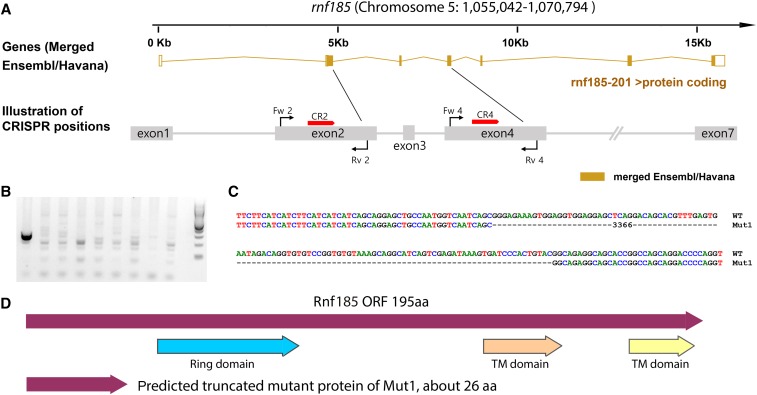
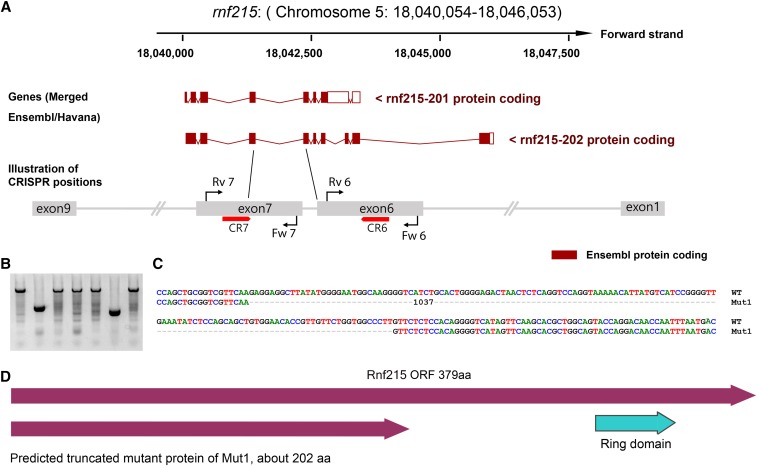
Due to the successful creation of LKOs in the zebrafish samarca2 gene, we tested the possibility of simultaneously targeting two genes, rnf185 and rnf215, which both are located on zebrafish chromosome 5, but well separated (∼18Mb). As these two genes are relatively small, we chose to target exon 2 and exon 4 of rnf185 (Figure 2A), and exon 6 and exon 7 of rnf215 (Figure 3A) by designing 1 gRNA for each exon. We then simultaneously injected all 4 gRNAs into 1-cell stage zebrafish embryos, and validated the effectiveness of the gRNAs (Supplementary Figure 2) by T7E1 assays on a portion of mixed injected fish embryos (20-30). We then raised the rest of these injected fish embryos to adulthood as F0 adult candidate founders. Nine F0 adult founders were randomly selected and the germline transmission rate of each gRNA induced mutation was evaluated by T7E1 assay on the mixed F1 embryos from the outcross with wildtype fish (Table 2). The rest of the F1 embryos from the same crosses were raised to adulthood as F1 fish for further LKO screening. As the expected deletions for these two genes are relatively smaller compared to the smarca2 gene, we randomly selected 48 F1 adult fish from the 9 F0 founders (Table 2). Adult F1 fish were screened for the presence of LKO deletion mutations for each gene by two independent PCRs using forward primers of earlier exons and reverse primers of later exons. As a result, 2 fish were found with LKO mutations between exon 2 and exon 4 in rnf185 and 3 fish between exon 6 and exon 7 in rnf215 out of 48 fish screened (Figures 2-3, Table 3). Interestingly, we identified a fish with LKOs in both genes, suggesting that it is possible to simultaneously target two genes in a single germline cell. Similar to the smarca2 LKO mutant rate, the overall efficiency was 4% for rnf185, and 6% for rnf215 (Table 3). To further validate the LKO mutations, we performed PCR by pairing the early and late exon primers and then cloned the products for sequencing. Deletions of >3kb between exon 2 and exon 4 in rnf185 (Figure 2C), and >1kb between exon 6 and exon 7 in rnf215 were confirmed (Figure 3C). ORF analysis suggests truncated proteins resulted from these LKOs (Figure 2D & 3D).
Figure 2.
LKO mutations induced by multiple gRNAs targeting the rnf185 gene in zebrafish. (A) Illustration of gRNAs that are against rnf185 exon 2 and exon 4 based on Ensemble annotation. Red bars with arrows indicate the positions of gRNAs against the rnf185 coding DNA. Red arrows match the gRNA directions. The lengths of illustrated exons are not proportional to the real size of corresponding exons. Black lines and arrows represent primers and their directions. (B) Representatives of positive adult F1 fish by PCR with primers (Fw 2 and Rv 4) that bind to exons 2 and 4 respectively. Among the 48 fish, 2 were identified to have large deletions between exon 2 and exon 4 (also see Table 1). Each lane is corresponding to an F1 adult fish. (C) The exon 2 - exon 4 deletion (3,366bp) was confirmed by Sanger sequencing. 3,366bp is the size of the omitted region including the dashes. (D) Predicted zebrafish Rnf185 wildtype and mutant protein based on the mutation positions. Protein domains were based on cBioPortal and SMART annotations. Mutant protein sizes were calculated by the ORF finder.
Figure 3.
LKO mutations induced by multiple gRNAs targeting the rnf215 gene in zebrafish. Illustration of gRNAs that are against rnf215 exon 6 and exon 7 based on Ensemble annotation. Red bars with arrows indicate the positions of gRNAs against the rnf215 coding DNA. Red arrows match the gRNA directions. The lengths of illustrated exons are not proportional to the real size of corresponding exons. Black lines and red arrows represent primers and their directions. (B) Representatives of positive adult F1 fish by PCR with primers Fw 6 and Rv 7 that bind to exon 6 and exon 7, respectively. The top band was amplified from wildtype and the lower small band was amplified from the LKO mutant. Among the 48 fish, 3 fish were identified to have large deletions between exon 6 and exon 7 (also see Table 1). Each lane is corresponding to an F1 adult fish. (C) The exon 6 – exon 7 deletion (1,037bp) was confirmed by Sanger sequencing. 1,037bp is the size of the omitted region including the dashes. (D) Predicted zebrafish Rnf215 wildtype and mutant proteins based on the mutation positions. Protein domains were based on cBioPortal and SMART annotations. Only wildtype protein from the longest transcript is shown here. Mutant protein sizes were calculated by ORF finder.
Table 2. The F0 to F1 transmission rates of induced mutations by rnf185 and rnf215 gRNAs.
| rnf185 F0 founder | CR2 % | CR4 % | LKO | # F1 adults analyzed | # F1 adults with an LKO deletion | % F1 adults with an LKO deletion |
|---|---|---|---|---|---|---|
| 1 | 9.67 | 16.90 | 5 | |||
| 2 | 9.08 | 11.08 | 5 | |||
| 3 | 15.56 | 26.76 | 5 | |||
| 4 | 9.13 | 12.30 | Yes | 5 | 1 | 20 |
| 5 | 15.57 | 20.24 | 5 | |||
| 6 | 3.19 | 25.36 | Yes | 5 | 1 | 20 |
| 7 | 18.31 | 27.49 | 5 | |||
| 8 | 3.84 | 6.75 | 5 | |||
| 9 | 25.65 | 19.06 | 8 |
| rnf215 F0 founder | CR6 % | CR7 % | LKO | # F1 adults analyzed | # F1 adults with an LKO deletion | % F1 adults with an LKO deletion |
|---|---|---|---|---|---|---|
| 1 | 3.48 | 5 | ||||
| 2 | 2.21 | 8.90 | Yes | 5 | 1 | 20 |
| 3 | 1.69 | 9.59 | 5 | |||
| 4 | 1.25 | 4.11 | 5 | |||
| 5 | 1.12 | 5 | ||||
| 6 | 0.93 | Yes | 5 | 1 | 20 | |
| 7 | 3.01 | 7.37 | Yes | 5 | 1 | 20 |
| 8 | 0.98 | 5.96 | 5 | |||
| 9 | 2.43 | 8.39 | 8 |
Note: Empty space indicates that no or uncertain activities detected. LKO: large knockout.
The percentage of mutant alleles at each gRNA target site were measured by analyzing T7E1 results from pools of F1 embryos, and the percentage of LKO deletion rate was calculated based on F1 adult fish.
Discussion
CRISPR has become an effective reverse genetic tool for zebrafish and other model organisms. Injecting multiple gRNAs simultaneously has been demonstrated to be more effective in mutagenizing zebrafish (Wu et al. 2018). Similarly, tRNA-based multiplex gRNA combined with Tol2 transposon has been reported for creating knockout zebrafish (Shiraki and Kawakami 2018). However, this has not been used for creating adult fish lines with LKOs yet, although there is a report with mouse models (Zhang et al. 2015). Additionally, a deletion of 1,423bps within the mir17a-mir92a region was reported in zebrafish embryos (Xiao et al. 2013). This ∼1.5kb deletion is still relatively small compared to the ∼1Mb deletion using TALEN in the same report, and this deletion was found in the injected F0 pooled fish embryos. Here, we demonstrate that LKOs up to 78kb can be created in adult F1 zebrafish by co-injection of multiple gRNAs that target different exons of the same gene. Moreover, two different genes on the same chromosome can be targeted simultaneously as exemplified by the rnf185 and rnf215 LKO zebrafish. Based on our data, these mutations can be identified in 5% of F1 fish. If our F0 founder fish were screened by PCR on mixed F1 fish embryos, the positive rate of LKOs could be further improved.
Due to the complex impacts of CRISPR-induced DNA mutations on mRNA transcription, splicing, and protein translation, generating indel mutations is not always an effective way to study loss-of-function phenotypes (Anderson et al. 2017; Tuladhar et al. 2019; Mou et al. 2017). In our laboratory, there are several cases where no obvious phenotypes can be observed in CRISPR generated zebrafish with various indel mutations (kcnj10a, kank1a, kank1b, mdm1, etc. unpublished data G. Zhang). Given the limited availability of zebrafish protein antibodies, it is difficult to distinguish whether single gRNA induced mutations do not lead to complete loss-of-function, or the targeted genes are simply not essential for zebrafish. The latter could be the case, since zebrafish contain paralogous genes, due to bony fish whole-genome duplication (Meyer and Van de Peer 2005; Hoegg et al. 2004). In the case of LKOs, most of the key coding exons are deleted, and this is more likely to generate severe loss-of-function mutants, especially when there are multiple RNA transcripts coded or alternative splicing events from the same DNA region. Thus, this approach is useful to solve the uncertainties surrounding a CRISPR-generated zebrafish without an obvious or strong phenotype. One caveat to this approach is there are potentially other coding elements (e.g., miRNAs) within the targeted regions. For example, the zebrafish omga gene is located within the 35th intron of the nf1b gene. Generally, co-deletion can be avoided if the genome reference is well annotated, however, this is not always the case. Another caveat is nonsense mutations caused by reading frame shift and RNA nonsense-mediated decay. Depending on the protein nature of the targeted gene, mutants can be simply null, dominant-negative, or constant-active. For example, N-terminal truncated Smarca2 may be dominant-negative since the N-terminal contains helicase and bromo domains. Thus, the LKO and indel mutants may provide us a variety of animal models to study targeted gene functions due to mutations that could lead to partial loss-of-function, null, or dominant-negative proteins. In addition, this approach is not only good for making null mutants but also can be an effective tool for studying gene promoters and other cis-regulatory DNA elements. For example, the upstream and downstream sequence of the start codon of smarca2 gene can be deleted for temporal and spatial expression change. Such gene expression changes can be examined with in situ hybridization or RT-PCR on zebrafish embryos or adult tissues.
This multiple gRNA based LKO method is relatively simple, and the directions of the gRNAs for Cas9 may not be important for generating LKOs, as both the same-orientation and opposite-orientation worked well in our experiments. Based on our experience, it seems that one effective gRNA per exon is good enough for LKO generation, but multiple gRNAs against the same DNA region may increase the cleavage rate. We did not thoroughly search for off-target mutations, but it should not be a major issue due to the following reasons: 1. All gRNAs with a single hit in the zebrafish genome were selected. 2. LKOs are a relatively low-frequency event (∼5%) and co-existence of rare off-target mutations is even less probable. 3. Out-crossing with wildtype fish for fish mutant line generation will also eliminate potential off-targets if they indeed exist. If the off-target mutation is key, Cas9 nickase could be another choice, though we did not examine it in this work.
Multiple gRNA cocktail injection into zebrafish embryos has been demonstrated for effective gene disruption in F0 fish embryos, and 4-gRNA sets have been computed for 21,386 genes (Wu et al. 2018). Interestingly, they indeed reported a low rate of small-sized site-spanning deletion for the hand2 and sox32 genes. They found that the frequency of this type of deletion decreases with increasing distance between gRNA target sites (Wu et al. 2018). However, no LKO deletion (>1kb) was reported although each gRNA showed high efficiency in the tested genes. Since generating LKO is not their primary goal, their injection method (yolk vs. cytoplasm) and mutation detecting method (high-throughput sequencing vs. PCR) may miss LKOs in their fish embryos. It is worth mentioning that, very recently, synthetic crRNA:tracrRNA duplex guide RNA was demonstrated highly efficient in zebrafish embryos (Hoshijima et al. 2019), and this could be a better choice for improving the LKO generation efficiency in the future.
Simultaneous multiple gene targeting in single microinjection is very useful for studying functionally related genes or genes in the same pathway. Evolutionarily conserved syntenies on chromosomes usually suggest there could be functional constraints, such as in the cases of CDKN2A and MTAP (Mavrakis et al. 2016), and TP53 and EIF5A-ALOX15b (Liu et al. 2016). Thus, this multiple gene LKO method could be an effective approach for cancer genetics. For example, rnf185 and rnf215 are located on the same chromosome in both human and zebrafish. Both chromosomal regions are underrepresented in human and zebrafish malignant peripheral nerve sheath tumors, suggesting they might be tumor suppressor genes. Our rnf185 and rnf215 zebrafish mutants may be used for cancer genetic studies in the future.
Acknowledgments
The research work reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under the Award Number R35GM124913. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agents. We thank Martin Silic for microinjection and manuscript proofreading and Han Han for zebrafish embryo collections.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11541960.
Communicating editor: D. Grunwald
Literature Cited
- Anderson J. L., Mulligan T. S., Shen M. C., Wang H., Scahill C. M. et al. , 2017. mRNA processing in mutant zebrafish lines generated by chemical and CRISPR-mediated mutagenesis produces unexpected transcripts that escape nonsense-mediated decay. PLoS Genet. 13: e1007105 10.1371/journal.pgen.1007105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody J. R., and Kern S. E., 2004. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. Biotechniques 36: 214–216. 10.2144/04362BM02 [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O. et al. , 2012. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2: 401–404 (erratum: Cancer Discov. doi: 10.1158/2159-8290.CD-12-0326). 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R. et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley K., and Zon L. I., 2000. Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 10: 252–256. 10.1016/S0959-437X(00)00074-5 [DOI] [PubMed] [Google Scholar]
- El-Brolosy M. A., Kontarakis Z., Rossi A., Kuenne C., Gunther S. et al. , 2019. Genetic compensation triggered by mutant mRNA degradation. Nature 568: 193–197. 10.1038/s41586-019-1064-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers G. P., Timberlake A. T., Mclean K. C., Monaghan J. R., and Crews C. M., 2014. Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development 141: 2165–2171. 10.1242/dev.105072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Akhmetova L. et al. , 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One 9: e98186 (erratum: PLoS One 9: e106396). 10.1371/journal.pone.0098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D. Y., Waite A. J., Katibah G. E., Miller J. C., Holmes M. C. et al. , 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649: 247–256. 10.1007/978-1-60761-753-2_15 [DOI] [PubMed] [Google Scholar]
- Hoegg S., Brinkmann H., Taylor J. S., and Meyer A., 2004. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J. Mol. Evol. 59: 190–203. 10.1007/s00239-004-2613-z [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Jurynec M.J., Klatt Shaw D., Jacobi A.M., Behlke M.A. et al. , 2019. Highly Efficient CRISPR-Cas9-Based Methods for Generating Deletion Mutations and F0 Embryos that Lack Gene Function in Zebrafish. Dev. Cell 51: 645–657.e4. 10.1016/j.devcel.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y. F., Reyon D., Maeder M. L., Tsai S. Q. et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius M. S., Hayes M. N., Moore F. E., Tang Q., Garcia S. P. et al. , 2018. tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. eLife 7 10.7554/eLife.37202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L. E., Wente S. R., and Chen W., 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110: 13904–13909. 10.1073/pnas.1308335110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke G. J., and Currie P. D., 2007. Animal models of human disease: zebrafish swim into view. Nat. Rev. Genet. 8: 353–367. 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- Liu K., Petree C., Requena T., Varshney P., and Varshney G. K., 2019. Expanding the CRISPR Toolbox in Zebrafish for Studying Development and Disease. Front. Cell Dev. Biol. 7: 13 10.3389/fcell.2019.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen C., Xu Z. M., Scuoppo C., Rillahan C. D. et al. , 2016. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531: 471–475. 10.1038/nature17157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D. Y., and Liu F., 2015. Genome Editing and Its Applications in Model Organisms. Genomics Proteomics Bioinformatics 13: 336–344. 10.1016/j.gpb.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Zhu P., Shi H., Guo L., Zhang Q. et al. , 2019. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568: 259–263. 10.1038/s41586-019-1057-y [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M. et al. , 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Serano J. M., Jarvis E., Bruce H. S., Wang J. et al. , 2016. CRISPR/Cas9 Mutagenesis Reveals Versatile Roles of Hox Genes in Crustacean Limb Specification and Evolution. Curr. Biol. 26: 14–26. 10.1016/j.cub.2015.11.021 [DOI] [PubMed] [Google Scholar]
- Mavrakis K. J., McDonald E. R. 3rd, Schlabach M. R., Billy E., Hoffman G. R. et al. , 2016. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science 351: 1208–1213. 10.1126/science.aad5944 [DOI] [PubMed] [Google Scholar]
- Meyer A., and Van de Peer Y., 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27: 937–945. 10.1002/bies.20293 [DOI] [PubMed] [Google Scholar]
- Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., and Valen E., 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research 42 (Web Server issue):W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H. W., Smith J. L., Peng L. T., Yin H., Moore J. et al. , 2017. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 18: 108 10.1186/s13059-017-1237-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Copley R. R., Doerks T., Ponting C. P., and Bork P., 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28: 231–234. 10.1093/nar/28.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T., and Kawakami K., 2018. A tRNA-based multiplex sgRNA expression system in zebrafish and its application to generation of transgenic albino fish. Sci. Rep. 8: 13366 10.1038/s41598-018-31476-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silic M. R., and Zhang G., 2018. Visualization of Cellular Electrical Activity in Zebrafish Early Embryos and Tumors. J. Vis. Exp. 134: e57330 10.3791/57330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Square T., Romasek M., Jandzik D., Cattell M. V., Klymkowsky M. et al. , 2015. CRISPR/Cas9-mediated mutagenesis in the sea lamprey Petromyzon marinus: a powerful tool for understanding ancestral gene functions in vertebrates. Development 142: 4180–4187. 10.1242/dev.125609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A. et al. , 2000. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29 (1):52, 54. [DOI] [PubMed] [Google Scholar]
- Tuladhar R., Yeu Y., Tyler Piazza J., Tan Z., Rene Clemenceau J. et al. , 2019. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 10: 4056 10.1038/s41467-019-12028-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M., 2000. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio), Univ. of Oregon Press, Eugene. [Google Scholar]
- Wu R.S., Lam I. I., Clay H., Duong D. N., Deo R. C. et al. , 2018. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell 46: 112–125.e4. 10.1016/j.devcel.2018.06.003 [DOI] [PubMed] [Google Scholar]
- Xiao A., Wang Z., Hu Y., Wu Y., Luo Z. et al. , 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res. 41: e141 10.1093/nar/gkt464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jia R., Palange N. J., Satheka A. C., Togo J. et al. , 2015. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One 10: e0120396 10.1371/journal.pone.0120396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25387/g3.11541960.