Abstract
The genus Oryzias consists of 35 medaka-fish species each exhibiting various ecological, morphological and physiological peculiarities and adaptations. Beyond of being a comprehensive phylogenetic group for studying intra-genus evolution of several traits like sex determination, behavior, morphology or adaptation through comparative genomic approaches, all medaka species share many advantages of experimental model organisms including small size and short generation time, transparent embryos and genome editing tools for reverse and forward genetic studies. The Java medaka, Oryzias javanicus, is one of the two species of medaka perfectly adapted for living in brackish/sea-waters. Being an important component of the mangrove ecosystem, O. javanicus is also used as a valuable marine test-fish for ecotoxicology studies. Here, we sequenced and assembled the whole genome of O. javanicus, and anticipate this resource will be catalytic for a wide range of comparative genomic, phylogenetic and functional studies. Complementary sequencing approaches including long-read technology and data integration with a genetic map allowed the final assembly of 908 Mbp of the O. javanicus genome. Further analyses estimate that the O. javanicus genome contains 33% of repeat sequences and has a heterozygosity of 0.96%. The achieved draft assembly contains 525 scaffolds with a total length of 809.7 Mbp, a N50 of 6,3 Mbp and a L50 of 37 scaffolds. We identified 21454 predicted transcripts for a total transcriptome size of 57, 146, 583 bps. We provide here a high-quality chromosome scale draft genome assembly of the euryhaline Javafish medaka (321 scaffolds anchored on 24 chromosomes (representing 97.7% of the total bases)), and give emphasis on the evolutionary adaptation to salinity.
Keywords: Medaka, evolution, whole genome sequencing, long reads, genetic map, transcriptome, adaptation, salinity
Medaka fishes belong to the genus Oryzias and are an emerging model system for studying the molecular basis of vertebrate evolution. This genus contains approximately 35 species, individually exhibiting numerous morphological, ecological and physiological differences and specificities (Inoue and Takei 2002, 2003; Parenti 2008; Mokodongan and Yamahira 2015). In addition, they all share many advantages of experimental model organisms, such as their small size, easy breeding, short generation time, transparent embryos, transgenic technology and genome-editing tools, with the “flag ship” species of this genus, the Japanese rice fish, Oryzias latipes (Wittbrodt et al. 2002; Kirchmaier et al. 2015). Such phenotypic variations, together with cutting edge molecular genetic tools make it possible to identify major loci that contribute to evolutionary differences, and to dissect the roles of individual genes and regulatory elements by functional tests. For example, a recent genetic mapping approach using interspecific hybrids identified the major chromosome regions that underlie the different hyperosmotic tolerance between species of the Oryzias genus (Myosho et al. 2018). Medaka fishes are also excellent models to study evolution of sex chromosomes and sex-determining loci among species (Takehana et al. 2007a, 2007b; Tanaka et al. 2007; Herpin and Schartl 2009), with the advantage of being also suitable models for providing functional evidences for these novel sex-determining genes by gain-of-function and/or loss-of-function experiments (Myosho et al. 2012; Takehana et al. 2014).
Among these species, the Java medaka, Oryzias javanicus (Figure 1), is unique as being the prototypic species of this genus with respect to adaptation to seawater. Previous phylogenetic studies divided the genus Oryzias into three monopyletic groups: (i) javanicus, (ii) latipes and (iii) celebensis species groups (Takehana et al. 2005; Mokodongan and Yamahira 2015). Most of the Oryzias species inhabit mainly freshwater biotopes, while only two species belonging to the javanicus group live in sea- or brackish water. One is O. javanicus, found in mangrove swamps from Thailand to Indonesia, and the other is O. dancena (previously named O. melastigma) living both in sea- and freshwaters from India to Malaysia. Although both species are highly adaptable to seawater, O. javanicus prefers hyperosmotic conditions while O. dancena favors hypoosmotic conditions at the west coast of Malaysian peninsula where their distribution ranges overlap (Yusof et al. 2012). In addition, O. javanicus is an important component of the mangrove ecosystem (Zulkifli et al. 2012), and has been used as a valuable marine test fish in several ecotoxicology studies (Koyama et al. 2008; Horie et al. 2018).
Figure 1.
A couple of Java medakas, Oryzias javanicus. Picture from K. Naruse, NBRP Medaka stock center (https://shigen.nig.ac.jp/medaka/top/top.jsp).
In this study, we sequenced and assembled the whole genome of O. javanicus, a model fish species for studying molecular mechanisms of seawater adaptation. In teleost fish, the major osmoregulatory organs i.e., gills, intestine and kidney, play different roles for maintaining body fluid homeostasis. Many genes encoding hormones, receptors, osmolytes, transporters, channels and cellular junction proteins are potentially involved in this osmotic regulation. In addition to osmoregulation, hatching enzyme activity dramatically fluctuates and adjusts at different salt conditions. At hatching stage, fish embryos secrete a specific cocktail of enzymes in order to dissolve the egg envelope, or chorion. In the medaka O. latipes, digestion of the chorion occurs through the cooperative action of two kinds of hatching enzymes, (i) the high choriolytic enzyme (HCE) and (ii) the low choriolytic enzyme (LCE) (Yasumasu et al. 2010). The HCE displays a higher activity in fresh- than in brackish waters (Kawaguchi et al. 2013). Thus, availability of a high-quality reference genome in O. javanicus would facilitate further research for investigating the molecular basis of physiological differences, including the osmotic regulation and the hatching enzyme activity, among Oryzias species.
Methods and Materials
Animal samplings
The wild stock of O. javanicus used in this study was supplied by the National Bio-Resource Project (NBRP) medaka in Japan. This stock (strain ID: RS831) was originally collected at Penang, Malaysia, and maintained in synthetic seawater (ca 3% of NaCl equivalent; although using half seawater is also possible) in aquaria under an artificial photoperiod of 14 hr light:10 hr darkness at 27 ± 2°. Genomic DNA was extracted from the whole body of a female (having ZW sex chromosome) using a conventional phenol/chloroform method, and was subjected to PacBio and 10X Genomics sequencings. For RNA-sequencing, total RNAs were extracted from nine female tissues (brain, bone, gill, heart, intestine, kidney, liver, muscle and ovary), and one male tissue (testis) using the RNeasy Mini Kit (Qiagen). For genetic mapping, we used a DNA panel consisting of 96 F1 progeny with their parents (originally described in a previous study (Takehana et al. 2008)). Phenotypic sex was determined by secondary sex characteristics of adult fish (six-month-old and sexually mature fish), namely, the shapes of dorsal and anal fins. All animal experiments performed in this study complied with the guideline of National Institute for Basic Biology, and have been approved by the Institutional Animal Care and Use Committee of National Institute of Natural Science (16A050 and 17A048).
PacBio genome sequencing:
Library construction and sequencing were performed according to the manufacturer’s instructions (Shared protocol-20kb Template Preparation Using BluePippin Size Selection system (15kb size Cutoff)). When required, DNA was quantified using the Qubit dsDNA HS Assay Kit (Life Technologies). DNA purity was assessed by spectrophotometry using the nanodrop instrument (Thermofisher), and size distribution and absence of degradation were monitored using the Fragment analyzer (AATI) (8–11). Purification steps were performed using 0.45X AMPure PB beads (PacBio). 80µg of DNA was purified and then sheared at 40kb using the megaruptor system (diagenode). DNA and END damage repair step was further performed for 5 libraries using the SMRTBell template Prep Kit 1.0 (PacBio). Blunt hairpin adapters were then ligated to the libraries. Libraries were subsequently treated with an exonuclease cocktail in order to digest unligated DNA fragments. Finally, a size selection step using a 15kb cutoff was performed on the BluePippin Size Selection system (Sage Science) using 0.75% agarose cassettes, Marker S1 high Pass 15-20kb. Conditioned sequencing primer V2 was annealed to the size-selected SMRTbell. The annealed libraries were then bound to the P6-C4 polymerase using a ratio of polymerase to SMRTbell set at 10:1. After performing a magnetic bead-loading step (OCPW), SMRTbell libraries were sequenced on 48 SMRTcells (RSII instrument at 0.25nM with a 360-min movie resulting in a total of 61.8Gb of sequence data (1.28Gb/SMRTcell).
10X Genomics genome sequencing:
Chromium library was prepared according to 10X Genomics’ protocol using the Genome Reagent Kits v1. Sample quantity and quality controls were further validated on Qubit, Nanodrop and Femto. Optimal performance has been characterized on input gDNA with a mean length greater than 50 kb. The library was prepared using 3 µg of high molecular weight (HMW) gDNA (cut off at 50kb using BluePippin system). In details, for the microfluidic Genome Chip, a library of Genome Gel Beads was combined with HMW template gDNA in Master Mix and partitioning oil in order to create Gel Bead-In-EMulsions (GEMs) in the Chromium. Each Gel Bead was functionalized with millions of copies of a 10x Barcoded primer. Upon dissolution of the Genome Gel Bead in the GEM, primers containing (i) an Illumina R1 sequence (Read 1 sequencing primer), (ii) a 16 bp 10x Barcode, and (iii) a 6 bp random primer sequence were released. Read 1 sequence and the 10x Barcode were added to the molecules during the GEM incubation. P5 and P7 primers, Read 2, and Sample Index were added during library construction. 8 cycles of PCR were performed for amplifying the library. Library quality was assessed using a Fragment analyzer. Finally, the library was sequenced on an Illumina HiSeq3000 using a paired-end read length of 2x150 pb with the Illumina HiSeq3000 sequencing kits resulting in 101.6Gb of raw sequence data.
Genome assembly and annotation:
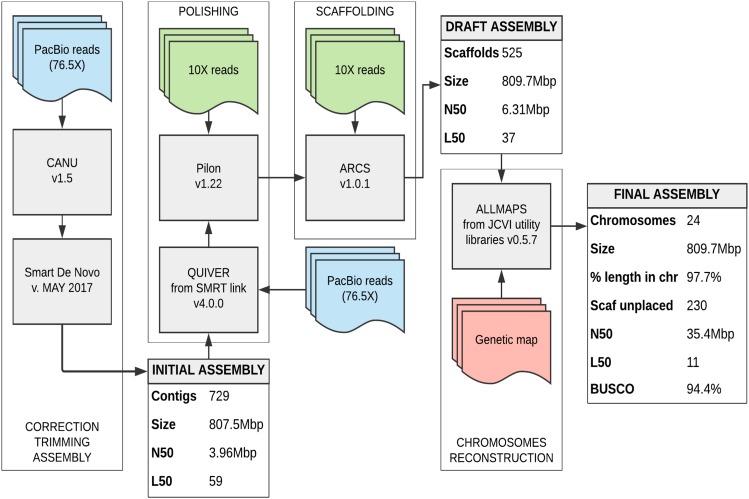
PacBio reads were corrected and trimmed using Canu v1.5 (Koren et al. 2017). Contigs were then assembled using SMARTdenovo version of May 2017 (Ruan 2019). The draft assembly produced contains 729 contigs with a total genome size of 807.5 Mbp, an N50 of 3,9 Mbp and a L50 of 59 contigs (Figure 2). To improve the assembly base pair quality two polishing steps were run. First, BLASR aligned PacBio reads were processed with Quiver from the Pacific Biosciences SMRT link software v.4.0.0. Second, 10X reads were realigned to the genome using Long Ranger v2.1.1 and the alignment file was processed with Pilon v1.22 (Walker et al. 2014). Third, the same 10X reads were aligned to the genome with BWA-MEM v0.7.12-r1039 (Li 2013) and the alignment file was processed with ARCS v1.0.1 (Yeo et al. 2018) to scaffold the genome. Both tools were run with default parameters. For genome annotation, the MAKER3 pipeline was employed ((Holt and Yandell 2011); Maker 3.01.02-beta in mpi mode to merge data from gene models and cDNA/protein evidences). Maker has been running with entries est_gff, protein_gff and pred_gff in run_evm = 1, est2genome = 0 and protein2genome = 0 mode. No AED cut-off was applied but AED scores have been used to select the best supported transcript for each gene.
Figure 2.
Oryzias javanicus assembly pipeline. Sequencing data are represented by colored rectangles with waved bases. Tools used are in gray rectangles. Assembly metrics are in gray and white rectangles. This pipeline is divided in stages symbolized by the frame.
Transcriptome RNA-seq sequencing and assembly:
RNA-seq libraries were prepared according to Illumina’s protocols using the Illumina TruSeq Stranded mRNA sample prep kit. Briefly, mRNAs were selected using poly-T beads, reverse-transcribed and fragmented. The resulting cDNAs were then subjected to adaptor ligation. 10 cycles of PCR were performed for amplifying the libraries. Quality of the libraries was assessed using a Fragment Analyzer. Quantification was performed by qPCR using the Kapa Library Quantification Kit. RNA-seq libraries were sequenced on an Illumina HiSeq3000 using a paired-end read length of 2x150 pb with the Illumina HiSeq3000 sequencing kits resulting in 95Gb of sequence data (28.9M reads pairs/library). The read quality of the RNA-seq libraries was evaluated using FastQC (Andrew S. 2010). De novo and reference-based transcriptome assemblies were produced. Reads were cleaned, filtered and de novo assembled using the DRAP pipeline v1.91 (Cabau et al. 2017) with the Oases assembler (Schulz et al. 2012). Assembled contigs were filtered in order to keep only those with at least one fragment per kilobase of transcript per million reads (FPKM). In the reference-based approach, all clean reads were mapped to the chromosomal assembly using STAR v2.5.1b (Dobin et al. 2013) with outWigType and outWigStrand options to output signal wiggle files. Cufflinks v2.2.1 (Trapnell et al. 2010) was used to assemble the transcriptome. All tissues have been de novo assembled separately and the 1 FPKM cut-off has been set on each library. The min, mean, median and max number of transcripts after the cufflinks assembly of each tissue are respectively 26223, 44992, 47804 and 59234.
RAD-library construction:
RAD-seq library was built following the Baird et al. (Baird et al. 2008) protocol with minor modifications. Briefly, between 400 to 500 ng of gDNA per fish were digested with SbfI-HF enzyme (R3642S, NEB). Digested DNA was purified using AMPure PX magnetic beads (Beckman Coulters) and ligated to indexed P1 adapters (1 index per sample) using concentrated T4 DNA ligase (M0202T, NEB). After quantification (Qubit dsDNA HS assay kit, Thermofisher) all samples were pooled in equal amounts. The pool was then fragmented on a S220 sonicator (Covaris) and purified with Minelute column (Qiagen). Finally, the sonicated DNA was size selected (250 to 450 bps) on a Pippin HT (Sage science) using a 2% agarose cassette, repaired using the End-It DNA-end repair kit (Tebu Bio) and adenylated at its 3′ ends using Klenow (exo-) (Tebu-Bio). P2 adapters were then ligated using concentrated T4 DNA ligase, and 50 ng of the ligation product were engaged in a 12 cycles PCR for amplification. After AMPure XP beads purification, the resulting library was checked on a Fragment Analyzer (Agilent) using the HS NGS kit (DNF-474-33) and quantified by qPCR using the KAPA Library Quantification Kit (Roche, ref. KK4824). Ultimately the whole library was denatured, diluted to 10 pM, clustered and sequenced using the rapid mode v2 SR100nt lane of a Hiseq2500 device (Illumina).
Identification of genes associated with osmoregulation
We listed known O. latipes genes that encode proteins associated with osmoregulation (e.g., hormones, pumps, transporters, channels, osmolytes-related and cellular junction proteins) based on literatures, and used HMMER version 3.1b2 (http://hmmer.org/) to identify specific Pfam domains (Pfam 32, (El-Gebali et al. 2019) included in these proteins (Supplemental Table S1). Using our gene model of O. javanicus together with Ensembl gene models of the O. dancena and O. latipes species complex (Hd-rR, HNI-II and HSOK), we counted the number of proteins containing the Pfam domains in each species (Supplemental Table S2).
Salt dependency of OjHCE:
The mature enzyme regions of OjHCE3 was amplified from their full-length cDNA using primers designed to contain suitable restriction enzyme sites (BamHI and NdeI) at the 5′ region. After digestion with BamHI and NdeI, the fragments were inserted into pET3c vector. The plasmid was transformed into E. coli BL21 (DE3) pLysE strain cells. The cells were cultivated, and recombinant protein was harvested as inclusion body as described in (Kawaguchi et al. 2013). After the inclusion body was dissolved in denaturing buffer (50 mM Tris-HCl (pH 8.0), 8 M urea, 0.1 M 2-mercaptoethanol and 1 mM EDTA), the recombinant protein was refolded as described in (Kawaguchi et al. 2013). The egg envelope digestion activity of OjHCE was determined by turbidimetric methods (Yamagami 1973; Kawaguchi et al. 2013). The isolated egg envelopes of medaka O. latipes were minced into fine fragments, and suspended in distilled water (DW). The suspension was allowed to stand overnight to remove rough fragments, and the supernatant containing fine fragments was used as substrate. The enzyme reaction was carried out in 400 μl of a reaction mixture containing 50 mmol/l Tris-HCl (pH 8.0), 0−0.75 mol/l NaCl, egg envelope suspension and recombinant HCE. The initial turbidity at 610 nm (T610) of the mixture was adjusted to approximately 55% when that of DW was 100%. Increment in transmission caused by the digestion of the fragmented envelopes was monitored for 3 min. The relative enzyme activity was expressed as the percentage of the highest activity under various salt concentrations.
Data availability
All genome and transcriptome information was deposited under the NCBI Bioproject number PRJNA505405. The Illumina sequencing data for the RAD-tag genetic map were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5326271 to SRX5326366. The genomic PacBio sequencing data were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5274121 to SRX5274138 and SRX5274139 to SRX5274169. The 10X genomics Illumina sequencing data were deposited in the Sequence Read Archive at NCBI with accession number SRX5274139. The transcriptome Illumina sequencing data were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5017469 to SRX5017479. The final chromosome assembly and genome annotation were deposited in GenBank at NCBI RWID00000000.1. Supplemental material available at figshare: https://doi.org/10.25387/g3.10310498.
Results and Discussion
Genome Characteristics
To estimate size and other genome characteristics, 10X reads were processed with Jellyfish v1.1.11 (Marçais and Kingsford 2011) to produce 21-mer distribution. The k-mer histogram was uploaded to GenomeScope (Vurture et al. 2017) with the max k-mer coverage parameter set to 10,000. Genome size was estimated around 908 Mbp, which is slightly higher than the 850 Mbp (0.87pg) estimated size reported on the Animal Genome Size Database (“Animal Genome Size Database:: Home”). Furthermore, this analysis estimates that the O. javanicus genome contains 33% of repeat sequences (around 303 Mbp) and has a heterozygosity of 0.96% (Table 1).
Table 1. GenomeScope outputs on O. javanicus genome statistics.
| Property | min | max |
|---|---|---|
| Heterozygosity | 0.960% | 0.964% |
| Genome Haploid Length | 908,146,324 bp | 908,641,143 bp |
| Genome Repeat Length | 303,610,795 bp | 303,776,222 bp |
| Genome Unique Length | 604,535,529 bp | 604,864,921 bp |
| Model Fit | 95.95% | 99.72% |
| Read Error Rate | 1.50% | 1.50% |
Genome assembly
Draft assembly contains 525 scaffolds with a total length of 809.7 Mbp, a N50 of 6,3 Mbp and a L50 of 37 scaffolds. This represents 89.1% of the k-mer estimated genome size. Given the high percentage of repeats in the O. javanicus genome (33%), it is possible that the PacBio assembly did not totally succeed in completing all repeated regions. The genome completeness was estimated using Benchmarking Universal Single-Copy Orthologs (BUSCO) v3.0 (Simão et al. 2015) based on 4,584 BUSCO orthologs derived from the Actinopterygii lineage leading to BUSCO scores of 4,327 (94.4%) complete BUSCOs, 176 (3.8%) fragmented BUSCOs and 81 (1.8%) missing BUSCOs.
Integration with the genetic map
RAD reads were trimmed by Trim Galore 0.4.3 (“Trim Galore”) with Cutadapt 1.12 (Martin 2011) and then mapped to the assembled scaffolds using BWA-MEM v0.7.17 (Li 2013). Uniquely mapped reads were extracted from the read alignments, and then called variant bases using uniquely mapped reads by samtools mpileup and bcftools call (Li 2011). Indels and variants with a low genotyping quality (GQ < 20), a low read depth (DP < 5), a low frequency of the minor allele (< 5%), more than four alleles in the family, no more than 5% individuals missing were removed by vcftools v0.1.15 (Danecek et al. 2011). After quality filtering, 6,375 variant sites were kept for the following analysis. Linkage map was constructed using this genotype information using Lep-MAP3 (Rastas 2017). Briefly, the filtered vcf file was loaded and the markers removed with high segregation distortion (Filtering2: dataTolerance = 0.001). Markers were then separated into 24 linkage groups with a LOD score threshold set at 9 and a fixed recombination fraction of 0.08 (SeparateChromosomes2: lodlimit = 9 and theta = 0.08). Two linkage groups were then excluded because of their small numbers of contained markers (less than 10). Classification of the markers was determined after maximum likelihood score indexing with 100 iterations (OrderMarkers2: numMergeIterations = 100) in each linkage group. The final map had 5,738 markers dispatched among 24 linkage groups spanning a total genetic distance of 1,221 cM.
The linkage map exhibited discrepancies between genomic scaffolds and genetic markers. Among 525 genomics scaffolds, 32 were linked to more than one linkage group. To split chimeric scaffolds with a higher precision and to rebuild chromosomes with a higher fidelity, we used a cross-species synteny map between the Java medaka (O. javanicus) scaffolds and the medaka (O. latipes) chromosomes in order to combine marker locations from genetic and synteny maps. To build the synteny map, medaka cDNAs were aligned to the Java medaka scaffolds using BLAT v36 (Kent 2002), and a list of pairwise correspondence of gene positions on Java medaka scaffolds and medaka chromosomes was established. 13,796 markers were added to the 5,738 markers of the genetic map. Java medaka chromosomes were then reconstructed using ALLMAPS from the JCVI utility libraries v0.5.7 (Tang et al. 2015). This package was used to combine genetic and synteny maps, to split chimeric scaffolds, to anchor, order and orient genomic scaffolds. The resulting chromosomal assembly consists of 321 scaffolds anchored on 24 chromosomes (97.7% of the total bases) and 231 unplaced scaffolds
Annotation results
The first annotation step was identifying repetitive DNA content using RepeatMasker v4.0.7 (“RepeatMasker Home Page”), Dust (Morgulis et al. 2006) and TRF v4.09 (Benson 1999). A species-specific de novo repeat library was built with RepeatModeler v1.0.11 (Smit and Hubley 2010). Repeated regions were located using RepeatMasker with the de novo and the Zebrafish (Danio rerio) libraries. Bedtools v2.26.0 (Quinlan and Hall 2010) was used to merge repeated regions identified with the three tools and to soft mask the genome. Repeats were estimated to account for 43.16% (349 Mbp) of our chromosomal assembly. The MAKER3 genome annotation pipeline v3.01.02-beta (Holt and Yandell 2011) combined annotations and evidences from three approaches: similarity with known fish proteins, assembled transcripts and de novo gene predictions. Protein sequences from 11 other fish species (Astyanax mexicanus, Danio rerio, Gadus morhua, Gasterosteus aculeatus, Lepisosteus oculatus, Oreochromis niloticus, Oryzias latipes, Poecilia formosa, Takifugu rubripes, Tetraodon nigroviridis, Xiphophorus maculatus) found in Ensembl were aligned to the masked genome using Exonerate v2.4 (Slater and Birney 2005). Previously assembled transcripts were used as RNA-seq evidence. A de novo gene model was built using Braker v2.0.4 (Hoff et al. 2016) with wiggle files provided by STAR as hints file for training GeneMark and Augustus. The best supported transcript for each gene was chosen using the quality metric Annotation Edit Distance (AED) (Eilbeck et al. 2009). The genome annotation gene completeness was assessed by BUSCO using the Actinopterygii group (Table 2). Finally, the predicted genes were subjected to similarity searches against the NCBI NR database using Diamond v0.9.22 (Buchfink et al. 2015). The top hit with a coverage over 70% and identity over 80% was retained.
Table 2. Java medaka assembly and annotation statistics.
| Gene annotation | |
|---|---|
| Number of genes | 21,454 |
| Number of transcripts | 21,454 |
| Transcriptome size | 57,146,583 bp |
| Mean transcript length | 2,663 bp |
| Longest transcript | 42,733 bp |
| Number of genes with significant hit against NCBI NR | 17,412 (81.2%) |
| Gene completeness | |
| Complete BUSCOs | 4,289 (93.6%) |
| Fragmented BUSCOs | 187 (4.1%) |
| Missing BUSCOs | 108 (2.3%) |
Mitochondrial genome and annotation
The previously sequenced Oryzias javanicus mitochondrial genome (NC_012981) (Setiamarga et al. 2009) was aligned to the chromosomal assembly using Blat. All hits were supported by a single scaffold. This scaffold was removed from the assembly, circularised and annotated using MITOS (Bernt et al. 2013). This new Oryzias javanicus mitochondrial genome is 16,789 bp long and encodes 13 genes, 2 rRNAs and 19 tRNAs.
Phylogenetic relationship:
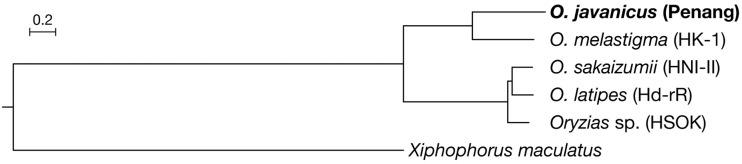
To precisely determine the phylogenetic position of O. javanicus within the genus Oryzias, we estimated the phylogenetic relationship using published whole genome datasets as references. Reference assemblies and annotations of O. latipes (Hd-rR: ASM223467v1), O. sakaizumii (HNI-II: ASM223471v1), Oryzias sp. (HSOK: ASM223469v1), O. dancena (Om_v0.7.RACA), and southern platyfish Xiphophorus maculatus (X_maculatus-5.0-male) were obtained from Ensembl Release 94 (http://www.ensembl.org/). Among the six genomes, orthologous groups were classified and 10,852 single-copy orthologous genes were identified using OrthoFinder 2.2.6 (Emms and Kelly 2015). For every single gene, codon alignment based on translated peptide sequences was generated by PAL2NAL (Suyama et al. 2006) and then trimmed by trimAl with ‘-autometed1’ option (Capella-Gutiérrez et al. 2009). All multi-sample fasta files were concatenated into a single file using AMAS concat by setting each gene as a separate partition (Borowiec 2016). A maximum likelihood tree was then inferred using IQ-TREE v1.6.6 (Nguyen et al. 2015) with the GTR+G substitution model for each codon, followed by an ultrafast bootstrap analysis of 1,000 replicates (Hoang et al. 2018). This tree (Figure 3) indicates that O. javanicus forms a monophyletic group with O. dancena but not with the O. latipes species complex (Hd-rR, HNI-II, and HSOK), being consistent with previous trees inferred from two mitochondrial genes and a nuclear gene (Takehana et al. 2005).
Figure 3.
Phylogenetic position of O. javanicus. Maximum likelihood tree was inferred from the concatenated codon-alignment of 10,852 single-copy genes among 5 reference assemblies of Oryzias species with Southern platyfish (Xiphophorus maculatus) as outgroup. All nodes were supported by 100% bootstrap values.
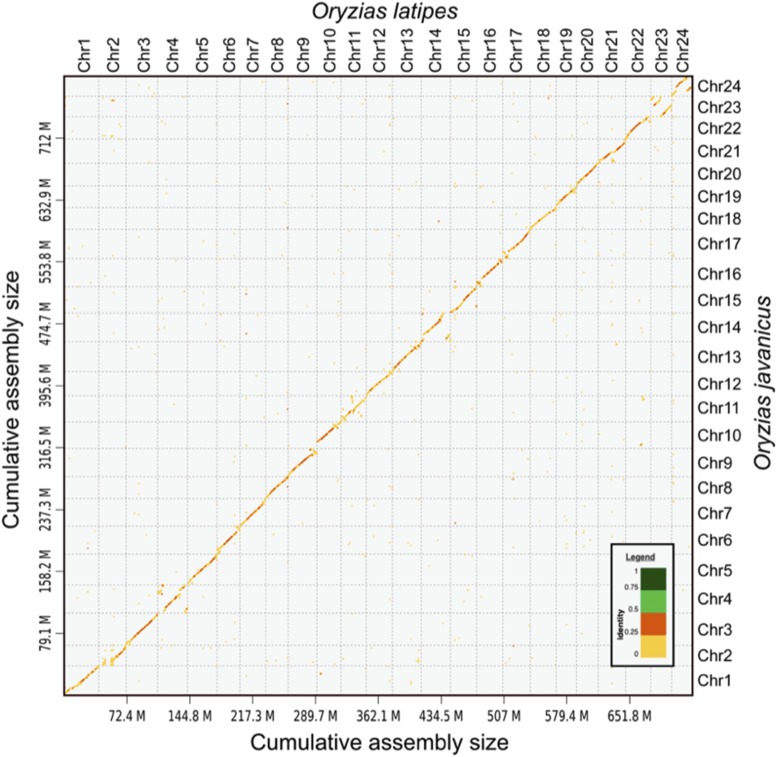
The D-GENIES (Cabanettes and Klopp 2018) genome-wide comparison of this O. javanicus genome compared to the O. latipes reference genome [Ensembl version ASM223467v1 (GCA_002234675.1)] shows that these two genomes are extremely colinear at the whole genome scale (Figure 4). At a chromosome scale the comparison with O. latipes shows that most of the O. javanicus chromosomes are strongly colinear with their single O. latipes chromosome counterparts (Supplemental Figure 1). Only a few O. Javanicus chromosomes are more deeply reorganized compared to their O. latipes chromosome counterparts with for instance, the O. javanicus LG04, LG14, LG23 and LG24 that display multiple intra chromosomal rearrangements and the O. javanicus LG10, LG11 and LG14 that show small inter chromosomal rearrangements (i.e., the insertion of a small region from an O. latipes different chromosome). With regards to their sex chromosomes O. latipes has a male heterogametic system (XX/XY) and the LG01 is the Y sex chromosome (see (Herpin and Schartl 2009) for review). O. javanicus has a female heterogametic sex determination system and LG16 is the W sex chromosome (Takehana et al. 2008). In O. javanicus both the LG01 (the O. latipes Y chromosome) and the LG16 (the O. javanicus W chromosome) display a strong chromosome collinearity with respectively the O. latipes LG01 and LG16. Indeed, and according to previous reports (Takehana et al. 2008) the dmrt1bY Y specific duplication/insertion on O. latipes LG01 is absent from the O. javanicus LG01 sequence.
Figure 4.
Genome-wide comparison of the Oryzias javanicus and Oryzias latipes genome assemblies. This dot-plot alignment of the O. javanicus genome with the O. latipes reference genome [Ensembl version ASM223467v1 (GCA_002234675.1)] was produced with the D-GENIES software (Cabanettes and Klopp 2018) available online at http://dgenies.toulouse.inra.fr/ with default visualization parameters and a slight filtering of the small matches. The different ranges of percentage of identity are shown in different colors (legend panel with the identity percentage color range).
Adaptation to salinity and hatching enzymes
To gain insight into gene family evolution associated with osmoregulation, we used HMMER version 3.1b2 to identify Pfam domain containing proteins in the O. javanicus genome. We used protein sequences based on our gene model of O. javanicus combined with Ensembl genes of the O. latipes species complex (Hd-rR, HNI-II and HSOK) and O. dancena for the Pfam search, and focused on 147 domains found in 224 proteins whose functions were related to osmoregulation (Supplemental Tables S1 and S2). Similar numbers of proteins were observed among species for each domain, suggesting that the osmoregulation gene repertoires are relatively conserved in Oryzias species. However, further detailed comparisons are required because gene annotation methods are different among data.
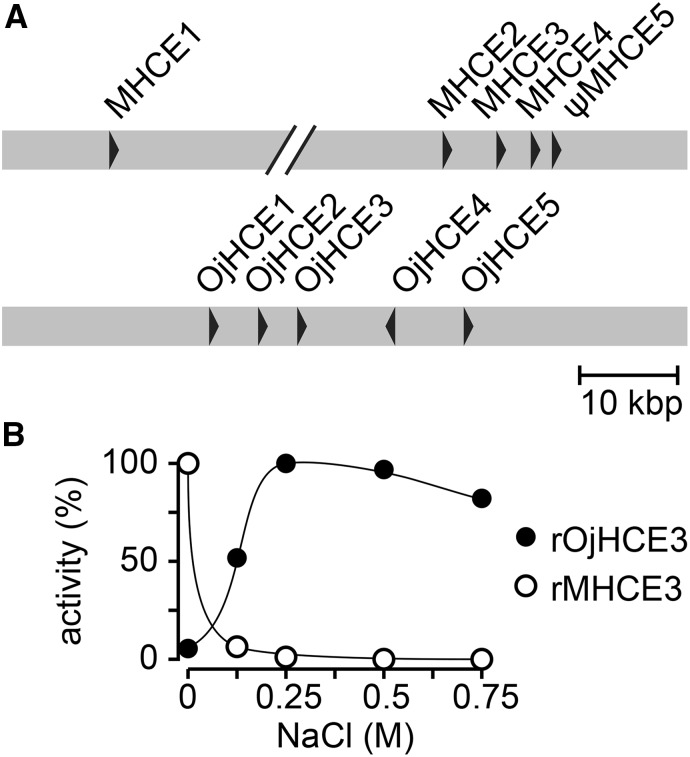
We then also focused on specific genes encoding hatching enzymes. In the genome of O. latipes, five copies of hce genes -including one pseudogene- are clustered tandemly with the same transcriptional direction on chromosome 3 (chr. 3), while only one single copy of the lce gene is located on chromosome 24 (chr. 24) (Kawaguchi et al. 2007). In O. javanicus 5 copies of the hce (Ojhce) gene are located on chromosome 3 and one lce (Ojlce) gene was found on chromosome 24. The amino acid sequence similarities in the mature enzyme region of the 5 Ojhce genes are between 89–99%. Only in comparison to O. latipes, within the five O. javanicus hce genes, the fourth one (Ojhce4) displays an opposite orientation compared to the others (Figure 5A) suggesting a re-arrangement within the hce gene cluster that has likely been occurring during the evolution of Oryzias lineage. Phylogenetic analyses indicated that all the cloned hce and lce genes were orthologous to other euteleosteans hce and lce respectively.
Figure 5.
Hatching enzyme of Oryzias javanicus. (A) hce gene cluster of O. latipes (mhce1-5) and O. javanicus (Ojhce1-5). Arrowheads indicate direction of transcription. (B) Salt dependency of O. javanicus hce (black circle) and O. latipes (white circle). Activities are shown as % of relative activity with respect to highest activity, which is considered as 100% in each species.
While LCE’s activity remains constant over various salinities, HCEs have been reported to show salt-dependent activity (Kawaguchi et al. 2013). In contrast to other Oryzias species, O. javanicus, being a euryhaline species, specifically adapted its physiology to higher water salinities. In order to test whether such adaptive evolution would translate at the level of HCE activity, recombinant OjHCE3 (rOjHCE3) was generated in an E. coli expression system, refolded, and its activity regarding to the digestion of the egg-envelope determined at various salt concentrations based on the method described in Kawaguchi et al. (Kawaguchi et al. 2013). Although rOjHCE3 showed virtually no activity at 0 M NaCl, an increased activity was apparent at elevated salt concentrations. Furtheron rOjHCE3 activity was recorded to be highest at 0.25 M NaCl, while still maintaining high activity up to 0.75 M NaCl (Figure 5B). In contrast, it has been reported that O. latipes HCEs show highest activity at 0 M NaCl, and drastically decrease when salt concentrations increase ((Kawaguchi et al. 2013), Figure 5B). These results suggest that salt preference of HCE enzymes is a species-specific adaptation to different salt environments at hatching.
The Java medaka, Oryzias javanicus, is one of the two species of medaka living in brackish/sea-waters. Being an important component of the mangrove ecosystem, O. javanicus is also used as a valuable marine test-fish for ecotoxicology studies. Here, we sequenced and assembled the whole genome of O. javanicus. Complementary sequencing approaches and data integration with a genetic map allowed the final assembly of the 908 Mbp of the O. javanicus genome. The final draft assembly contains 525 scaffolds with a total length of 809.7 Mbp, a N50 of 6,3 Mbp and a L50 of 37 scaffolds. Providing here a high-quality draft genome assembly of the euryhaline Javafish medaka, we anticipate this resource will be catalytic for a wide range of comparative genomic, phylogenetic and functional studies within the genus Oryzias and beyond.
Acknowledgments
This work was supported by a “Projet Incitatif PHASE department 2015” grant (Grant ID ACI_PHASE, INRAE) to AH, a NIBB Collaborative Research Initiative to KN, NIBB individual Collaboration Research Project to MK, and Grants-in-Aid for Young Scientists to YT (Grant ID 16K18590) and MK (Grant ID 16K18593). AH was additionally funded by the project AquaCRISPR (ANR-16-COFA-0004-01). The GeT and MGX core facilities were supported by France Génomique National infrastructure, funded as part of “Investissement d’avenir” program managed by Agence Nationale pour la Recherche (contract ANR-10-INBS-09). The GeT core facility was also supported by the GET-PACBIO program (« Programme operationnel FEDER-FSE MIDI-PYRENEES ET GARONNE 2014-2020 »).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.10310498.
Communicating editor: D. J. Grunwald
Literature Cited
- Andrew S., 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Animal Genome Size Database Available at: http://www.genomesize.com/index.php. Accessed: September 13, 2019.
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L. et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 3: e3376 10.1371/journal.pone.0003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt M., Donath A., Jühling F., Externbrink F., Florentz C. et al. , 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- Borowiec M. L., 2016. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4: e1660 10.7717/peerj.1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B., Xie C., and Huson D. H., 2015. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12: 59–60. 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- Cabanettes F., and Klopp C., 2018. D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6: e4958 10.7717/peerj.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabau C., Escudié F., Djari A., Guiguen Y., Bobe J. et al. , 2017. Compacting and correcting Trinity and Oases RNA-Seq de novo assemblies. PeerJ 5: e2988 10.7717/peerj.2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J. M., and Gabaldón T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C. et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K., Moore B., Holt C., and Yandell M., 2009. Quantitative measures for the management and comparison of annotated genomes. BMC Bioinformatics 10: 67 10.1186/1471-2105-10-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S., Mistry J., Bateman A., Eddy S. R., Luciani A. et al. , 2019. The Pfam protein families database in 2019. Nucleic Acids Res. 47: D427–D432. 10.1093/nar/gky995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157 10.1186/s13059-015-0721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herpin A., and Schartl M., 2009. Molecular mechanisms of sex determination and evolution of the Y-chromosome: insights from the medakafish (Oryzias latipes). Mol. Cell. Endocrinol. 306: 51–58. 10.1016/j.mce.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., and Vinh L. S., 2018. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 35: 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff K. J., Lange S., Lomsadze A., Borodovsky M., and Stanke M., 2016. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. 10.1093/bioinformatics/btv661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., and Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie Y., Kanazawa N., Yamagishi T., Yonekura K., and Tatarazako N., 2018. Ecotoxicological Test Assay Using OECD TG 212 in Marine Java Medaka (Oryzias javanicus) and Freshwater Japanese Medaka (Oryzias latipes). Bull. Environ. Contam. Toxicol. 101: 344–348. 10.1007/s00128-018-2398-1 [DOI] [PubMed] [Google Scholar]
- Inoue K., and Takei Y., 2003. Asian medaka fishes offer new models for studying mechanisms of seawater adaptation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 136: 635–645. 10.1016/S1096-4959(03)00204-5 [DOI] [PubMed] [Google Scholar]
- Inoue K., and Takei Y., 2002. Diverse adaptability in oryzias species to high environmental salinity. Zool. Sci. 19: 727–734. 10.2108/zsj.19.727 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Yasumasu S., Hiroi J., Naruse K., Suzuki T. et al. , 2007. Analysis of the exon-intron structures of fish, amphibian, bird and mammalian hatching enzyme genes, with special reference to the intron loss evolution of hatching enzyme genes in Teleostei. Gene 392: 77–88. 10.1016/j.gene.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Yasumasu S., Shimizu A., Kudo N., Sano K. et al. , 2013. Adaptive evolution of fish hatching enzyme: one amino acid substitution results in differential salt dependency of the enzyme. J. Exp. Biol. 216: 1609–1615. 10.1242/jeb.069716 [DOI] [PubMed] [Google Scholar]
- Kent W. J., 2002. BLAT–the BLAST-like alignment tool. Genome Res. 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier S., Naruse K., Wittbrodt J., and Loosli F., 2015. The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). Genetics 199: 905–918. 10.1534/genetics.114.173849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S., Walenz B. P., Berlin K., Miller J. R., Bergman N. H. et al. , 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27: 722–736. 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama J., Kawamata M., Imai S., Fukunaga M., Uno S. et al. , 2008. Java medaka: a proposed new marine test fish for ecotoxicology. Environ. Toxicol. 23: 487–491. 10.1002/tox.20367 [DOI] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., 2013 Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN]
- Marçais G., and Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M., 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI]
- Mokodongan D. F., and Yamahira K., 2015. Origin and intra-island diversification of Sulawesi endemic Adrianichthyidae. Mol. Phylogenet. Evol. 93: 150–160. 10.1016/j.ympev.2015.07.024 [DOI] [PubMed] [Google Scholar]
- Morgulis A., Gertz E. M., Schäffer A. A., and Agarwala R., 2006. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J. Comput. Biol. 13: 1028–1040. 10.1089/cmb.2006.13.1028 [DOI] [PubMed] [Google Scholar]
- Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y. et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. 10.1534/genetics.111.137497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myosho T., Takahashi H., Yoshida K., Sato T., Hamaguchi S. et al. , 2018. Hyperosmotic tolerance of adult fish and early embryos are determined by discrete, single loci in the genus Oryzias. Sci. Rep. 8: 6897 10.1038/s41598-018-24621-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H. A., von Haeseler A., and Minh B. Q., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti L. R., 2008. A phylogenetic analysis and taxonomic revision of ricefishes, Oryzias and relatives (Beloniformes, Adrianichthyidae). Zool. J. Linn. Soc. 154: 494–610. 10.1111/j.1096-3642.2008.00417.x [DOI] [Google Scholar]
- Quinlan A. R., and Hall I. M., 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastas P., 2017. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics 33: 3726–3732. 10.1093/bioinformatics/btx494 [DOI] [PubMed] [Google Scholar]
- RepeatMasker Home Page Available at: http://www.repeatmasker.org/. Accessed: September 13, 2019.
- Ruan, J., 2019 Ultra-fast de novo assembler using long noisy reads: Ruanjue/smartdenovo. Available at: https://github.com/ruanjue/smartdenovo. Accessed: September 13, 2019.
- Schulz M. H., Zerbino D. R., Vingron M., and Birney E., 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. 10.1093/bioinformatics/bts094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiamarga D. H. E., Miya M., Yamanoue Y., Azuma Y., Inoue J. G. et al. , 2009. Divergence time of the two regional medaka populations in Japan as a new time scale for comparative genomics of vertebrates. Biol. Lett. 5: 812–816. 10.1098/rsbl.2009.0419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Slater G. S. C., and Birney E., 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. F. A., and R. Hubley, 2010 RepeatModeler Open-1.0.
- Suyama M., Torrents D., and Bork P., 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y., Demiyah D., Naruse K., Hamaguchi S., and Sakaizumi M., 2007a Evolution of different Y chromosomes in two medaka species, Oryzias dancena and O. latipes. Genetics 175: 1335–1340. 10.1534/genetics.106.068247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana Y., Hamaguchi S., and Sakaizumi M., 2008. Different origins of ZZ/ZW sex chromosomes in closely related medaka fishes, Oryzias javanicus and O. hubbsi. Chromosome Res. 16: 801–811. 10.1007/s10577-008-1227-5 [DOI] [PubMed] [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K. et al. , 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157 10.1038/ncomms5157 [DOI] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., Hamaguchi S., and Sakaizumi M., 2007b Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116: 463–470. 10.1007/s00412-007-0110-z [DOI] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., and Sakaizumi M., 2005. Molecular phylogeny of the medaka fishes genus Oryzias (Beloniformes: Adrianichthyidae) based on nuclear and mitochondrial DNA sequences. Mol. Phylogenet. Evol. 36: 417–428. 10.1016/j.ympev.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Takehana Y., Naruse K., Hamaguchi S., and Sakaizumi M., 2007. Evidence for different origins of sex chromosomes in closely related Oryzias fishes: substitution of the master sex-determining gene. Genetics 177: 2075–2081. 10.1534/genetics.107.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Zhang X., Miao C., Zhang J., Ming R. et al. , 2015. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 16: 3 10.1186/s13059-014-0573-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G. et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trim Galore Available at: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. Accessed: September 13, 2019.
- Vurture G. W., Sedlazeck F. J., Nattestad M., Underwood C. J., Fang H. et al. , 2017. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33: 2202–2204. 10.1093/bioinformatics/btx153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B. J., Abeel T., Shea T., Priest M., Abouelliel A. et al. , 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt J., Shima A., and Schartl M., 2002. Medaka–a model organism from the far East. Nat. Rev. Genet. 3: 53–64. 10.1038/nrg704 [DOI] [PubMed] [Google Scholar]
- Yamagami K., 1973. Some enzymological properties of a hatching enzyme (chorionase) isolated from the fresh-water teleost, Oryzias latipes. Comp. Biochem. Physiol. B 46: 603–616. 10.1016/0305-0491(73)90100-4 [DOI] [PubMed] [Google Scholar]
- Yasumasu S., Kawaguchi M., Ouchi S., Sano K., Murata K. et al. , 2010. Mechanism of egg envelope digestion by hatching enzymes, HCE and LCE in medaka, Oryzias latipes. J. Biochem. 148: 439–448. [DOI] [PubMed] [Google Scholar]
- Yeo S., Coombe L., Warren R. L., Chu J., and Birol I., 2018. ARCS: scaffolding genome drafts with linked reads. Bioinformatics 34: 725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof S., Ismail A., Koito T., Kinoshita M., and Inoue K., 2012. Occurrence of two closely related ricefishes, Javanese medaka (Oryzias javanicus) and Indian medaka (O. dancena) at sites with different salinity in Peninsular Malaysia. Environ. Biol. Fishes 93: 43–49. 10.1007/s10641-011-9888-x [DOI] [Google Scholar]
- Zulkifli, S. Z., F. Mohamat-Yusuff, A. Ismail, and N. Miyazaki, 2012 Food preference of the giant mudskipper Periophthalmodon schlosseri (Teleostei : Gobiidae). Knowl. Managt. Aquatic Ecosyst. 07.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genome and transcriptome information was deposited under the NCBI Bioproject number PRJNA505405. The Illumina sequencing data for the RAD-tag genetic map were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5326271 to SRX5326366. The genomic PacBio sequencing data were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5274121 to SRX5274138 and SRX5274139 to SRX5274169. The 10X genomics Illumina sequencing data were deposited in the Sequence Read Archive at NCBI with accession number SRX5274139. The transcriptome Illumina sequencing data were deposited in the Sequence Read Archive at NCBI with accession numbers SRX5017469 to SRX5017479. The final chromosome assembly and genome annotation were deposited in GenBank at NCBI RWID00000000.1. Supplemental material available at figshare: https://doi.org/10.25387/g3.10310498.