Abstract
Bacillus firmus nematicidal bacterial strains are used to control plant parasitic nematode infestation of crops in agricultural production. Proteases are presumed to be the primary nematode virulence factors in nematicidal B. firmus degrading the nematode cuticle and other organs. We determined and compared the whole genome sequences of two nematicidal strains. Comparative genomics with a particular focus on possible virulence determinants revealed a wider range of possible virulence factors in a B. firmus isolate from a commercial bionematicide and a wild type Bacillus sp. isolate with nematicidal activity. The resulting 4.6 Mb B. firmus I-1582 and 5.3 Mb Bacillus sp. ZZV12-4809 genome assemblies contain respectively 18 and 19 homologs to nematode-virulent proteases, two nematode-virulent chitinase homologs in ZZV12-4809 and 28 and 36 secondary metabolite biosynthetic clusters, projected to encode antibiotics, small peptides, toxins and siderophores. The results of this study point to the genetic capability of B. firmus and related species for nematode virulence through a range of direct and indirect mechanisms.
Keywords: Bacillus firmus, complete genomes, bioinformatics, biological control, nematicidal activity, virulence factors
Bacillus firmus (Bacillales; Bacillaceae) is a species of Gram-positive, rod-shaped bacteria capable of producing endospores under adverse conditions. B. firmus is a cosmopolitan species that can be isolated primarily from soil, but also from a variety of different environments including wastewater and marine sediments (Geng et al. 2014). B. firmus has the potential for biotechnological exploitation, since various strains have been studied for bioremediation applications, such as: enzyme production from agro-industrial waste (Husseiny et al. 2008); heavy metals detoxification (Bachate et al. 2013; Noroozi et al. 2017); textile dye discoloration in wastewater discharge (Rathod and Pathak 2018); and microbial-enhanced heavy oil recovery (Shibulal et al. 2018), among others. Importantly, B. firmus is an agriculturally useful bacteria, as it has been demonstrated to promote plant growth and alleviate abiotic saline stress (El-Esawi et al. 2018), as well as offering plants protection against plant-parasitic nematodes, including root-knot nematodes (RKNs) (Keren-Zur et al. 2000; Giannakou et al. 2004). RKNs belonging to the Meloidogyne genus (Nematoda, Tylenchida, Meloidogynidae) are endoparasites and among the most important plant pests (Sasser 1977; Trudgill and Blok 2001; Bebber et al. 2014). RKNs infestations lead to physiologically stressed, low-yielding plants (Abad et al. 2003) and 5% crop yield loss on average (Carter and Sasser 1985), significantly contributing to about 110 billion EUR per year in economic damage due to plant-parasitic nematodes in agricultural systems across the world (Danchin et al. 2013).
Various broad-spectrum chemical pesticides have been used in crop production for decades to minimize the damage caused by soil-borne plant parasitic nematodes, but many were banned or phased out due to the associated toxicities (Regulation (EC) No. 1107/2009). Biopesticides based on microbial biocontrol agents can be used to control RKNs infestation as a safer alternative to agrochemicals and many candidate bacterial and fungal species have been studied for their potential use as biological nematicides (Wilson and Jackson 2013). Different biopesticides against RKNs are also commercially available, but preparations based on B. firmus have seen the widest commercial use - with the B. firmus strain I-1582 in the bionematicide preparation BioNem-WP (AgroGreen) being deployed on the market in the early 2000s (Keren-Zur et al. 2000; Giannakou et al. 2004). Despite commercial use, little is known about the exact mode of action of B. firmus against RKNs (Tian et al. 2007; Geng et al. 2016; Valencia and Kotcon 2016). Bacterial proteases are considered to be the primary virulence factor in nematicidal B. firmus strains (Lian et al. 2007; Tian et al. 2007; Geng et al. 2016). However, Bacillus spp. are known to produce various secondary metabolites, enzymes and toxins, which have never been studied for nematicidal activity. One example is a cereulide-like emetic toxin purified from B. firmus ATCC 14575T and ATCC 8247 strains (Taylor et al. 2005). Additionally, Bacillus spp. may inhibit RKNs or alleviate their effects on plants indirectly by strengthening plant defense mechanisms (Kloepper et al. 2004), release of repellents (Valencia and Kotcon 2016), and plant growth-promotion (El-Esawi et al. 2018). Screening for secondary metabolite production potential in nematicidal bacteria could thus be warranted since there are various secondary metabolite compounds from other organisms with substantial nematotoxic properties (Khalil 2013). With the accessibility of next-generation sequencing, bacterial genomes can be assessed through bioinformatics analysis for the genetic potential to produce nematicidal substances, including novel secondary metabolites and virulence factors useful in agriculture. This approach has been used by Zheng et al. (2016) to screen the genomic sequences of 120 Bacillus strains exhibiting nematicidal activity against bacterivorous nematode Caenorhabditis elegans for the presence of various virulence factors. The whole-genome sequence analysis of B. firmus strain DS-1 was also the first step in the determination of nematicidal serine protease Sep1, capable of inhibiting the growth and development of the nematodes C. elegans and Meloidogyne incognita (Geng et al. 2014; 2016). Bioinformatics investigation of the nature of B. firmus virulence against plant parasitic nematodes like RKNs is however still lacking – especially assessment of the range of possible virulence factors that could be found in B. firmus genomes.
The aim of this study was to compare the range of possible virulence determinants in the genomes of two geographically and phylogenetically distinct nematicidal Bacillus strains – a B. firmus isolate from widely used commercial bionematicide and a local wild-type Bacillus sp. isolate with nematicidal activity.
Materials and Methods
Bacterial strains
Two Bacillus strains with nematicidal potential were used in this study. Strain B. firmus I-1582 was isolated from the commercially available biological seed treatment preparation VOTiVO FS (Bayer CropScience, Germany), which is used to protect against soil-borne plant parasitic nematodes. Strain Bacillus sp. ZZV12-4809 was isolated from the pea (Pisum sativum L.) rhizosphere, in Maribor, Slovenia. Both strains were cultivated in LB liquid and solid media at 23°. The nematicidal activity of both isolates has been demonstrated in prior in vitro and pot experiments (Susič et al. 2019). For endospore differential staining, bacterial cultures in exponential growth phase were inoculated into liquid Difco Sporulation Medium (DSM) (Monteiro et al. 2014) for 48h at 37°, stained according to Schaeffer and Fulton (1933) and visualized under a microscope.
Species identification
For bacterial identification, total bacterial DNA was isolated from a single bacterial colony grown on an LB agar plate at 23° for 48h. Bacteria were identified according to the 16S rRNA gene sequence, which was amplified using the forward 27f (5′-GAGAGTTTGATCCTGGCTCAG-3′) and reverse 1495r (5′-CTACGGCTACCTTGTTACGA-3′) primers and with cycling conditions as described by Bandi et al. (1994). 16S rRNA sequences were first searched against quality-controlled databases of 16S rRNA sequences in the EzBioCloud Identify service (Yoon et al. 2017). Second, the average nucleotide identity (ANI) was calculated based on the genomes of B. firmus I-1582, Bacillus sp. ZZV12-4809 and five other B. firmus sequences available in GenBank: B. firmus DS1 (GenBank assembly accession: GCA_000565285.1), B. firmus LK28 (GCA_001038755.1), B. firmus NBRC 15306 (GCA_001591465.1), B. firmus NCTC10335 (GCA_900445365.1), B. firmus 14_TX (GCA_003315495.1) and B. oceanisediminis 2691 (GCA_000294775.2), using JSpeciesWS (Richter et al. 2016; http://jspecies.ribohost.com/jspeciesws/#home), implementing the algorithms as described by Goris et al. (2007). JSpeciesWS was also used to calculate tetra-nucleotide signatures (Tetra) correlation indexes and to search the associated database for related genomes using the Tetra Correlation Search (TCS) function. For general phylogenetic positioning of B. firmus I-1582 and Bacillus sp. ZZV12-4809 within the genus, we included the two genomes in phylogenetic analysis of 222 assembled Bacillus genomes from type material available in GenBank. Phylogenetic analysis was carried out with GToTree v1.1.10 (Lee 2019; https://github.com/AstrobioMike/GToTree), using the hidden Markov model (HMM) single-copy gene set (119 HMMs) for Firmicutes and the maximum likelihood (ML) method for tree reconstruction. The best-fitting substitution model was selected according to the Akaike information criterion (AIC) and Bayesian information criterion (BIC) scores, with consensus trees calculated from 1000 bootstrap replicates. Heatmaps visualizing the ANI values (%) and phylogenetic trees were rendered in R with the packages ‘Superheat’ (Barter and Yu 2018), ‘ggtree’ (Yu et al. 2017) and ‘ggplot2’ (Wickham 2016).
Genome sequencing, assembly and annotation
For sequencing, total DNA was isolated from overnight cultures of B. firmus I-1582 and Bacillus sp. ZZV12-4809 with the QIAamp DNA Mini Kit (Qiagen, Germany). Paired-end libraries were prepared following the manufacturer’s instructions with the Nextera XT DNA Library Preparation Kit (Illumina, USA) and then sequenced on a MiSeq System (Illumina) using the MiSeq Reagent Kit v3 (600-cycle) (Illumina). Raw sequence reads were adapter-trimmed (Trimmomatic) and quality filtered (FastQC). Genome sequences were assembled de novo and annotated using the Comprehensive Genome Analysis Service at the Pathosystems Resource Integration Center (PATRIC, https://www.patricbrc.org) bioinformatics utility (Wattam et al. 2017). The assembly and annotation pipeline was optimized for Illumina MiSeq reads and involved a Velvet (Zerbino and Birney 2008) run with hash length 35, error correction of sequenced reads with BayesHammer (Nikolenko et al. 2013), and assembly with SPAdes (Bankevich et al. 2012), with a k-mer value of up to 99. Assembly Rapid Annotation using Subsystem Technology (ARAST) quality score was used to sort the assembly results. Assemblies were additionally assessed with BlobTools (Laetsch and Blaxter 2017) to detect contaminant DNA sequences. Genome assemblies were annotated with the RAST tool kit (RASTtk) (Brettin et al. 2015). The annotation included functional assignments of predicted protein coding sequences (CDS) to Enzyme Commission (EC) numbers (Schomburg et al. 2004), Gene Ontology (GO) assignments (Ashburner et al. 2000), and mapping to KEGG pathways (Kanehisa et al. 2016), genus-specific protein families (PLFams) and cross-genus protein families (PGFams) (Davis et al. 2016). Additionally, predicted protein sequences were separately assigned to clusters of orthologous groups (COG) using eggNOG-mapper v4.5.1 (Huerta-Cepas et al. 2017). Genome assemblies and syntenic regions were visualized as circular maps using plotMyGBK (https://github.com/microgenomics/plotMyGBK) and GGisy (https://github.com/Sanrrone/GGisy) python scripts that depend on the R environment (R Core Team 2018), with the packages ‘data.table’ (Dowle et al. 2019), ‘Rsamtools’ (Morgan et al. 2019) and ‘OmicCircos’ (Hu et al. 2014). Where applicable, various graphic elements were arranged with Inkscape v0.92.4 (https://inkscape.org).
Comparative genomics and bioinformatics analysis
The OrthoVenn web server with default parameters (E-value 1e−5 and inflation value 1.5) (Wang et al. 2015; http://www.bioinfogenome.net/OrthoVenn/) was used to identify orthologous gene clusters in the assembled genomes of B. firmus I-1582 and Bacillus sp. ZZV12-4809, and five other B. firmus sequences available in GenBank as described above. Homologs to known transporters, virulence factors, drug targets and antibiotic resistance genes, were identified using PATRIC by homology to known sequences in the following databases: Transporter Classification Database (TCDB) (Saier et al. 2016), Virulence Factor Database (VFDB) (Chen et al. 2016), PATRIC_VF (Mao et al. 2015), Therapeutic Target Database (TTD) (Zhu et al. 2010), DrugBank 4.0 (Law et al. 2014), Comprehensive Antibiotic Resistance Database (CARD) (McArthur et al. 2013) and the National Database of Antibiotic Resistant Organisms (NDARO). Antimicrobial resistance (AMR) genes were also annotated by using the k-mer-based AMR gene detection method, which utilizes PATRIC’s curated collection of representative AMR gene sequence variants (Wattam et al. 2017). Putative nematode-virulent proteases were revealed by protein BLAST search on the assembled genomes, by sequence similarity with an e-value of 10−5 and minimum 30% sequence identity over 60% of both protein lengths (Rost 1999; Galagan et al. 2003). Selected protein sequences were aligned with Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) to calculate the percent (%) identity matrix and presented in a heatmap. The annotated genome assembly sequence files, including information for contigs and ORFs, were analyzed with antiSMASH v4.2.0 (Blin et al. 2017) and RIPPMiner with pairwise BLAST (Agrawal et al. 2017) to predict and analyze various types of secondary metabolite gene clusters.
Data availability
The complete genome sequence data, including raw sequence reads, genome assemblies and annotations of B. firmus I-1582 and Bacillus sp. ZZV12-4809 used in this study were submitted to NCBI, GenBank under the BioProject accession ID: PRJNA533096. Supplemental material available at figshare: https://doi.org/10.25387/g3.11522544.
Results and Discussion
Microbial features and identification of bacterial species
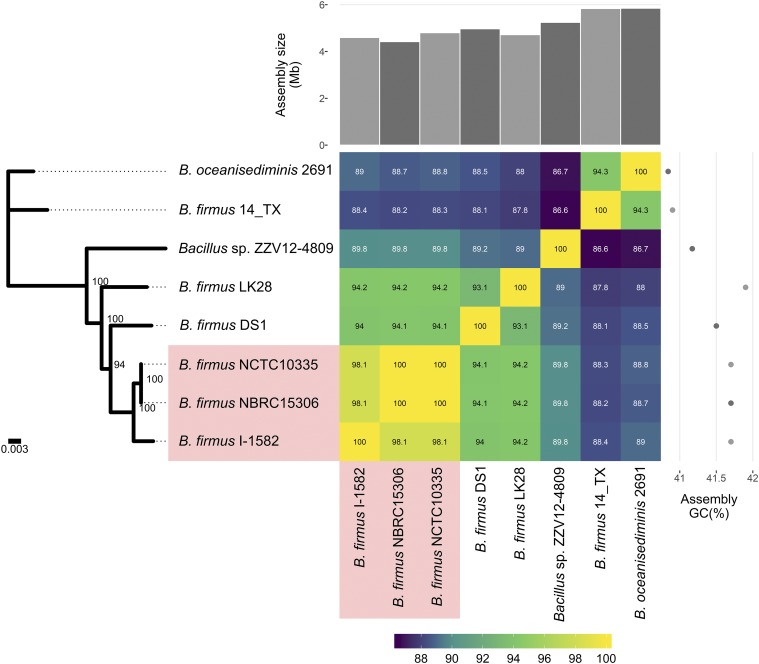
Colony and cell morphology as well as 16S rRNA gene and whole genome sequence analysis were performed to determine species identity of the two studied nematicidal bacterial strains. Strains I-1582 and ZZV12-4809 generally formed round, pale-cream colonies on LB agar medium under the described growth conditions, with 2 µm-long rod-shaped cells able to form endospores (Supplementary Figure 1B,C). B. firmus I-1582 and Bacillus sp. ZZV12-4809 16S rRNA gene sequence amplification (Supplementary Figure 1A) and EzBioCloud analysis showed 100% and 99.23% similarity (Supplementary Table 1), respectively, to the 16S rRNA sequence of B. firmus strain NBRC 15306T (GenBank accession: BCUY01000205). Tetra analysis was in accordance with 16S rRNA results, and a TCS search for related genomes returned B. firmus DS1 as the closest genome in the database to I-1582 and ZZV12-4809, with Z-scores of 0.99866 and 0.99709, respectively (Supplementary Table 2B,C). Average nucleotide identity values (ANI %) were calculated between B. firmus I-1582, Bacillus sp. ZZV12-4809 and all 5 B. firmus genome assemblies currently publicly available in GenBank, with Bacillus oceanisediminis 2691 as an outgroup (see Materials and Methods). I-1582 and ZZV12-4809 shared moderate to high (ANIb/ANIm) identity with each other, as well as with the type B. firmus strain NBRC 15306 (Figure 1; Supplementary Table 2). According to the strict 95% cut-off value to delineate species boundary (Richter and Rosselló-Móra 2009; Chun et al. 2018), strain I-1582, but not ZZV12-4809, could be considered as B. firmus. Phylogenetic analysis within the genus Bacillus to discern the phylogenetic position of this strain accompanied by the ANI values showed that ZZV12-4089 was still most closely related to the B. firmus group (Supplementary Figure 2). ANI calculations and phylogenetic analysis indicated previously unreported variability within the strains of B. firmus that points to incorrect species circumscription in some strains. Therefore, description of a novel species could be warranted (Figure 1).
Figure 1.
Maximum likelihood phylogenetic tree based on the 8 available genomic assemblies of Bacillus firmus and Bacillus oceanisediminis 2691 with heatmap annotation. Nodes annotated with bootstrap support. Average nucleotide identity (ANI) values (%) are presented in a heatmap, ranging from lower (violet) to higher sequence identity (green-yellow), and clustered according to the phylogenetic tree. ANI analysis showed 100% sequence identity between B. firmus NBRC 15306 and B. firmus NCTC 10335, which is in accordance with information from the NCBI GenBank (data not shown). B. firmus I-1582 exhibited 98.14% sequence identity with strains NBRC 15306 and NCTC 10335 (highlighted in red), and 89.83% sequence identity with Bacillus sp. ZZV12-4809. The heatmap was annotated with a bar chart showing the varying sizes (Mb) of all 8 assemblies (at the top) and their respective GC (%) content (right-hand side).
B. firmus I-1582 and Bacillus sp. ZZV12-4809 share general genome characteristics
The genome assemblies of B. firmus I-1582 and Bacillus sp. ZZV12-4809 were 4,597,711 and 5,245,841 bp long, and yielded 199 and 90 contigs, respectively (Table 1; Supplementary Figures 3,4). The genomes of the two strains have slightly different G+C content and similar numbers of tRNA and rRNA genes. In B. firmus I-1582, 75.4% of predicted coding sequences (CDSs) were assigned to the clusters of orthologous groups (COG) categories, and in Bacillus sp. ZZV12-4809 74% of predicted CDSs were assigned to COGs, although the functional category of many CDS could not be defined (Table 2). Multiple syntenic blocks were found to be shared with B. firmus I-1582 and Bacillus sp. ZZV12-4809 assemblies. High-synteny blocks (above 90% nucleotide identity), equal to or larger than 15 kbp, were present in 11 contigs in ZZV12-4809, aligning to 15 contigs in I-1582 (Supplementary Figure 5). Gene assignment to the COG categories indicated similar fractions of CDS assigned to different categories in both strains. Some differences in COG categories were observed between I-1582 and ZZV12-4809; certain categories contain a larger fraction of genes in the first while others in the second corresponding genomes (Table 2).
Table 1. Sequencing, assembly and annotation information for the Bacillus firmus I-1582 and Bacillus sp. ZZV12-4809 genomes.
| B. firmus I-1582 | Bacillus sp. ZZV12-4809 | |
|---|---|---|
| Assembly information | ||
| Assembly size (bp) | 4,597,711 | 5,245,841 |
| Number of contigs | 199 | 90 |
| G+C content (%) | 41.70 | 41.17 |
| Largest contig (bp) | 277,315 | 587,391 |
| N50 (bp) | 55,490 | 172,048 |
| L50 | 24 | 9 |
| Estimated coverage (times ×) | 158 | 58 |
| Annotation information | ||
| CDS | 5,048 | 5,671 |
| rRNA genes | 23 | 15 |
| tRNA genes | 103 | 102 |
| Hypothetical proteins | 1,600 | 1,953 |
| Proteins with functional assignments | 3,448 | 3,718 |
| Proteins with EC number assignments | 1,110 | 1,135 |
| Proteins with GO assignments | 934 | 957 |
| Proteins with Pathway assignments | 848 | 863 |
| Proteins with PATRIC genus-specific family (PLfam) assignments | 4,498 | 4,562 |
| Proteins with PATRIC cross-genus family (PGfam) assignments | 4,518 | 4,611 |
Table 2. The number and percentage (%) of genes assigned to COG categories in the genomes of Bacillus firmus I-1582 and Bacillus sp. ZZV12-4809.
| B. firmus I-1582 | Bacillus sp. ZZV12-4809 | |||
|---|---|---|---|---|
| Gene number | Gene percent (%) | Gene number | Gene percent (%) | |
| CDS assigned to COG categories | 3,805 | 100 | 4,194 | 100 |
| COG category | ||||
| [n] Not assigned to any COG category | 365 | 8.75 | 424 | 9.18 |
| [m] Assigned to multiple COG categories | 55 | 1.32 | 58 | 1.26 |
| [B] Chromatin structure and dynamics | 1 | 0.02 | 1 | 0.02 |
| [C] Energy production and conversion | 230 | 5.52 | 224 | 4.85 |
| [D] Cell cycle control, cell division, chromosome partitioning | 39 | 0.94 | 36 | 0.78 |
| [E] Amino acid transport and metabolism | 340 | 8.15 | 381 | 8.25 |
| [F] Nucleotide transport and metabolism | 88 | 2.11 | 94 | 2.04 |
| [G] Carbohydrate transport and metabolism | 189 | 4.53 | 245 | 5.31 |
| [H] Coenzyme transport and metabolism | 130 | 3.12 | 113 | 2.45 |
| [I] Lipid transport and metabolism | 118 | 2.83 | 124 | 2.69 |
| [J] Translation, ribosomal structure and biogenesis | 172 | 4.13 | 174 | 3.77 |
| [K] Transcription | 253 | 6.07 | 306 | 6.63 |
| [L] Replication, recombination and repair | 239 | 5.73 | 223 | 4.83 |
| [M] Cell wall / membrane / envelope biogenesis | 175 | 4.20 | 183 | 3.96 |
| [N] Cell motility | 40 | 0.96 | 38 | 0.82 |
| [O] Posttranslational modification, protein turnover, chaperones | 113 | 2.71 | 121 | 2.62 |
| [P] Inorganic ion transport and metabolism | 217 | 5.20 | 254 | 5.50 |
| [Q] Secondary metabolites biosynthesis, transport and catabolism | 55 | 1.32 | 69 | 1.49 |
| [S] Function unknown | 1081 | 25.92 | 1257 | 27.22 |
| [T] Signal transduction mechanisms | 163 | 3.91 | 174 | 3.77 |
| [U] Intracellular trafficking, secretion, and vesicular transport | 29 | 0.70 | 31 | 0.67 |
| [V] Defense mechanisms | 78 | 1.87 | 88 | 1.91 |
Comparative genomics reveals high overall similarity and certain strain-specific gene clusters
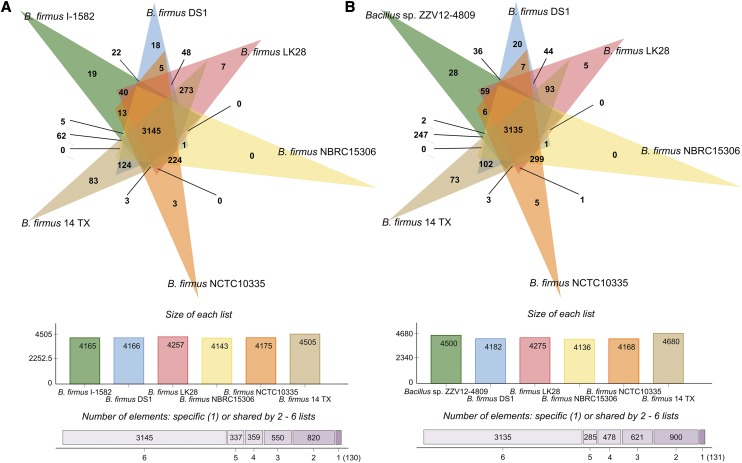
Genome-wide analysis of orthologous clusters is frequently used in comparative genomics studies – commonly in whole genome analysis across species, as orthologous genes are clusters of genes in different species, originating by vertical descent from a single gene in the last common ancestor (Wang et al. 2015). Genome-wide analysis of orthologous clusters revealed that the I-1582 and ZZV12-4809 genomes shared the vast majority of gene families / orthologous clusters (3850), while there were 1.03 and 2.33% specific clusters in I-1582 and ZZV12-4809 cluster lists, respectively. Expanding on the ANI results, the I-1582 and ZZV12-4809 proteomes were separately compared with five other B. firmus in the GenBank database (Figure 2). The strains formed 5341 clusters, 5211 orthologous clusters (listed in at least two strains) and 3116 single-copy gene clusters, when I-1582 was compared with other strains (Figure 2A); or 5550 clusters, 5419 orthologous clusters and 3096 single-copy gene clusters when ZZV12-4809 was compared with others (Figure 2B). Apart from the orthologous clusters shared by all the strains, some strains shared a proportionally higher number of clusters with each other. 14_TX shared almost 4 times more additional orthologous clusters with ZZV12-4809 than with I-1582, despite the fact that 14_TX and I-1582 share a higher nucleotide identity. This result could be due to the fact that there were simply more clusters formed by the ZZV12-4809 list than I-1582. Comparison of I-1582 and ZZV12-4809 lists identified 40 and 92 clusters respectively specific to I-1582 or ZZV12-4809. These strain-specific clusters could potentially be involved in important biological processes, so affiliations across GO terms were checked for differences between the strains. Genes in these clusters seemed to be involved in different molecular functions in I-1582 or ZZV12-4809 (Supplementary Table 4).
Figure 2.
Venn diagrams generated by OrthoVenn showing orthologous clusters shared separately by A) Bacillus firmus I-1582 (green) and B) Bacillus sp. ZZV12-4809 (green) and five other Bacillus species, specifically B. firmus DS1 (blue), LK28 (red), NBRC 15306 (yellow), NCTC10335 (orange), and 14_TX (brown). When A) B. firmus I-1582 was compared with other B. firmus strains, there were 820, 550, 359, 337 and 3145 clusters shared between 2, 3, 4, 5 and 6 strains, respectively, while 130 clusters (clusters of singleton genes) were found in only 1 of the 6 Bacillus strains compared. Comparison of B) ZZV12-4809 with other strains resulted in 900, 621, 478, 285 and 3135 clusters shared between 2, 3, 4, 5 and 6 strains, respectively, while 131 clusters of singleton genes were found in only 1 of the 6 strains compared.
Antimicrobial resistance genes, drug targets, transporters and virulence factors
Groups of genes that could contribute toward virulent activity of studied bacterial strains were investigated. The B. firmus I-1582 and Bacillus sp. ZZV12-4809 genomes were respectively annotated with 75 and 87 genes homologous to known transporters, virulence factors, drug targets and antibiotic resistance genes (Table 3). The two genomes shared more than 80% of these genes. Most of the shared genes were associated with resistance to triclosan, phenicol, peptide, aminoglycoside and tetracycline classes of antibiotics. However, strain ZZV12-4809 possessed 14 unique genes not present in I-1582 (Supplementary Table 5) – most of which seemed to be involved in resistance to various antibiotics. Among these, there were genes vgbA and vatD involved in resistance to streptogramin antibiotics; bcrC, bacitracin resistance; vanR, glycopeptide antibiotic resistance; fosB, fosfomycin resistance; catA9, resistance to chloramphenicol; rlmAII, resistance to the macrolide antibiotics tylosin and telithromycin (Takaya et al. 2013); and genes blaA and blaD, involved in resistance to β-lactam antibiotics. Of the genes present in strain ZZV12-4809 but not in strain I-1582, there were also capB and capC, which were classified as virulence factors involved in anti-phagocytosis (Candela and Fouet 2006); and aapA encoding an Amino-acid permease AapA transporter. Genes for putative gap-family peptidoglycolipid transporter and hemH for coproporphyrin ferrochelatase (EC 4.99.1.9) were found in I-1582 but not in ZZV12-4809. The latter gene product was classified as a drug target for N-Methylmesoporphyrin (Supplementary Table 5).
Table 3. Number of genes associated with antimicrobial resistance, drug targets, transporters, various virulence factors and secondary metabolites found in Bacillus firmus I-1582 and Bacillus sp. ZZV12-4809 genome assemblies.
| Number of determinants in the genome | ||
|---|---|---|
| Associated process | B. firmus I-1582 | Bacillus sp. ZZV12-4809 |
| Antimicrobial resistance genes | 40 | 48 |
| Drug targets | 12 | 11 |
| Transporters | 13 | 13 |
| Human / animal virulence factors | 6 | 9 |
| Nematode virulence factors | ||
| Proteases | 18 | 19 |
| Chitinases | 0 | 2 |
| Secondary metabolite clusters | 28 | 36 |
Multiple nematode virulence factors in I-1582 and ZZV12-4809 genomes
In order to assess the virulence potential of the two sequenced Bacillus strains, we performed a BLASTP search of the previously described virulence factors of bacterial, fungal, plant and animal origin surveyed by Zheng et al. (2016), bacterial proteases identified by Geng et al. (2016), and whole-genome biosynthetic cluster prediction. The candidate virulence factors found were classified into classes such as proteases (Supplementary Table 6), chitinases (Supplementary Table 8), peptides, and other secondary metabolites – such as putative siderophore compounds and toxins (Supplementary Table 9).
Proteases:
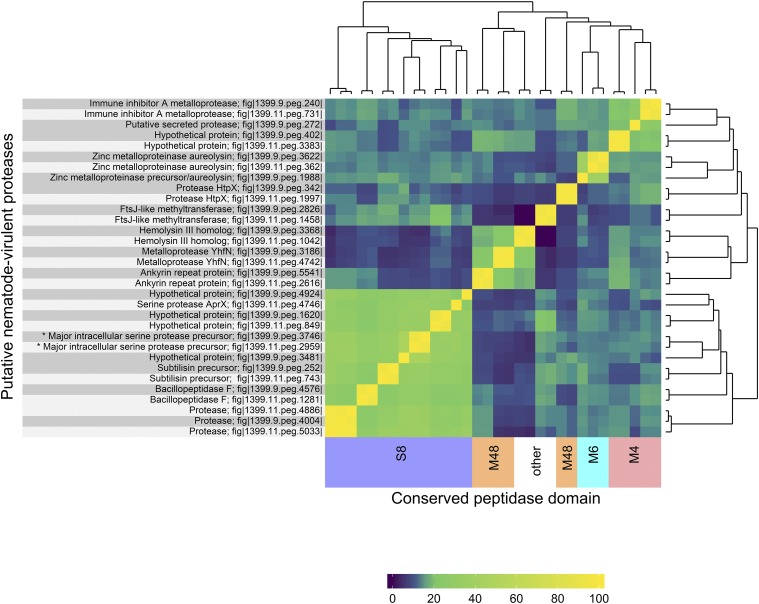
Using known nematode-virulent protease sequences as queries (Supplementary Table 6), respectively 18 and 19 different homologous sequences were found in B. firmus I-1582 and Bacillus sp. ZZV12-4809 (Supplementary Table 7). The sequences clustered loosely into groups based on the conserved family domains, as determined with InterProScan. Most of them belonged to the peptidase S8 domain family (Figure 3). Five, including Immune inhibitor A metalloproteases (InhA), hypothetical proteins and one putative secreted protease found only in ZZV12-4809, belonged to the M6 peptidase family. Three sequences annotated as Zn-metalloproteinase aureolysin (EC 3.4.24.29), were found to have conserved peptidase M4 domain. Six protein sequences were found to contain M48 peptidase domains, although two of these, annotated as HtpX proteases, had substantially shorter amino-acid sequences and did not cluster with the rest of the group. Haemolysin III homologs clustered more readily with peptidase M48 family proteins, while haemolysin A homologs clustered better with peptidase S8 family proteins. Homologs of haemolysin A in B. firmus genomes had similar domains to TlyA from Mycobacterium tuberculosis, which is known to exhibit hemolytic exotoxin activity in vitro (Rahman et al. 2010). Haemolysin III homologs from I-1582 and ZZV12-4809 showed high sequence identity with the haemolysin III family protein from B. firmus DS1, which exhibited some nematicidal activity in vitro against C. elegans N2 (Geng et al. 2016). Most of the putative nematode-virulent proteases had direct homologous counterparts in either the I-1582 or ZZV12-4809 genome, as indicated by high identity correlation in amino-acid sequences (Figure 3). One protease gene ‘fig|1399.11.peg.4746’ in the I-1582 did not have a counterpart in the ZZV12-4809 genome while 4 genes (‘fig|1399.9.peg.4924’, ‘fig|1399.9.peg.3481’, ‘fig|1399.9.peg.1988’ and ‘fig|1399.9.peg.272’) in ZZV12-4809 did not have a counterpart in the I-1582. This study revealed numerous putative nematode-virulent proteases in the B. firmus I-1582 and Bacillus sp. ZZV12-4809 genomes. However, previous work with B. firmus DS1 has demonstrated that most of the tested putative proteases did not exhibit any nematicidal effects and only recombinant protease EWG10090 (Serine protease 1; Sep1) led to significant 73.2% mortality of C. elegans N2 when expressed (Geng et al. 2016). Homologs to Sep1 were present in the B. firmus I-1582 and Bacillus sp. ZZV12-4809 genomes. Protein sequences ‘fig|1399.11.peg.2959’ in I-1582 and ‘fig|1399.9.peg.3746’ in ZZV12-4809 showed 98% and 93% identity respectively with Sep1 from B. firmus DS1.
Figure 3.
Clustering of putative nematode-virulent proteases found in Bacillus firmus I-1582 (light-gray) and Bacillus sp. ZZV12-4809 (dark-gray) genomes. All-against-all comparison of amino-acid sequences is based on percent (%) identity values calculated with Clustal Omega and presented in a heatmap, ranging from lower (violet) to higher sequence identity (green-yellow). A dendrogram was created using hierarchical clustering by Euclidean distance. The identities of analyzed proteins are given as genomic annotations together with PATRIC IDs (left-hand margin). Colored highlights (bottom margin) represent clustering of sequences according to the conserved peptidase family domains: S8 (blue), M4 (magenta), M6 (cyan), M48 (orange), or other (no color). Asterisks (*) denote high-similarity protein sequences to the nematicidal protease Sep1 from B. firmus DS1.
Chitinases:
Chitinases are considered to be putative nematode-virulence factors, since they may digest chitinous components of nematode eggs. Additionally, chitinases can enhance the deleterious effects of proteases, as postulated previously. Zheng et al. (2016) found that Bacillus strains with genes encoding putative virulent proteases and chitinases had higher nematode mortality rates when compared to strains with only protease genes. Through an initial BLAST search, two putative chitinase sequences were found only in the Bacillus sp. ZZV12-4809 genome, but were below the selection threshold (see Materials and Methods). Upon searching the NCBI Conserved Domain Database (CDD), both protein sequences were found to contain the chitin-hydrolyzing GH18-chitinase domain and could thus be considered putative nematode-virulent chitinases. Sequence ‘fig|1399.9.peg.2658’ showed the highest sequence identity with chitinase ABW96521.1 (33%; E-value = 1.0e-31) from the fungus Pochonia chlamydosporia, while sequence ‘fig|1399.9.peg.3724’ showed highest sequence identity with chitinase ABP37997.1 (26%; E-value = 1.0e-17) from the fungus Purpureocillium lilacinum. Both fungi are nematophagous and have been shown to parasitize eggs of the RKN Meloidogyne enterolobii (Silva et al. 2017). Through a recursive BLAST search, two additional sequences were found in the I-1582 assembly and one in ZZV12-4809 but appeared to lack the catalytic chitinase domain (Supplementary Table 8).
Secondary metabolites:
The secondary-metabolite gene clusters (antiSMASH) analysis revealed the presence of 28 and 36 predicted clusters in strains I-1582 and ZZV12-4809, respectively. Of these, only 9 and 12 were assigned to specific types, and five predicted clusters in each genome assembly (clusters 11, 21, 24, 25 and 28 in strain I-1582, and clusters 10, 11, 16, 17 and 35 in strain ZZV12-4809) were found on the contig borders and were likely to be incomplete (Supplementary Table 9). One gene cluster in strain I-1582 and two gene clusters in the ZZV12-4809 genome were predicted to be involved in the synthesis of the siderophore petrobactin which is considered a virulence factor of Bacillus anthracis (Wilson et al. 2010) and is the primary siderophore produced by this bacterium under iron starvation (Koppisch et al. 2005). Due to the biological importance of iron (Beneduzi et al. 2012), siderophore production by rhizobacteria can indirectly inhibit the growth of phytopathogenic soil microorganisms by limiting the bioavailability of iron, or promote plant growth through plant-microbe interactions (Singh et al. 2011). In I-1582, one lanthipeptide was predicted with 43.5% sequence identity (e = 5e-04) with paenicidin A. Two lanthipeptides were predicted in ZZV12-4809, the first showed very weak sequence identity with plantazolicin (44%, e = 2.9) and the second showed 38.9% identity (e = 6e-10) with cerecidin A7. Bacillus spp. are known to produce class I and II lanthipeptides with antibacterial activity – lantibiotics (Barbosa et al. 2015). Clusters 10 and 24 in the ZZV12-4809 genome were found to contain genes associated with bacteriocin-terpene (cluster 10) or bacteriocin (cluster 24) synthesis, while in I-1582, one cluster associated with terpene synthesis was predicted. Bacteriocins are antimicrobial peptides produced by bacteria, which have the ability to kill or inhibit closely related bacterial strains without the negative effects on bacteriocin-producing bacterium (Yang et al. 2014). In ZZV12-4809, cluster 19 was predicted to contain a hybrid type I polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) system, with 35% of genes similar to paenilamicin biosynthetic cluster. Paenilamicin has previously been identified as an antibiotic compound produced by the honey bee pathogen Paenibacillus larvae (Garcia-Gonzalez et al. 2014). Additionally, polysaccharide-type cluster 5 in I-1582 was found to contain two genes similar to aveBIII and aveBII from the avermectin oleandrose (GenBank: AB032523.1) biosynthetic cluster (Supplementary Table 9). Avermectins are considered to be potent anthelmintics and insecticides (Wohlert et al. 2001), one example being the widely commercialized insecticide / acaricide / nematicide / anthelmintic abamectin from Streptomyces avermitilis (Khalil 2013). The results of this analysis indicated an array of possible secondary metabolites potentially produced by the two studied Bacillus strains. This information is useful for further experimental evaluation of these secondary metabolites for nematicidal activity, especially since the exact mechanism(s) of action by B. firmus against nematodes are not fully understood. Although the cuticle-degrading proteases are thought the primary virulence factor in Bacillus (Lian et al. 2007; Geng et al. 2016), it has been observed previously that various B. firmus strains elicited high nematode mortality rates, while having relatively few homologs or gene copies to known nematode-virulence factors in their genomes compared to other bacteria (Zheng et al. 2016). This could be due to high expression rates, novel interaction by known virulence factors, or the existence of as yet uncharacterized nematotoxic compounds.
Conclusions
Through whole genome sequencing, comparative genomics and bioinformatics analysis it was possible to determine differences between the B. firmus I-1582 and Bacillus sp. ZZV12-4809 genomes and to assess their genetic capacity for nematode virulence. Although the B. firmus group appears to be relatively homogenous according to 16S rDNA, ANI analysis of whole genome assemblies available in GenBank highlighted previously unreported variability within the B. firmus group that probably mirrors the diverse habitats from which these strains were originally isolated. The differences in ANI% were sufficient to suggest incorrect species circumscription of some strains in GenBank. Various homologs to known nematode-virulent proteases from different organisms were found, specifically 18 and 19 different homologous sequences were found respectively in the B. firmus I-1582 and Bacillus sp. ZZV12-4809 strains. Additionally, two putative nematode-virulent chitinases were found in ZZV12-4809, containing the GH18-chitinase domain necessary to hydrolyze chitin and degrade chitinous structures in nematode eggs, thus contributing to their mortality. Additionally, 28 and 36 secondary metabolite clusters that could express an array of compounds belonging to siderophores, toxins, bacteriocins, lanthipeptides and antibiotics were predicted respectively in the genomes of B. firmus I-1582 and Bacillus sp. ZZV12-4809. The activity and nematotoxic potential of these putative secondary metabolites has yet to be experimentally characterized, but the resulting body of information points to a range of possible direct and indirect mechanisms of B. firmus and related species that could contribute to nematode virulence and could thus be used for targeted studies on virulence mechanisms, gene expression, and proteomic and metabolic studies of B. firmus, widely used as a biocontrol agent in crop production.
Acknowledgments
We thank Tanja Zidarič for her contribution during strain isolation and Marko Verce for his help with the program code written in Python.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25387/g3.11522544.
Communicating editor: M.-A. Félix
Literature Cited
- Abad P., Favery B., Rosso M.-N., and Castagnone-Sereno P., 2003. Root-knot nematode parasitism and host response: molecular basis of a sophisticated interaction. Mol. Plant Pathol. 4: 217–224. 10.1046/j.1364-3703.2003.00170.x [DOI] [PubMed] [Google Scholar]
- Agrawal P., Khater S., Gupta M., Sain N., and Mohanty D., 2017. RiPPMiner: a bioinformatics resource for deciphering chemical structures of RiPPs based on prediction of cleavage and cross-links. Nucleic Acids Res. 45: W80–W88. 10.1093/nar/gkx408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachate S. P., Nandre V. S., Ghatpande N. S., and Kodam K. M., 2013. Simultaneous reduction of Cr(VI) and oxidation of As(III) by Bacillus firmus TE7 isolated from tannery effluent. Chemosphere 90: 2273–2278. 10.1016/j.chemosphere.2012.10.081 [DOI] [PubMed] [Google Scholar]
- Bandi C., Damiani G., Magrassi L., Grigolo A., Fani R. et al. , 1994. Flavobacteria as intracellular symbionts in cockroaches. Proc. R. Soc. Lond. B Biol. Sci. 257: 43–48. 10.1098/rspb.1994.0092 [DOI] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M. et al. , 2012. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa J., Caetano T., and Mendo S., 2015. Class I and Class II lanthipeptides produced by Bacillus spp. J. Nat. Prod. 78: 2850–2866. 10.1021/np500424y [DOI] [PubMed] [Google Scholar]
- Barter R. L., and Yu B., 2018. Superheat: An R package for creating beautiful and extendable heatmaps for visualizing complex data. J. Comput. Graph. Stat. 27: 910–922. 10.1080/10618600.2018.1473780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebber D. P., Holmes T., and Gurr S. J., 2014. The global spread of crop pests and pathogens. Glob. Ecol. Biogeogr. 23: 1398–1407. 10.1111/geb.12214 [DOI] [Google Scholar]
- Beneduzi A., Ambrosini A., and Passaglia L. M. P., 2012. Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35: 1044–1051. 10.1590/S1415-47572012000600020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Wolf T., Chevrette M. G., Lu X., Schwalen C. J. et al. , 2017. antiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45: W36–W41. 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S. et al. , 2015. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5: 8365 10.1038/srep08365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela T., and Fouet A., 2006. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 60: 1091–1098. 10.1111/j.1365-2958.2006.05179.x [DOI] [PubMed] [Google Scholar]
- Carter C. C., and Sasser J. N., 1985. Overview of the international Meloidogyne project 1975–1984, pp. 19–24 in An Advanced Treatise on Meloidogyne, Volume I: Biology and Control, edited by Carter C. C. and Sasser J. N.. Department of Plant Pathology, North Carolina State University and the United States Agency for International Development, Raleigh, NC. [Google Scholar]
- Chen L., Zheng D., Liu B., Yang J., and Jin Q., 2016. VFDB 2016: hierarchical and refined dataset for big data analysis - 10 years on. Nucleic Acids Res. 44: D694–D697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Oren A., Ventosa A., Christensen H., Arahal D. R. et al. , 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68: 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Danchin E. G. J., Arguel M.-J., Campan-Fournier A., Perfus-Barbeoch L., Magliano M. et al. , 2013. Identification of novel target genes for safer and more specific control of root-knot nematodes from a pan-genome mining. PLoS Pathog. 9: e1003745 10.1371/journal.ppat.1003745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. J., Gerdes S., Olsen G. J., Olson R., Pusch G. D. et al. , 2016. PATtyFams: Protein families for the microbial genomes in the PATRIC database. Front. Microbiol. 7: 118 10.3389/fmicb.2016.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowle, M., A. Srinivasan, J. Gorecki, M. Chirico, P. Stetsenko et al., 2019 Package ‘data.table’ R package version 1.12.0. https://cran.r-project.org/web/packages/data.table/data.table.pdf
- El-Esawi M. A., Alaraidh I. A., Alsahli A. A., Alamri S. A., Ali H. M. et al. , 2018. Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 132: 375–384. 10.1016/j.plaphy.2018.09.026 [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D. et al. , 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. 10.1038/nature01554 [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalez E., Müller S., Hertlein G., Heid N., Süssmuth R. D. et al. , 2014. Biological effects of paenilamicin, a secondary metabolite antibiotic produced by the honey bee pathogenic bacterium Paenibacillus larvae. MicrobiologyOpen 3: 642–656. 10.1002/mbo3.195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C., Nie X., Tang Z., Zhang Y., Lin J. et al. , 2016. A novel serine protease, Sep1, from Bacillus firmus DS-1 has nematicidal activity and degrades multiple intestinal-associated nematode proteins. Sci. Rep. 6: 25012 10.1038/srep25012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C., Tang Z., Peng D., Shao Z., Zhu L. et al. , 2014. Draft genome sequence of Bacillus firmus DS1. J. Biotechnol. 177: 20–21. 10.1016/j.jbiotec.2014.02.012 [DOI] [PubMed] [Google Scholar]
- Giannakou I. O., Karpouzas D. G., and Prophetou-Athanasiadou D., 2004. A novel non-chemical nematicide for the control of root-knot nematodes. Appl. Soil Ecol. 26: 69–79. 10.1016/j.apsoil.2003.09.002 [DOI] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P. et al. , 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57: 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Hu Y., Yan C., Hsu C.-H., Chen Q.-R., Niu K. et al. , 2014. OmicCircos: A simple-to-use R package for the circular visualization of multidimensional omics data. Cancer Inform. 13: 13–20. 10.4137/CIN.S13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L. P., Szklarczyk D., Jensen L. J. et al. , 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34: 2115–2122. 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husseiny S. M., Bayoumi R. A., El-Gamal M. S., Ahmad A. I., and Khashaba H. M., 2008. Production and purification of pectinase, avicelase and carboxymethyl cellulase by fermentation of agro-industrial wastes using B. firmus and B. laterosporus. N. Egypt. J. Microbiol. 19: 326–352. [Google Scholar]
- Kanehisa M., Sato Y., Kawashima M., Furumichi M., and Tanabe M., 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44: D457–D462. 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Zur M., Antonov J., Bercovitz A., Feldman K., Husid A. et al. , 2000. Bacillus firmus formulations for the safe control of root-knot nematodes, pp. 47–52 in Proceedings of the Brighton Crop Protection Conference on Pests and Diseases, Farnham, Surrey: : British Crop Protection Council, Brighton. [Google Scholar]
- Khalil M. S., 2013. Abamectin and azadirachtin as eco-friendly promising biorational tools in integrated nematodes management programs. J. Plant Pathol. Microbiol. 4: 174 10.4172/2157-7471.1000174 [DOI] [Google Scholar]
- Kloepper J. W., Ryu C.-M., and Zhang S., 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94: 1259–1266. 10.1094/PHYTO.2004.94.11.1259 [DOI] [PubMed] [Google Scholar]
- Koppisch A. T., Browder C. C., Moe A. L., Shelley J. T., Kinkel B. A. et al. , 2005. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals 18: 577–585. 10.1007/s10534-005-1782-6 [DOI] [PubMed] [Google Scholar]
- Laetsch D. R., and Blaxter M. L., 2017. BlobTools: Interrogation of genome assemblies. F1000 Res. 6: 1287 10.12688/f1000research.12232.1 [DOI] [Google Scholar]
- Law V., Knox C., Djoumbou Y., Jewison T., Guo A. C. et al. , 2014. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42: D1091–D1097. 10.1093/nar/gkt1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. D., 2019. GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35: 4162–4164. 10.1093/bioinformatics/btz188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian L. H., Tian B. Y., Xiong R., Zhu M. Z., Xu J. et al. , 2007. Proteases from Bacillus: a new insight into the mechanism of action for rhizobacterial suppression of nematode populations. Lett. Appl. Microbiol. 45: 262–269. 10.1111/j.1472-765X.2007.02184.x [DOI] [PubMed] [Google Scholar]
- Mao C., Abraham D., Wattam A. R., Wilson M. J. C., Shukla M. et al. , 2015. Curation, integration and visualization of bacterial virulence factors in PATRIC. Bioinformatics 31: 252–258. 10.1093/bioinformatics/btu631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur A. G., Waglechner N., Nizam F., Yan A., Azad M. A. et al. , 2013. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57: 3348–3357. 10.1128/AAC.00419-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro S. M. S., Clemente J. J., Carrondo M. J. T., and Cunha A. E., 2014. Enhanced spore production of Bacillus subtilis grown in a chemically defined medium. Adv. Microbiol. 4: 444–454. 10.4236/aim.2014.48049 [DOI] [Google Scholar]
- Morgan, M., H. Pagès, V. Obenchain, and N. Cahoon, 2019 Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 3.8. 10.18129/B9.bioc.Rsamtools [DOI]
- Nikolenko S. I., Korobeynikov A. I., and Alekseyev M. A., 2013. BayesHammer: Bayesian clustering for error correction in single-cell sequencing. BMC Genomics 14: S7 10.1186/1471-2164-14-S1-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noroozi M., Amoozegar M. A., Pourbabaei A. A., Naghavi N. S., and Nourmohammadi Z., 2017. Isolation and characterization of mercuric reductase by newly isolated halophilic bacterium, Bacillus firmus MN8. Global J. Environ. Sci. Manage. 3: 427–436. [Google Scholar]
- R Core Team , 2018. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rahman A., Srivastava S. S., Sneh A., Ahmed N., and Krishnasastry M. V., 2010. Molecular characterization of tlyA gene product, Rv1694 of Mycobacterium tuberculosis: a non-conventional hemolysin and a ribosomal RNA methyl transferase. BMC Biochem. 11: 35 10.1186/1471-2091-11-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod M. G., and Pathak A. P., 2018. Efficient decolorization of textile dyes by alkaline protease producing bacterial consortia. Indian J. Geo-Mar. Sci. 47: 1468–1477. [Google Scholar]
- Regulation (EC) No 1107/2009 of the European Parliament and of the Council of October 21, 2009 concerning the placing of plant protection products on the market and repealing Council Directives 79/117/EEC and 91/414/EEC, 2009. https://eur-lex.europa.eu/eli/reg/2009/1107/oj/eng
- Richter M., and Rosselló-Móra R., 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106: 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R., Oliver Glöckner F., and Peplies J., 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32: 929–931. 10.1093/bioinformatics/btv681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., 1999. Twilight zone of protein sequence alignments. Protein Eng. 12: 85–94. 10.1093/protein/12.2.85 [DOI] [PubMed] [Google Scholar]
- Saier M. H., Reddy V. S., Tsu B. V., Ahmed M. S., Li C. et al. , 2016. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 44: D372–D379. 10.1093/nar/gkv1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasser J. N., 1977. Worldwide dissemination and importance of the root-knot nematodes, Meloidogyne spp. J. Nematol. 9: 26–29. [PMC free article] [PubMed] [Google Scholar]
- Schaeffer A. B., and Fulton M. D., 1933. A Simplified Method of Staining Endospores. Science 77: 194 10.1126/science.77.1990.194 [DOI] [PubMed] [Google Scholar]
- Schomburg I., Chang A., Ebeling C., Gremse M., Heldt C. et al. , 2004. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 32: D431–D433. 10.1093/nar/gkh081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibulal B., Al-Bahry S., Al-Wahaibi Y., Elshafie A., Al-Bemani A. et al. , 2018. Microbial-enhanced heavy oil recovery under laboratory conditions by Bacillus firmus BG4 and Bacillus halodurans BG5 isolated from heavy oil fields. Colloids Interfaces 2: 1–18. 10.3390/colloids2010001 [DOI] [Google Scholar]
- Silva S. D., Carneiro R. M. D. G., Faria M., Souza D. A., Monnerat R. G. et al. , 2017. Evaluation of Pochonia chlamydosporia and Purpureocillium lilacinum for suppression of Meloidogyne enterolobii on tomato and banana. J. Nematol. 49: 77–85. 10.21307/jofnem-2017-047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J. S., Pandey V. C., and Singh D. P., 2011. Efficient soil microorganisms: A new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 140: 339–353. 10.1016/j.agee.2011.01.017 [DOI] [Google Scholar]
- Susič, N., S. Širca, P. Strajnar, and B. Gerič Stare, 2019 Assessing the nematicidal activity of Bacillus firmus strains, pp. 134 in Abstract volume of the 14th Slovenian Conference on Plant Protection with International Participation, Maribor, Slovenia. [Google Scholar]
- Takaya A., Sato Y., Shoji T., and Yamamoto T., 2013. Methylation of 23S rRNA nucleotide G748 by RlmAII methyltransferase renders Streptococcus pneumoniae telithromycin susceptible. Antimicrob. Agents Chemother. 57: 3789–3796. 10.1128/AAC.00164-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. W., Sutherland A. D., Aidoo K. E., and Logan N. A., 2005. Heat-stable toxin production by strains of Bacillus cereus, Bacillus firmus, Bacillus megaterium, Bacillus simplex and Bacillus licheniformis. FEMS Microbiol. Lett. 242: 313–317. 10.1016/j.femsle.2004.11.022 [DOI] [PubMed] [Google Scholar]
- Tian B., Yang J., and Zhang K.-Q., 2007. Bacteria used in the biological control of plant-parasitic nematodes: populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 61: 197–213. 10.1111/j.1574-6941.2007.00349.x [DOI] [PubMed] [Google Scholar]
- Trudgill D. L., and Blok V. C., 2001. Apomictic, polyphagous root-knot nematodes: exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 39: 53–77. 10.1146/annurev.phyto.39.1.53 [DOI] [PubMed] [Google Scholar]
- Valencia L., and Kotcon J. B., 2016. Efficacy and Mode of Action of Bacillus firmus as a bionematicide for the northern root-knot nematode, Meloidogyne hapla, and dagger nematode, Xiphinema americanum. J. Nematol. 48: 378–379. [Google Scholar]
- Wang Y., Coleman-Derr D., Chen G., and Gu Y. Q., 2015. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43: W78–W84. 10.1093/nar/gkv487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A. R., Davis J. J., Assaf R., Boisvert S., Brettin T. et al. , 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45: D535–D542. 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing, New York. [Google Scholar]
- Wilson M. K., Abergel R. J., Arceneaux J. E. L., Raymond K. N., and Byers B. R., 2010. Temporal production of the two Bacillus anthracis siderophores, petrobactin and bacillibactin. Biometals 23: 129–134. 10.1007/s10534-009-9272-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. J., and Jackson T. A., 2013. Progress in the commercialisation of bionematicides. BioControl 58: 715–722. 10.1007/s10526-013-9511-5 [DOI] [Google Scholar]
- Wohlert S.-E., Lomovskaya N., Kulowski K., Fonstein L., Occi J. L. et al. , 2001. Insights about the biosynthesis of the avermectin deoxysugar L-oleandrose through heterologous expression of Streptomyces avermitilis deoxysugar genes in Streptomyces lividans. Chem. Biol. 8: 681–700. 10.1016/S1074-5521(01)00043-6 [DOI] [PubMed] [Google Scholar]
- Yang S.-C., Lin C.-H., Sung C. T., and Fang J.-Y., 2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 5: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S.-H., Ha S.-M., Kwon S., Lim J., Kim Y. et al. , 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67: 1613–1617. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Smith D. K., Zhu H., Guan Y., and Lam T. T.-Y., 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8: 28–36. 10.1111/2041-210X.12628 [DOI] [Google Scholar]
- Zerbino D. R., and Birney E., 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Zheng J., Zhang Z., Peng D., and Sun M., 2016. Nematicidal spore-forming Bacilli share similar virulence factors and mechanisms. Sci. Rep. 6: 31341 10.1038/srep31341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Han B., Kumar P., Liu X., Ma X. et al. , 2010. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 38: D787–D791. 10.1093/nar/gkp1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence data, including raw sequence reads, genome assemblies and annotations of B. firmus I-1582 and Bacillus sp. ZZV12-4809 used in this study were submitted to NCBI, GenBank under the BioProject accession ID: PRJNA533096. Supplemental material available at figshare: https://doi.org/10.25387/g3.11522544.