Abstract
Background
Large‐cell neuroendocrine carcinoma of the lung (LCNEC) and small‐cell lung carcinoma (SCLC) are neuroendocrine neoplasms. However, the underlying mechanisms of common and distinct genetic characteristics between LCNEC and SCLC are currently unclear. Herein, protein expression profiles and possible interactions with miRNAs were provided by integrated bioinformatics analysis, in order to explore core genes associated with tumorigenesis and prognosis in SCLC and LCNEC.
Methods
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037 gene expression profiles were obtained from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) in LCNEC and SCLC, as compared with normal lung tissues, were selected using the GEO2R online analyzer and Venn diagram software. Gene ontology (GO) analysis was performed using Database for Annotation, Visualization and Integrated Discovery. The biological pathway analysis was performed using the FunRich database. Subsequently, a protein–protein interaction (PPI) network of DEGs was generated using Search Tool for the Retrieval of Interacting Genes and displayed via Cytoscape software. The PPI network was analyzed by the Molecular Complex Detection app from Cytoscape, and 16 upregulated hub genes were selected. The Oncomine database was used to detect expression patterns of hub genes for validation. Furthermore, the biological pathways of these 16 hub genes were re‐analyzed, and potential interactions between these genes and miRNAs were explored via FunRich.
Results
A total of 384 DEGs were identified. A Venn diagram determined 88 common DEGs. The PPI network was constructed with 48 nodes and 221 protein pairs. Among them, 16 hub genes were extracted, 14 of which were upregulated in SCLC samples, as compared with normal lung specimens, and 10 were correlated with the cell cycle pathway. Furthermore, 57 target miRNAs for 8 hub genes were identified, among which 31 miRNAs were correlated with the progression of carcinoma, drug‐resistance, radio‐sensitivity, or autophagy in lung cancer.
Conclusion
This study provided effective biomarkers and novel therapeutic targets for diagnosis and prognosis of SCLC and LCNEC.
Keywords: bioinformatics analysis, biomarker, LCNEC, lung cancer, SCLC
Common and distinct features of DEGs of SCLC and LCNEC were identified by integrated bioinformatics methods. Genes related to the cell cycle pathway were discussed. Target miRNAs for hub genes were identified and discussed.
1. INTRODUCTION
High‐grade pulmonary neuroendocrine neoplasms (NENs), small‐cell lung cancer (SCLC), and large‐cell neuroendocrine carcinoma of the lung (LCNEC) are among the malignancies with the highest mortality rates. Since the revised 2015 World Health Organization classification of lung tumors classified LCNEC as a NEN (Travis, Brambilla, Burke, Marx, & Nicholson, 2015), debates and investigations of the new classification were ceaseless. LCNEC was grouped with NENs due to its similarities with SCLC in terms of morphology, DNA methylation, protein levels, mutational aspects, and transcriptional levels (Aly et al., 2019; Karlsson et al., 2015, 2014; Miyoshi et al., 2017; Rossi et al., 2005; Simbolo et al., 2017), in spite of a certain extent of heterogeneity (Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM), 2013; Rossi et al., 2005; Simbolo et al., 2017). In addition, despite the differences in histopathological characteristics between SCLC and LCNEC, their clinical characteristics are similar, including a generally poor outcome, greater incidence in men than in women, and advanced stage diagnosis (Asamura et al., 2006; Cerilli, Ritter, Mills, & Wick, 2001; Xu et al., 2019). With regard to treatment, individuals with SCLC and LCNEC may manifest different sensitivities to different types of treatment, owing to gene alterations between SCLC‐like LCNEC and NSCLC‐like LCNEC (Rekhtman et al., 2016).
Genome‐based medicine, which has been used in oncology for >10 years, can rapidly detect differentially expressed genes and provide greater chances of reducing malignancy‐related morbidity and mortality (Vogelstein et al., 2013). Therefore, a number of valuable clues obtained from the SCLC regulation and LCNEC development could be investigated from different perspectives using gene chips. However, the understanding of the genetic expression of these two types of NEN is limited. Genetic analysis was therefore conducted to clarify the genomic characteristics of NENs and determine the similarities between SCLC and LCNEC, consequently identifying more potential genetic candidates for treatment.
In this study, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037 expression microarray data were downloaded from the Gene Expression Omnibus (GEO) database. DEGs were analyzed and their interaction patterns in this dataset were revealed through a protein–protein interaction (PPI) network. In addition, possible DEG functions were explored by gene and biological pathway enrichment analysis. The functions of the significant genes involved in major function terms were discussed, and a number of them might be novel candidates for SCLC and LCNEC treatment. The workflow of the present study is depicted in Figure 1 (free images were obtained from Servier Medical Art).
Figure 1.
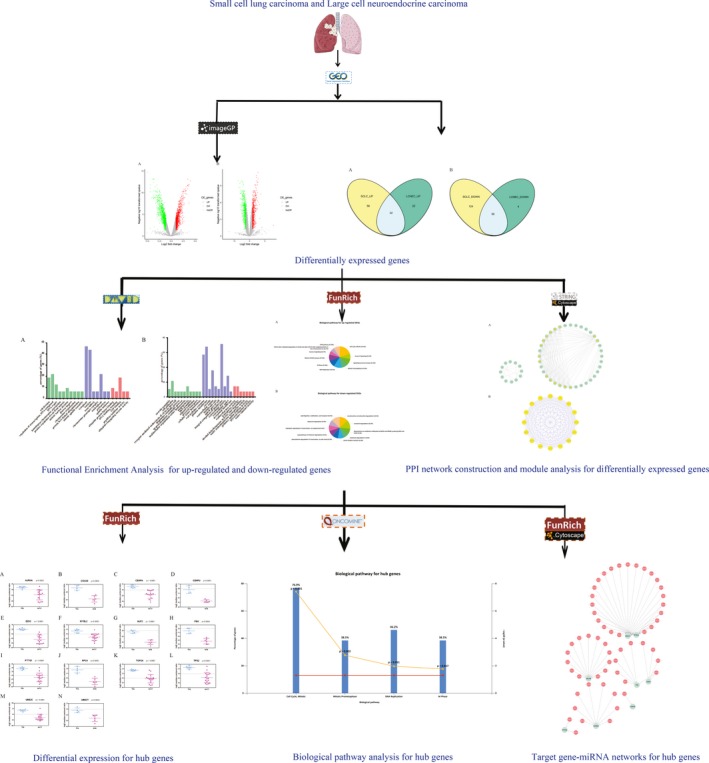
Workflow for identification of hub genes and pathways in small cell lung carcinoma and large cell neuroendocrine carcinoma of the lung. Step 1: We identified the differentially expressed genes commonly in small cell lung carcinoma and large cell neuroendocrine carcinoma of the lung. Step 2: The GO and biological pathway analysis were performed and the PPI network was constructed. Next, we identified the hub genes from the PPI network. Step 3: We validated expression patterns and re‐analysis biological pathway of hub genes. Then, the target miRNAs associated with hub genes were identified and gene‐miRNA networks were constructed. Free images were obtained from Servier Medical Art
2. MATERIALS AND METHODS
2.1. Ethical compliance
This article does not contain any studies with human participants or animals performed by any of the authors, and therefore no ethical compliance is required.
2.2. Gene expression profile data
Gene expression profiles (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037) were obtained from the GEO database, based on the GPL962 CHUGAI 41K platform. Data including 18 primary normal lung tissues, 8 primary LCNEC tissues, 15 primary small‐cell lung carcinoma tissues, and 9 small‐cell lung carcinoma cell lines were downloaded. The profiles of differentially expressed genes in SCLC and LCNEC are presented in a volcano plot online http://www.ehbio.com/ImageGP/.
2.3. Data processing of DEGs
The mRNA expression raw data were analyzed by GEO2R online tools (Davis & Meltzer, 2007). To identify DEGs in SCLC and LCNEC, as compared with normal lung specimens, |logFC| > 2 and adjusted p < .05 were used as the threshold. A Venn diagram was drawn to visualize shared genes in SCLC and LCNEC specimens.
2.4. Functional enrichment analysis of DEGs
Annotation, Visualization and Integrated Discovery (DAVID) (Huang da, Sherman, & Lempicki, 2009a, 2009b), a bioinformatics online tool, provides functional annotations and gene analysis. Purpose genes were integrated using Gene ontology (GO) analysis in DAVID, to identify their biological properties. Biological pathway analysis of DEGs and hub genes was performed using FunRich software (Pathan et al., 2017), a self‐contained software tool used for function enrichment and interaction network evaluation of sets of genes and proteins.
2.5. PPI network construction and module analysis
Search Tool for the Retrieval of Interacting Genes (STRING) (Szklarczyk et al., 2015)) is a database of known and potential PPIs. DEGs were evaluated by STRING and a combined score of ≥0.4 was set as the cut‐off value. The network was visualized by Cytoscape (Shannon et al., 2003) and the Cytoscape app Molecular Complex Detection (MCODE) was then used to examine marked modules of the PPI network, with the advanced option set to a max. depth of 100, degree cut‐off of 2, k‐core of 2, and node score cut‐off of 0.2.
2.6. Oncomine analysis
To validate the hub genes, the Oncomine website (http://www.oncomine.org) was used to obtain mRNA expression data to compare SCLC and normal lung tissues from Bhattacharjee et al. (2001) and Garber et al. (2001).
2.7. Prediction of target miRNAs for DEGs
To detect predicted miRNAs corresponding to core genes selected from MCODE, miRNA enrichment in FunRich software was performed and miRNAs found to interact with hub genes were defined. The results were input into and visualized by Cytoscape.
2.8. Statistical analysis
In this study, a student's t‐test was carried out using GraghPad Prism 5 to generate a p‐value for the comparison of cancerous specimens and normal control samples. p < .05 was considered to indicate a statistically significant difference.
3. RESULTS
3.1. Identification of DEGs
DEGs in the SCLC and LCNEC samples, as compared with normal ones, were determined using GEO2R online analyzer (|logFC| > 2 and adjusted p < .05) (Davis & Meltzer, 2007). A total of 270 and 114 DEGs were extracted from SCLC and LCNEC samples, respectively. A “volcano plot” between p‐values and fold changes was established to plot the differentially expressed genes (Figure 2). A Venn diagram was then drawn to confirm the shared DEGs in the two subtypes (Figure 3). Finally, 88 common DEGs were confirmed, including 56 downregulated and 32 upregulated genes in the SCLC and LCNEC samples (Table 1).
Figure 2.
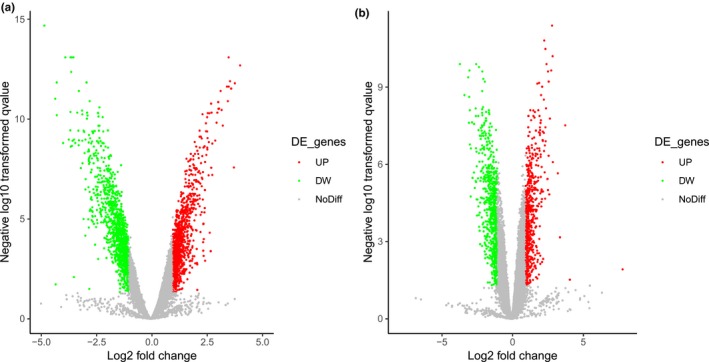
Volcano plot of the DEGs. (a) Volcano plot for the DEGs in small cell lung carcinoma, as compared with normal lung samples. (b) Volcano plot for the DEGs in large cell neuroendocrine carcinoma of the lung, as compared with normal lung samples. The x‐axis demonstrates the log 2 fold change, and the y‐axis demonstrates the log10 (adjusted p‐value). The red dots indicate up‐regulated genes, and the green dots indicate down‐regulated genes. The DEGs were filtered on the basis of a |log 2 fold change|> 1.0 and an adjusted p‐value of < .05. The grey dots represent genes with no significant difference. DEGs, differentially expressed genes
Figure 3.
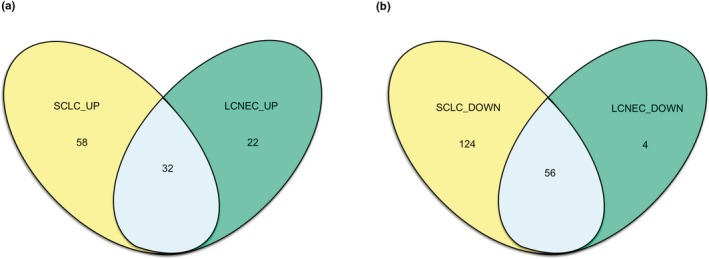
Identification of 88 common DEGs in two types of NENs (SCLC and LCNEC) via Venn diagrams. (a) 32 DEGs are upregulated in the two types of NENs (logFC > 0). (b) 56 DEGs are downregulated in the two types of NENs (logFC < 0). Different color meant different types. SClC, small cell lung carcinoma. LCNEC, large cell neuroendocrine carcinoma of the lung
Table 1.
Commonly DEGs in the SCLC and LCNEC samples
| DEGs | Genes name |
|---|---|
| Upregulated | ZNF829 BCCIP PTTG1 CENPA FAM123A SESN2 CSE1L LOC256676///CDCA5 PBK UBE2TC5orf13 ZNF587 UNQ353///CSNK1G1///KIAA0101 HMGB3 SOX4 FARP1 C1orf215///STMN1 MLF1IP AURKA MYBL2 ZWINTAS///ZWINT UBE2C EZH2 RFC4 CDCA8 SLC2A1 LOC253012 TPX2 TTKCDCA7 TOP2A NUF2 |
| Downregulated | ADH1B CAV1 WIF1 SFTPC FMO2 TSHZ2///ADAMTS8 USP42 MFAP4 SCGB1A1 IGSF10 TIMP3 CLDN18 OGN CAV2 MAOA FABP4 ZNF521 SVEP1 EPAS1 TPSB2///TPSAB1 FHL1 FAT4 CAB39L ADH1C PI4KAP1 C2orf34 AGER LOC144571///PZP///A2M AQP4 FAM75A1 SFTPH AQP1 CAMP HBE1 EMCN HNRNPH1 SLC39A8 SUSD2 FLJ44379 LIMCH1 SLPI CYBRD1 HOPX C13orf15 LOC100131277///TACC1 LYVE1 GPR116 MAOB SELENBP1 PGM5 EMP2 PMP22 S1PR1 ABCA8 ANGPTL1 RGS2 |
Abbreviations: DEGs, differentially expressed genes; LCNEC, large cell neuroendocrine carcinoma; SCLC, small cell lung carcinoma.
3.2. Enrichment analysis
All 88 DEGs were analyzed using DAVID and FunRich software. The GO analysis results were as follows: (a) For biological processes (BP), the significantly enriched terms of upregulated DEGs were cell division, regulation of transcription, DNA repair, homologous chromosome segregation, positive regulation of exit from mitosis, mitotic nuclear division, glucose import, chromosome organization, protein K11‐linked ubiquitination, mitotic spindle organization and mitotic spindle assembly, and the significant terms of downregulated DEGs were caveola assembly, cell adhesion, receptor‐mediated endocytosis of virus by host cell, dopamine catabolic process, neurotransmitter catabolic process, multicellular organismal water homeostasis, angiogenesis, bleb assembly, ethanol oxidation, glycerol transport, and cellular water homeostasis; (b) for molecular function (MF), the notably enriched terms of upregulated DEGs were ubiquitin protein ligase binding, DNA binding, adenosine triphosphate (ATP) binding, peptidase inhibitor activity and ubiquitin conjugating enzyme activity, and those for downregulated DEGs were oxidoreductase, transporter, alcohol dehydrogenase (zinc‐dependent), water transmembrane transporter, primary amine oxidase, glycerol channel and water channel activities; (c) for GO cell component (CC), the upregulated DEGs were particularly enriched in cytoplasm, nucleus, axon hillock, chromosome passenger complex, nucleoplasm, pronucleus and spindle midzone, and downregulated DEGs in extracellular exosome, plasma membrane, caveola, extracellular region, membrane raft, apical part of cell, integral component of membrane, extracellular matrix, extracellular space, and acrosomal membrane (Table 2). These results are displayed in bar graphs produced on GraghPad Prism 5 (Figure 4).
Table 2.
Enriched Go terms of the DEGs
| Category | Term | Count | % | p‐Value | FDR | |
|---|---|---|---|---|---|---|
| Up regulated | GOTERM_BP_DIRECT | GO:0051301~cell division | 6 | 0.142993327 | 4.04E‐06 | 0.004971483 |
| GOTERM_BP_DIRECT | GO:0006355~regulation of transcription DNA‐templated | 7 | 0.166825548 | 3.53E‐04 | 0.432578524 | |
| GOTERM_BP_DIRECT | GO:0006281~DNA repair | 4 | 0.095328885 | 8.15E‐04 | 0.997007591 | |
| GOTERM_BP_DIRECT | GO:0045143~homologous chromosome segregation | 2 | 0.047664442 | .006639221 | 7.86341138 | |
| GOTERM_BP_DIRECT | GO:0031536~positive regulation of exit from mitosis | 2 | 0.047664442 | .008292426 | 9.731016783 | |
| GOTERM_BP_DIRECT | GO:0007067~mitotic nuclear division | 3 | 0.071496663 | .013030094 | 14.89218414 | |
| GOTERM_BP_DIRECT | GO:0046323~glucose import | 2 | 0.047664442 | .018156528 | 20.17047326 | |
| GOTERM_BP_DIRECT | GO:0051276~chromosome organization | 2 | 0.047664442 | .023053317 | 24.9298377 | |
| GOTERM_BP_DIRECT | GO:0070979~protein K11‐linked ubiquitination | 2 | 0.047664442 | .037603917 | 37.57710332 | |
| GOTERM_BP_DIRECT | GO:0007052~mitotic spindle organization | 2 | 0.047664442 | .037603917 | 37.57710332 | |
| GOTERM_BP_DIRECT | GO:0090307~mitotic spindle assembly | 2 | 0.047664442 | .045597774 | 43.66110836 | |
| GOTERM_CC_DIRECT | GO:0005737~cytoplasm | 15 | 0.357483317 | 2.21E‐05 | 0.022280174 | |
| GOTERM_CC_DIRECT | GO:0005634~nucleus | 14 | 0.333651096 | 5.40E‐05 | 0.054460588 | |
| GOTERM_CC_DIRECT | GO:0043203~axon hillock | 2 | 0.047664442 | .00741693 | 7.237425098 | |
| GOTERM_CC_DIRECT | GO:0032133~chromosome passenger complex | 2 | 0.047664442 | .00741693 | 7.237425098 | |
| GOTERM_CC_DIRECT | GO:0005654~nucleoplasm | 7 | 0.166825548 | .010502317 | 10.10650713 | |
| GOTERM_CC_DIRECT | GO:0045120~pronucleus | 2 | 0.047664442 | .013312558 | 12.64989615 | |
| GOTERM_CC_DIRECT | GO:0051233~spindle midzone | 2 | 0.047664442 | .022093237 | 20.18460143 | |
| GOTERM_MF_DIRECT | GO:0031625~ubiquitin protein ligase binding | 3 | 0.076628352 | .011662791 | 10.93819391 | |
| GOTERM_MF_DIRECT | GO:0008301~DNA binding bending | 2 | 0.051085568 | .015851015 | 14.59560577 | |
| GOTERM_MF_DIRECT | GO:0005524~ATP binding | 6 | 0.153256705 | .027091606 | 23.75384229 | |
| GOTERM_MF_DIRECT | GO:0030414~peptidase inhibitor activity | 2 | 0.051085568 | .030054212 | 26.01571812 | |
| GOTERM_MF_DIRECT | GO:0061631~ubiquitin conjugating enzyme activity | 2 | 0.051085568 | .038482685 | 32.12464677 | |
| Down regulated | GOTERM_BP_DIRECT | GO:0070836~caveola assembly | 3 | 0.046260601 | 6.99E−05 | 0.098559412 |
| GOTERM_BP_DIRECT | GO:0007155~cell adhesion | 6 | 0.092521203 | .007419687 | 9.977332032 | |
| GOTERM_BP_DIRECT | GO:0019065~receptor‐mediated endocytosis of virus by host cell | 2 | 0.030840401 | .008018493 | 10.74080337 | |
| GOTERM_BP_DIRECT | GO:0042420~dopamine catabolic process | 2 | 0.030840401 | .013329193 | 17.25342044 | |
| GOTERM_BP_DIRECT | GO:0042135~neurotransmitter catabolic process | 2 | 0.030840401 | .021243157 | 26.14357095 | |
| GOTERM_BP_DIRECT | GO:0050891~multicellular organismal water homeostasis | 2 | 0.030840401 | .021243157 | 26.14357095 | |
| GOTERM_BP_DIRECT | GO:0001525~angiogenesis | 4 | 0.061680802 | .021777665 | 26.71078904 | |
| GOTERM_BP_DIRECT | GO:0032060~bleb assembly | 2 | 0.030840401 | .026484616 | 31.53390357 | |
| GOTERM_BP_DIRECT | GO:0006069~ethanol oxidation | 2 | 0.030840401 | .031698626 | 36.53140253 | |
| GOTERM_BP_DIRECT | GO:0015793~glycerol transport | 2 | 0.030840401 | .034295382 | 38.89185575 | |
| GOTERM_BP_DIRECT | GO:0009992~cellular water homeostasis | 2 | 0.030840401 | .034295382 | 38.89185575 | |
| GOTERM_CC_DIRECT | GO:0070062~extracellular exosome | 16 | 0.246723207 | .001317986 | 1.408901211 | |
| GOTERM_CC_DIRECT | GO:0005886~plasma membrane | 19 | 0.292983809 | .003743762 | 3.955034143 | |
| GOTERM_CC_DIRECT | GO:0005901~caveola | 3 | 0.046260601 | .010295194 | 10.53630766 | |
| GOTERM_CC_DIRECT | GO:0005576~extracellular region | 10 | 0.154202005 | .01150877 | 11.70950819 | |
| GOTERM_CC_DIRECT | GO:0045121~membrane raft | 4 | 0.061680802 | .012603718 | 12.75603307 | |
| GOTERM_CC_DIRECT | GO:0045177~apical part of cell | 3 | 0.046260601 | .013532583 | 13.63498448 | |
| GOTERM_CC_DIRECT | GO:0016021~integral component of membrane | 20 | 0.308404009 | .018955008 | 18.60768595 | |
| GOTERM_CC_DIRECT | GO:0031012~extracellular matrix | 4 | 0.061680802 | .032469722 | 29.89178143 | |
| GOTERM_CC_DIRECT | GO:0005615~extracellular space | 8 | 0.123361604 | .036771948 | 33.17391345 | |
| GOTERM_CC_DIRECT | GO:0002080~acrosomal membrane | 2 | 0.030840401 | .043912815 | 38.31524409 | |
| GOTERM_MF_DIRECT | GO:0016491~oxidoreductase activity | 4 | 0.061680802 | .014279333 | 14.99791487 | |
| GOTERM_MF_DIRECT | GO:0005215~transporter activity | 4 | 0.061680802 | .014662761 | 15.37073704 | |
| GOTERM_MF_DIRECT | GO:0004024~alcohol dehydrogenase activity zinc‐dependent | 2 | 0.030840401 | .015188693 | 15.87969632 | |
| GOTERM_MF_DIRECT | GO:0005372~water transmembrane transporter activity | 2 | 0.030840401 | .015188693 | 15.87969632 | |
| GOTERM_MF_DIRECT | GO:0008131~primary amine oxidase activity | 2 | 0.030840401 | .015188693 | 15.87969632 | |
| GOTERM_MF_DIRECT | GO:0015254~glycerol channel activity | 2 | 0.030840401 | .030151975 | 29.24210187 | |
| GOTERM_MF_DIRECT | GO:0015250~water channel activity | 2 | 0.030840401 | .040003884 | 36.95130237 |
Abbreviations: DEGs, differentially expressed genes; Go, Gene ontology.
Figure 4.
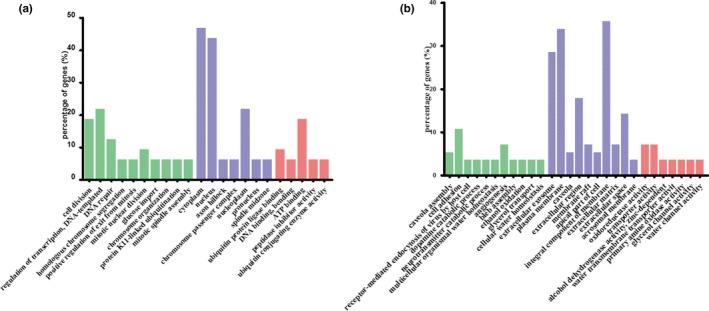
GO terms of overlapped differentially expressed genes. (a) Up‐regulated genes. (b) Down‐regulated genes. Green bars, biological process; blue bars, cellular component; red bars, molecular function. GO, Gene ontology
The biological pathway analysis results are shown in Figure 5, indicating that upregulated DEGs were notably enriched in cell cycle (mitotic), DNA replication, ataxia telangiectasia‐mutated (ATM) pathway, mitotic M‐M/G1 phases, signaling by Aurora kinases, M phase, mitotic prometaphase, while downregulated DEGs in lipid digestion, mobilization and transport, noradrenaline and adrenaline degradation, and serotonin degradation (Figure 5).
Figure 5.
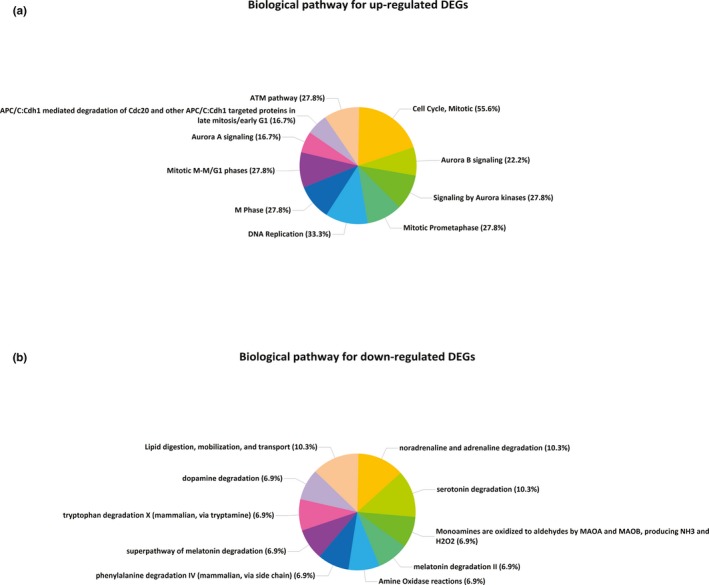
Biological pathway analysis for DEGs. (a) Biological pathways for upregulated DEGs. (b) Biological pathways for downregulated DEGs. DEGs, differentially expressed genes
3.3. PPI network and module analysis
Using STRING, a PPI network containing 48 nodes and 221 protein pairs was constructed, with a combined score of >0.4. As shown in Figure 6, most nodes in the PPI network were upregulated DEGs in SCLC and LCNEC specimens. Next, the MCODE app in Cytoscape was used for further analysis, which showed that 16 of 48 nodes were hub nodes (Figure 6). These core nodes were all upregulated genes: Enhancer of zeste homolog 2 (EZH2, OMIM:601573), cell division cycle‐associated 1 (CDCA1/NUF2, OMIM:611772), PDZ binding kinase, CDCA8(OMIM:609977), CDCA7(OMIM:609937), aurora kinase A (AURKA, OMIM:603072), replication factor C4 (RFC4, OMIM:102577), ubiquitin conjugating enzyme E2 C (UBE2C, OMIM:605574), centromere protein A (CENPA, OMIM:117139), targeting protein for xklp2 (TPX2, OMIM:605917), TTK protein kinase (TTK, OMIM:604092), V‐Myb avian myeloblastosis viral oncogene homolog‐like 2 (MYBL2, OMIM:601415), pituitary tumor transforming gene 1 (PTTG1, OMIM:604147), DNA topoisomerase II α (TOP2A, OMIM:126430), centromere protein U (CENPU, OMIM:611511), and UBE2T( OMIM:610538).
Figure 6.
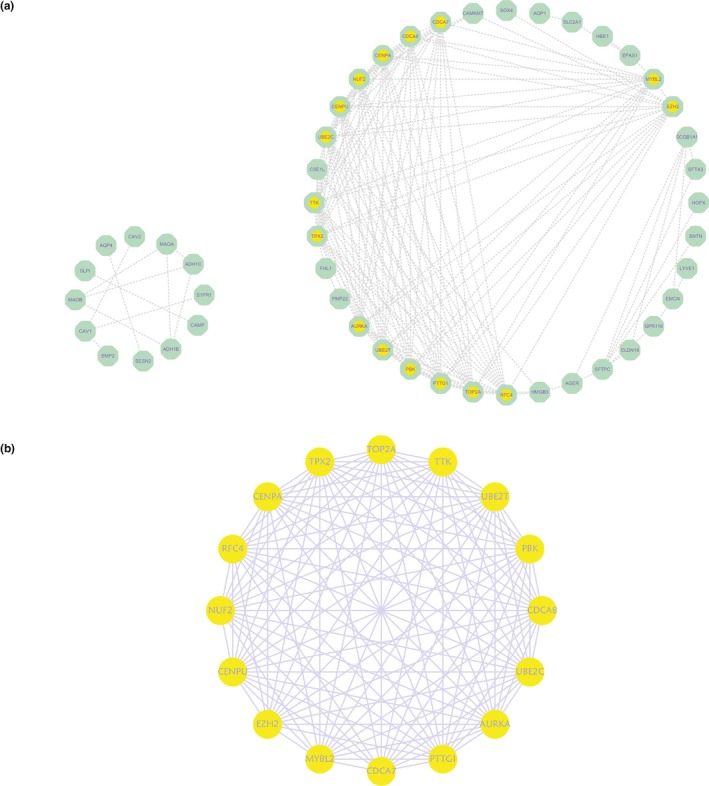
Common DEGs PPI network constructed by STRING online database and Module analysis. (a) There were a total of 48 DEGs in the DEGs PPI network complex. (b) The most significant module in the PPI network of the DEGs Module analysis of Cytoscape software (degree cutoff = 2, node score cutoff = 0.2, k‐core = 2, and max. depth = 100). The nodes meant proteins; the edges meant the interaction of proteins; yellow nodes meant DEGs with high marks. DEGs, differentially expressed genes
3.4. Analysis of core genes by the Oncomine database
The Oncomine database (http://www.oncomine.org) was used to determine the expression patterns of 16 hub genes between normal and cancerous lung tissues. Due to the rarity of LCNEC, the expression pattern was only obtained in SCLC samples and compared with normal lung samples. The results showed that, except for TTK (deficiency in expression data), 14 of 15 genes were highly expressed in SCLC specimens, as compared with normal ones, which was consistent with the http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037 data (Figure 7).
Figure 7.
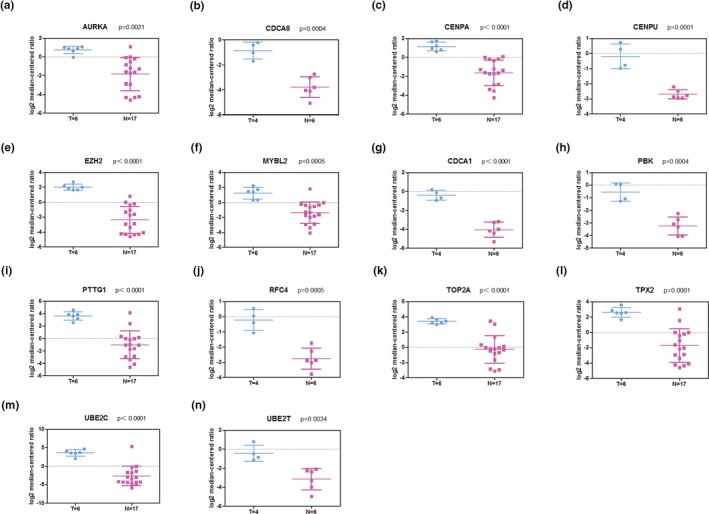
Significantly expressed 14 genes in SCLC tissues, as compared with normal tissues. SCLC, small cell lung carcinoma
3.5. Re‐analysis of hub genes by biological pathway analysis
To better apprehend the pathway correlated with these 16 selected DEGs, biological pathway analysis was performed again by FunRich. The results showed that these 16 core genes were markedly enriched in the cell cycle pathway, mitotic prometaphase, DNA replication and M phase (Figure 8). Among them the cell cycle pathway was the most markedly enriched, containing 10 genes (CDCA1, CDCA8, AURKA, RFC4, UBE2C, CENPA, CENPU, MYBL2, PTTG1, and TOP2A) (p = 2.30182e‐11).
Figure 8.
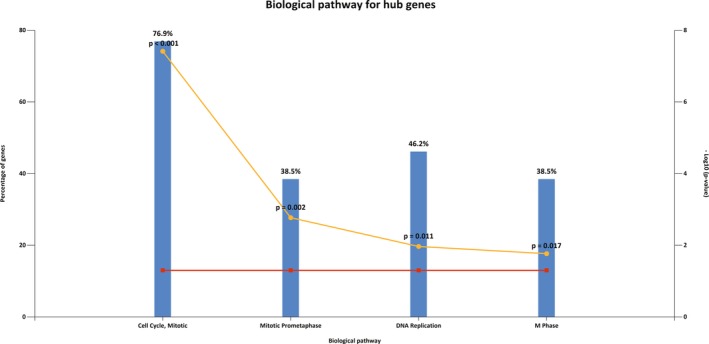
Re‐analysis of 16 hub genes by biological pathway enrichment. X axis represents significant biological pathway terms; Y axis represents percentage of genes or –log10 (p‐value)
3.6. Prediction of target miRNAs for hub genes
The target human miRNAs for hub genes were determined by FunRich software, and the mRNA‐miRNA network was displayed by Cytoscape (Figure 9). The genes found to be connected with miRNAs were TTK, PTTG1, MYBL2, EZH2, CENPA, CDCA8, CDCA7, and AURKA. Hub genes and their corresponded target miRNAs are presented in Table 3. Finally, 31 miRNAs, whose functions were correlated with tumor progression (Chang et al., 2019; Chang et al., 2017; Chen et al., 2015; Gao et al., 2018; Gu et al., 2014; Han et al., 2019; He et al., 2019; Kucuksayan et al., 2019; Li, Ma, Zhang, Ji, & Jin, 2014; Li, Ran, et al., 2018; Li & He, 2014; Li, Fu, et al., 2019; Li, Cui, Li, Zhang, & Li, 2018; Liu et al., 2017; Liu, Liu, Deng, et al., 2019; Liu, Liu, & Lu, 2019; Liu, Liu, Zhang, Tong, & Gan, 2019; Liu, Miao, et al., 2016; Luo et al., 2018; Tang et al., 2018; Wang et al., 2017; Wang, Cao, Su, Li, & Yan, 2019; Wu et al., 2015; Wu, Mo, Wan, Dan, & Hu, 2019; Xia, Jing, Li, & Lv, 2018; Xu et al., 2018; Zeng et al., 2018; Zhang, Hao, et al., 2018; Zhang et al., 2014, 2017; Zhu et al., 2017), drug resistance (Bian et al., 2019; Gao et al., 2014; Lin, Xie, Lu, Hu, & Chang, 2018; Pan, Chen, Shen, & Tantai, 2019; Su et al., 2016; Xu et al., 2017; Zhou, Wang, & Feng, 2014), radiosensitivity (Liu, Li, & Gao, 2016), recurrence (Sim et al., 2018) or autophagy (Pan et al., 2019) in lung cancer, were identified (Table 4).
Figure 9.
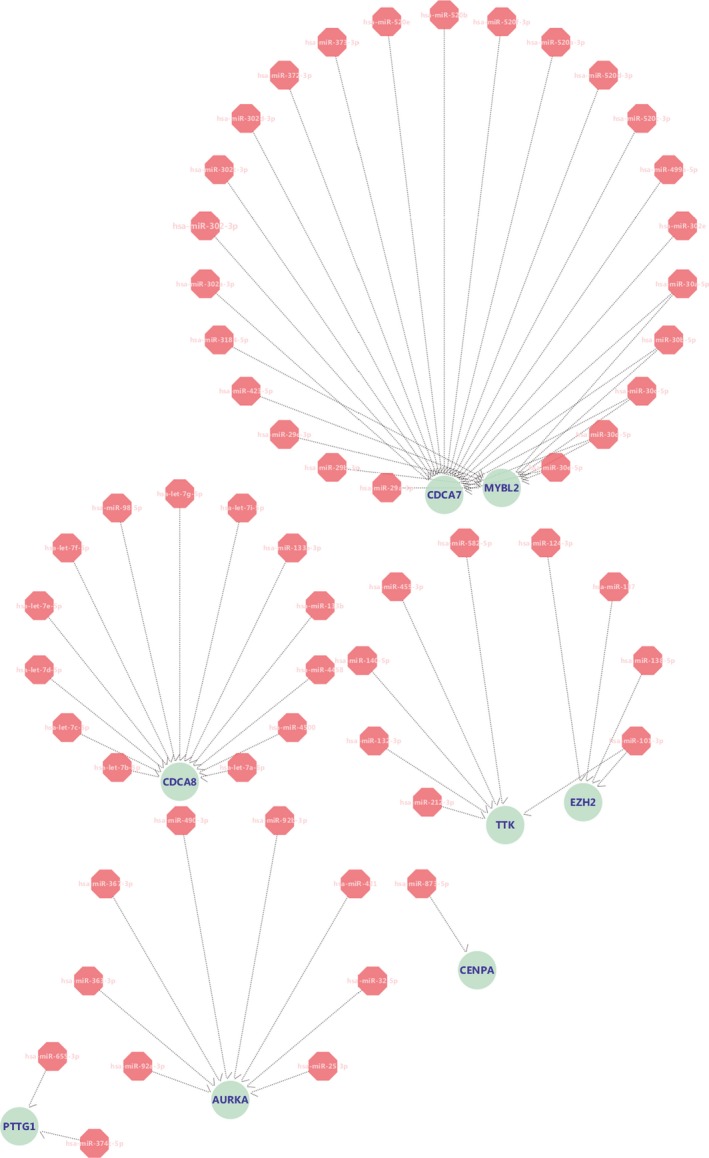
Target gene‐miRNA networks of hub genes. Blue node stands for 8 hub genes, pink node stands for miRNA. The network was visualized by Cytoscape
Table 3.
Hub genes and corresponded target miRNAs
| Gene | miRNA |
|---|---|
| TTK | hsa‐miR‐101‐3p hsa‐miR‐212‐3p hsa‐miR‐132‐3p hsa‐miR‐140‐5p hsa‐miR‐455‐3p hsa‐miR‐582‐5p |
| PTTG1 | hsa‐miR‐655‐3p hsa‐miR‐374c‐5p |
| MYBL2 | hsa‐miR‐29a‐3p hsa‐miR‐29b‐3p hsa‐miR‐30c‐5p hsa‐miR‐30a‐5p hsa‐miR‐30b‐5p hsa‐miR‐30d‐5p hsa‐miR‐29c‐3p hsa‐miR‐30e‐5p hsa‐miR‐423‐5p hsa‐miR‐3184‐5p |
| EZH2 | hsa‐miR‐101‐3p hsa‐miR‐124‐3p hsa‐miR‐137 hsa‐miR‐138‐5p |
| CENPA | hsa‐miR‐873‐5p |
| CDCA8 | hsa‐let‐7a‐5p hsa‐let‐7b‐5p hsa‐let‐7c‐5p hsa‐let‐7d‐5p hsa‐let‐7e‐5p hsa‐let‐7f‐5p hsa‐miR‐98‐5p hsa‐let‐7g‐5p hsa‐let‐7i‐5p hsa‐miR‐133a‐3p hsa‐miR‐133b hsa‐miR‐4458 hsa‐miR‐4500 |
| CDCA7 | hsa‐miR‐30a‐5p hsa‐miR‐30b‐5p hsa‐miR‐30c‐5p hsa‐miR‐30d‐5p hsa‐miR‐30e‐5p hsa‐miR‐302a‐3p hsa‐miR‐302‐3p hsa‐miR‐302c‐3p hsa‐miR‐302d‐3p hsa‐miR‐372‐3p hsa‐miR‐373‐3p hsa‐miR‐520e hsa‐miR‐520b hsa‐miR‐520f‐3p hsa‐miR‐520a‐3p hsa‐miR‐520d‐3p hsa‐miR‐520c‐3p hsa‐miR‐499a‐5p hsa‐miR‐302e |
| AURKA | hsa‐miR‐25‐3p hsa‐miR‐92a‐3p hsa‐miR‐363‐3p hsa‐miR‐367‐3p hsa‐miR‐490‐3p hsa‐miR‐92b‐3p hsa‐miR‐421 hsa‐miR‐32‐5p |
Table 4.
Functions of miRNAs in lung cancer
| Gene | miRNA |
|---|---|
| TTK | hsa‐miR‐101‐3p hsa‐miR‐212‐3p hsa‐miR‐132‐3p hsa‐miR‐140‐5p hsa‐miR‐455‐3p hsa‐miR‐582‐5p |
| PTTG1 | hsa‐miR‐655‐3p hsa‐miR‐374c‐5p |
| MYBL2 | hsa‐miR‐29a‐3p hsa‐miR‐29b‐3p hsa‐miR‐30c‐5p hsa‐miR‐30a‐5p hsa‐miR‐30b‐5p hsa‐miR‐30d‐5p hsa‐miR‐29c‐3p hsa‐miR‐30e‐5p hsa‐miR‐423‐5p hsa‐miR‐3184‐5p |
| EZH2 | hsa‐miR‐101‐3p hsa‐miR‐124‐3p hsa‐miR‐137 hsa‐miR‐138‐5p |
| CENPA | hsa‐miR‐873‐5p |
| CDCA8 | hsa‐let‐7a‐5p hsa‐let‐7b‐5p hsa‐let‐7c‐5p hsa‐let‐7d‐5p hsa‐let‐7e‐5p hsa‐let‐7f‐5p hsa‐miR‐98‐5p hsa‐let‐7g‐5p hsa‐let‐7i‐5p hsa‐miR‐133a‐3p hsa‐miR‐133b hsa‐miR‐4458 hsa‐miR‐4500 |
| CDCA7 | hsa‐miR‐30a‐5p hsa‐miR‐30b‐5p hsa‐miR‐30c‐5p hsa‐miR‐30d‐5p hsa‐miR‐30e‐5p hsa‐miR‐302a‐3p hsa‐miR‐302‐3p hsa‐miR‐302c‐3p hsa‐miR‐302d‐3p hsa‐miR‐372‐3p hsa‐miR‐373‐3p hsa‐miR‐520e hsa‐miR‐520b hsa‐miR‐520f‐3p hsa‐miR‐520a‐3p hsa‐miR‐520d‐3p hsa‐miR‐520c‐3p hsa‐miR‐499a‐5p hsa‐miR‐302e |
| AURKA | hsa‐miR‐25‐3p hsa‐miR‐92a‐3p hsa‐miR‐363‐3p hsa‐miR‐367‐3p hsa‐miR‐490‐3p hsa‐miR‐92b‐3p hsa‐miR‐421 hsa‐miR‐32‐5p |
4. DISCUSSION
To determine more possible targets for the exploration of SCLC and LCNEC biomarkers and molecular mechanisms, integrated bioinformatics methods based on the http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1037 profile dataset were performed in the present study. Eighteen primary normal lung tissues, 8 primary LCNEC tissues, 15 primary SCLC tissues, and 9 SCLC cell lines were enrolled and analyzed. Using GEO2R and Venn tools, 88 common DEGs were detected (|logFC| > 2, adjusted p < .05) in SCLC and LCNEC, including 32 up‐ (|logFC < 0) and 56 downregulated DEGs (|logFC < 0). Subsequently, functional enrichment and GO analysis were performed by DAVID, which showed the following: (a) For BP, the significantly enriched terms of up‐regulated DEGs were cell division, regulation of transcription, DNA repair, homologous chromosome segregation, positive regulation of exit from mitosis, mitotic nuclear division, glucose import, chromosome organization, protein K11‐linked ubiquitination, mitotic spindle organization and mitotic spindle assembly, and the significantly enriched terms of downregulated DEGs were caveola assembly, cell adhesion, receptor‐mediated endocytosis of virus by host cell, dopamine catabolic process, neurotransmitter catabolic process, multicellular organismal water homeostasis, angiogenesis, bleb assembly, ethanol oxidation, glycerol transport, and cellular water homeostasis; (b) for MF, the notably enriched terms of upregulated DEGs were ubiquitin protein ligase binding, DNA binding, ATP binding, peptidase inhibitor activity and ubiquitin conjugating enzyme activity, and those of downregulated DEGs were oxidoreductase, transporter, alcohol dehydrogenase (zinc‐dependent), water transmembrane transporter, primary amine oxidase, glycerol channel and water channel activities; (c) for CC, upregulated DEGs were particularly enriched in the cytoplasm, nucleus, axon hillock, chromosome passenger complex, nucleoplasm, pronucleus and spindle midzone, and down‐regulated DEGs in extracellular exosome, plasma membrane, caveola, extracellular region, membrane raft, apical part of cell, integral component of membrane, extracellular matrix, extracellular space and acrosomal membrane. To further explore the role of DEGs in biological pathways, the FunRich database was used. It was revealed that upregulated DEGs were notably enriched in the cell cycle pathway (mitotic), DNA replication, ATM pathway, mitotic M‐M/G1 phases, signaling by Aurora kinases, M phase, and mitotic prometaphase, while downregulated DEGs in lipid digestion, mobilization transport, noradrenaline and adrenaline degradation, and serotonin degradation. A PPI network complex of 48 nodes with 221 edges was then generated using STRING and Cytoscape. Next, 16 key upregulated genes were selected by MCODE analysis. Furthermore, following a re‐analysis of biological pathways by FunRich, 10 genes (CDCA1, CDCA8, AURKA, RFC4, UBE2C, CENPA, CENPU, MYBL2, PTTG1, and TOP2A) enriched in the cell cycle pathway were screened. In the meantime, target miRNAs associated with hub genes were predicted, and 8 genes (TTK, PTTG1, MYBL2, EZH2, CENPA, CDCA8, CDCA7, and AURKA) and 51 miRNAs involved in the mRNA‐miRNA regulatory network of SCLC and LCNEC were identified.
CDCA1, is a centromeric protein (Nabetani, Koujin, Tsutsumi, Haraguchi, & Hiraoka, 2001), also known as NUF2. The gene has been reported to be co‐expressed with cell cycle‐related genes, including CDC2, cyclin, TOP2A, and others (Walker, 2001), which was in line with the present results. It has been identified that the coactivation of KNTC2 and CDCA1 and their cognate interactions had an impact on pulmonary carcinogenesis, and that the inhibition of the KNTC2‐CDCA1 complex would provide a novel strategy for therapy of lung cancer treatment. CDCA1 was also regarded as a potential interacting protein for ZW10, a prognostic marker for lung cancer (Hayama et al., 2006; Yuan et al., 2018). In addition, CDCA1 is projected to be a candidate of tumor‐associated antigens (TAA) in SCLC. CDCA1, with increasing levels in SCLC, could act as a TAA in inducing cytotoxic T lymphocytes (CTLs) which killed tumor cell lines, and therefore made differences in immunotherapy.(Harao et al., 2008).
CDCA8, a member of the chromosomal passenger complex, is indispensable for the transmission of the genome in the process of cell division (Hindriksen, Meppelink, & Lens, 2015). Bidkhori et al constructed a co‐expression network and identified CDCA8 as a crucial gene in lung adenocarcinoma (Bidkhori et al., 2013). Another study implied that the CDCA8‐AURKB pathway participated in the process of lung carcinogenesis (Hayama et al., 2007). However, we failed to obtain studies of the impact of CDCA8 neither in SCLC nor in LCNEC.
AURKA, a serine‐threonine protein kinase, was found to be involved in several crucial mitotic events and was regarded as an oncogene in multiple types of cancer (Vader & Lens, 2008). In SCLC cell lines, a decreased level of AURKA inhibited cell proliferation and promoted apoptosis, and the inhibition of AURKA may be involved in the G2/M phase arrest of the cell cycle (Lu et al., 2014). Apart from that way, AURKA was involved in immunotherapy in SCLC by being regarded as a TAA to induce CTLs.(Babiak et al., 2014) Recently, more and more studies are concentrating on Aurora Kinase Inhibitors, PF‐03814735 and AZD1152, which establishes its pivotal status of SCLC in vitro and vivo.(Helfrich et al., 2016; Hook et al., 2012) Studies have also underlined the role of AURKA in platinum‐resistance as well as acquired resistance to third‐generation EGFR inhibitors (both osimertinib and rociletinib) (Gay et al., 2019; Shah et al., 2019). In addition, AURKA was reportedly correlated with poor prognosis in smoking‐related lung adenocarcinoma (Zhang, Hao, et al., 2018).
RFC4 is a member of the RFC family and has been found to be involved in DNA replication (Arai et al., 2009). In colorectal cancer, the forced expression of RFC4 was associated with tumor progression, poor prognosis (Xiang et al., 2014), and radioresistance (X. C. Wang, Yue, et al., 2019). RFC4 knockdown in liver cancer cells could suppress proliferation and induce apoptosis (Arai et al., 2009). In lung adenocarcinoma, RFC4 was regulated by an oncogene, protein kinases C (PKC)‐iota, and correlated with the PKC‐iota expression in several more tumor types (Erdogan, Klee, Thompson, & Fields, 2009).
UBE2C, a ubiquitin‐conjugating enzyme, works with the ubiquitin‐activating enzyme E1 and ubiquitin protein ligase E3 to degradate key regulatory molecules in cell cycle progression (Mayer, 2000). In addition, UBE2C intervened in the cell cycle, apoptosis and transcription processes and controlled tumoriogenesis. Previous studies have implied that UBE2C promotes cell growth (Jin et al., 2019; van Ree, Jeganathan, Malureanu, & van Deursen, 2010; Sivakumar et al., 2017) in NSCLC and is correlated with poor prognosis (Chen et al., 2011; Guo et al., 2017; Psyrri et al., 2012). In addition, UBE2C participated in the miR 495‐UBE2C‐ABCG2/ERCC1 axis regulating cisplatin resistance and mediating autophagy in NSCLC (Guo, Jin, et al., 2018; Guo, Wu, et al., 2018).UBE2C were in similarly high level in small‐cell prostate carcinoma (SCPC) and LCNEC of the prostate with xenografts, and the expression of UBE2C in SCLC and LCNEC of the lung might bear a strong resemblance to that of SCPC and LCNEC of the prostate (Tzelepi et al., 2012).
CENPA, a centromere‐specific 17‐kDa protein, is a crucial histone H3 variant that acts as a vital epigenetic mark for the identity and propagation of the centromere (Allshire & Karpen, 2008). CENPA played an essential role in cell cycle regulation and tumor cell survival (Takada et al., 2017). Recent studies have identified the impacts of CENPA in lung cancer. Liu et al. (2018) used integrated microarray analysis and confirmed CENPA, cyclin‐dependent kinase 1, and cell‐division cycle protein 20 as a cluster of prognostic biomarkers for patients with lung adenocarcinoma. It has also been revealed that CENPA intensifies the aggressive phenotype of lung adenocarcinoma and is associated with patient prognosis (Cheng et al., 2019; Wu et al., 2014, 2012).
CENPU, also known as PBIP1/KLIP1/CENP‐50/MLF1IP, is indispensable for kinetochore‐microtubule attachment, chromosome segregation, and recovery from spindle damage (Kagawa et al., 2014; Walter et al., 2015). Mounting evidence (Li, Zhang, Zhang, & Wang, 2018; Wang et al., 2017; Zhang et al., 2015) has indicated that CENPU influences several types of cancer, such as bladder and ovarian cancer, and prostate carcinoma. Furthermore, CENPU predicted poor survival in patients with lung carcinoma and affected lung tumor growth (Li, Wang, Pang, Zhang, & Wang, 2019; Wang et al., 2018; Zhang, Li, Zhang, & Shi, 2018).
MYBL2 participated in cell cycle regulation, cell survival, and EMT‐associated processes in carcinoma (Musa, Aynaud, Mirabeau, Delattre, & Grunewald, 2017). Accumulating evidence has indicated that MYBL2 regulated tumorigenesis and cancer progression in multiple types of cancer. The high expression of MYBL2 disrupted the DREAM complex and then hindered the cell cycle process in breast and ovarian cancer (Iness et al., 2019). In liver cancer cells, YAP supported cell proliferation in an MYBL2‐dependant manner (Wei et al., 2019). In addition, Fan et al. (2018) and Jin et al. (2017) stressed the oncogenic impacts of MYBL2 and its capacity to serve as a therapeutic target in NSCLC. Moreover the overexpression of MYBL2 was found to be related to that of the ubiquitin carboxyterminal hydrolase UCHL1 (an enzyme expressed in neurons and neuroendocrine cells in the lung) in murine, because of the reciprocal action of the UCHL1 enzyme and MYBL2 (Long, Long, Tsirigotis, & Gray, 2003).
PTTG1 is a novel proto‐oncogene that is overexpressed in different types of human cancers, including endocrine‐ and nonendocrine‐related types (Vlotides, Eigler, & Melmed, 2007). Wu, Zhang, et al. (2019)) revealed the importance of PTTG1 as a potential biomarker in NSCLC. Accordingly, Wang, Liu, and Chen (2016) emphasized the prognostic value of PTTG1 in lung cancer, showing the level of PGGT1 as an optimal parameter to predict malignant status and prognosis in NSCLC patients.
TOP2A is a type of isozyme type II topoisomerases (Li & Liu, 2001). A dysregulated TOP2A has been observed in multiple types of cancer and been used in the treatment of several carcinomas (Lan et al., 2014; Li et al., 2015; Wesierska‐Gadek & Skladanowski, 2012). TOP2A has been shown to be involved not only in NSCLC (Chien et al., 2019; Olaussen & Postel‐Vinay, 2016), but also in SCLC (Chiappori et al., 2010). Chiappori et al. (2010) recommended TOP2A as a biomarker of chemotherapeutic efficacy in SCLC. Chang et al. (2017)) and Gray et al. (2017) considered TOP2A as a target of anti‐small cell lung cancer agents. Furthermore, in silico analyses showed TOP2A as target gene of miR‐27a‐5p and miR‐34b‐3p, both of whom were tumor suppressors in SCLC (Mizuno et al., 2017). A current research showed that compared with lung adenocarcinoma, TOP2A was a potential drug target of the therapy in LCNEC due to its overexpression examined by immunohistochemistry (Makino et al., 2016). Although TOP2A was highly expressed in NENs, no evident difference in expression levels was found between SCLC and LCNEC (Neubauer et al., 2016).
The results of re‐analysis of biological pathway revealed that most key genes were enriched in cell cycle (mitotic), mitotic prometaphase, DNA replication, and M phase. Accordingly, because of the high genome instability as a result of high level replication stress in SCLC, the inhibition of DNA‐replication stress‐response, especially in combination with cisplatin, was one of the vulnerabilities of SCLC, which provided a novel therapy for this type of cancer (Bian & Lin, 2019; Nagel et al., 2019). Prometaphase arrest caused by mitotic spindle formation defects that resulting from the depletion of Aurora A, was also involved with the inhibition of cell growth in SCLC (Du et al., 2019). Additionally, the inhibition of a pivotal mitotic regulator led to the increase in the population of cells in the G2/M phase (Wang et al., 2018), while treatment with etoposide in TP53 gene abnormalities ones resulted in the lack of G1/S arrest (Soues, Wiltshire, & Smith, 2001), all of which were associated with pathways of mitotic cell cycle or M phase in SCLC cell lines.
To sum up, CDCA1 and AURK4 might be characteristic especially to SCLC, while MYBL2 is the biomarker of LCNEC. The biological pathway of DNA replication, mitotic cell cycle, mitotic prometaphase, and M phase might be characteristic to SCLC. In contrast, no evidence has been shown that any pathway enrichment is related to LCNEC, and the genes of CDCA8, RFC4, UBE2C, CENPA, CENPU, PTTG1, and TOP2A are not predicted to participate particularly in SCLC or LCNEC. Although further researches should be performed focusing on more functions of these key genes and pathways, our discoveries provide potential candidates useful for the diagnosis and discrimination between SCLC and LCNEC.
miRNAs play essential roles in the genesis, development, and progression of carcinoma (Rupaimoole & Slack, 2017). Aberrant miRNA expression profiles have been identified in multiple subtypes of lung cancer (Lai et al., 2019; Mao et al., 2019; Wang et al., 2018). In addition to DEGs and their functions, miRNAs can influence multiple processes of lung cancer, including drug resistance, tumor progression and recurrence, autophagy and radio‐sensitivity. The role of miRNAs in tumor growth is shown in Table 4. Recent studies have also revealed the impacts of miRNAs, including miR‐363‐3p, miR‐30a‐5p, miR‐133b, miR‐98‐5p, miR‐138‐5p, miR‐137, and miR‐138‐5p, on drug resistance in NSCLC, showing that miRNAs participated in the mechanisms of traditional chemotherapeutic drugs [gemcitabine (Bian et al., 2019), paclitaxel (Xu et al., 2017), and cisplatin (Lin et al., 2018; Pan et al., 2019; Su et al., 2016; Zhou et al., 2014)] as well as targeted drugs [gefitinib (Gao et al., 2014)]. In addition, Sim et al demonstrated that let‐7g‐5p was associated with recurrence in stage I lung adenocarcinoma (Sim et al., 2018). Other studies also elucidated the roles of miRNAs in autophagy and radio‐sensitivity (Liu, Miao, et al., 2016; Pan et al., 2019).
However, the present study had several limitations. First, due to the rarity of LCNEC and SCLC, we failed to provide meaningful prognosis information from public databases. Second, the present data were obtained from the GEO database, so the reliability and quality of the statistics cannot be evaluated. Third, this study lacks more convincing evidence, such as qPCR and immunohistochemistry results.
5. CONCLUSIONS
In conclusion, common DEGs of SCLC and LCNEC were identified by integrated bioinformatics methods and 16 hub genes were selected from PPI network, among which 10 genes were shown to be related to the most significantly enriched pathway, cell cycle pathway. The roles of these genes and pathways in lung cancer were further discussed and the application of these genes in treatment of SCLC and LCNEC is worthy of further study. In addition, the functions in lung cancer of miRNAs corresponding to 16 core genes were discussed. Several studies have tried to explore novel strategies (Christopoulos et al., 2017; Kujtan et al., 2018; Saunders et al., 2015) for the treatment of SCLC and LCNEC. Attempts at identifying the values of molecular targets, such as spread through air spaces, and immune therapy, such as the expression of PD‐L1 and mutation burden, are ongoing (Aly et al., 2019; Kim et al., 2018). To determine the various roles of emerging molecules in the process of tumorigenesis and progression of SCLC and LCNEC, further research should be conducted.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of Shandong Province (No. ZR2017MH062) and the Science and Technology for People's Livelihood Project of Qingdao (No. 17‐3‐3‐33‐nsh).
Dong S, Liang J, Zhai W, Yu Z. Common and distinct features of potentially predictive biomarkers in small cell lung carcinoma and large cell neuroendocrine carcinoma of the lung by systematic and integrated analysis. Mol Genet Genomic Med. 2020;8:e1126 10.1002/mgg3.1126
Shenghua Dong and Jun Liang contributed equally to this work.
Funding information
Natural Science Foundation of Shandong Province, Grant number: ZR2017MH062; Science and Technology for People's Livelihood Project of Qingdao, Grant number: 17‐3‐3‐33‐nsh.
REFERENCES
- Allshire, R. C. , & Karpen, G. H. (2008). Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nature Reviews Genetics, 9(12), 923–937. 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aly, R. G. , Rekhtman, N. , Li, X. , Takahashi, Y. , Eguchi, T. , Tan, K. S. , … Travis, W. D. (2019). Spread through air spaces (STAS) is prognostic in atypical carcinoid, large cell neuroendocrine carcinoma, and small cell carcinoma of the lung. Journal of Thoracic Oncology, 14(9), 1583–1593. 10.1016/j.jtho.2019.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai, M. , Kondoh, N. , Imazeki, N. , Hada, A. , Hatsuse, K. , Matsubara, O. , & Yamamoto, M. (2009). The knockdown of endogenous replication factor C4 decreases the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Liver International: Official Journal of the International Association for the Study of the Liver, 29(1), 55–62. 10.1111/j.1478-3231.2008.01792.x [DOI] [PubMed] [Google Scholar]
- Asamura, H. , Kameya, T. , Matsuno, Y. , Noguchi, M. , Tada, H. , Ishikawa, Y. , … Nagai, K. (2006). Neuroendocrine neoplasms of the lung: A prognostic spectrum. Journal of Clinical Oncology, 24(1), 70–76. 10.1200/jco.2005.04.1202 [DOI] [PubMed] [Google Scholar]
- Babiak, A. , Steinhauser, M. , Gotz, M. , Herbst, C. , Dohner, H. , & Greiner, J. (2014). Frequent T cell responses against immunogenic targets in lung cancer patients for targeted immunotherapy. Oncology Reports, 31(1), 384–390. 10.3892/or.2013.2804 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee, A. , Richards, W. G. , Staunton, J. , Li, C. , Monti, S. , Vasa, P. , … Meyerson, M. (2001). Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America, 98(24), 13790–13795. 10.1073/pnas.191502998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian, W. G. , Zhou, X. N. , Song, S. , Chen, H. T. , Shen, Y. , & Chen, P. (2019). Reduced miR‐363‐3p expression in non‐small cell lung cancer is associated with gemcitabine resistance via targeting of CUL4A. European Review for Medical and Pharmacological Sciences, 23(2), 649–659. 10.26355/eurrev_201901_16879 [DOI] [PubMed] [Google Scholar]
- Bian, X. , & Lin, W. (2019). Targeting DNA replication stress and DNA double‐strand break repair for optimizing SCLC Treatment. Cancers (Basel), 11(9), 10.3390/cancers11091289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidkhori, G. , Narimani, Z. , Hosseini Ashtiani, S. , Moeini, A. , Nowzari‐Dalini, A. , & Masoudi‐Nejad, A. (2013). Reconstruction of an integrated genome‐scale co‐expression network reveals key modules involved in lung adenocarcinoma. PLoS ONE, 8(7), e67552 10.1371/journal.pone.0067552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerilli, L. A. , Ritter, J. H. , Mills, S. E. , & Wick, M. R. (2001). Neuroendocrine neoplasms of the lung. American Journal of Clinical Pathology, 116(Suppl), S65–S96. 10.1309/bpy4-vgtb-patf-vuxu [DOI] [PubMed] [Google Scholar]
- Chang, J. , Gao, F. , Chu, H. , Lou, L. , Wang, H. , & Chen, Y. (2019). miR‐363‐3p inhibits migration, invasion, and epithelial‐mesenchymal transition by targeting NEDD9 and SOX4 in non‐small‐cell lung cancer. Journal of Cellular Physiology, 235(2), 1808–1820. 10.1002/jcp.29099 [DOI] [PubMed] [Google Scholar]
- Chang, T.‐H. , Tsai, M.‐F. , Gow, C.‐H. , Wu, S.‐G. , Liu, Y.‐N. , Chang, Y.‐L. , … Shih, J.‐Y. (2017). Upregulation of microRNA‐137 expression by Slug promotes tumor invasion and metastasis of non‐small cell lung cancer cells through suppression of TFAP2C. Cancer Letters, 402, 190–202. 10.1016/j.canlet.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Chen, D. I. , Guo, W. , Qiu, Z. , Wang, Q. , Li, Y. , Liang, L. , … He, X. (2015). MicroRNA‐30d‐5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non‐small cell lung cancer. Cancer Letters, 362(2), 208–217. 10.1016/j.canlet.2015.03.041 [DOI] [PubMed] [Google Scholar]
- Chen, Z. , Zhang, C. , Wu, D. , Chen, H. , Rorick, A. , Zhang, X. , & Wang, Q. (2011). Phospho‐MED1‐enhanced UBE2C locus looping drives castration‐resistant prostate cancer growth. EMBO Journal, 30(12), 2405–2419. 10.1038/emboj.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z. , Yu, C. , Cui, S. , Wang, H. , Jin, H. , Wang, C. , … Qin, W. (2019). circTP63 functions as a ceRNA to promote lung squamous cell carcinoma progression by upregulating FOXM1. Nature Communications, 10(1), 3200 10.1038/s41467-019-11162-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappori, A. A. , Zheng, Z. , Chen, T. , Rawal, B. , Schell, M. J. , Mullaney, B. P. , & Bepler, G. (2010). Features of potentially predictive biomarkers of chemotherapeutic efficacy in small cell lung cancer. Journal of Thoracic Oncology, 5(4), 484–490. 10.1097/JTO.0b013e3181ccb27b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien, C.‐M. , Yang, J.‐C. , Wu, P.‐H. , Wu, C.‐Y. , Chen, G.‐Y. , Wu, Y.‐C. , … Chiu, C.‐C. (2019). Phytochemical naphtho[1,2‐b] furan‐4,5dione induced topoisomerase II‐mediated DNA damage response in human non‐small‐cell lung cancer. Phytomedicine, 54, 109–119. 10.1016/j.phymed.2018.06.025 [DOI] [PubMed] [Google Scholar]
- Christopoulos, P. , Engel‐Riedel, W. , Grohé, C. , Kropf‐Sanchen, C. , von Pawel, J. , Gütz, S. , … Thomas, M. (2017). Everolimus with paclitaxel and carboplatin as first‐line treatment for metastatic large‐cell neuroendocrine lung carcinoma: A multicenter phase II trial. Annals of Oncology, 28(8), 1898–1902. 10.1093/annonc/mdx268 [DOI] [PubMed] [Google Scholar]
- Clinical Lung Cancer Genome Project (CLCGP); Network Genomic Medicine (NGM) . (2013). A genomics‐based classification of human lung tumors. Science Translational Medicine, 5(209), 209ra153 10.1126/scitranslmed.3006802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S. , & Meltzer, P. S. (2007). GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics, 23(14), 1846–1847. 10.1093/bioinformatics/btm254 [DOI] [PubMed] [Google Scholar]
- Du, J. , Yan, L. , Torres, R. , Gong, X. , Bian, H. , Marugán, C. , … Campbell, R. M. (2019). Aurora A‐selective inhibitor LY3295668 leads to dominant mitotic arrest, apoptosis in cancer cells, and shows potent preclinical antitumor efficacy. Molecular Cancer Therapeutics, 18(12), 2207–2219. 10.1158/1535-7163.mct-18-0529 [DOI] [PubMed] [Google Scholar]
- Erdogan, E. , Klee, E. W. , Thompson, E. A. , & Fields, A. P. (2009). Meta‐analysis of oncogenic protein kinase Ciota signaling in lung adenocarcinoma. Clinical Cancer Research, 15(5), 1527–1533. 10.1158/1078-0432.ccr-08-2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, X. , Wang, Y. , Jiang, T. , Cai, W. , Jin, Y. , Niu, Y. , Bu, Y. (2018). B‐Myb mediates proliferation and migration of non‐small‐cell lung cancer via suppressing IGFBP3. International Journal of Molecular Sciences, 19(5), 1479 10.3390/ijms19051479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, X. , Zhao, H. , Diao, C. , Wang, X. , Xie, Y. , Liu, Y. , … Zhang, M. (2018). miR‐455‐3p serves as prognostic factor and regulates the proliferation and migration of non‐small cell lung cancer through targeting HOXB5. Biochemical and Biophysical Research Communications, 495(1), 1074–1080. 10.1016/j.bbrc.2017.11.123 [DOI] [PubMed] [Google Scholar]
- Gao, Y. , Fan, X. , Li, W. , Ping, W. , Deng, Y. , & Fu, X. (2014). miR‐138‐5p reverses gefitinib resistance in non‐small cell lung cancer cells via negatively regulating G protein‐coupled receptor 124. Biochemical and Biophysical Research Communications, 446(1), 179–186. 10.1016/j.bbrc.2014.02.073 [DOI] [PubMed] [Google Scholar]
- Garber, M. E. , Troyanskaya, O. G. , Schluens, K. , Petersen, S. , Thaesler, Z. , Pacyna‐Gengelbach, M. , … Petersen, I. (2001). Diversity of gene expression in adenocarcinoma of the lung. Proceedings of the National Academy of Sciences of the United States of America, 98(24), 13784–13789. 10.1073/pnas.241500798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay, C. M. , Tong, P. , Cardnell, R. J. , Sen, T. , Su, X. , Ma, J. , … Byers, L. A. (2019). Differential sensitivity analysis for resistant malignancies (DISARM) identifies common candidate therapies across platinum‐resistant cancers. Clinical Cancer Research, 25(1), 346–357. 10.1158/1078-0432.ccr-18-1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, J. E. , Heist, R. S. , Starodub, A. N. , Camidge, D. R. , Kio, E. A. , Masters, G. A. , … Goldenberg, D. M. (2017). Therapy of small cell lung cancer (SCLC) with a topoisomerase‐I‐inhibiting antibody‐drug conjugate (ADC) targeting trop‐2. Sacituzumab Govitecan. Clin Cancer Res, 23(19), 5711–5719. 10.1158/1078-0432.ccr-17-0933 [DOI] [PubMed] [Google Scholar]
- Gu, H. , Yang, T. , Fu, S. , Chen, X. , Guo, L. , & Ni, Y. (2014). MicroRNA‐490‐3p inhibits proliferation of A549 lung cancer cells by targeting CCND1. Biochemical and Biophysical Research Communications, 444(1), 104–108. 10.1016/j.bbrc.2014.01.020 [DOI] [PubMed] [Google Scholar]
- Guo, J. , Jin, D. , Wu, Y. , Yang, L. , Du, J. , Gong, K. , … Xi, S. (2018). The miR 495‐UBE2C‐ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin‐resistant non‐small cell lung cancer cells. EBioMedicine, 35, 204–221. 10.1016/j.ebiom.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Guo, J. , Wu, Y. , Du, J. , Yang, L. , Chen, W. , Gong, K. , … Xi, S. (2018). Deregulation of UBE2C‐mediated autophagy repression aggravates NSCLC progression. Oncogenesis, 7(6), 49 10.1038/s41389-018-0054-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Ding, Z. , Huang, N. , Huang, Z. , Zhang, N. , & Xia, Z. (2017). Forkhead Box M1 positively regulates UBE2C and protects glioma cells from autophagic death. Cell Cycle, 16(18), 1705–1718. 10.1080/15384101.2017.1356507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, C. , Kawata, M. , Hamada, Y. , Kondo, T. , Wada, J. , Asano, K. , … Narita, M. (2019). Analyses of the possible anti‐tumor effect of yokukansan. Journal of Natural Medicines, 73(3), 468–479. 10.1007/s11418-019-01283-x [DOI] [PubMed] [Google Scholar]
- Harao, M. , Hirata, S. , Irie, A. , Senju, S. , Nakatsura, T. , Komori, H. , … Nishimura, Y. (2008). HLA‐A2‐restricted CTL epitopes of a novel lung cancer‐associated cancer testis antigen, cell division cycle associated 1, can induce tumor‐reactive CTL. International Journal of Cancer, 123(11), 2616–2625. 10.1002/ijc.23823 [DOI] [PubMed] [Google Scholar]
- Hayama, S. , Daigo, Y. , Kato, T. , Ishikawa, N. , Yamabuki, T. , Miyamoto, M. , … Nakamura, Y. (2006). Activation of CDCA1‐KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Research, 66(21), 10339–10348. 10.1158/0008-5472.can-06-2137 [DOI] [PubMed] [Google Scholar]
- Hayama, S. , Daigo, Y. , Yamabuki, T. , Hirata, D. , Kato, T. , Miyamoto, M. , … Nakamura, Y. (2007). Phosphorylation and activation of cell division cycle associated 8 by aurora kinase B plays a significant role in human lung carcinogenesis. Cancer Research, 67(9), 4113–4122. 10.1158/0008-5472.can-06-4705 [DOI] [PubMed] [Google Scholar]
- He, S. , Li, Z. , Yu, Y. , Zeng, Q. , Cheng, Y. , Ji, W. , … Lu, S. (2019). Exosomal miR‐499a‐5p promotes cell proliferation, migration and EMT via mTOR signaling pathway in lung adenocarcinoma. Experimental Cell Research, 379(2), 203–213. 10.1016/j.yexcr.2019.03.035 [DOI] [PubMed] [Google Scholar]
- Helfrich, B. A. , Kim, J. , Gao, D. , Chan, D. C. , Zhang, Z. , Tan, A. C. , & Bunn, P. A. (2016). Barasertib (AZD1152), a small molecule aurora B inhibitor, inhibits the growth of SCLC cell lines in vitro and in vivo. Molecular Cancer Therapeutics, 15(10), 2314–2322. 10.1158/1535-7163.mct-16-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindriksen, S. , Meppelink, A. , & Lens, S. M. (2015). Functionality of the chromosomal passenger complex in cancer. Biochemical Society Transactions, 43(1), 23–32. 10.1042/bst20140275 [DOI] [PubMed] [Google Scholar]
- Hook, K. E. , Garza, S. J. , Lira, M. E. , Ching, K. A. , Lee, N. V. , Cao, J. , … Pavlicek, A. (2012). An integrated genomic approach to identify predictive biomarkers of response to the aurora kinase inhibitor PF‐03814735. Molecular Cancer Therapeutics, 11(3), 710–719. 10.1158/1535-7163.MCT-11-0184 [DOI] [PubMed] [Google Scholar]
- Huang da, W. , Sherman, B. T. , & Lempicki, R. A. (2009a). Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research, 37(1), 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da, W. , Sherman, B. T. , & Lempicki, R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols, 4(1), 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Iness, A. N. , Felthousen, J. , Ananthapadmanabhan, V. , Sesay, F. , Saini, S. , Guiley, K. Z. , … Litovchick, L. (2019). The cell cycle regulatory DREAM complex is disrupted by high expression of oncogenic B‐Myb. Oncogene, 38(7), 1080–1092. 10.1038/s41388-018-0490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, D. , Guo, J. , Wu, Y. , Du, J. , Wang, X. , An, J. , … Wang, W. (2019). UBE2C, directly targeted by miR‐548e‐5p, increases the cellular growth and invasive abilities of cancer cells interacting with the EMT marker protein zinc finger E‐box binding homeobox 1/2 in NSCLC. Theranostics, 9(7), 2036–2055. 10.7150/thno.32738 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jin, Y. , Zhu, H. , Cai, W. , Fan, X. , Wang, Y. , Niu, Y. , … Bu, Y. (2017). B‐Myb is up‐regulated and promotes cell growth and motility in non‐small cell lung Cancer. International Journal of Molecular Sciences, 18(6), 860 10.3390/ijms18060860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa, N. , Hori, T. , Hoki, Y. , Hosoya, O. , Tsutsui, K. , Saga, Y. , … Fukagawa, T. (2014). The CENP‐O complex requirement varies among different cell types. Chromosome Research, 22(3), 293–303. 10.1007/s10577-014-9404-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, A. , Brunnström, H. , Lindquist, K. E. , Jirström, K. , Jönsson, M. , Rosengren, F. , … Staaf, J. (2015). Mutational and gene fusion analyses of primary large cell and large cell neuroendocrine lung cancer. Oncotarget, 6(26), 22028–22037. 10.18632/oncotarget.4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson, A. , Jonsson, M. , Lauss, M. , Brunnstrom, H. , Jonsson, P. , Borg, A. , … Staaf, J. (2014). Genome‐wide DNA methylation analysis of lung carcinoma reveals one neuroendocrine and four adenocarcinoma epitypes associated with patient outcome. Clinical Cancer Research, 20(23), 6127–6140. 10.1158/1078-0432.ccr-14-1087 [DOI] [PubMed] [Google Scholar]
- Kim, H. S. , Lee, J. H. , Nam, S. J. , Ock, C.‐Y. , Moon, J.‐W. , Yoo, C. W. , … Han, J.‐Y. (2018). Association of PD‐L1 expression with tumor‐infiltrating immune cells and mutation burden in high‐grade neuroendocrine carcinoma of the lung. Journal of Thoracic Oncology, 13(5), 636–648. 10.1016/j.jtho.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Kucuksayan, H. , Akgun, S. , Ozes, O. N. , Alikanoglu, A. S. , Yildiz, M. , Dal, E. , & Akca, H. (2019). TGF‐beta‐SMAD‐miR‐520e axis regulates NSCLC metastasis through a TGFBR2‐mediated negative‐feedback loop. Carcinogenesis, 40(5), 695–705. 10.1093/carcin/bgy166 [DOI] [PubMed] [Google Scholar]
- Kujtan, L. , Muthukumar, V. , Kennedy, K. F. , Davis, J. R. , Masood, A. , & Subramanian, J. (2018). The Role of systemic therapy in the management of stage I large cell neuroendocrine carcinoma of the lung. Journal of Thoracic Oncology, 13(5), 707–714. 10.1016/j.jtho.2018.01.019 [DOI] [PubMed] [Google Scholar]
- Lai, J. , Yang, H. , Zhu, Y. , Ruan, M. , Huang, Y. , & Zhang, Q. (2019). MiR‐7‐5p‐mediated downregulation of PARP1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer, 19(1), 602 10.1186/s12885-019-5798-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, J. , Huang, H.‐Y. , Lee, S.‐W. , Chen, T.‐J. , Tai, H.‐C. , Hsu, H.‐P. , … Li, C.‐F. (2014). TOP2A overexpression as a poor prognostic factor in patients with nasopharyngeal carcinoma. Tumour Biology, 35(1), 179–187. 10.1007/s13277-013-1022-6 [DOI] [PubMed] [Google Scholar]
- Li, H. , Zhang, H. , & Wang, Y. (2018). Centromere protein U facilitates metastasis of ovarian cancer cells by targeting high mobility group box 2 expression. American Journal of Cancer Research, 8(5), 835–851. [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Wang, Z. G. , Pang, L. B. , Zhang, R. H. , & Wang, Y. Y. (2019). Reduced CENPU expression inhibits lung adenocarcinoma cell proliferation and migration through PI3K/AKT signaling. Bioscience, Biotechnology, and Biochemistry, 83(6), 1077–1084. 10.1080/09168451.2019.1588094 [DOI] [PubMed] [Google Scholar]
- Li, P. , Ma, L. , Zhang, Y. , Ji, F. , & Jin, F. (2014). MicroRNA‐137 down‐regulates KIT and inhibits small cell lung cancer cell proliferation. Biomedicine and Pharmacotherapy, 68(1), 7–12. 10.1016/j.biopha.2013.12.002 [DOI] [PubMed] [Google Scholar]
- Li, Q. , Ran, P. , Zhang, X. , Guo, X. , Yuan, Y. , Dong, T. , … Xiao, C. (2018). Downregulation of N‐Acetylglucosaminyltransferase GCNT3 by miR‐302b‐3p decreases non‐small cell lung cancer (NSCLC) cell proliferation, migration and invasion. Cellular Physiology and Biochemistry, 50(3), 987–1004. 10.1159/000494482 [DOI] [PubMed] [Google Scholar]
- Li, T. K. , & Liu, L. F. (2001). Tumor cell death induced by topoisomerase‐targeting drugs. Annual Review of Pharmacology and Toxicology, 41, 53–77. 10.1146/annurev.pharmtox.41.1.53 [DOI] [PubMed] [Google Scholar]
- Li, W. , & He, F. (2014). Monocyte to macrophage differentiation‐associated (MMD) targeted by miR‐140‐5p regulates tumor growth in non‐small cell lung cancer. Biochemical and Biophysical Research Communications, 450(1), 844–850. 10.1016/j.bbrc.2014.06.075 [DOI] [PubMed] [Google Scholar]
- Li, X. , Fu, Q. , Li, H. , Zhu, L. , Chen, W. , Ruan, T. , … Yu, X. (2019). MicroRNA‐520c‐3p functions as a novel tumor suppressor in lung adenocarcinoma. FEBS Journal, 286(14), 2737–2752. 10.1111/febs.14835 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Cui, X. , Li, Y. , Zhang, T. , & Li, S. (2018). Upregulated expression of miR‐421 is associated with poor prognosis in non‐small‐cell lung cancer. Cancer Management and Research, 10, 2627–2633. 10.2147/cmar.s167432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Shen, X. , Wang, X. , Li, A. , Wang, P. , Jiang, P. , … Feng, Q. (2015). EGCG regulates the cross‐talk between JWA and topoisomerase IIalpha in non‐small‐cell lung cancer (NSCLC) cells. Scientific Reports, 5, 11009 10.1038/srep11009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C. , Xie, L. , Lu, Y. , Hu, Z. , & Chang, J. (2018). miR‐133b reverses cisplatin resistance by targeting GSTP1 in cisplatin‐resistant lung cancer cells. International Journal of Molecular Medicine, 41(4), 2050–2058. 10.3892/ijmm.2018.3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C. H. , Lv, D. S. , Li, M. , Sun, G. , Zhang, X. F. , & Bai, Y. (2017). MicroRNA‐4458 suppresses the proliferation of human lung cancer cells in vitro by directly targeting Lin28B. Acta Pharmacologica Sinica, 38(9), 1297–1304. 10.1038/aps.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G. , Li, Y. I. , & Gao, X. (2016). Overexpression of microRNA‐133b sensitizes non‐small cell lung cancer cells to irradiation through the inhibition of glycolysis. Oncology Letters, 11(4), 2903–2908. 10.3892/ol.2016.4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Liu, S. , Deng, X. , Rao, J. , Huang, K. , Xu, G. , & Wang, X. (2019). MicroRNA‐582‐5p suppresses non‐small cell lung cancer cells growth and invasion via downregulating NOTCH1. PLoS ONE, 14(6), e0217652 10.1371/journal.pone.0217652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Liu, L. , & Lu, S. (2019). lncRNA H19 promotes viability and epithelial‐mesenchymal transition of lung adenocarcinoma cells by targeting miR‐29b‐3p and modifying STAT3. International Journal of Oncology, 54(3), 929–941. 10.3892/ijo.2019.4695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. T. , Wang, Y. , Zhang, J. , Ye, F. , Huang, X. H. , Li, B. , & He, Q. Y. (2018). A novel strategy of integrated microarray analysis identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic biomarkers in lung adenocarcinoma. Cancer Letters, 425, 43–53. 10.1016/j.canlet.2018.03.043 [DOI] [PubMed] [Google Scholar]
- Liu, X. , Liu, J. , Zhang, X. , Tong, Y. , & Gan, X. (2019). MiR‐520b promotes the progression of non‐small cell lung cancer through activating Hedgehog pathway. Journal of Cellular and Molecular Medicine, 23(1), 205–215. 10.1111/jcmm.13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Miao, L. , Ni, R. , Zhang, H. , Li, L. , Wang, X. , … Wang, J. (2016). microRNA‐520a‐3p inhibits proliferation and cancer stem cell phenotype by targeting HOXD8 in non‐small cell lung cancer. Oncology Reports, 36(6), 3529–3535. 10.3892/or.2016.5149 [DOI] [PubMed] [Google Scholar]
- Long, E. M. , Long, M. A. , Tsirigotis, M. , & Gray, D. A. (2003). Stimulation of the murine Uchl1 gene promoter by the B‐Myb transcription factor. Lung Cancer, 42(1), 9–21. 10.1016/s0169-5002(03)00279-4 [DOI] [PubMed] [Google Scholar]
- Lu, Y. , Liu, Y. , Jiang, J. , Xi, Z. , Zhong, N. , Shi, S. , … Wei, X. (2014). Knocking down the expression of aurora‐A gene inhibits cell proliferation and induces G2/M phase arrest in human small cell lung cancer cells. Oncology Reports, 32(1), 243–249. 10.3892/or.2014.3194 [DOI] [PubMed] [Google Scholar]
- Luo, J. , Zhu, H. , Jiang, H. , Cui, Y. , Wang, M. , Ni, X. , & Ma, C. (2018). The effects of aberrant expression of LncRNA DGCR5/miR‐873‐5p/TUSC3 in lung cancer cell progression. Cancer Medicine, 7(7), 3331–3341. 10.1002/cam4.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino, T. , Mikami, T. , Hata, Y. , Otsuka, H. , Koezuka, S. , Isobe, K. , … Iyoda, A. (2016). Comprehensive biomarkers for personalized treatment in pulmonary large cell neuroendocrine carcinoma: A comparative analysis with adenocarcinoma. Annals of Thoracic Surgery, 102(5), 1694–1701. 10.1016/j.athoracsur.2016.04.100 [DOI] [PubMed] [Google Scholar]
- Mao, Y. , Xue, P. , Li, L. , Xu, P. , Cai, Y. , Chu, X. , … Zhu, S. (2019). Bioinformatics analysis of mRNA and miRNA microarray to identify the key miRNAgene pairs in smallcell lung cancer. Mol Med Rep, 20(3), 2199–2208. 10.3892/mmr.2019.10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, R. J. (2000). The meteoric rise of regulated intracellular proteolysis. Nature Reviews Molecular Cell Biology, 1(2), 145–148. 10.1038/35040090 [DOI] [PubMed] [Google Scholar]
- Miyoshi, T. , Umemura, S. , Matsumura, Y. , Mimaki, S. , Tada, S. , Makinoshima, H. , … Tsuchihara, K. (2017). Genomic profiling of large‐cell neuroendocrine carcinoma of the lung. Clinical Cancer Research, 23(3), 757–765. 10.1158/1078-0432.ccr-16-0355 [DOI] [PubMed] [Google Scholar]
- Mizuno, K. , Mataki, H. , Arai, T. , Okato, A. , Kamikawaji, K. , Kumamoto, T. , … Seki, N. (2017). The microRNA expression signature of small cell lung cancer: Tumor suppressors of miR‐27a‐5p and miR‐34b‐3p and their targeted oncogenes. Journal of Human Genetics, 62(7), 671–678. 10.1038/jhg.2017.27 [DOI] [PubMed] [Google Scholar]
- Musa, J. , Aynaud, M. M. , Mirabeau, O. , Delattre, O. , & Grunewald, T. G. (2017). MYBL2 (B‐Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death and Disease, 8(6), e2895 10.1038/cddis.2017.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabetani, A. , Koujin, T. , Tsutsumi, C. , Haraguchi, T. , & Hiraoka, Y. (2001). A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma, 110(5), 322–334. 10.1007/s004120100153 [DOI] [PubMed] [Google Scholar]
- Nagel, R. , Avelar, A. T. , Aben, N. , Proost, N. , van de Ven, M. , van der Vliet, J. , … Berns, A. (2019). Inhibition of the replication stress response is a synthetic vulnerability in SCLC that acts synergistically in combination with cisplatin. Molecular Cancer Therapeutics, 18(4), 762–770. 10.1158/1535-7163.mct-18-0972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, E. , Wirtz, R. M. , Kaemmerer, D. , Athelogou, M. , Schmidt, L. , Sanger, J. , & Lupp, A. (2016). Comparative evaluation of three proliferation markers, Ki‐67, TOP2A, and RacGAP1, in bronchopulmonary neuroendocrine neoplasms: Issues and prospects. Oncotarget, 7(27), 41959–41973. 10.18632/oncotarget.9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaussen, K. A. , & Postel‐Vinay, S. (2016). Predictors of chemotherapy efficacy in non‐small‐cell lung cancer: A challenging landscape. Annals of Oncology, 27(11), 2004–2016. 10.1093/annonc/mdw321 [DOI] [PubMed] [Google Scholar]
- Pan, X. , Chen, Y. , Shen, Y. , & Tantai, J. (2019). Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR‐138‐5p/ATG7. Cell Death and Disease, 10(6), 429 10.1038/s41419-019-1660-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan, M. , Keerthikumar, S. , Chisanga, D. , Alessandro, R. , Ang, C.‐S. , Askenase, P. , … Mathivanan, S. (2017). A novel community driven software for functional enrichment analysis of extracellular vesicles data. Journal of Extracellular Vesicles, 6(1), 1321455 10.1080/20013078.2017.1321455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psyrri, A. , Kalogeras, K. T. , Kronenwett, R. , Wirtz, R. M. , Batistatou, A. , Bournakis, E. , … Fountzilas, G. (2012). Prognostic significance of UBE2C mRNA expression in high‐risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Annals of Oncology, 23(6), 1422–1427. 10.1093/annonc/mdr527 [DOI] [PubMed] [Google Scholar]
- Rekhtman, N. , Pietanza, M. C. , Hellmann, M. D. , Naidoo, J. , Arora, A. , Won, H. , … Ladanyi, M. (2016). Next‐generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma‐like and non‐small cell carcinoma‐like subsets. Clinical Cancer Research, 22(14), 3618–3629. 10.1158/1078-0432.ccr-15-2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, G. , Cavazza, A. , Marchioni, A. , Longo, L. , Migaldi, M. , Sartori, G. , … Brambilla, E. (2005). Role of chemotherapy and the receptor tyrosine kinases KIT, PDGFRalpha, PDGFRbeta, and Met in large‐cell neuroendocrine carcinoma of the lung. Journal of Clinical Oncology, 23(34), 8774–8785. 10.1200/jco.2005.02.8233 [DOI] [PubMed] [Google Scholar]
- Rupaimoole, R. , & Slack, F. J. (2017). MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nature Reviews Drug Discovery, 16(3), 203–222. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- Saunders, L. R. , Bankovich, A. J. , Anderson, W. C. , Aujay, M. A. , Bheddah, S. , Black, K. A. , … Dylla, S. J. (2015). A DLL3‐targeted antibody‐drug conjugate eradicates high‐grade pulmonary neuroendocrine tumor‐initiating cells in vivo. Science Translational Medicine, 7(302), 302ra136 10.1126/scitranslmed.aac9459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, K. N. , Bhatt, R. , Rotow, J. , Rohrberg, J. , Olivas, V. , Wang, V. E. , … Bandyopadhyay, S. (2019). Aurora kinase A drives the evolution of resistance to third‐generation EGFR inhibitors in lung cancer. Nature Medicine, 25(1), 111–118. 10.1038/s41591-018-0264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P. , Markiel, A. , Ozier, O. , Baliga, N. S. , Wang, J. T. , Ramage, D. , … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim, J. , Kim, Y. , Kim, H. , Shin, S. J. , Kim, D. H. , Paik, S. S. , & Jang, K. (2018). Identification of recurrence‐associated microRNAs in stage I lung adenocarcinoma. Medicine (Baltimore), 97(25), e10996 10.1097/md.0000000000010996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbolo, M. , Mafficini, A. , Sikora, K. O. , Fassan, M. , Barbi, S. , Corbo, V. , … Scarpa, A. (2017). Lung neuroendocrine tumours: Deep sequencing of the four World Health Organization histotypes reveals chromatin‐remodelling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. The Journal of Pathology, 241(4), 488–500. 10.1002/path.4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar, S. , Lucas, F. A. S. , McDowell, T. L. , Lang, W. , Xu, L. I. , Fujimoto, J. , … Kadara, H. (2017). Genomic landscape of atypical adenomatous hyperplasia reveals divergent modes to lung adenocarcinoma. Cancer Research, 77(22), 6119–6130. 10.1158/0008-5472.can-17-1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soues, S. , Wiltshire, M. , & Smith, P. J. (2001). Differential sensitivity to etoposide (VP‐16)‐induced S phase delay in a panel of small‐cell lung carcinoma cell lines with G1/S phase checkpoint dysfunction. Cancer Chemotherapy and Pharmacology, 47(2), 133–140. 10.1007/s002800000227 [DOI] [PubMed] [Google Scholar]
- Su, T. J. , Ku, W. H. , Chen, H. Y. , Hsu, Y. C. , Hong, Q. S. , Chang, G. C. , … Chen, J. J. (2016). Oncogenic miR‐137 contributes to cisplatin resistance via repressing CASP3 in lung adenocarcinoma. American Journal of Cancer Research, 6(6), 1317–1330. [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk, D. , Franceschini, A. , Wyder, S. , Forslund, K. , Heller, D. , Huerta‐Cepas, J. , … von Mering, C. (2015). STRING v10: Protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43(D1), D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, M. , Zhang, W. , Suzuki, A. , Kuroda, T. S. , Yu, Z. , Inuzuka, H. , … Zhang, Q. (2017). FBW7 loss promotes chromosomal instability and tumorigenesis via cyclin E1/CDK2‐mediated phosphorylation of CENP‐A. Cancer Research, 77(18), 4881–4893. 10.1158/0008-5472.can-17-1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, L. X. , Chen, G. H. , Li, H. , He, P. , Zhang, Y. , & Xu, X. W. (2018). Long non‐coding RNA OGFRP1 regulates LYPD3 expression by sponging miR‐124‐3p and promotes non‐small cell lung cancer progression. Biochemical and Biophysical Research Communications, 505(2), 578–585. 10.1016/j.bbrc.2018.09.146 [DOI] [PubMed] [Google Scholar]
- Travis, W. D. , Brambilla, E. , Burke, A. P. , Marx, A. , & Nicholson, A. G. (2015). Introduction to The 2015 World Health Organization Classification of tumors of the lung, pleura, thymus, and heart. Journal of Thoracic Oncology, 10(9), 1240–1242. 10.1097/jto.0000000000000663 [DOI] [PubMed] [Google Scholar]
- Tzelepi, V. , Zhang, J. , Lu, J.‐F. , Kleb, B. , Wu, G. , Wan, X. , … Aparicio, A. M. (2012). Modeling a lethal prostate cancer variant with small‐cell carcinoma features. Clinical Cancer Research, 18(3), 666–677. 10.1158/1078-0432.CCR-11-1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader, G. , & Lens, S. M. (2008). The Aurora kinase family in cell division and cancer. Biochimica et Biophysica Acta, 1786(1), 60–72. 10.1016/j.bbcan.2008.07.003 [DOI] [PubMed] [Google Scholar]
- van Ree, J. H. , Jeganathan, K. B. , Malureanu, L. , & van Deursen, J. M. (2010). Overexpression of the E2 ubiquitin‐conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. Journal of Cell Biology, 188(1), 83–100. 10.1083/jcb.200906147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlotides, G. , Eigler, T. , & Melmed, S. (2007). Pituitary tumor‐transforming gene: Physiology and implications for tumorigenesis. Endocrine Reviews, 28(2), 165–186. 10.1210/er.2006-0042 [DOI] [PubMed] [Google Scholar]
- Vogelstein, B. , Papadopoulos, N. , Velculescu, V. E. , Zhou, S. , Diaz, L. A. Jr , & Kinzler, K. W. (2013). Cancer genome landscapes. Science, 339(6127), 1546–1558. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, M. G. (2001). Drug target discovery by gene expression analysis: Cell cycle genes. Current Cancer Drug Targets, 1(1), 73–83. [DOI] [PubMed] [Google Scholar]
- Walter, R. B. , Walter, D. J. , Boswell, W. T. , Caballero, K. L. , Boswell, M. , Lu, Y. , … Savage, M. G. (2015). Exposure to fluorescent light triggers down regulation of genes involved with mitotic progression in Xiphophorus skin. Comparative Biochemistry and Physiology Part C: Toxicology and Pharmacology, 178, 93–103. 10.1016/j.cbpc.2015.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Liu, Y. , & Chen, Y. (2016). Pituitary tumor transforming gene‐1 in non‐small cell lung cancer: Clinicopathological and immunohistochemical analysis. Biomedicine and Pharmacotherapy, 84, 1595–1600. 10.1016/j.biopha.2016.10.047 [DOI] [PubMed] [Google Scholar]
- Wang, S. , Liu, B. , Zhang, J. , Sun, W. , Dai, C. , Sun, W. , & Li, Q. (2017). Centromere protein U is a potential target for gene therapy of human bladder cancer. Oncology Reports, 38(2), 735–744. 10.3892/or.2017.5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Cao, R. , Su, W. , Li, Y. , & Yan, H. (2019). miR‐655‐3p inhibits cell migration and invasion by targeting pituitary tumor‐transforming 1 in non‐small cell lung cancer. Bioscience, Biotechnology, and Biochemistry, 1–6, 10.1080/09168451.2019.1617109 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Chen, D. , Gao, J. , Long, H. , Zha, H. , Zhang, A. , … Wu, W. (2018). Centromere protein U expression promotes non‐small‐cell lung cancer cell proliferation through FOXM1 and predicts poor survival. Cancer Management and Research, 10, 6971–6984. 10.2147/cmar.s182852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.‐C. , Yue, X. , Zhang, R.‐X. , Liu, T.‐Y. , Pan, Z.‐Z. , Yang, M.‐J. , … Liu, R.‐Y. (2019). Genome‐wide RNAi screening identifies RFC4 as a factor that mediates radioresistance in colorectal cancer by facilitating nonhomologous end joining repair. Clinical Cancer Research, 25(14), 4567–4579. 10.1158/1078-0432.ccr-18-3735 [DOI] [PubMed] [Google Scholar]
- Wei, T. , Weiler, S. M. E. , Tóth, M. , Sticht, C. , Lutz, T. , Thomann, S. , … Breuhahn, K. (2019). YAP‐dependent induction of UHMK1 supports nuclear enrichment of the oncogene MYBL2 and proliferation in liver cancer cells. Oncogene, 38(27), 5541–5550. 10.1038/s41388-019-0801-y [DOI] [PubMed] [Google Scholar]
- Wesierska‐Gadek, J. , & Skladanowski, A. (2012). Therapeutic intervention by the simultaneous inhibition of DNA repair and type I or type II DNA topoisomerases: One strategy, many outcomes. Future Med Chem, 4(1), 51–72. 10.4155/fmc.11.175 [DOI] [PubMed] [Google Scholar]
- Wu, A. , Li, J. , Wu, K. , Mo, Y. , Luo, Y. , Ye, H. , … Yang, Z. (2015). MiR‐373‐3p promotes invasion and metastasis of lung adenocarcinoma cells. Zhongguo Fei Ai Za Zhi, 18(7), 427–435. 10.3779/j.issn.1009-3419.2015.07.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Mo, Q. , Wan, X. , Dan, J. , & Hu, H. (2019). NEAT1/hsa‐mir‐98‐5p/MAPK6 axis is involved in non‐small‐cell lung cancer development. Journal of Cellular Biochemistry, 120(3), 2836–2846. 10.1002/jcb.26442 [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Chen, Y.‐F. , Fu, J. , You, Q.‐H. , Wang, S.‐M. , Huang, X. , … Zhang, S.‐H. (2014). Short hairpin RNA‐mediated down‐regulation of CENP‐A attenuates the aggressive phenotype of lung adenocarcinoma cells. Cellular Oncology (Dordr), 37(6), 399–407. 10.1007/s13402-014-0199-z [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Qian, Y. M. , Zhao, X. L. , Wang, S. M. , Feng, X. J. , Chen, X. F. , & Zhang, S. H. (2012). Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer, 77(2), 407–414. 10.1016/j.lungcan.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Wu, Q. , Zhang, B. , Sun, Y. , Xu, R. , Hu, X. , Ren, S. , … Wang, Z. (2019). Identification of novel biomarkers and candidate small molecule drugs in non‐small‐cell lung cancer by integrated microarray analysis. OncoTargets and Therapy, 12, 3545–3563. 10.2147/ott.s198621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, H. , Jing, H. , Li, Y. , & Lv, X. (2018). Long noncoding RNA HOXD‐AS1 promotes non‐small cell lung cancer migration and invasion through regulating miR‐133b/MMP9 axis. Biomedicine and Pharmacotherapy, 106, 156–162. 10.1016/j.biopha.2018.06.073 [DOI] [PubMed] [Google Scholar]
- Xiang, J. , Fang, L. , Luo, Y. , Yang, Z. , Liao, Y. I. , Cui, J. I. , … Wang, J. (2014). Levels of human replication factor C4, a clamp loader, correlate with tumor progression and predict the prognosis for colorectal cancer. Journal of Translational Medicine, 12, 320 10.1186/s12967-014-0320-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Chen, K. E. , Lu, C. , Gu, J. , Zeng, H. , Xu, Y. , … Ge, D. I. (2019). Large cell neuroendocrine carcinoma shares similarity with small cell carcinoma on the basis of clinical and pathological features. Translational Oncology, 12(4), 646–655. 10.1016/j.tranon.2019.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Cai, J. , Wang, L. , Jiang, L. , Huang, J. , Hu, R. , & Ding, F. (2018). MicroRNA‐30e‐5p suppresses non‐small cell lung cancer tumorigenesis by regulating USP22‐mediated Sirt1/JAK/STAT3 signaling. Experimental Cell Research, 362(2), 268–278. 10.1016/j.yexcr.2017.11.027 [DOI] [PubMed] [Google Scholar]
- Xu, X. , Jin, S. , Ma, Y. , Fan, Z. , Yan, Z. , Li, W. , … Ye, Q. (2017). miR‐30a‐5p enhances paclitaxel sensitivity in non‐small cell lung cancer through targeting BCL‐2 expression. Journal of Molecular Medicine (Berlin), 95(8), 861–871. 10.1007/s00109-017-1539-z [DOI] [PubMed] [Google Scholar]
- Yuan, W. , Xie, S. , Wang, M. , Pan, S. , Huang, X. , Xiong, M. , … Shao, L. (2018). Bioinformatic analysis of prognostic value of ZW10 interacting protein in lung cancer. OncoTargets and Therapy, 11, 1683–1695. 10.2147/ott.s149012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z. , Li, Y. , Pan, Y. , Lan, X. , Song, F. , Sun, J. , … Liang, L. I. (2018). Cancer‐derived exosomal miR‐25‐3p promotes pre‐metastatic niche formation by inducing vascular permeability and angiogenesis. Nature Communications, 9(1), 5395 10.1038/s41467-018-07810-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Hao, C. , Zhai, R. , Wang, D. I. , Zhang, J. , Bao, L. , … Yao, W. U. (2018). Downregulation of exosomal let‐7a‐5p in dust exposed‐ workers contributes to lung cancer development. Respiratory Research, 19(1), 235 10.1186/s12931-018-0949-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Ji, G. , Shao, Y. , Qiao, S. , Jing, Y. , Qin, R. , … Shao, C. (2015). MLF1 interacting protein: A potential gene therapy target for human prostate cancer? Medical Oncology, 32(2), 454 10.1007/s12032-014-0454-1 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Qian, J. , Qiang, Y. , Huang, H. , Wang, C. , Li, D. , & Xu, B. (2014). Down‐regulation of miR‐4500 promoted non‐small cell lung cancer growth. Cellular Physiology and Biochemistry, 34(4), 1166–1174. 10.1159/000366329 [DOI] [PubMed] [Google Scholar]
- Zhang, Q. , Li, Y. D. , Zhang, S. X. , & Shi, Y. Y. (2018). Centromere protein U promotes cell proliferation, migration and invasion involving Wnt/beta‐catenin signaling pathway in non‐small cell lung cancer. European Review for Medical and Pharmacological Sciences, 22(22), 7768–7777. 10.26355/eurrev_201811_16400 [DOI] [PubMed] [Google Scholar]
- Zhang, X. , He, X. , Liu, Y. , Zhang, H. , Chen, H. , Guo, S. , & Liang, Y. (2017). MiR‐101‐3p inhibits the growth and metastasis of non‐small cell lung cancer through blocking PI3K/AKT signal pathway by targeting MALAT‐1. Biomedicine and Pharmacotherapy, 93, 1065–1073. 10.1016/j.biopha.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Zhou, D. H. , Wang, X. , & Feng, Q. (2014). EGCG enhances the efficacy of cisplatin by downregulating hsa‐miR‐98‐5p in NSCLC A549 cells. Nutrition and Cancer, 66(4), 636–644. 10.1080/01635581.2014.894101 [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Zeng, Y. , Li, W. , Qin, H. , Lei, Z. , Shen, D. , … Liu, Z. (2017). CD73/NT5E is a target of miR‐30a‐5p and plays an important role in the pathogenesis of non‐small cell lung cancer. Molecular Cancer, 16(1), 34 10.1186/s12943-017-0591-1 [DOI] [PMC free article] [PubMed] [Google Scholar]


