Abstract
In this review of the application of proteomics and metabolomics to kidney disease research, we review key concepts, highlight illustrative examples, and outline future directions. The proteome and metabolome reflect the influence of environmental exposures in addition to genetic coding. Circulating levels of proteins and metabolites are dynamic and modifiable, and thus amenable to therapeutic targeting. Design and analytic considerations in proteomics and metabolomics studies should be tailored to the investigator’s goals. For the identification of clinical biomarkers, adjustment for all potential confounding variables, particularly GFR, and strict significance thresholds are warranted. However, this approach has the potential to obscure biologic signals and can be overly conservative given the high degree of intercorrelation within the proteome and metabolome. Mass spectrometry, often coupled to up-front chromatographic separation techniques, is a major workhorse in both proteomics and metabolomics. High-throughput antibody- and aptamer-based proteomic platforms have emerged as additional, powerful approaches to assay the proteome. As the breadth of coverage for these methodologies continues to expand, machine learning tools and pathway analyses can help select the molecules of greatest interest and categorize them in distinct biologic themes. Studies to date have already made a substantial effect, for example elucidating target antigens in membranous nephropathy, identifying a signature of urinary peptides that adds prognostic information to urinary albumin in CKD, implicating circulating inflammatory proteins as potential mediators of diabetic nephropathy, demonstrating the key role of the microbiome in the uremic milieu, and highlighting kidney bioenergetics as a modifiable factor in AKI. Additional studies are required to replicate and expand on these findings in independent cohorts. Further, more work is needed to understand the longitudinal trajectory of select protein and metabolite markers, perform transomics analyses within merged datasets, and incorporate more kidney tissue–based investigation.
Keywords: Metabolomics, proteomics, proteome, diabetic nephropathy, diabetic nephropathies, metabolome, membranous glomerulonephritis, research personnel, confounding factors (epidemiology), prognosis, goals, kidney, mass spectrometry, biomarkers, microbiota, peptides, machine learning, environmental exposure, energy metabolism, chronic renal insufficiency, biological products, albumins, Kidney Genomics Series
Introduction
As detailed in other reviews in this series, there has been tremendous progress in our ability to analyze the approximately 3 billion DNA base pairs of the human genome (or informative subsets of the genome) in large patient populations, with cascading benefits on our understanding of kidney disease pathophysiology, risk prediction, and treatment. By interrogating molecular domains downstream from the genome, proteomics and metabolomics have the potential to build on these advances, as well as add novel insights. Because the proteome and metabolome represent the summative effects of gene function, they are sometimes classified within the overarching field of “functional genomics,” which aims to understand the relationship between genotype and phenotype on a genome-wide scale (1). All of these “omics” approaches embrace an unbiased examination of human health and disease, but assay methodologies can vary widely. Further, although there are shared analytic approaches to large datasets regardless of molecule type, there are also areas of conceptual and operational divergence.
Key Concepts in Metabolomics and Proteomics in Kidney Disease
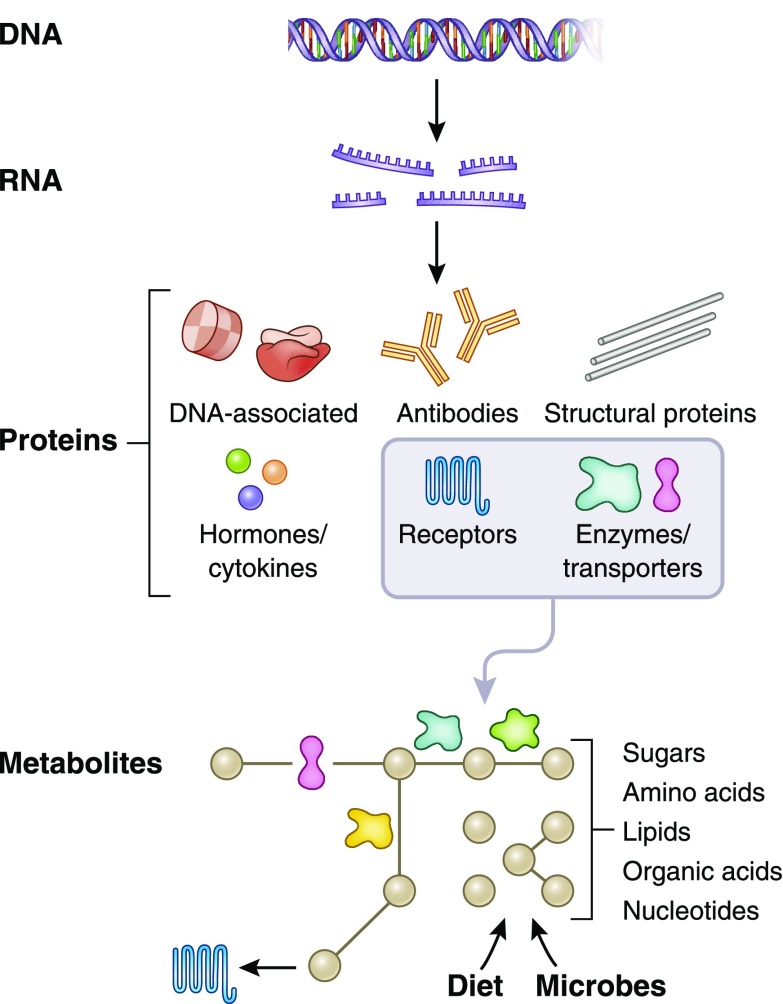
The fundamental flow of information in biologic systems is from DNA (genome) to RNA (transcriptome) to proteins (proteome) to metabolites (metabolome) (Figure 1). Although somatic mutations certainly occur (and are of critical importance in cancers), from the standpoint of kidney disease, DNA sequence can largely be viewed as static and uniform throughout the body. Thus, an assessment of the kidney genome does not require sequencing of kidney DNA, but rather can be inferred from DNA analysis of any other source (2). By contrast, the proteome and metabolome are highly dynamic—indeed, this is a major reason why an examination of these domains is critical for understanding the “functional” consequences of the genome. Further, the proteome and metabolome differ throughout the body, such that proteomic or metabolomic analysis of liver, muscle, kidney, blood, urine, etc. will yield different results (3,4). Heterogeneity also exists within tissues, for example across glomerular, endothelial, and tubular cells in the kidney (5). Thus, examination of the proteome and metabolome has clear advantages: compared with the genome, the proteome and metabolome provide information about biology in a specific time and place, more “proximal” to disease phenotype. Further, both can be applied to blood and urine, accessible biofluids central to current diagnostic and prognostic approaches in clinical nephrology—indeed, metabolites such as urea, creatinine, glucose, and uric acid, and proteins such as albumin, cystatin C, complement, and parathyroid hormone, are indispensable to the practicing nephrologist. Perhaps most importantly, proteins and metabolites are modifiable, and thus amenable to therapeutic targeting.
Figure 1.
The conceptual relationship between the proteome and metabolome and their upstream counterparts. DNA transcription and subsequent RNA translation yield proteins with a broad range of structure and function. A subset of proteins, particularly enzymes and transporters, plays a prominent role in modulating metabolites (black dots in Figure), which are derived from endogenous metabolism and exogenous sources such as diet and the microbiome.
Although proteomics and metabolomics have some distinct strengths, they suffer from one clear disadvantage relative to genomics—the inability to infer causality, for example when associations are observed between specific proteins or metabolites and disease. In genomics, whether for Mendelian disorders or more complex conditions with polygenic contributions to disease risk, the germ-line change(s) in DNA sequence by definition precedes disease onset. A genome-wide association study may show that a disease is associated with single-nucleotide polymorphisms of uncertain function, the large majority of which are noncoding and/or intronic; although these studies are unable to pinpoint the exact genetic change(s) driving the association, they nevertheless implicate the specific genomic regions tagged by the single-nucleotide polymorphisms in disease pathogenesis (6). Importantly, subsequent experimental work is required to prove causal inferences generated by genomic analyses; nevertheless, the temporal relationship between germ-line genotype and disease onset is unidirectional. By contrast, the dynamic nature of the proteome and metabolome mean that the direction of influence, i.e., from protein/metabolite ←→ disease, may be challenging to discern.
Confounding needs to be thoughtfully addressed in any proteomic or metabolomic study. In proteomics, particularly analysis of urine, accounting for underlying differences in albuminuria/proteinuria is essential because protein appearance in the urine could reflect a specific biologic process in the kidney or simply nonspecific passage through a damaged glomerular filtration barrier. In metabolomics, accounting for differences in body mass index, diabetes, and fasting status at sample collection are essential given the extensive effect of insulin action on systemic metabolism and the blood metabolome (7). The proteome and metabolome are also influenced by age, sex, diet, and diurnal variations, although our understanding of these interactions is incomplete (8–10).
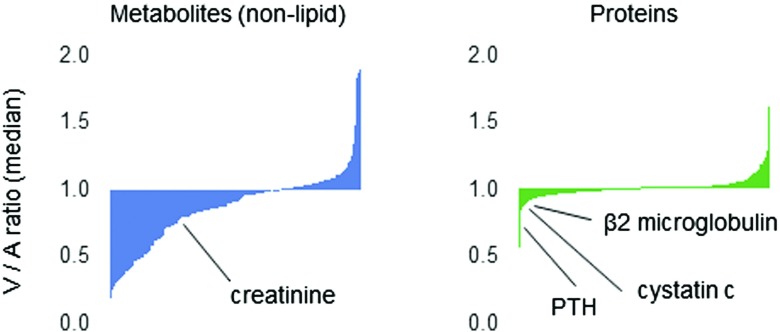
Perhaps more than any other potential confounder, GFR requires thoughtful consideration in the application of proteomics and metabolomics to kidney disease (11,12). Figure 2 shows the results of proteomic and metabolomic profiling of blood sampled from the aorta (A) and kidney vein (V) of ten human subjects. Creatinine and cystatin C are highlighted as metabolite and protein indices of GFR, respectively. Setting aside lipid metabolites, which are predominantly circulating within lipoproteins, the majority of blood metabolites undergo some degree of kidney clearance—because they are small and polar, many presumably undergo glomerular filtration, but others also undergo net tubular secretion or metabolism within the organ, as we have previously shown for select metabolites that have a greater relative A-V reduction than creatinine (13). Because they are larger, the majority of circulating proteins do not undergo glomerular filtration and most of the proteome in this analysis does not demonstrate significant A-V reductions. There are notable exceptions, however, including proteins such as β2-microglobulin and parathyroid hormone which are known to undergo some kidney clearance. Some metabolites and proteins also demonstrate an A-V increase, raising the possibility of net release from the kidney.
Figure 2.
Kidney arteriovenous gradients for the circulating metabolome and proteome. Panels show median venous-to-arterial (V/A) ratio for approximately 300 nonlipid metabolites measured by liquid chromatography–mass spectrometry and approximately 1000 proteins measured by aptamer in samples obtained from ten individuals. PTH, parathyroid hormone.
In considering the effect of kidney function on the proteome and metabolome, one must also understand the limitations of adjusting for GFR in statistical analyses. On one hand, GFR (even if directly measured) does not account for the full breadth of kidney function, such as tubular absorption or secretion. Thus, adjusting for GFR does not exclude residual confounding attributable to differences in underlying kidney health. At the same time, adjusting for GFR has the potential to obscure biologically meaningful signals. For example, a molecule might have an inverse correlation with GFR because it is nephrotoxic and causes GFR decline. This possibility is arguably more likely for circulating proteins than metabolites, as the proteome subsumes a broad range of biologic domains including inflammation, thrombosis, metabolism, cell growth, and differentiation, etc. Circulating enzymes, cytokines, and hormones are all attractive as potential participants in disease pathogenesis. Notably, recent studies have begun to assign unanticipated functional roles for select circulating metabolites, sometimes acting through previously “orphan” G-protein–coupled receptors (14). Many of these findings appear anchored around the crosstalk of gut microbes and host immunity and metabolism; given their relevance to inflammation, BP, and energy utilization, the metabolites in this subset are potential causal mediators of kidney disease and its complications.
Assay Methodology
Whereas nucleic acids comprise four to five similar chemical motifs, proteins and metabolites span a broad range of size, polarity, and structure, raising considerable analytic challenges. For a detailed review of proteomic and metabolomic methodologies, the reader is referred to other in-depth reviews (15–18). Mass spectrometry (MS), which differentiates ions—for example peptide fragments or metabolites—on the basis of mass-to-charge ratio, has been a major workhorse for both proteomics and metabolomics. Typically, MS is coupled to up-front separation techniques, such as liquid chromatography (LC) or gas chromatography, that can have a substantial effect on which analytes are ultimately measured. Two-dimensional gel electrophoresis has also been used extensively in proteomics, whereas nuclear magnetic resonance spectrometry has been an important alternative for metabolomics. For MS, proteins need to be enzymatically digested into multiple smaller peptides and peptide fragments before analysis. This requirement for complex preanalytic sample preparation has historically limited the throughput of MS-based proteomics. More recently, highly multiplexed methods that capture and detect hundreds or thousands of proteins have been developed (15,19). Specific protein capture is achieved using nucleotide-labeled antibodies or aptamers (nucleic acid–based), and then detection and quantitation are on the basis of nucleic acid oligomerization. The breadth, sensitivity, and throughput of these approaches make them powerful research tools that can be applied to large study populations. For both proteomics and metabolomics, assay validation is important. Traditionally, this has involved using an orthogonal approach to confirm top hits, for example ELISA for proteins or tandem MS for metabolites. Because proteomic and metabolomic approaches usually provide information on relative protein or metabolite levels, rather than absolute concentrations, orthogonal methods may also be required for absolute quantitation.
Analytic Considerations
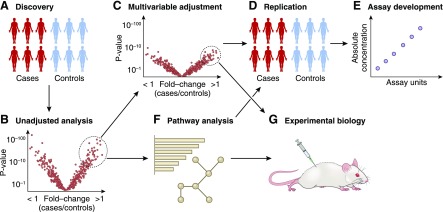
As with genomics, correction for multiple hypothesis testing is required in proteomics and metabolomics to reduce the risk of false discovery. However, there is no uniformly applied threshold analogous to the P<5×10–8 genome-wide threshold, in part because the actual sizes of the proteome and metabolome are uncertain (and contingent on whether one includes post-translational modifications, xenobiotics, etc.) and different platforms provide incomplete and only partially overlapping coverage of these molecular domains. This latter limitation has made independent replication across cohorts and meta-analysis more challenging in proteomics and metabolomics than genomics. For discovery analyses, most investigators have favored using either Bonferroni adjustment or false discovery rate, for example using the Benjamini–Hochberg procedure (20). Given the intercorrelation of many analytes in the context of biologic pathways, however, these approaches may be overly conservative and, as with adjustment for GFR, have the potential to obscure biologic signals. If the goal is to identify individual biomarkers with robust disease associations above and beyond other established predictors, then fully adjusted models and strict significance thresholds are ideal. A focus on biologic discovery may warrant more flexibility (Figure 3).
Figure 3.
Outline of common analytic approaches in human proteomics and metabolomics studies. First (A), proteomic or metabolomic analysis is performed on samples obtained from individuals with or without a phenotype, e.g., CKD progression (an alternative is to examine the association of proteins and metabolites with a continuous phenotype, such as eGFR slope). A “volcano plot” (B) can be used to visualize the fold-change and associated P value across all analytes in the case versus control comparison. The strengths of associations are often significantly stronger in unadjusted analysis. Bonferroni adjustment or false discovery rate are often used to account for multiple hypothesis testing. For the identification of clinical biomarkers, (C) it is important to adjust for all potential confounding variables (particularly eGFR and proteinuria), (D) followed by replication in an independent cohort and (E) assay development and validation, i.e., using an orthogonal assay to confirm specificity and permit absolute quantitation. A focus on biologic discovery may warrant less stringent significant thresholds, consideration of unadjusted results, and the use of (F) pathway analysis and machine learning. Ultimately, (G) mechanistic interrogation at the bench is required to test the causal role for select proteins and metabolites in disease.
Machine learning and pathway analyses are potent methods to help organize and decode omics datasets. Machine learning methods such as least absolute shrinkage and selection operator, elastic net, and random forest regression each have different strengths. In omics datasets, many variables may be collinear, and random forest regression may handle this better than other methods. If the goal is to create a parsimonious risk model, least absolute shrinkage and selection operator or elastic net may be more suitable. Support-vector machines or clustering are methods that use algorithms to categorize variables in supervised or unsupervised machine learning. These methods can be tuned to select the number of “top hits” appropriate to the research goal. A handful of top hits may be ideal when designing a biomarker risk panel, whereas several hundred proteins or metabolites will be more amenable to pathway analyses. Pathway analysis is a broad term encompassing several methods to organize large datasets into functionally distinct biologic themes (21,22). These methods are more established and have been used more widely in proteomics (and genomics) than in metabolomics. There are many publicly available software programs for pathway analysis, including Gene Set Enrichment Analysis; Database for Annotation, Visualization, and Integrated Discovery; and Search Tool for the Retrieval of Interacting Genes/Proteins. Ingenuity Pathway Analysis is also widely used and requires a private license.
Illustrative Examples in the Literature
Application of Proteomics to Understanding Biology of Membranous Nephropathy
The story of the M-type phospholipase A(2) receptor (PLA2R) illustrates the potential for proteomic discovery to inform pathobiology and guide diagnosis and treatment. On the basis of its histopathology, primary membranous nephropathy was long recognized as an antibody-mediated autoimmune glomerular disease, but the target antigen was unknown. Beck et al. performed western blots of protein extracts of normal human glomeruli with serum from patients with primary membranous nephropathy and secondary membranous nephropathy and from normal controls. The investigators identified a 185-kDa reactive protein band specific to primary membranous nephropathy and then applied liquid chromatography–mass spectrometry (LC-MS) to the reactive protein band to detect PLA2R. Further, they showed that PLA2R was expressed in podocytes from healthy kidneys and that antibodies to PLA2R were found in the glomerular immune deposits of patients with primary membranous nephropathy (23). A similar approach, western blotting followed by LC-MS, was used to identify thrombospondin type-1 domain–containing 7A (THSD7A) as an additional target antigen for autoantibodies in some patients with anti-PLA2R–negative membranous nephropathy (24). Since these landmark findings, autoantibodies to PLA2R have been found to account for approximately 70%–80% of cases of primary membranous nephropathy, with autoantibodies to THSD7A accounting for a much smaller fraction (25). These discoveries have transformed clinical care, because serum autoantibody levels have been found to correlate with membranous nephropathy disease activity and response to immunosuppression (26,27). A recent exploratory study applied LC-MS–based proteomics to serum from patients with idiopathic membranous nephropathy who did or did not achieve complete remission on calcineurin inhibitors, and identified serum amyloid A1 protein as a candidate marker that may provide further prognostic information on top of PLA2R antibody levels (28).
Utility of Proteomics for Predicting CKD Incidence and Prognosis
Whereas the example of membranous nephropathy demonstrates how proteomic discovery can focus interest on one or two specific proteins, other studies have considered a wider array of molecules. Mischak and colleagues (29) have endorsed the value of combining multiple peptide or protein biomarkers, on the premise that the high variability of single biomarkers limits accuracy. Using capillary electrophoresis–MS, these investigators compared urine from 230 patients with a range of CKD causes and 379 controls with normal kidney function. Cluster analysis by support vector machines was used to combine 273 urinary peptides that differed between CKD and healthy controls into an aggregate urinary peptide marker termed CKD273 (30). Subsequent studies that included several thousand patient samples have shown that CKD273 may be of higher utility than standard metrics for detection and prediction of CKD (31,32). The US Food and Drug Administration has issued a letter of support encouraging further studies with CKD273 as a biomarker panel in early phase clinical trials “to determine its clinically utility for prognostic enrichment, drug development decisions, and study design considerations,” but also noting that CKD273 does not appear to perform well in prediction of progression to ESKD or doubling of serum creatinine in patients with more advanced disease (29).
Major components of CKD273 include collagen fragments, perhaps indicative of accumulation of intrakidney extracellular matrix, along with fragments of blood-derived proteins involved in inflammation and tissue repair. Notably, chronic inflammation has drawn considerable interest as a risk marker and potential etiologic factor in diabetic kidney disease progression. For example, Niewczas and colleagues have highlighted a strong association between circulating TNF receptors 1 and 2 (TNF-R1 and TNF-R2) and the risk of ESKD in diabetic kidney disease (33). To more comprehensively assess inflammation in diabetic kidney disease, these investigators recently used an aptamer-based approach to measure a panel of 194 proteins that includes most of the circulating inflammatory proteins known in the literature and most proteins previously studied in diabetic kidney disease (34). Discovery analysis was performed in 219 subjects with type 1 diabetes from the Joslin Kidney Study, followed by validation in 144 subjects with type 2 diabetes from the Joslin Kidney Study and replication in 162 subjects with type 2 diabetes in the Pima Indian Study. This study identified a Kidney Risk Inflammatory Signature (KRIS), comprised of 17 inflammatory proteins enriched for TNF-R superfamily members, significantly associated with the 10-year risk of ESKD. The investigators also assessed KRIS proteins in urine from a small case-control analysis of rapid GFR decline and found that most were elevated in cases and none were reduced. Further, they found no significant correlation between the majority of circulating KRIS protein levels and their corresponding gene expression in kidney biopsy specimens obtained from 56 Pima Indians. On the basis of these observations, the authors posit that nonkidney sources (such as leukocytes) play an important role in generating circulating KRIS proteins, and perhaps as etiologic drivers of diabetic kidney disease.
Metabolomics and the Contribution of the Gut Microbiome to Uremia
Many metabolomics studies have examined blood or urine with the goal to identify metabolite alterations associated with CKD or to identify metabolite predictors of CKD progression. To date, no single metabolite has emerged as an unequivocal causal factor in the pathogenesis of CKD or its complications; this relates in part to the nonoverlap across platforms noted above and the heterogeneity of study populations, but also to the sheer number of metabolomic perturbations that manifest as kidney clearance is lost. However, one major theme that has emerged is the fundamental contribution made by the gut microbiome to the blood metabolome in CKD. For example, Meyer and colleagues (35) used LC-MS to profile blood obtained from nine patients on hemodialysis with intact colons and six patients on hemodialysis who had previously undergone colectomy. Compared with patients with ESKD with intact colons, patients with ESKD who had undergone colectomy had markedly lower levels of at least 35 metabolites, including indoxyl sulfate and p-cresol sulfate. Importantly, the levels of these colon-derived metabolites were substantially higher in the patients with ESKD than in individuals with normal kidney function, and the majority were not effectively removed by conventional hemodialysis. Abe and colleagues (36) have raised specific interest in phenyl sulfate, a tyrosine metabolite derived from gut microbes, as a potential causal factor in diabetic kidney disease. In patients with diabetes, they found that phenyl sulfate levels correlate with albuminuria, and, further, that phenyl sulfate administration and inhibition of the bacterial enzyme responsible for its synthesis induce and ameliorate albuminuria in mice, respectively.
Hazen and colleagues have focused on a specific gut-derived metabolite, trimethylamine-N-oxide (TMAO), as a potential causal factor in cardiovascular disease. Initial interest in TMAO was motivated by an LC-MS metabolomics analysis of blood samples obtained from individuals with prevalent cardiovascular disease, with subsequent studies showing that elevated TMAO is associated with new-onset cardiovascular events (37,38). The requirement for gut flora on blood TMAO was demonstrated with elegant feeding studies before and after gut flora suppression using antibiotics (37,38), and a functional role for TMAO in atherogenesis has been posited on the basis of its ability to inhibit reverse cholesterol transport, increase platelet hyperresponsiveness, and promote macrophage foam cell formation (39,40). In mice, dietary TMAO loading can also cause tubular injury and kidney fibrosis (41). Because TMAO is primarily excreted via the kidney and levels increase with CKD, several groups have examined its association with cardiovascular disease among individuals with CKD and ESKD, with mixed results (41–44). Whether or not molecules such as phenyl sulfate and TMAO prove to be important causal factors in CKD, these studies underscore the ability of metabolomics to integrate both endogenous and exogenous outputs, with the latter including contributions from diet and the microbiome.
Application of Metabolomics for Translational AKI Research
Parikh and colleagues (45) applied LC-MS–based metabolomics to examine endogenous kidney metabolism in mice with loss- and gain-of-function of Peroxisome proliferator-activated receptor γ coactivator 1-α, which they have shown are sensitized and protected from AKI, respectively. They found that niacinamide levels were lower in kidneys from PGC1α-deficient animals and that niacinamide levels fell in response to AKI. By contrast, niacinamide levels were higher in transgenic PGC1α animals and niacinamide supplementation was renoprotective in different injury models, in part by rescuing AKI-induced impairment in lipid metabolism. Niacinamide is a precursor in the salvage pathway of NAD+, an essential cofactor for energy metabolism that is a key determinant of cellular health across a range of tissues. Parikh and colleagues (46) have used LC-MS to demonstrate that urinary metabolites indicative of reduced de novo NAD+ biosynthesis are associated with AKI risk in humans, and, further, have generated pilot data raising the possibility that NAD+ boosting with oral niacinamide supplementation may attenuate kidney injury in the context of cardiac surgery. These promising initial findings underscore an important advantage of metabolomics-based discovery, the potential for rapid translation. More specifically, many metabolites are readily delivered in vivo, and their cognate enzymes, transporters, and receptors are all potential targets for small-molecule agonism or antagonism.
Future Directions
Although progress has been made, more work is required to harness the full potential of proteomics and metabolomics in the study of kidney disease. In one example, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) CKD Biomarkers Consortium (www.ckdbiomarkersconsortium.org) is actively generating blood proteomic and metabolomic data in >3000 individuals with CKD, spanning several different study cohorts, including both adults and children (47). In addition to enhancing statistical power compared with prior CKD studies, this coordinated effort will enable examination of different outcomes such as CKD progression, cardiovascular events, and cognitive development, as well as different “subphenotypes,” i.e., looking at patient subsets on the basis of diagnosis (48), comorbidity, or other differentiating features. Because some proteomics and metabolomics data will be generated in a common sample set that has been genotyped, there will also be an opportunity for integrative analyses; paradigms exist for genome-wide association studies of the blood proteome or metabolome, whereas efforts to integrate the proteome and metabolome are less developed.
Even as proteomic and metabolomic studies in kidney disease continue to expand, important opportunities remain. To date, there is a relative paucity of data in large cohorts with repeated measurements over time that show the longitudinal trajectory of the proteome and metabolome. Further, the spatial heterogeneity of the proteome and metabolome mean that an examination of blood or urine provides only limited information about intraorgan biology. The NIDDK Kidney Precision Medicine Project (www.kpmp.org) is anticipated to provide much-needed human kidney tissues for biomarker research, and this program and more mechanistic investigation in model systems will be critical to determine both why select proteins or metabolites are altered in kidney disease and whether they play a causal role in disease pathogenesis. Recent advances in identifying post-translational modifications of proteins will enhance our understanding of the proteome in kidney disease (49). In parallel, the integration of genomics with proteomics and metabolomics may permit Mendelian randomization–based approaches to test whether select markers lie in causal pathways (50). These efforts will require vigorous collaboration across disciplines, and, taken together, will build on the momentum of genomic discovery in nephrology research to further advance our understanding and therapeutic approaches to kidney disease.
Disclosures
Dr. Dubin and Dr. Rhee have nothing to disclose.
Funding
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants U01DK108809 and U01DK106981 and National Institute of Nursing Research grant R01NR017399.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Sauer S, Lange BM, Gobom J, Nyarsik L, Seitz H, Lehrach H: Miniaturization in functional genomics and proteomics. Nat Rev Genet 6: 465–476, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Heather JM, Chain B: The sequence of sequencers: The history of sequencing DNA. Genomics 107: 1–8, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F: Proteomics. Tissue-based map of the human proteome. Science 347: 1260419, 2015 [DOI] [PubMed] [Google Scholar]
- 4.Sugimoto M, Ikeda S, Niigata K, Tomita M, Sato H, Soga T: MMMDB: Mouse multiple tissue metabolome database. Nucleic Acids Res 40: D809–D814, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoyer KJR, Dittrich S, Bartram MP, Rinschen MM: Quantification of molecular heterogeneity in kidney tissue by targeted proteomics. J Proteomics 193: 85–92, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Hirschhorn JN, Daly MJ: Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95–108, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Newgard CB: Metabolomics and metabolic diseases: Where do we stand? Cell Metab 25: 43–56, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao C, Zhao M, Chen X, Sun H, Yang Y, Xiao X, Guo Z, Liu X, Lv Y, Chen X, Sun W, Wu D, Gao Y: Comprehensive analysis of individual variation in the urinary proteome revealed significant gender differences. Mol Cell Proteomics 18: 1110–1122, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaeva S, Ansermet C, Centeno G, Pradervand S, Bize V, Mordasini D, Henry H, Koesters R, Maillard M, Bonny O, Tokonami N, Firsov D: Nephron-specific deletion of circadian clock gene Bmal1 alters the plasma and renal metabolome and impairs drug disposition. J Am Soc Nephrol 27: 2997–3004, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebholz CM, Zheng Z, Grams ME, Appel LJ, Sarnak MJ, Inker LA, Levey AS, Coresh J: Serum metabolites associated with dietary protein intake: Results from the Modification of Diet in Renal Disease (MDRD) randomized clinical trial. Am J Clin Nutr 109: 517–525, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christensson A, Ash JA, DeLisle RK, Gaspar FW, Ostroff R, Grubb A, Lindström V, Bruun L, Williams SA: The impact of the glomerular filtration rate on the human plasma proteome. Proteomics Clin Appl 12: e1700067, 2018 [DOI] [PubMed] [Google Scholar]
- 12.Sekula P, Goek ON, Quaye L, Barrios C, Levey AS, Römisch-Margl W, Menni C, Yet I, Gieger C, Inker LA, Adamski J, Gronwald W, Illig T, Dettmer K, Krumsiek J, Oefner PJ, Valdes AM, Meisinger C, Coresh J, Spector TD, Mohney RP, Suhre K, Kastenmüller G, Köttgen A: A metabolome-wide association study of kidney function and disease in the general population. J Am Soc Nephrol 27: 1175–1188, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee EP, Clish CB, Ghorbani A, Larson MG, Elmariah S, McCabe E, Yang Q, Cheng S, Pierce K, Deik A, Souza AL, Farrell L, Domos C, Yeh RW, Palacios I, Rosenfield K, Vasan RS, Florez JC, Wang TJ, Fox CS, Gerszten RE: A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol 24: 1330–1338, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW: GPCR-mediated signaling of metabolites. Cell Metab 25: 777–796, 2017 [DOI] [PubMed] [Google Scholar]
- 15.Gramolini A, Lau E, Liu PP: Identifying low-abundance biomarkers: Aptamer-based proteomics potentially enables more sensitive detection in cardiovascular diseases. Circulation 134: 286–289, 2016 [DOI] [PubMed] [Google Scholar]
- 16.Aebersold R, Mann M: Mass spectrometry-based proteomics. Nature 422: 198–207, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Kalim S, Rhee EP: An overview of renal metabolomics. Kidney Int 91: 61–69, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sas KM, Karnovsky A, Michailidis G, Pennathur S: Metabolomics and diabetes: Analytical and computational approaches. Diabetes 64: 718–732, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JG, Gerszten RE: Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation 135: 1651–1664, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300, 1995 [Google Scholar]
- 21.Kelder T, Conklin BR, Evelo CT, Pico AR: Finding the right questions: Exploratory pathway analysis to enhance biological discovery in large datasets. PLoS Biol 8: e1000472, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatri P, Sirota M, Butte AJ: Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput Biol 8: e1002375, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck LH Jr, Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, Klein JB, Salant DJ: M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med 361: 11–21, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomas NM, Beck LH Jr, Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, Dolla G, Hoxha E, Helmchen U, Dabert-Gay AS, Debayle D, Merchant M, Klein J, Salant DJ, Stahl RAK, Lambeau G: Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med 371: 2277–2287, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Couser WG: Primary membranous nephropathy. Clin J Am Soc Nephrol 12: 983–997, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoxha E, Thiele I, Zahner G, Panzer U, Harendza S, Stahl RA: Phospholipase A2 receptor autoantibodies and clinical outcome in patients with primary membranous nephropathy. J Am Soc Nephrol 25: 1357–1366, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bomback AS: Management of membranous nephropathy in the PLA2R Era. Clin J Am Soc Nephrol 13: 784–786, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Cai J, Jiao X, Zhang S, Liu H, Ding X: Response predictors to calcineurin inhibitors in patients with primary membranous nephropathy. Am J Nephrol 47: 266–274, 2018 [DOI] [PubMed] [Google Scholar]
- 29.Pontillo C, Mischak H: Urinary peptide-based classifier CKD273: Towards clinical application in chronic kidney disease. Clin Kidney J 10: 192–201, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good DM, Zürbig P, Argilés A, Bauer HW, Behrens G, Coon JJ, Dakna M, Decramer S, Delles C, Dominiczak AF, Ehrich JH, Eitner F, Fliser D, Frommberger M, Ganser A, Girolami MA, Golovko I, Gwinner W, Haubitz M, Herget-Rosenthal S, Jankowski J, Jahn H, Jerums G, Julian BA, Kellmann M, Kliem V, Kolch W, Krolewski AS, Luppi M, Massy Z, Melter M, Neusüss C, Novak J, Peter K, Rossing K, Rupprecht H, Schanstra JP, Schiffer E, Stolzenburg JU, Tarnow L, Theodorescu D, Thongboonkerd V, Vanholder R, Weissinger EM, Mischak H, Schmitt-Kopplin P: Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics 9: 2424–2437, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schanstra JP, Zürbig P, Alkhalaf A, Argiles A, Bakker SJ, Beige J, Bilo HJ, Chatzikyrkou C, Dakna M, Dawson J, Delles C, Haller H, Haubitz M, Husi H, Jankowski J, Jerums G, Kleefstra N, Kuznetsova T, Maahs DM, Menne J, Mullen W, Ortiz A, Persson F, Rossing P, Ruggenenti P, Rychlik I, Serra AL, Siwy J, Snell-Bergeon J, Spasovski G, Staessen JA, Vlahou A, Mischak H, Vanholder R: Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol 26: 1999–2010, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontillo C, Zhang ZY, Schanstra JP, Jacobs L, Zürbig P, Thijs L, Ramírez-Torres A, Heerspink HJL, Lindhardt M, Klein R, Orchard T, Porta M, Bilous RW, Charturvedi N, Rossing P, Vlahou A, Schepers E, Glorieux G, Mullen W, Delles C, Verhamme P, Vanholder R, Staessen JA, Mischak H, Jankowski J: Prediction of chronic kidney disease stage 3 by CKD273, a urinary proteomic biomarker. Kidney Int Rep 2: 1066–1075, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niewczas MA, Gohda T, Skupien J, Smiles AM, Walker WH, Rosetti F, Cullere X, Eckfeldt JH, Doria A, Mayadas TN, Warram JH, Krolewski AS: Circulating TNF receptors 1 and 2 predict ESRD in type 2 diabetes. J Am Soc Nephrol 23: 507–515, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, Park J, Nair V, Schlafly A, Saulnier PJ, Satake E, Simeone CA, Shah H, Qiu C, Looker HC, Fiorina P, Ware CF, Sun JK, Doria A, Kretzler M, Susztak K, Duffin KL, Nelson RG, Krolewski AS: A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25: 805–813, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aronov PA, Luo FJ, Plummer NS, Quan Z, Holmes S, Hostetter TH, Meyer TW: Colonic contribution to uremic solutes. J Am Soc Nephrol 22: 1769–1776, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikuchi K, Saigusa D, Kanemitsu Y, Matsumoto Y, Thanai P, Suzuki N, Mise K, Yamaguchi H, Nakamura T, Asaji K, Mukawa C, Tsukamoto H, Sato T, Oikawa Y, Iwasaki T, Oe Y, Tsukimi T, Fukuda NN, Ho HJ, Nanto-Hara F, Ogura J, Saito R, Nagao S, Ohsaki Y, Shimada S, Suzuki T, Toyohara T, Mishima E, Shima H, Akiyama Y, Akiyama Y, Ichijo M, Matsuhashi T, Matsuo A, Ogata Y, Yang CC, Suzuki C, Breeggemann MC, Heymann J, Shimizu M, Ogawa S, Takahashi N, Suzuki T, Owada Y, Kure S, Mano N, Soga T, Wada T, Kopp JB, Fukuda S, Hozawa A, Yamamoto M, Ito S, Wada J, Tomioka Y, Abe T: Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat Commun 10: 1835, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL: Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL: Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 368: 1575–1584, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL: Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, Hazen SL: Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell 165: 111–124, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang WH, Wang Z, Kennedy DJ, Wu Y, Buffa JA, Agatisa-Boyle B, Li XS, Levison BS, Hazen SL: Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res 116: 448–455, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stubbs JR, House JA, Ocque AJ, Zhang S, Johnson C, Kimber C, Schmidt K, Gupta A, Wetmore JB, Nolin TD, Spertus JA, Yu AS: Serum Trimethylamine-N-Oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J Am Soc Nephrol 27: 305–313, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalim S, Clish CB, Wenger J, Elmariah S, Yeh RW, Deferio JJ, Pierce K, Deik A, Gerszten RE, Thadhani R, Rhee EP: A plasma long-chain acylcarnitine predicts cardiovascular mortality in incident dialysis patients. J Am Heart Assoc 2: e000542, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafi T, Powe NR, Meyer TW, Hwang S, Hai X, Melamed ML, Banerjee T, Coresh J, Hostetter TH: Trimethylamine N-Oxide and cardiovascular events in hemodialysis patients. J Am Soc Nephrol 28: 321–331, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tran MT, Zsengeller ZK, Berg AH, Khankin EV, Bhasin MK, Kim W, Clish CB, Stillman IE, Karumanchi SA, Rhee EP, Parikh SM: PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature 531: 528–532, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poyan Mehr A, Tran MT, Ralto KM, Leaf DE, Washco V, Messmer J, Lerner A, Kher A, Kim SH, Khoury CC, Herzig SJ, Trovato ME, Simon-Tillaux N, Lynch MR, Thadhani RI, Clish CB, Khabbaz KR, Rhee EP, Waikar SS, Berg AH, Parikh SM: De novo NAD+ biosynthetic impairment in acute kidney injury in humans. Nat Med 24: 1351–1359, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee EP, Waikar SS, Rebholz CM, Zheng Z, Perichon R, Clish CB, Evans AM, Avila J, Denburg MR, Anderson AH, Vasan RS, Feldman HI, Kimmel PL, Coresh J; CKD Biomarkers Consortium: Variability of two metabolomic platforms in CKD. Clin J Am Soc Nephrol 14: 40–48, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grams ME, Tin A, Rebholz CM, Shafi T, Köttgen A, Perrone RD, Sarnak MJ, Inker LA, Levey AS, Coresh J: Metabolomic alterations associated with cause of CKD. Clin J Am Soc Nephrol 12: 1787–1794, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaffer LV, Millikin RJ, Miller RM, Anderson LC, Fellers RT, Ge Y, Kelleher NL, LeDuc RD, Liu X, Payne SH, Sun L, Thomas PM, Tucholski T, Wang Z, Wu S, Wu Z, Yu D, Shortreed MR, Smith LM: Identification and quantification of proteoforms by mass spectrometry. Proteomics 19: e1800361, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekula P, Del Greco M F, Pattaro C, Köttgen A: Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 27: 3253–3265, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]