Abstract
Background:
More than 30% of estrogen receptor-positive breast cancers are resistant to primary hormone therapy, and about 40% that initially respond to hormone therapy eventually acquire resistance. Although the mechanisms of hormone therapy resistance remain unclear, aberrant DNA methylation has been implicated in oncogenesis and drug resistance.
Purpose:
We investigated the relationship between methylome variations in circulating tumor DNA and exemestane resistance, to track hormone therapy efficacy.
Methods:
We prospectively recruited 16 patients who were receiving first-line therapy in our center. All patients received exemestane-based hormone therapy after enrollment. We collected blood samples at baseline, first follow-up (after 2 therapeutic cycles) and at detection of disease progression. Disease that progressed within 6 months under exemestane treatment was considered exemestane resistance but was considered relatively exemestane-sensitive otherwise. We obtained circulating tumor DNA-derived methylomes using the whole-genome bisulfide sequencing method. Methylation calling was done by BISMARK software; differentially methylated regions for exemestane resistance were calculated afterward.
Results:
Median follow-up for the 16 patients was 19.0 months. We found 7 exemestane resistance-related differentially methylated regions, located in different chromosomes, with both significantly different methylation density and methylation ratio. Baseline methylation density and methylation ratio of chromosome 6 [32400000-32599999] were both high in exemestane resistance. High baseline methylation ratios of chromosome 3 [67800000-67999999] (P = .013), chromosome 3 [140200000-140399999] (P = .037), and chromosome 12 [101200000-101399999] (P = .026) could also predict exemestane resistance. During exemestane treatment, synchronized changes in methylation density and methylation ratio in chromosome 6 [32400000-32599999] could accurately stratify patients in terms of progression-free survival (P = .000033). Cutoff values of methylation density and methylation ratio for chromosome 6 [149600000-149799999] were 0.066 and 0.076, respectively.
Conclusion:
Methylation change in chromosome 6 [149600000-149799999] is an ideal predictor of exemestane resistance with great clinical potential.
Keywords: advanced breast cancer, exemestane resistance, methylomes, circulating tumor DNA
Introduction
Breast cancer (BC) is the second leading cause of cancer-related death among women worldwide.1 However, as the use of companion diagnostics has gained popularity among physicians and researchers,2 a growing body of prognostic biomarkers have been identified for use in BC,3-5 such as Oncotype Dx and PAM50.
DNA methylation has been shown to be a valuable biomarker.6,7 DNA methylation occurs mostly at 5′-C-phosphate-G-3′ (CpG) dinucleotides and is catalyzed by DNA methyltransferases (DNMTs), including DNMT1, DNMT3α, and DNMT3β.8 Hypermethylation of CpG islands associated with tumor suppressor genes is rare in normal cells, but common in cancer cells.8 The effects of DNA methylation on BC have been widely studied in the past 3 decades.7,9-12
Although hormone receptor (HR) overexpression is predictive of hormone therapy (HT) response in BC, nearly 40% of patients eventually develop resistance.13 This is partly due to the loss of estrogen receptor (ER) expression during HT.14,15 Several predictors of HT sensitivity are available besides ER expression. Widschwendter et al 16 found that ARHI methylation in tumor samples predicted survival in nontamoxifen-treated HR+ patients with BC. They also found that CYP1B1 methylation in tumor samples predicted tamoxifen response. Because of their easy accessibility, circulating biomarkers related to HT sensitivity have also gained much attention. Martínez-Galán and her colleagues17 observed a significant inverse correlation between hypermethylation of ESR1 in circulating tumor DNA (ctDNA) and ER expression status in primary BC tumors. They proposed that ESR1 methylation status can predict poor prognosis and HT resistance in luminal BC. The relationship between methylation of the RASSF1A promoter region and ER expression could indicate prognosis during HT.18 Although hypermethylation of certain genes is predictive for HT response, HT resistance eventually develops anyway, due to the multiple factors that affect response.10,19,20 Thus, investigating methylation status at the single-gene level is insufficient. In the present study, we aimed to monitor and predict exemestane resistance (EXEr) using ctDNA-based methylomes. Our findings may provide a clue to the relationship between whole-genome ctDNA methylation profiles and EXEr in advanced HR+ BC.
Material and Method
Patients and Samples
The study was approved by the Medical Ethical Committee of Peking University Cancer Hospital & Institute (Approval No. 2016KT47). We recruited 16 patients with HR+ BC who were treated at the Peking Cancer Hospital & Institute between April 2009 and June 2017. Their tumors’ ER status was evaluated by applying antibodies to ER to archival formalin-fixed paraffin-embedded tissue. A sample was considered ER+ if at least 1% of its tumor cells showed positive staining.21 All scoring was performed by pathologists, according to the American Society of Clinical Oncology/College of American Pathologists guidelines.22
Inclusion criteria for every patient were (1) pathologically ER+ primary tumor; (2) treatment with EXE, with or without chemotherapy and/or target therapy after relapse; (3) complete clinical, pathological, and follow-up data available; and (4) provision of written and informed consent.
In the present study, cancers that progressed within 6 months of starting EXE treatment were defined as EXEr; otherwise, tumors were defined as relatively exemestane sensitive (EXEs). Plasma DNA samples from each patient were collected at (1) the beginning of EXE therapy (baseline); (2) first follow-up (after 2 therapeutic cycles); and (3) when disease progression (PD) was diagnosed. Such blood draws collected 8.0 mL peripheral blood in BD Vacutainer tubes (Becton, Dickinson and Co, Franklin Lakes, New Jersey). Samples were stored at room temperature and analyzed within 24 hours. Progression-free survival (PFS) was defined as the time from EXE interference to PD or death.
Cell Lines and Reagents
The BC cell line MCF7 was purchased from the American Type Culture Collection (Manassas, Virginia) and cultured in Dulbecco’s modified Eagle medium/high-glucose medium (Gibco-BRL, Rockville, Indiana) supplemented with 10% fetal bovine serum (Corning CellGro, Virginia), 100 U/mL penicillin, and 100 μg/mL streptomycin in 5% CO2 at 37°C. Exemestane (high-performance liquid chromatography purity ≥ 98%) was purchased from Energy Chemical Co, Ltd (Energy Chemical, Shanghai, China). CellTiter 96 Aqueous One Solution Reagent was purchased from Biotech Co, Ltd (Promega, Beijing, China). Wild-type MCF-7 cells were exposed stepwise to increased concentrations of EXE (100-1000 μM) to develop the resistant cell line, MCF-7/ADM. Resistance index (RI) was determined by cell viability assays and calculated as (IC50·R)/(IC50·W) = RI, where IC50 R is IC50 of resistant cells and IC50 W is IC50 of wild-type cells. Cells for which RI >5 were considered resistant. Resistant cells were not used after more than 2 passages after being cultured.
Cell Viability Assay
The IC50 for each cell line was measured using CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS). Breast cancer cells (2 × 103 cells/well) were cultured overnight in 96-well plates. The medium was then replaced with medium that contained various concentrations of EXE (0, 30, 60, 90, or 120 μM). After incubation for 72 hours, 20 μL of MTS + 100 μL complete medium were added to each well and the cells were incubated at 37°C for 3 to 4 hours in a humidified, 5% CO2 atmosphere. Supernatants were carefully discarded, and 100 μL dimethyl sulfoxide was added to each well. Absorbance values were read in a Microplate Reader (Bio-Rad, Hercules, California) at 490 nm. Each data point represents the mean ± standard deviation (SD) of 4 replications.
DNA Preparation and Whole-Genome Bisulfide Sequencing Library Preparation
The 8-mL specimens of patients’ peripheral blood were spun into plasma and frozen at −80°C until extraction. Cell-free DNA (cfDNA) was extracted from 600 µL plasma with the MagMAX Cell-Free DNA Isolation Kit (A29319; ThermoFisher, Massachusetts, USA), following the manufacturer’s protocols. The cfDNA was then quantified by quantitative polymerase chain reaction with iTaq Universal SYBR Green Supermix #1725121 (Bio-Rad, California, USA) with primers ALU115 and ALU247. Genomic DNA of MCF-7 and of MCF-7/EXE was extracted using QIAamp DNA Mini Kit 50T (Lot 51304 QIAGEN, Düsseldorf, Germany).
Bisulfite conversion of DNA was performed with an EZ DNA Methylation-gold kit (D5005; Zymo Research, California, USA), according to the manufacturer’s protocols. The methyl-seq lib preparation was followed by the cfDNA protocol of Accel-NGS Methyl-Seq DNA Library Kit (Cat No. 30096; Swift Biosciences, Michigan, USA). We conducted high-throughput sequencing using an Illumina HiSeq XTEN (Illumina, San Diego, California, USA).
Sequence Analysis Methods
Cutadapt (version 1.14) was used to trim the sequence barcode from the original FastQC sequence data. FastQC (version 0.11.5) was then used to check the whole sequence quality and the GC content. Bismark (version 0.18.1) was used to align the trimmed reads to the human genome (version hg19) with the recommended parameters. A customized C++ program was used to summarize the methylation ratio (MR) and methylation density (MD). A sliding window of 200 kB was used to calculate methylation index across the genome. Methylation indices for specified blocks of all the samples were determined using the methods described above. They were then combined into a matrix. Regions with large amounts of missing data and 0 values were removed from the matrix. All the pathological parameters of 16 patients were converted to the standard numerical format. The linear mixed model of the R software package (version 3.3.1) was used to associate methylation indices with methylation data. Customized Perl scripts were used to draw association maps and related heatmaps. The hclust package of the R software (version 3.3.1) was used to cluster the samples. The default complete linkage method was used in this clustering process. The amounts of sequencing data and the percentages of uniquely mapped reads were both similar in different samples. The quality of all sequencing data was Q20 > 95%, and Q30 ≈ 90%, indicating that the data were of sufficiently high quality to provide stable results for the intended analysis.
Statistical Analysis
Associations between different pathological features were examined by contingency tables. Correlations between the methylation status (MD or MR) and patients’ clinical parameters was evaluated by Spearman rank correlation coefficient. Differences in MD and MR between the EXEr and EXEs groups were evaluated using Student t test or the Mann-Whitney U test. Kaplan-Meier survival curves were calculated according to baseline methylation status of differentially methylated regions (DMRs) or changes in methylation status during EXE treatment. The PFS of each group was compared using log-rank tests. For all analyses, P <.05 was considered significant. Area under the curve (AUC) was calculated by receiver operating characteristic (ROC) curve analysis. Cutoff values for relevant parameters were based on the Youden index, calculated as (sensitivity + specificity) − 1. Data are expressed as mean ± SD. All analyses were performed on SPSS version 22.0. All statistical tests were 2 sided.
Results
Patient Characteristics
We carried out ctDNA methylation analyses on blood samples from 16 patients who received EXE-based therapy until their relapses. Of these patients, 15 (93.8%) had more than 2 blood samples collected at baseline, first follow-up, and/or PD, respectively, and 1 (6.2%) patient had only 1 sample at baseline. Their median age at diagnosis was 53.0 years (range: 31.0-70.0 years). Their median follow-up period was 19.0 months (range: 2.0-35.6 months). Four (25.0%) patients were confirmed to have EXEr disease. Their clinicopathologic characteristics are shown in Table 1.
Table 1.
Clinicopathological Characteristics of 16 Patients.
| Characteristic | n (%) |
|---|---|
| Age at diagnosis (years) | |
| >45 | 11 (68.8) |
| ≤45 | 5 (31.2) |
| Unknown | 0 |
| Whether received adjuvant HT | |
| Yes | 11 (68.8) |
| No | 5 (31.2) |
| Unknown | 0 |
| Therapeutic status of adjuvant HT | |
| Relapse within 2 years while on adjuvant HT | 0 (0.0) |
| Relapse after 2 years while on adjuvant HT | 4 (36.4) |
| Relapse within 1 year of completing adjuvant HT | 0 (0.0) |
| Relapse after 1 year of completing adjuvant HT | 7 (63.6) |
| Not applicable | 5 |
| Regimens prior to exemestane in first line | |
| Antharcycline and/or Taxanes | 9 (56.25) |
| Trastuzumab and/or pertuzumab | 2 (12.5) |
| None | 7 (43.75) |
| Unknown | 0 |
| Histology of primary tumor | |
| Ductal | 15 (93.8) |
| Lobular | 1 (6.2) |
| Others | 0 (0.0) |
| Unknown | 0 |
| Initial HER2 status | |
| Positive | 3 (20.0) |
| Negative | 12 (80.0) |
| Unknown | 1 |
| Initial TNM stage | |
| I | 1 (6.2) |
| II | 8 (50.0) |
| III | 2 (12.5) |
| IV | 5 (31.3) |
| Unknown | 0 |
| Sites of metastases at relapse | |
| Liver | 3 (18.8) |
| Lung | 8 (50.0) |
| Brain | 1 (6.3) |
| Bone | 5 (31.3) |
| Lymph node | 7 (43.8) |
| Chest wall | 1 (6.3) |
| Unknown | 0 |
Abbreviation: HT, hormone therapy; TNM, Tumor Node Metastasis.
Methylomes Variation During the Course of EXE Treatment
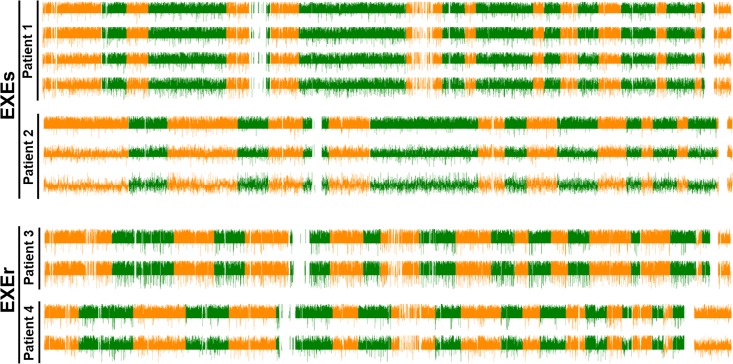
We first focused on DNA methylome variations (among all 23 chromosomes) during EXE treatment. Whole-genomic DNA methylation levels at baseline, first follow-up (if any), and progression were measured for each patient. We found ongoing genome-wide demethylation to occur during EXE treatment in all 16 patients, including large-scale genome-wide demethylation in both the EXEs group and the EXEr group. Therefore, overall DNA demethylation seems to be associated with EXE treatment rather than EXE response (Figure 1).
Figure 1.
Methylome variations during exemestane (EXE) treatment. Four of the 16 patients were exemestane resistant (EXEr), and 12 were exemestane sensitive (EXEs). Differential variations in methylation regions between samples taken at different time points (baseline [upper], first follow-up [middle], and diagnosis of disease progression [lower]) are shown. Each value is calculated using the formula: 2 × ([methylation rate] − methylome variations during EXE treatment. Four of the 16 patients were EXEr and depicted in different colors (orange or green).
Selection of Primary EXEr-Related DMRS Using Bismark Analysis
As large-scale variations in patients’ methylomes occurred during the course of EXE treatment, we next sought to identify variations related to EXEr tumors. Using Bismark analysis software, we evaluated correlations between methylation status and EXEr. Differential MD and MR regions for which P < .05 are summarized in Supplementary Tables 1 and 2, respectively. We found 79 differential MD regions (covering 175 genes) and 70 differential MR regions (covering 223 genes) that were correlated with EXEr. The respective differential MD and MR regions both included 7 DMRs in common. These 7 DMRs covered genes such as the human leukocyte antigens (HLA), which affect antitumor immune response, and tripartite motif containing 42 (TRIM42), which affects apoptosis and cell-cycle regulation (Table 2).
Table 2.
Summary of the EXE Resistance-Related DMRs in Terms of Methylation Density and Methylation Ratio.
| Genomic Location of DMRs | Methylation Density (P Value) | Methylation Ratio (P Value) | Overlap Genes |
|---|---|---|---|
| Chr1[149600000-149799999] | .026 | .050 | LINC00623; LINC00869; LOC103091866 |
| Chr3 [67800000-67999999] | .026 | .049 | SUCLG2-AS1 |
| Chr3 [140200000-140399999] | .029 | .038 | CLSTN2; CLSTN2-AS1; TRIM42 |
| Chr6 [32400000-32599999] | .005 | .013 | HLA-DRA; HLA-DRB5; HLA-DRB6; HLA-DRB1 |
| Chr9 [46800000-46999999] | .019 | .002 | FAM74A1 |
| Chr10 [46400000-46599999] | .019 | .034 | PTPN20 |
| Chr12 [101200000-101399999] | .016 | .041 | ANO4 |
Abbreviations: DMRs, differentially methylated regions; EXE, exemestane.
Baseline Methylation Status of the 7 Selected DMRs in the EXEr and EXEs Groups
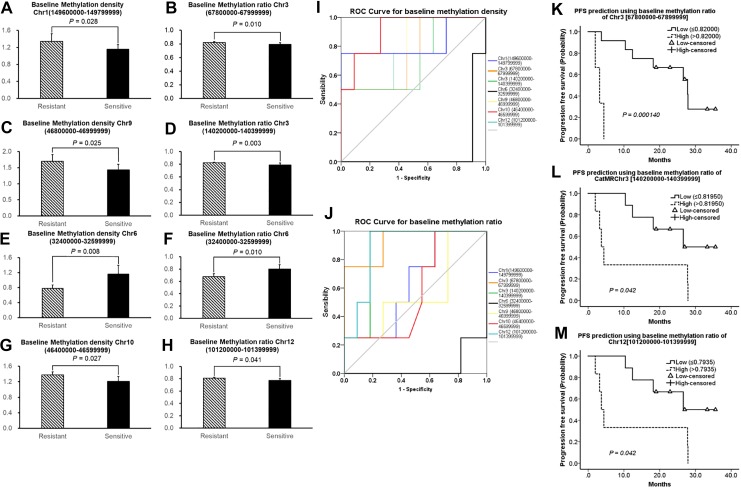
To validate the results of methylation profile analysis, we compared the methylation status of the 7 selected DMRs in ctDNA between baseline blood samples of EXEr and EXEs patients. In these DMRs, baseline MDs in Chr1 [149600000-149799999] (P = .028), Chr9 [46800000-46999999] (P = .025), and Chr10 [46400000-46599999] (P = .027) were higher in the EXEr samples than in EXEs samples (Figure 2A, C, and G); and baseline MRs in Chr3 [67800000-67999999] (P = .01), Chr3 [140200000-140399999] (P = .003), and Chr12 [101200000-101399999] were higher in EXEr samples than in EXEs samples (Figure 2B, D, and H). Most of these MD and MR regions did not overlap. However, the MD (P = .008) and MR (P = .010) of Chr6 [32400000-32599999] were lower in EXEr samples than in EXEs samples (Figure 2E and F). Receiver operating characteristic analysis showed that the baseline MD of Chr10 [46400000-46599999] (AUC = 0.909, P = .019, 95% confidence interval [CI], 0.752-1.000) and baseline MRs of Chr3 [67800000-67999999] (AUC = 0.932, P = .013, 95% CI, 0.787-1.000), Chr3 [140200000-140399999] (AUC = 0.864, P = .037, 95% CI, 0.670-1.000), and Chr12 [101200000-101399999] (AUC = 0.886, P = .026, 95% CI, 0.714-1.000) were predictive for EXEr (Figure 2I and J). Using ROC curves, we calculated the Youden index of these parameters and set corresponding threshold values. We then stratified patients by high and low MD or MR values and found that baseline MRs of Chr3 [67800000-67999999] (P = .00014), Chr3 [140200000-140399999] (P = .042), and Chr12 [101200000-101399999] (P = .042) constituted a prognostic marker for PFS after EXE treatment (Figure 2K-M). However, we did not find a similar result for baseline MD of Chr10 [46400000-46599999] (P = .189, data not shown).
Figure 2.
Comparison of baseline methylation status of 7 selected DMRs between EXEr and EXEs patients. Comparison of baseline MD of certain DMRs between EXEr and EXEs group (A, C, E, G); comparison of baseline MR of certain DMRs between EXEr and EXEs groups (B, D, F, H). Receiver operating characteristic curve analysis for 7 selected DMRs in terms of MD (I) and MR (J). Kaplan-Meier survival analysis for baseline MR of Chr3 [67800000-67999999] (K), Chr3 [140200000-140399999] (L), and Chr12 [101200000-101399999] (M). “Resistant” indicates EXEr patients, “sensitive” indicates EXEs patients. Chr indicates chromosome; DMRs, differential methylation regions; EXEr, exemestane resistance; EXEs, exemestane sensitive; MD, methylation density; MR, methylation ratio.
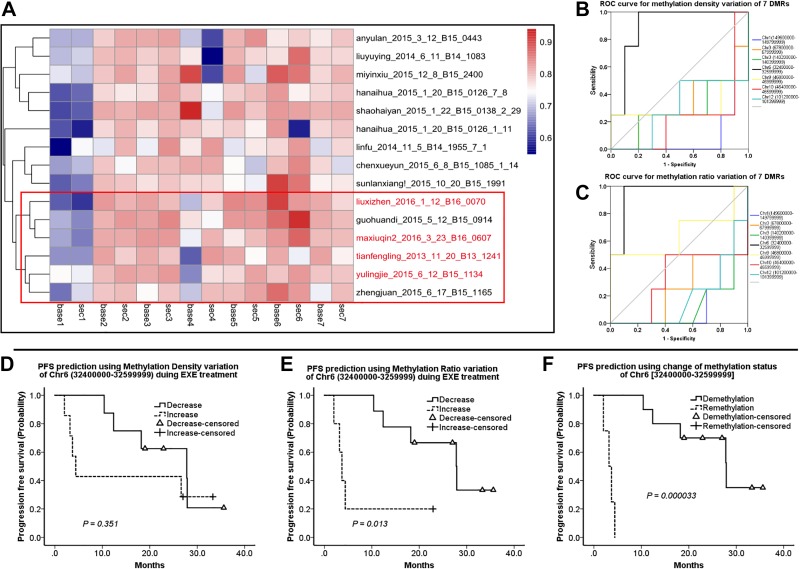
Methylation Changes in 7 Selected DMRs During Therapeutic EXE Courses
As baseline MR was more relevant to primary EXEr, we investigated whether methylation changes during EXE treatment were also predictive for acquired EXEr. We clustered the 15 patients by their MR variations for 7 DMRs. As shown in the heatmap, the EXEr patients were largely clustered together and shared a similar MR variation pattern in the 7 DMRs (Figure 3A). Interestingly, only changes of MD (AUC = 0.900, P = .024, 95% CI, 0.732-1.000) and MR (AUC = 0.950, P = .011, 95% CI, 0.835-1.000) in Chr6 [32400000-32599999] were predictive for acquired EXEr; the other 6 DMRs did not have similar prognostic power (Figure 3B and C). Using the cutoff value calculated by Youden index, we classified the 15 patients into those with greater or less methylation of the Chr6 DMR [149600000-149799999]. Kaplan-Meier survival analysis showed that greater MR of Chr6 [149600000-149799999] was significantly associated with shorter PFS (7.24 ± 3.519 months vs 25.71 ± 3.182 months, P = .013, Figure 3E). However, similar predictive power was not found for MD variations in Chr6 [149600000-149799999] (13.32 ± 5.776 months vs 24.15 ± 3.238 months, P = .351, Figure 3D). As ROC curve analysis suggested that both MD and MR variations in Chr6 [149600000-149799999] could identify EXEr, we checked if combining them could enhance the prediction power for prognosis. As expected, the synchronized increase in MD and MR in Chr6 [149600000-149799999] was a strong predictor for shorter PFS (3.33 ± 0.506 vs 26.31 ± 3.003, P = .000033, Figure 2F). The threshold values of MD and MR for Chr6 [149600000-149799999] were 0.066 and 0.076, respectively.
Figure 3.
Methylation status changes in EXE-resistant (EXEr) and EXE-sensitive (EXEs) groups during EXE treatment. (A), Red: Hierarchical clustering analysis of methylation ratio (MR) variation in all 15 patients and EXEr patients. (B) and (C), Receiver operating characteristic curve analysis shows changes in methylation density (MD) and MR for 7 selected differentially methylated regions. (D-F) Kaplan-Meier survival analyses for MD variation (D) and MR variation (E) and synchronized MD and MR variation (F) of Chr6 [149600000-149799999]. Demethylation: synchronized decrease in MD and MR, Remethylation: synchronized increase in MD and MR. Chr indicates chromosome.
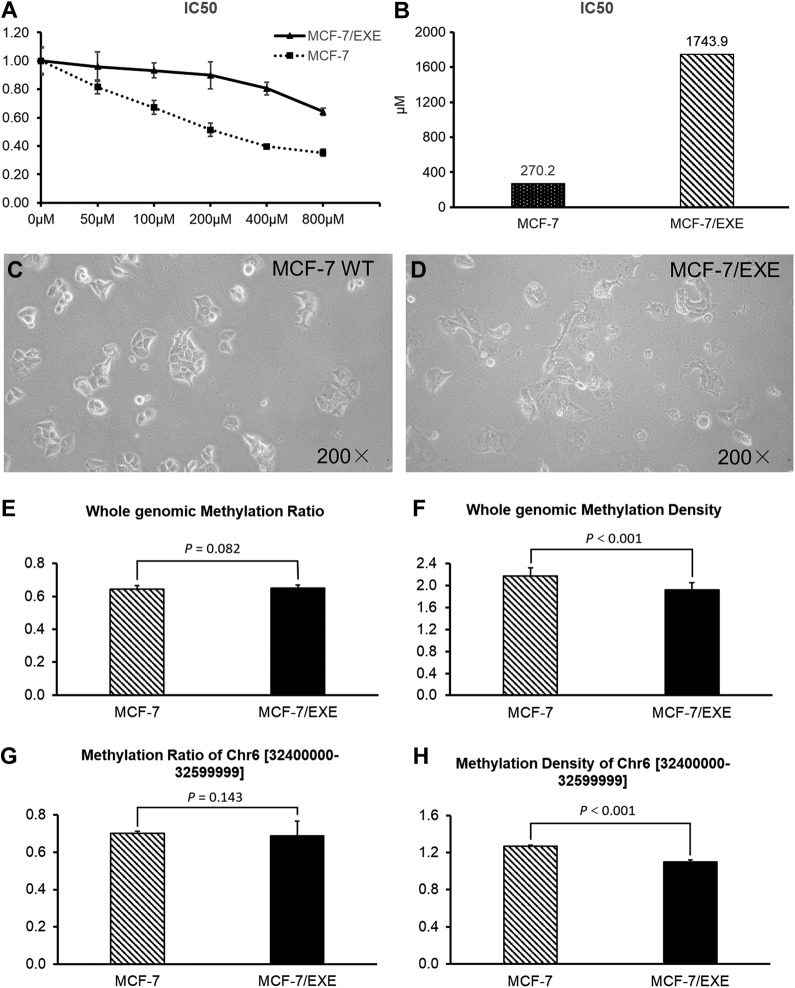
Methylation Changes in EXEr MCF-7
To validate the 7 EXEr-related MDRs found in blood samples, we developed an EXEr MCF-7 cell line (MCF-7/EXE) by subjecting wild-type MCF-7 to increasing concentrations of EXE. As the cell viability assay showed, the IC50s of wild-type MCF-7 and MCF-7/EXE were 270.2 and 1743.9 μM, respectively (Figure 4A and B). As the IC50 of the EXE-treated MCF-7 was about 5 times that of wild-type MCF-7, it was considered MCF-7/EXE. Morphologically, the MCF-7/EXE cells were slenderer and had more pseudopodia than did wild-type MCF-7 (Figure 4C and D). We next explored differences in methylomes between wild-type MCF-7 and MCF-7/EXE cells. We found that although genomic MRs of MCF-7 and MCF-7/EXE were similar (P = .082, Figure 4E), MCF-7/EXE showed significantly lower genomic MD (P < .001, Figure 4F), notably in Chr6 [32400000-32599999] (P < .001, Figure 4H), compared to MCF-7. This result was consistent with our clinical evidence: Demethylation of Chr6 [32400000-32599999] may be a predictor of EXEr in ER+ BC.
Figure 4.
Genomic methylation status in MCF-7 human breast cancer cells and in exemestane-resistant MCF-7 (MCF-7/EXE) cells. (A) Viability assay of MCF-7 and MCF-7/EXE cells subjected to increasing concentrations of exemestane. (B) IC50 of MCF-7 and MCF-7/EXE were calculated using cell viability curve. Morphologies of (C) MCF-7 and (D) MCF-7/EXE (magnification: ×200). Comparisons between MCF-7 and MCF-7/EXE for (E) average genomic methylation ratio (MR) and (F) average genomic methylation density (MD). Comparisons between MCF-7 and MCF-7/EXE for (G) MR and (H) MD of chromosome 6 [32400000-32599999].
Discussion
This is the first study to interrogate whole-methylome variations in advanced BC to predict therapeutic response. Abnormal DNA methylation is common in many cancer types and has emerged as potential prognostic marker in advanced BC.17,18,23,24 However, little is known about the mechanism of EXEr other than the mTOR pathway25,26 and ESR1 mutation.27 For example, mTOR pathway has been shown to be involved in autophagy-mediated EXE resistance.28 Several studies suggested that global methylation changes in peripheral blood DNA of patients with BC is associated with cancer development, progression, and metastasis.29-31 A prospective biomarker study (TBCRC005) showed that high cumulative methylation index is a predictor of worse PFS and OS for patients with systemically treated BC.32 He et al 11 also found that BC cell lines that developed chemoresistance in vitro all showed massive changes in methylation patterns compared with chemosensitive controls. Epigenetic dysregulation of the ER signaling pathway is crucial for tumor response to HT.33,34 For instance, ectopic methylation of ER co-regulators (such as SRC1, SRC3, TIF2, CBP, and NCOR1) is reportedly involved in tamoxifen resistance.35,36
As DNA bisulfite treatment followed by next-generation sequencing technology allowed us to analyze ctDNA methylation patterns at the genome-wide scale, we aim to find the crucial DNA methylation factors that are responsible for EXEr. Our data suggest that massive demethylation after EXE treatment is inevitable in both EXEr and EXEs patients and might contribute to the development of primary and/or acquired EXEr. At baseline, the MD or MR of most DMRs were lower in EXEs samples than in EXEr samples. These DMRs contained coding genes that are involved in many cellular processes such as actin polymerization (protein tyrosine phosphatase, nonreceptor type 20), Ca2+-activated Cl− ion channel control (anoctamin-4), apoptosis, or cell-cycle regulation (TRIM42). We also found the baseline MRs—but not baseline MDs—of Chr3 [67800000-67999999], Chr3 [140200000-140399999], and Chr12 [101200000-101399999] could predict PFS after EXE treatment. Although MR and MD are both important measures of DNA methylation status, either factor alone does not wholly represent DNA methylation status. Thus, methylation of Chr3 [67800000-67999999], Chr3 [140200000-140399999], and Chr12 [101200000-101399999] cannot be feasibly associated with PFS. In contrast, Chr6 [149600000-149799999] is the only region that showed lower MD and MR in both EXEr samples (Figure 2E and F). Moreover, in vitro data also showed lower MD for Chr6 [149600000-149799999] in the MCF-7/EXE cell line (Figure 4H). We, therefore, believe that low methylation in Chr6 [149600000-149799999] is a particular feature of EXEr. Interestingly, upregulated MD and MR of Chr6 [149600000-149799999] during EXE treatment were also predictive for EXEr (Figure 3B and C). When considered simultaneously, MD and MR become an ideal prognostic factor for PFS after EXE treatment (Figure 3F). The overlapped genes in Chr6 [149600000-149799999] all belonged to the HLA class II α chain paralogue family (HLA-DR); they therefore play a central role in the immune system37 and are reportedly associated with ER+ BC risk38 and survival.39 In 2 independent cohorts of patients with anti-PD-1-treated melanoma, MHC-II positivity in tumor cells was associated with therapeutic response and PFS and OS.40 Positive MHC-II expression, including HLA-DR in tumor cells, was associated with better disease-free survival in patients who had triple-negative BC with lymph node metastasis.41 Meanwhile, estradiol (E2) and ERα signaling pathway was found to help regulate interferon-γ-inducible HLA-II in BC cells. These findings indicate a prognostically important interaction between the ER pathway and HLA-II expression.42 Our data indicate an association between EXEr and HLA-II methylation. We believe that remethylation of Chr6 [149600000-149799999] during EXE therapy could be a very useful predictor for EXEr.
Our study found, for the first time, that methylation changes during EXE treatment correlate with EXEr status. The methylation status of certain DNA region is an ideal predictor for EXEr. However, this study was limited by its small cohort. Our findings and methods should be validated in a larger patient group.
Supplemental Material
Supplemental Material, Copy_of_Suplementary_Table_4 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Copy_of_Supplementary_Table_3 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Figure_1 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Table_1_EXEr_related_differential_methylation_density_regions for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Table_2_EXEr_related_differential_methylation_ratio_regions for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Acknowledgments
The authors thank all the patients for their volunteered participation in the present study. The authors also thank Marla Brunker, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Abbreviations
- AUC
area under the curve; BC, breast cancer
- cfDNA
cell-free DNA; CI, confidence interval
- CMI
cumulative methylation index
- CpG
5′-C-phosphate-G-3′
- ctDNA
circulating tumor DNA
- Chr
chromosome
- DMRs
differentially methylated regions
- DNMTs
DNA methyltransferases
- ER
estrogen receptor
- EXE
exemestane
- EXEr
exemestane resistance
- EXEs
exemestane sensitive
- HLA
human leukocyte antigen
- HR
hormone receptor
- HT
hormone therapy
- MD
methylation density
- MR
methylation ratio
- OS
overall survival
- PD
disease progression
- PFS
progression-free survival
- ROC
receiver operating characteristic
- TRIM42
tripartite motif containing 42
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (Grant Nos. 81502269 and 21273051).
ORCID iD: Xiao-ran Liu  https://orcid.org/0000-0003-0912-3210
https://orcid.org/0000-0003-0912-3210
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. [DOI] [PubMed] [Google Scholar]
- 2. Rosenbaum JN, Weisman P. The evolving role of companion diagnostics for breast cancer in an era of next-generation omics. Am J Pathol. 2017;187(10):2185–2198. [DOI] [PubMed] [Google Scholar]
- 3. Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163(2):506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fang F, Turcan S, Rimner A, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3(75):75ra25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davalos V, Martinez-Cardus A, Esteller M. The epigenomic revolution in breast cancer: from single-gene to genome-wide next-generation approaches. Am J Pathol. 2017;187(10):2163–2174. [DOI] [PubMed] [Google Scholar]
- 8. Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Hao DP, Li JJ, Wang L, Di LJ. Genome-wide methylome and chromatin interactome identify abnormal enhancer to be risk factor of breast cancer. Oncotarget. 2017;8(27):44705–44719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abdel-Hafiz HA. Epigenetic mechanisms of tamoxifen resistance in luminal breast cancer. Diseases. 2017;5(3):E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He DX, Gu F, Gao F, Hao JJ, Gong D, Gu XT, et al. Genome-wide profiles of methylation, microRNAs, and gene expression in chemoresistant breast cancer. Sci Rep. 2016;6:24706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fukagawa A, Ishii H, Miyazawa K, Saitoh M. Deltaef1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 2015;4(1):125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Early Breast Cancer Trialists’ Collaborative Group. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365(9472):1687–1717. [DOI] [PubMed] [Google Scholar]
- 14. Dalmau E, Armengol-Alonso A, Munoz M, Segui-Palmer MA. Current status of hormone therapy in patients with hormone receptor positive (HR+) advanced breast cancer. Breast 2014;23(6):710–720. [DOI] [PubMed] [Google Scholar]
- 15. Li HP, Ji JF, Hou KY, Jia TZ, et al. Clinical study of aromatase inhibitors in advanced breast cancer. Beijing Da Xue Xue Bao Yi Xue Ban. 2007;39(2):193–196. [PubMed] [Google Scholar]
- 16. Widschwendter M, Siegmund KD, Muller HM, et al. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res. 2004;64(11):3807–3813. [DOI] [PubMed] [Google Scholar]
- 17. Martinez-Galan J, Torres-Torres B, Nunez MI, et al. ESR1 gene promoter region methylation in free circulating DNA and its correlation with estrogen receptor protein expression in tumor tissue in breast cancer patients. BMC Cancer. 2014;14:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kajabova V, Smolkova B, Zmetakova I, et al. RASSF1A promoter methylation levels positively correlate with estrogen receptor expression in breast cancer patients. Transl Oncol. 2013;6(3):297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang MJ, Professor Hope S. Rugo: reversing resistance and hormone therapy for metastatic breast cancer. Chin Clin Oncol. 2014;3(4):51. [DOI] [PubMed] [Google Scholar]
- 20. Yue W, Fan P, Wang J, Li Y, Santen RJ. Mechanisms of acquired resistance to endocrine therapy in hormone-dependent breast cancer cells. J Steroid Biochem Mol Biol. 2007;106(1-5):102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Allred DC, Harvey JM, Berardo M, Clark GM. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod Pathol. 1998;11(2):155–168. [PubMed] [Google Scholar]
- 22. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buchegger K, Riquelme I, Viscarra T, et al. Reprimo, a potential p53-dependent tumor suppressor gene, is frequently hypermethylated in estrogen receptor alpha-positive breast cancer. Int J Mol Sci. 2017;18(8):E1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Harbeck N, Nimmrich I, Hartmann A, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26(31):5036–5042. [DOI] [PubMed] [Google Scholar]
- 25. Chumsri S, Sabnis G, Tkaczuk K, Brodie A. MTOR inhibitors: changing landscape of endocrine-resistant breast cancer. Future Oncol. 2014;10(3):443–456. [DOI] [PubMed] [Google Scholar]
- 26. Chen S, Masri S, Wang X, Phung S, Yuan YC, Wu X. What do we know about the mechanisms of aromatase inhibitor resistance? J Steroid Biochem Mol Biol. 2006;102(1-5):232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fribbens C, O’Leary B, Kilburn L, et al. Plasma ESR1 mutations and the treatment of estrogen receptor-positive advanced breast cancer. J Clin Oncol. 2016;34(25):2961–2968. [DOI] [PubMed] [Google Scholar]
- 28. Amaral C, Augusto TV, Tavares-da-Silva E, Roleira FMF, Correia-da-Silva G, Teixeira N. Hormone-dependent breast cancer: targeting autophagy and PI3K overcomes exemestane-acquired resistance. J Steroid Biochem Mol Biol. 2018;183:51–61. [DOI] [PubMed] [Google Scholar]
- 29. Khan SI, Aumsuwan P, Khan IA, Walker LA, Dasmahapatra AK. Epigenetic events associated with breast cancer and their prevention by dietary components targeting the epigenome. Chem Res Toxicol. 2012;25(1):61–73. [DOI] [PubMed] [Google Scholar]
- 30. Zhao Z, Wang L, Di LJ. Compartmentation of metabolites in regulating epigenome of cancer. Mol Med. 2016;22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuchiba A, Iwasaki M, Ono H, et al. Global methylation levels in peripheral blood leukocyte DNA by LUMA and breast cancer: a case–control study in Japanese women. Br J Cancer. 2014;110(11):2765–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Visvanathan K, Fackler MS, Zhang Z, et al. Monitoring of serum DNA methylation as an early independent marker of response and survival in metastatic breast cancer: TBCRC 005 prospective biomarker study. J Clin Oncol. 2017;35(7):751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mann M, Cortez V, Vadlamudi RK. Epigenetics of estrogen receptor signaling: role in hormonal cancer progression and therapy. Cancers (Basel). 2011;3(3):1691–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hervouet E, Cartron PF, Jouvenot M, Delage-Mourroux R. Epigenetic regulation of estrogen signaling in breast cancer. Epigenetics. 2013;8(3):237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fleming FJ, Myers E, Kelly G, et al. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004;57(10):1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Girault I, Bieche I, Lidereau R. Role of estrogen receptor alpha transcriptional coregulators in tamoxifen resistance in breast cancer. Maturitas. 2006;54(4):342–351. [DOI] [PubMed] [Google Scholar]
- 37. Yang XX, Pan HZ, Li PY, et al. HLA class II variants in Chinese breast cancer patients. Asian Pac J Cancer Prev. 2011;12(11):3075–3079. [PubMed] [Google Scholar]
- 38. Quan L, Gong Z, Yao S, et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int J Can. 2014;134(6):1408–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Calabro A, Beissbarth T, Kuner R, et al. Effects of infiltrating lymphocytes and estrogen receptor on gene expression and prognosis in breast cancer. Breast Cancer Res Treat. 2009;116(1):69–77. [DOI] [PubMed] [Google Scholar]
- 40. Johnson DB, Estrada MV, Salgado R, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park IA, Hwang SH, Song IH, et al. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS One. 2017;12(8):e0182786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mostafa AA, Codner D, Hirasawa K, et al. Activation of ERalpha signaling differentially modulates IFN-gamma induced HLA-class II expression in breast cancer cells. PLoS One. 2014;9(1):e87377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Copy_of_Suplementary_Table_4 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Copy_of_Supplementary_Table_3 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Figure_1 for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Table_1_EXEr_related_differential_methylation_density_regions for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment
Supplemental Material, Supplementary_Table_2_EXEr_related_differential_methylation_ratio_regions for Methylome Variation Predicts Exemestane Resistance in Advanced ER+ Breast Cancer by Xiao-ran Liu, Ru-yan Zhang, Hao Gong, Hope S. Rugo, Ling-bo Chen, Yuan Fu, Jian-wei Che, Jian Tie, Bin Shao, Feng-ling Wan, Wei-yao Kong, Guo-hong Song, Han-fang Jiang, Guo-bing Xu and Hui-ping Li in Technology in Cancer Research & Treatment