Abstract
Immunotherapy is a rapidly growing field for cancer treatment. In contrast to conventional cancer therapies, immunotherapeutic strategies focus on reactivating the immune system to mount an antitumor response. Despite the encouraging outcome in clinical trials, a large proportion of patients still do not respond to treatment and many experience different degrees of immune-related adverse events. Furthermore, it is now increasingly appreciated that even many conventional cancer therapies such as radiotherapy could have a positive impact on the host immune system for better clinical response. Hence, there is a need to better understand tumor immunity in order to design immunotherapeutic strategies, especially evidence-based combination therapies, for improved clinical outcomes. With this aim, cancer research turned its attention to profiling the immune contexture of either the tumor microenvironment (TME) or peripheral blood to uncover mechanisms and biomarkers which might aid in precision immunotherapeutics. Conventional technologies used for this purpose were limited by the depth and dimensionality of the data. Advances in newer techniques have, however, greatly improved the breadth and depth, as well as the quantity and quality of data that can be obtained. The result of these advances is a wealth of new information and insights on how the TME could be affected by various immune cell-types, and how this might in turn impact the clinical outcome of cancer patients . We highlight herein some of the high-dimensional technologies currently employed in immune profiling in cancer and summarize the insights and potential benefits they could bring in designing better cancer immunotherapies.
Keywords: immunology, tumors, functional imaging, genetics, oncology
Introduction
The tumor microenvironment (TME) is complex, comprising of tumor cells, infiltrating immune cells, fibroblasts, tumor vasculature and extracellular matrix.1 These various compartments are now understood to play an interactive and collective role in tumorigenesis, cancer progression, metastasis and even response to therapy.2 The immune contexture of the cancer, defined as the location, density and functional orientation of the different immune cell populations,3 has been shown to determine the disease prognosis and efficacy of various cancer therapies, especially the rapidly progressing cancer immunotherapy.2 4 5 Unlike conventional cancer therapies, immunotherapy targets the immune system or the TME of the patient to induce an antitumor immune response.1 Immunotherapeutic strategies include cytokine administration, cancer vaccines, adoptive T cell therapy and immune checkpoint blockades (ICB).6 While results from clinical trials using immunotherapy, particularly the ICBs, have been promising, a large proportion of patients still do not respond to treatment.6 Many patients also experience treatment-related autoimmune toxicities or immune-related adverse events (irAEs).6 Therefore, in order to understand the mechanisms behind response and irAEs (or the lack of them) following immunotherapy, attempts have been made to profile the immune contexture of the TME or peripheral blood, with the aim to discover novel strategies and biomarkers that might guide treatment decisions for an enhanced clinical outcome.7
The various technologies used for immune profiling can be divided into two broad categories: transcriptomic-based and proteomic-based technologies.8 Many limitations posed by conventional technologies, such as fewer analysis parameters available,9 10 larger quantities of sample required11 and overlapping detection signals,9 10 were significantly improved by the availability of newer and higher-dimensional technologies. These current cutting-edge technologies provide researchers with unprecedented capability to conduct deep immunophenotyping of the TME or peripheral blood of patients with cancer. This review highlights the latest high-dimensional technologies used for immunoprofiling in cancer and examines the insights and potential benefits they could bring, especially to advance the progress of cancer immunotherapy (figure 1).
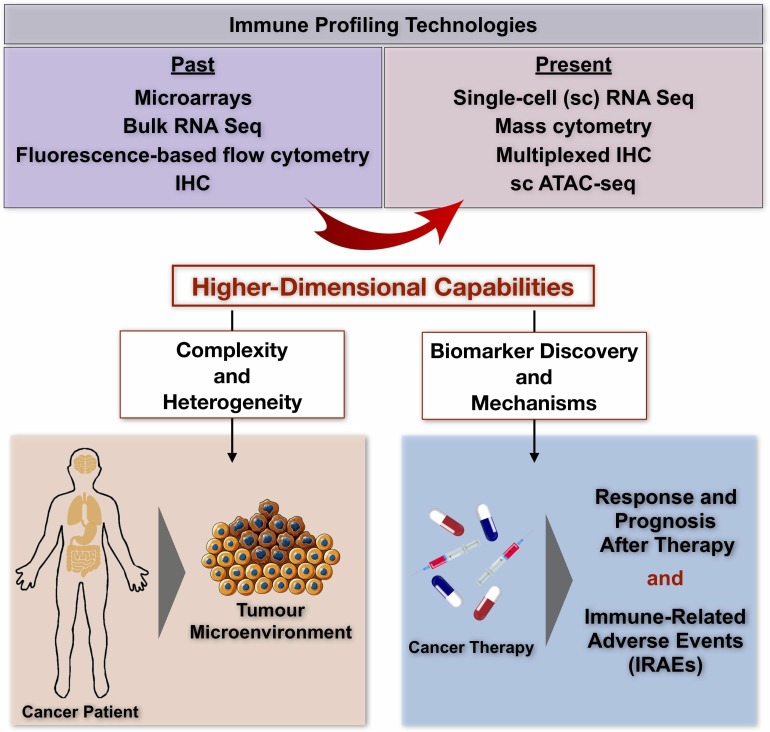
Figure 1.
Recent development of high-dimensional immune profiling technologies. Transition of immunoprofiling technologies from conventional tools in the past, to cutting-edge, high-dimensional technologies in the present which have overcome previous limitations and are equipped with enhanced capabilities for a deeper understanding of the complex and heterogeneous tumor microenvironment as well as for the identification of potential biomarkers for immunotherapy. ATAC, assay for transposase-accessible chromatin; Seq, sequencing; sc, single-cell.
Conventional immune profiling tools in cancer
The earlier generation of immune profiling technologies have provided valuable insights into the mechanisms behind tumor-immune cell interactions and facilitated breakthroughs in biomarker discovery for disease prognosis as well as prediction of therapeutic response. These technologies include bulk transcriptome assays like microarrays8 11 12 and bulk RNA Sequencing (RNA-Seq),8 13–15 as well as lower-dimensional protein-based platforms such as early-generation fluorescence-based flow cytometry8 16–18 and conventional immunohistochemistry (IHC).19 20
Nonetheless, these technologies have inherent limitations which restrict a deeper understanding of the tumor-immune landscapes. For instance, conventional bulk transcriptomic analyses such as microarrays and RNA-seq rely on the extraction of RNA from a pooled population of cells from tumor tissue.21 Consequently, this bulk transcriptomic approach precludes the identification of rare cell types and the resolution between cells with similar expression patterns or between tumor versus immune cells.22 It also limits the understanding of the function and/or phenotype of individual cell types.8
For proteomic-based analyses, advances in technology and the development of new fluorochromes and laser detectors have enabled the most advanced flow cytometers to routinely measure up to 18 different protein markers simultaneously.9 Conventional fluorescence-based flow cytometry is, however, limited by the overlapping emission spectra of the fluorochromes, a problem which is exacerbated by the use of more fluorochromes.23 This renders a detailed examination of cell phenotypes difficult on this platform as more parameters are required to distinguish the complex immune subsets in biological systems like tumors.23
Finally, conventional IHC or immunofluorescent microscopy enables the detection of cellular antigens within tissue sections using enzyme-labeled or fluorochrome-labeled antibodies and is useful for identifying cell types and their spatial location within tissues.24 However, no more than four markers can be used simultaneously due to chromogenic or fluorescent spectra overlap.10 19 As such, conventional IHC techniques would also fall short in capturing the degree of heterogeneity and complexity of immune phenotypes that exist in tumor-infiltrating immune cells.8
Thus, while the above-mentioned conventional technologies provide a good global picture of the immune landscape of tumor, they may not be effective in capturing the complexity and depth of immune phenotypes and potential functional cues in a dynamic system within TME.25
High-dimensional technologies for next-generation immune profiling in cancer
In the attempt to address the need to capture cellular heterogeneity and detect rare immune subsets, technologies have advanced toward single-cell capabilities and high-dimensional analyses with new technologies such as single-cell RNA-Seq (scRNA-Seq), cytometry by time of flight (CyTOF) and multiplex IHC (mIHC). These technologies have overcome many limitations posed by conventional immune profiling technologies and provided breakthroughs in biomarker discovery and the understanding of immune mechanisms, particularly in the field of tumor immunology.
The development of scRNA-Seq addressed the limitations of bulk transcriptomic data, rendering them more useful for dissecting phenotypic and functional heterogeneity among single cells.21 It built on the concept of RNA-Seq (which has been well-described previously26–28), with the additional steps of isolating single cells, followed by a powerful amplification process to generate cDNA from minute cellular RNA quantities.21 This enables the analysis of the transcriptome at single-cell level, while preserving the accuracy and complexity of bulk analysis methods.29 Recent advances complemented with tissue imaging capabilities provided additional and valuable spatial information of these scRNA-seq data.30 31
Beyond scRNA-seq, the single-cell assay for transposase-accessible chromatin sequencing (scATAC-Seq) could also be applied to study epigenetic alterations.32 scATAC-Seq identifies active DNA regulatory elements and allows rapid and sensitive profiling of genome-wide chromatin accessibility, at a single cell level.32
CyTOF, on the other hand, addressed the limited dimensionality in conventional fluorescence-based flow cytometry23 by combining mass spectrometry and flow cytometry, using antibodies conjugated with stable isotopes of rare earth metals (lanthanide metals) instead of fluorophores.9 Samples are nebulized into single-cell droplets, vaporized by the high temperature plasma to generate an ion cloud with heavy metal probe ions which are then quantified using mass-to-charge ratio.23 33 Little background signal is produced because these metal reporters are uncommon in biological specimens.9 Finally, the discrete readout of an isotope presents little or no overlapping spectra. More than 40 markers can be detected on a single cell without the need for combining smaller, separate panels of antibodies, thus greatly enhancing the dimensionality of cytometric analysis.23
Finally, advancements in dye-cycling techniques, where staining, imaging and dye inactivation are done repeatedly, have enabled the detection of up to 61 different antigens on the same tissue sample using fluorescence microscopy.34 At the same time, new technologies incorporating mass spectrometry have been explored for mIHC, enabling the imaging of 32 proteins simultaneously using laser ablation of tissue coupled with CyTOF,35 and up to 100 proteins simultaneously using multiplexed ion beam imaging (MIBI).36 Mass cytometry-based mIHC techniques are promising technologies that are still under development. Furthermore, advances in fluorescence imaging hardware and software as well as fluorescent dyes have enabled routine simultaneous imaging of up to seven fluorescent markers on a single section.24 This breakthrough overcame the restriction of two to four markers by conventional IHC or immunofluorescent confocal systems.19
The data generated by these new technologies enable researchers to conduct deep immunophenotyping at a single-cell level. More importantly, combinations of these advanced multidimensional technologies are being used increasingly in the study of the complex immune response, yielding insights which would have been challenging to obtain using the older generation of technologies. Some of the highlights of these studies will be described in the next sections.
Multidimensional immune profiling reveals phenotypic and functional complexity of immune subsets in the TME
With the ability to conduct deep profiling of immune cells at a single-cell level, researchers have now begun to better appreciate the complexity of the immune response, the interactions between immune subsets and the interaction between host and stroma cells in the TME. These studies, as summarized in table 1, highlight the ability of single-cell, high-dimensional analyses to capture the complex network and relationships which might exist among cells in the TME. They enable the identification of novel immune cell-types and also help us to understand how their presence or recruitment o might affect the local antitumor immune response (figure 2). Such complexity and heterogeneity would not have been revealed without the ability to profile individual cells, as with the bulk analysis tools.
Table 1.
Phenotypic and functional complexity of tumor-immune landscapes revealed by high-dimensional and single-cell analysis technologies
| Cancer type | High dimensional technologies | Key observations | References |
| Clear cell renal cell carcinoma | CyTOF, mIHC |
|
Chevrier et al, 201837 |
| Hepatocellular carcinoma | CyTOF, mIHC |
|
Chew et al, 201738 |
| Hepatocellular carcinoma | scRNA-Seq, mIHC |
|
Zheng et al, 201739 |
| Breast carcinoma | scRNA-Seq |
|
Azizi et al, 201840 |
| Basal cell carcinoma | scATAC-Seq |
|
Satpathy et al, 201942 |
| Triple-negative breast cancer | MIBI |
|
Keren et al, 201843 |
| Colorectal cancer | scRNA-Seq, mIHC |
|
Zhang et al, 201841 |
| Lung adenocarcinoma | CyTOF, scRNA-Seq, mIHC |
|
Lavin et al, 201744 |
CyTOF, cytometry by time of flight; DC, dendritic cell; HCC, hepatocellular carcinoma; IDO, indoleamine-pyrrole 2,3-dioxygenase; MAIT cells, mucosal-associated invariant T cells; MIBI, multiplexed ion beam imaging; mIHC, multiplex immunohistochemistry; NK, natural killer; PBMC, peripheral blood mononuclear cell; scATAC-Seq, single-cell assay for transposase-accessible chromatin sequencing; TAM, tumor-associated macrophage; TCR, T cell receptor; TLS, tertiary lymphoid structures; TME, tumor microenvironment.
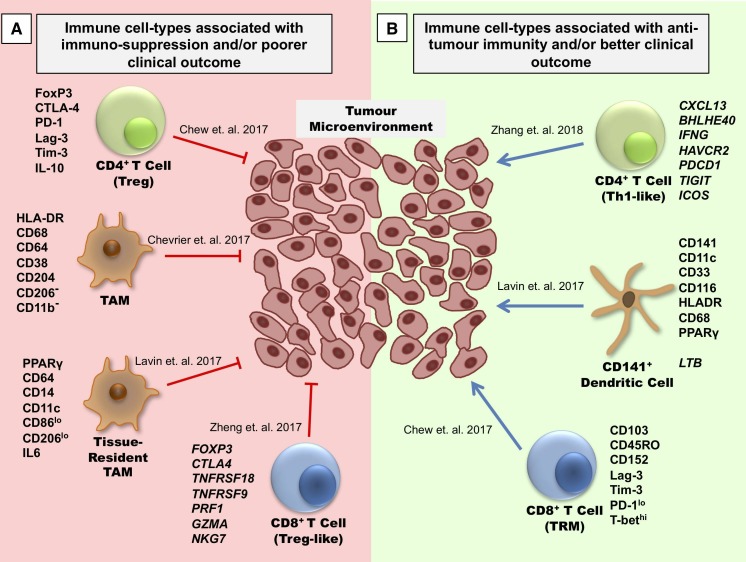
Figure 2.
Characteristics of selective immune subsets important for antitumor immune activity as revealed by multidimensional immunoprofiling. (A) Immune cell-types correlated with immunosuppressive characteristics in the TME and poorer clinical outcome in patients with cancer. (B) Immune cell-types correlated with antitumor immunity and better clinical outcome in patients with cancer. TAM, tumor-associated macrophage; Th1, T-helper type 1 cells; TME, tumor microenvironment; Treg, regulatory T cells; TRM, tissue-resident memory cells.
For instance, in a study on clear cell renal cell carcinoma (CCRCC), CyTOF was used to interrogate the T cell and tumor-associated macrophage (TAM) populations.37 The expression of exhaustion markers on T cells, such as TIM3 and CTLA4, was found to be heterogeneous, whereby PD-1 was broadly expressed. This suggests that anti-PD-1 immunotherapy might be effective in CCRCC. Moreover, a population of CD38+CD204+CD206- TAMs was associated with the immunosuppressive T cell phenotype, indicated by its positive association with PD-1+CD8+ T cells and regulatory T cells (Tregs).37 This finding was validated by the observation with mIHC (4-color) where these TAMs could colocalize with CD8+ T cells in the tumor. In characterizing both the T cell and TAM populations, this study provided an immune atlas of CCRCC which would aid in understanding the mechanisms behind immunotherapy.
Similarly, a study done by Chew et al employed CyTOF to conduct deep immune profiling of the TME, the adjacent non-TME and the peripheral blood mononuclear cells (PBMCs) of hepatocellular carcinoma (HCC).38 It was found that key immune subsets such as T cells, natural killer (NK) cells and myeloid cells were differentially enriched across these three compartments. The functional phenotype of each subset was investigated using markers for exhaustion (PD-1, LAG-3), immunosuppression (IL-10) and inflammatory response (TNFa, IFNy, GzB). It was found that immune cells within the TME are more exhausted and immunosuppressive, even when compared with adjacent non-tumor liver tissues with the background of chronic inflammation due to hepatitis infection.38 mIHC was also used to validate CyTOF findings on formalin-fixed paraffin-embedded (FFPE) tissues, showing that Tregs and exhausted CD8+ T cells were indeed enriched in the TME. Furthermore, the abundance of PD-1-expressing memory CD8+ T cells was reduced in advanced-stage tumors, implying their role in tumor progression.38 This study provided an in-depth understanding on the different phenotypes and functions of infiltrating immune cells when exposed to different microenvironments and concluded that the more exhausted state of immune cells in the TME provided a rationale for the successful application of ICB therapies in HCC.38
The development of single-cell transcriptomic technology also enables researchers to capture the heterogeneity of immune subsets within the TME more comprehensively and trace their developmental relationships. In a study on six patients with HCC by Zheng et al, scRNA-Seq was performed on 5063 individual CD4+ and CD8+ T cells isolated from the TME, non-TME and peripheral blood to dissect their transcriptomic profiles and T cell receptor (TCR) sequences.39 Unique subpopulations such as CD8+FoxP3+ regulatory-like cells were identified within the HCC TME, and this was validated by mIHC. It was also found that clonal expansion of CD8+ T cells and Tregs occurred to a greater extent within the TME as opposed to non-TME or PBMC.39 Finally, the ability to define the TCR sequence at a single-cell level enabled the study of developmental relationships and activation states of these T cell clusters. It was inferred that exhausted T cell populations were more closely related to populations of intermediate development rather than fully-differentiated effector populations, making these intermediate populations attractive checkpoint therapeutic targets.39 This large amount of data detailing the development and composition of the HCC immune landscape would be a valuable resource in understanding and developing new therapeutic strategies.
A similar scRNA seq study by Azizi et al was done on breast carcinoma, but on a larger scale of 47 016 single immune cells collected from eight patients.40 The authors found that the heterogeneity of T cells and myeloid cells greatly increased in tumor tissue as compared with normal breast tissue, with about double the number of unique cell-subsets. Genes which contributed most to this diversity were related to signaling pathways such as inflammation, hypoxia and nutrient supply, suggesting complex responses and modifications in adaptation to the hostile TME.40 Paired scRNA-Seq and TCR sequencing also enabled the authors to map gene expression to TCR clone in each cell, and their data suggested that the phenotypic states of T cells were likely to be affected by both TCR stimulation and signals from the TME. Moreover, in macrophages, both M1 and M2 associated genes were often expressed by the same cells, challenging the conventional model of polarized macrophage phenotypes.40 The ability to extract such detailed information would help to improve the understanding in the protumor or antitumor roles of the immune cells in breast cancer.
Another study by Zhang et al on colorectal cancer (CRC) also used scRNA-Seq and TCR sequencing to map the distribution, clonal expansion, migration and developmental transitions of sorted T cells from tumor tissue, adjacent normal tissue and peripheral blood.41 Similar to previous studies described above, they found that tumor tissue consisted of CD8+ and CD4+ T cells with more exhausted, clonally expanded and less mobile phenotypes while normal tissue and peripheral blood were associated with a more naive or recently activated phenotype. The authors were also able to analyze developmental patterns between these immune subsets across tissue types based on their TCR clonotypes and identify a population of CX3CL13+BHLHE40+IFNG+ Th1-like cells which were developmentally linked to T-effector memory cells.41 These cells were enriched in CRC tumors which demonstrated microsatellite instability, a tumor phenotype associated with response to anti-PD1 immunotherapy. Thus, single-cell technology enabled the authors to better explain the mechanisms behind successful immunotherapy and provided a basis to focus on these novel T cells as a therapeutic target.41
The study of open, actively-transcribed chromatin regions can also bring insights into immune cell heterogeneity in the TME. Satpathy et al used scATAC-Seq to examine the chromatin profile of primary tumor biopsies from patients with basal cell carcinoma receiving anti-PD1 immunotherapy.42 It was found that CD8+ exhausted T cells as well as T follicular helper cells were expanded post-therapy. Moreover, these two cell-types shared a set of transcription factor motifs associated with their differentiation, suggesting a common gene regulatory program driving their development following anti-PD1 immunotherapy.42
An mIHC technology called MIBI described by Angelo et al holds great potential for future multiplex TME studies of tumor tissues.36 This method uses an oxygen primary ion beam to rasterize the tissue sample surface and is technically capable of detecting up to 100 different markers in repeated scan cycles of the same field of view.36 First described in 2014, this technique has now been improved for full-automation and was used to interrogate the TME in triple-negative breast cancer (TNBC).43 With a panel of 36 markers targeted at tumor-related proteins, functional markers, immune-related and immunoregulatory proteins, the authors were able to construct a tumor-immune landscape of TNBC and correlate features such as tissue architecture and cellular phenotype to the clinical parameters of the patients.43 For example, they classified the tumor-immune composition and organization in TNBC into three archetypes: cold (with few immune cells), mixed (with tumor and immune cells mixed together) and compartmentalized (with tumor and immune cells spatially separated) and found that the compartmentalized organization correlated with better survival.43
Finally, a combination of multidimensional technologies was used by Lavin et al to dissect the immune microenvironment in early lung adenocarcinoma.44 CyTOF and scRNA-Seq analyses uncovered a distinct tumor macrophage signature when compared with their normal lung counterparts and a high ratio of tumor-specific macrophage to lung-resident macrophage gene expression was correlated with poorer patient survival.44 Distinct changes in the phenotypes of lymphocytes and antigen-presenting cells (APCs) were also found in the tumor as compared with normal lung tissue. For instance, NK cells, CD16+ monocytes and CD141+ dendritic cells (DCs) were found to be reduced whereas PPARγhi macrophages were enriched in tumor tissue.44 CD141+ DCs in particular has been implicated in T cell recruitment and the formation of tertiary lymphoid structures. Moreover, this immune signature characteristic of the tumor lesions was independent of TNM staging, suggesting that immunotherapy targeting this innate immune pathway, particularly tumor-infiltrating myeloid subsets, might benefit patients even at an early stage.44 Thus, the combination of scRNA-Seq, CyTOF and mIHC enabled the authors to understand and explain how immune cell interactions might contribute to tumorigenesis and use this information to formulate potentially novel therapeutic strategies for the patients.
High-dimensional immune profiling for biomarkers discovery in immunotherapy
Apart from enabling a deeper understanding of the local immune response in the TME, these advanced, high-dimensional technologies have also been highly valuable in screening for immune-related gene and/or protein signatures which might serve as potential biomarkers in predicting response to cancer therapy (table 2).
Table 2.
Biomarker discovery and disease prognosis after cancer therapy
| Cancer type | High dimensional technology | Key observations | References |
| Hepatocellular carcinoma | CyTOF |
|
Chew et al, 201845 |
| Melanoma | CyTOF |
|
Krieg et al, 201746 |
| Melanoma | scRNA-Seq, mIHC |
|
Sade-Feldman et al, 201847 |
| Melanoma | scRNA-Seq, mIHC |
|
Jerby-Arnon et al, 201848 |
| Melanoma | CyTOF, scRNA-Seq |
|
Das et al, 201749 |
CyTOF, cytometry by time of flight; ICB, immune checkpoint blockade; irAE, immune-related adverse event; mIHC, multiplex immunohistochemistry; PBMC, peripheral blood mononuclear cell; scRNA-seq, single-cell RNA sequencing; Y90-RE, Yttrium-90 radioembolization.
One particular study in HCC investigated the immune response to Yttrium-90 radioembolization (Y90-RE) using CyTOF on tumor-infiltrating leucocytes and PBMCs.45 The authors tracked the immune response pre-Y90-RE and post-Y90-RE treatment and detected increased immune subsets such as TNFα-expressing CD8+ and CD4+ T cells and APCs in post-treatment PBMCs, indicating an active systemic immune response elicited by Y90-RE therapy. They also discovered that patients who demonstrated a sustained therapeutic response to Y90-RE treatment showed higher percentages of CD8+ T cells expressing PD-1 (suggesting preactivation) and chemokine receptors, CCR5 and CXCR6 (suggesting homing to the tumor), in both the pretreatment and post-treatment PBMCs.45 Based on this differential expression of immune markers in pretreatment PBMCs, a predictive model for sustained clinical response to Y90-RE treatment was built and validated.45 Thus, the ability to conduct deep immunophenotyping could guide the discovery of potential predictive biomarkers for clinical response to aid in treatment decisions in patients.
Similar studies to predict response to immunotherapy have also been done in melanoma. Krieg et al also employed CyTOF to characterize the lymphocytes and myeloid cells in patients with melanoma before and after anti-PD1 immunotherapy and found that the frequency of CD4+ and CD8+ T cells was lower, while that of the myeloid cells was higher in the PBMCs of responders.46 This was also reflected in the TME where responders had higher frequencies of tumor-infiltrating CD4+ and CD8+ T cells. Moreover, a higher frequency of CD14+CD16-CD33+HLA-DRhi monocytes capable of antigen-presentation was found in the PBMCs of patients before anti-PD-1 therapy, and hence, predictive of a more favorable response to treatment and survival.46 These monocytes also expressed ICAM-1, suggesting a more activated status. Thus, this study provided a rationale to validate this immune-cell signature for clinical stratification of patients before anti-PD1 therapy.
Besides CyTOF, scRNA-Seq has also proven to be very useful in the search for potential biomarkers predictive of response to therapy. Sade-Feldman et al conducted single-cell analysis on tumor biopsies taken at baseline and during treatment, from patients with metastatic melanoma who underwent immune checkpoint therapy.47 They found that CD8+ T cells from the biopsies could be clustered into two major states based on gene expression: one associated with memory, activation and survival (including Wnt transcription factor-7, TCF7) and the other with cell exhaustion. The ratio of the activated to exhausted CD8+ T cells in the tumor correlated with the patients’ response to immunotherapy. This was then validated by IHC staining whereby a higher density of memory and activated TCF7+CD8+ T cells in tumor tissue, both at baseline and post-treatment, was indeed associated with better response to immunotherapy.47 Thus, this study highlights the capability of scRNA-Seq in screening for potential biomarkers in tumor infiltrating immune cells of the patients who might benefit from immunotherapy.
Biomarker discovery is not limited to immune cell types; potential biomarkers might also be found in malignant cells. Jerby-Arnon et al, using scRNA-Seq, reported an immune resistance molecular signature expressed by melanoma tumor cells which was associated with lower levels of CD8+ T cell infiltration and resistance to immune checkpoint therapy.48 The authors validated this with mIHC, showing that the expression pattern of this resistance program (p53, Myc, DLL3, HLA-A, c-Jun, SQSTM1 and LAMP2) was associated with T cell exclusion in tumor tissue. Moreover, it was shown that this resistance program could potentially be regulated by CDK4/6 inhibitors.48 Thus, scRNA-Seq provided the capability to identify potential biomarkers on malignant cells which could predict response to immunotherapy and to rationally formulate potential drug targets to enhance this response.
High-dimensional immunomonitoring could also be used to identify potential predictive biomarkers for immunotherapy-induced irAEs. For instance, a combination of flow cytometry, CyTOF and scRNA-Seq was employed to analyze changes (pretreatment and post-treatment) in circulating B cells of patients with advanced melanoma who received combination ICB therapy or mono-ICB (anti-CTLA-4 or anti-PD-1).49 It was found that a significant decline in overall B cell numbers postimmunotherapy correlated with a shorter time to onset and higher grade of irAEs following combination ICB, possibly providing an early biomarker for immunotherapy-induced autoimmunity. Moreover, this study identified an increase in circulating CD21lo B cells with enhanced activation and IFN-γ signaling post-immunotherapy, suggesting that this population might be an unintended target of combination ICB.49 Thus, these high-dimensional technologies demonstrate the potential for the identification of biomarkers to predict immunotherapy-induced irAEs, for better therapeutic strategies in the clinics.
It should be acknowledged that in many of these studies, especially those employing scRNA-Seq, the cohort of patients enrolled is small, which raises the question of small sample bias. Nonetheless, they show the capability of these high-dimensional technologies in screening for potential biomarkers predictive of clinical outcome after immunotherapy (table 2) and provide a rational basis for validating these signatures in larger cohorts of patients.
Future direction of multidimensional immune profiling technology
The development of single-cell, multi-parametric technology has revolutionized the field of immune profiling. Nonetheless, there is still much room for improvement in terms of profiling capacity, scalability and cost. Newer technologies and methods of analysis are currently being developed, which will enable researchers to conduct profiling at an even higher dimensionality and depth.
The use of lanthanide-labeled antibodies and detection via mass spectrometry has been explored in tissue imaging, harnessing the high-dimensional capability of CyTOF for spatially-resolved proteomic measurements.35 Besides MIBI (as mentioned above), another technique conducted laser ablation of FFPE tissue coupled with a CyTOF mass cytometer could achieve a 32-plex IHC analysis on proteins and their modifications.35 This technique was later extended to include mRNA analysis, enabling the simultaneous detection of 3 mRNAs and 16 proteins at a single-cell level in breast cancer tissue.50 In this study, the authors were able to study single-cell correlations between the mRNA and protein levels for HER2 and CK19 genes. They also found that cells which expressed CXCL10 (a T cell chemoattractant) often clustered together and were correlated with a higher frequency of T cells in the sample.50 With improvements in multiplexing capability, this technology holds great potential in furthering the study of the relationships between mRNA, protein and signaling networks in the TME. These mass cytometry-based tissue proteomic imaging techniques hold several advantages over conventional IHC. One, would be the reduced background or sample autofluorescence signal with little or no spectral overlap between the lanthanide metal reporters.35 36 Moreover, mIHC could provide spatial or localization information of immune cells within TME. Finally, the ability to maximize the amount of information obtained using mIHC techniques enables us to take full advantage of the abundant source of routinely archived FFPE clinical samples with long patient follow-up history.51
RNA sequencing is also currently being explored as a tool to conduct spatial studies. In a method termed Spatial Transcriptomics (ST),31 oligonucleotides were immobilized on glass slides with arrays of spots, each of which contained a positional barcode. Cryopreserved tissue sections were mounted on these slides and imaged, followed by permeabilization and reverse transcription of the mRNA from the tissue. Following probe release from the slide and sequencing, the positional barcodes allowed matching of the genes expression to their original position in the tissue. This enabled the spatial, albeit bulk, expression of genes to be determined within the particular spot from the tissue section.31 The main limitation of this technology was the lack of single-cell resolution as each of these spots captured the transcriptomes of at least 10 or more cells from the same region. As an extension of this method, scRNA-seq was integrated with ST by separately performing scRNA seq and ST from one half of the same tissue and inferring the enrichment of specific cell subsets by comparing the gene signature defined by scRNA seq with ST genes enriched for that same region.52 The specific cell types would be inferred using a statistical deconvolution method which had been used previously to estimate cell-type proportions in bulk tissue RNA seq data.53 This method allowed the identification of three populations of cancer cells in pancreatic ductal adenocarcinoma and their distinct locations in the tumor.52
The imaging of RNA has since been further expanded by using a method called multiplexed error-robust fluorescence in situ hybridization (MERFISH), capable of imaging up to 10,000 RNA species at the single cell level.30 54 Cellular RNA was labeled with encoding probes containing a target sequence and two readout sequences, followed by successive rounds of hybridization and imaging using readout probes. Thus, the expression of RNA at a single-molecule level could be spatially located within a single cell.30 Moreover, using an RNA velocity approach modified from La Manno et al,55 a pseudotime ordering of cell-cycle phases was constructed.30 Finally, scRNA-seq was combined with MERFISH to investigate the hypothalamic preoptic region in mice.56 With such advanced tools, it is conceivable that the characteristics, phenotype and localization of tumor and immune cells within the TME could be more clearly defined so that the mechanisms of tumor progression and response to therapy could be better understood.
Increasingly, sequencing technologies are being harnessed to increase multiplexing capability of immune proteomic profiling. For instance, a recently developed technique called antibody-sequencing (Abseq) has increased the dimensionality of single-cell protein profiling to an even greater extent, providing theoretically limitless multiplexing capability.57 Abseq uses DNA oligo-tagged antibodies which are identified at the single-cell level using droplet microfluidic barcoding and DNA sequencing technology. With a short tag length of 10 bases, Abseq provides more than a million unique sequences for epitope identification and could theoretically label every single member of the human proteome, detecting as low an expression as a single antibody per cell.57 As an extension to Abseq, another technique called Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq) enables the simultaneous, unbiased, multiplex profiling of both the proteome and transcriptome of a single cell.58 Protein epitopes are detected via oligonucleotide-conjugated antibodies (similar to Abseq). After separation into single cell droplets, cell lysis releases cellular mRNA and the oligonucleotides bound to the cell-surface epitopes and allows for separate analyses.58 This technique provides the ability to study gene and protein expression within a single cell simultaneously and would enable researchers to understand how the cell might regulate its transcriptome and protein phenotype under different biological conditions. These technologies are currently only suitable for the detection of surface protein markers, a limitation which could be overcome with future improvements in RNA-seq technology.
The huge volume of data generated in a scRNA-Seq experiment also provides a wealth of information which can be analyzed in various ways, allowing researchers to conduct their investigations from different angles. One such dimension has been called pseudotime analysis, referring to computational models which order cells along a trajectory based on similarities in their expression patterns.59 Also known as trajectory inference (TI) methods, these models operate under the assumption that single-cell data is a snapshot of a continuous process of different cells at different time points along a common developmental process.60 A multitude of TI methods are available, as reviewed in Salens et al (2019).59 Such analyses could be useful in understanding how immune cell development is affected by tumor progression and vice versa. For example, the study done by Zheng et al mentioned above also conducted a pseudotime analysis on CD8+ T cells from the TME, placing the CD8 cell clusters in an order from a naïve, to an effector and finally to an exhausted phenotype, demonstrating a transition from activation to exhaustion.39
Many of these single-cell technologies currently stand in their infancy: protocols and techniques are still being optimized. Consequently, the cost of conducting large-scale screening of patients in a clinical setting would likely be too high to be feasible. Nonetheless, as these technologies improve, and with commercialization, increased efficiency and throughput aas well as lower costs could make them available to clinical laboratories in the future.
Implications for immunotherapy and conclusion
The TME is heterogeneous and complex in its molecular and immunological features.61 Multiple immune subsets and factors in the TME greatly influence the outcome of immunotherapy2 and disease prognosis.62 As described in the review above, the balance between immune evasion and antitumor activity, as well as a patient’s response to therapy, is greatly affected by the recruitment or accumulation of specific immune cell types, which in turn influence the immune response in TME. Importantly, the variation in these cellular subsets, both within and across cancer types, precludes the development of a single effective treatment for all patients. The success of cancer immunotherapy, therefore, likely hinges on the ability to tailor personalized courses of therapy to each individual patient, based on their immune profiles. In order to achieve such personalized treatment, it is necessary to be able to capture the heterogeneity of TME within each cancer type and to gain a mechanistic understanding of how each network interacts and functions. Such knowledge will help to predict the clinical response to treatment as well as to design better therapeutic strategies for the patients. The ability to conduct deep immunophenotyping at a single-cell level and to simultaneously extract transcriptomic, proteomic and spatial information propels us closer to the goal of designing more effective personalized immunotherapy.
Footnotes
Contributors: SC researched and drafted the manuscript. VC conceptualized the ideas, guided SC in his research, wrote and edited the manuscript.
Funding: This work was supported by the National Medical Research Council (NMRC), Singapore (ref numbers: TCR15Jun006, CIRG16may048, CSAS16Nov006, CSASI17may003 and LCG17MAY003).
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Yu Y, Cui J. Present and future of cancer immunotherapy: a tumor microenvironmental perspective (review). Oncol Lett 2018;16:4105–13. 10.3892/ol.2018.9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klemm F, Joyce JA. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol 2015;25:198–213. 10.1016/j.tcb.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fridman WH, Pagès F, Sautès-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 2012;12:298–306. 10.1038/nrc3245 [DOI] [PubMed] [Google Scholar]
- 4. Zhang H, Chen J. Current status and future directions of cancer immunotherapy. J Cancer 2018;9:1773–81. 10.7150/jca.24577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346–54. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- 6. Farkona S, Diamandis EP, Blasutig IM. Cancer immunotherapy: the beginning of the end of cancer? BMC Med 2016;14:73 10.1186/s12916-016-0623-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beatty GL, Gladney WL. Immune escape mechanisms as a guide for cancer immunotherapy. Clin Cancer Res 2015;21:687–92. 10.1158/1078-0432.CCR-14-1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lyons YA, Wu SY, Overwijk WW, et al. Immune cell profiling in cancer: molecular approaches to cell-specific identification. NPJ Precis Oncol 2017;1:26. 10.1038/s41698-017-0031-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bendall SC, Nolan GP, Roederer M, et al. A deep profiler's guide to cytometry. Trends Immunol 2012;33:323–32. 10.1016/j.it.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stack EC, Wang C, Roman KA, et al. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 2014;70:46–58. 10.1016/j.ymeth.2014.08.016 [DOI] [PubMed] [Google Scholar]
- 11. Russo G, Zegar C, Giordano A. Advantages and limitations of microarray technology in human cancer. Oncogene 2003;22:6497–507. 10.1038/sj.onc.1206865 [DOI] [PubMed] [Google Scholar]
- 12. Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. 10.1038/35000501 [DOI] [PubMed] [Google Scholar]
- 13. Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the juncture of clinical translation. Nat Rev Genet 2018;19:93–109. 10.1038/nrg.2017.96 [DOI] [PubMed] [Google Scholar]
- 14. Foerster F, Hess M, Gerhold-Ay A, et al. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci Rep 2018;8:5351 10.1038/s41598-018-21937-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau D, Bobe AM, Khan AA. Rna sequencing of the tumor microenvironment in precision cancer immunotherapy. Trends Cancer 2019;5:149–56. 10.1016/j.trecan.2019.02.006 [DOI] [PubMed] [Google Scholar]
- 16. Martens A, Wistuba-Hamprecht K, Geukes Foppen M, Foppen MG, et al. Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res 2016;22:2908–18. 10.1158/1078-0432.CCR-15-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barlogie B, Raber MN, Schumann J, et al. Flow cytometry in clinical cancer research. Cancer Res 1983;43:3982–97. [PubMed] [Google Scholar]
- 18. Danova M, Torchio M, Comolli G, et al. The role of automated cytometry in the new era of cancer immunotherapy. Mol Clin Oncol 2018;9:355–61. 10.3892/mco.2018.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gnjatic S, Bronte V, Brunet LR, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 2017;5:44. 10.1186/s40425-017-0243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbst RS, Soria J-C, Kowanetz M, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–7. 10.1038/nature14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potter SS. Single-Cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 2018;14:479–92. 10.1038/s41581-018-0021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newell EW, Becht E. High-Dimensional Profiling of Tumor-Specific Immune Responses: Asking T Cells about What They "See" in Cancer. Cancer Immunol Res 2018;6:2–9. 10.1158/2326-6066.CIR-17-0519 [DOI] [PubMed] [Google Scholar]
- 23. Bjornson ZB, Nolan GP, Fantl WJ. Single-Cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol 2013;25:484–94. 10.1016/j.coi.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsurui H, Nishimura H, Hattori S, et al. Seven-color fluorescence imaging of tissue samples based on Fourier spectroscopy and singular value decomposition. J Histochem Cytochem. 2000;48:653–62. 10.1177/002215540004800509 [DOI] [PubMed] [Google Scholar]
- 25. Greenplate AR, Johnson DB, Ferrell PB, et al. Systems immune monitoring in cancer therapy. Eur J Cancer 2016;61:77–84. 10.1016/j.ejca.2016.03.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ozsolak F, Milos PM. Rna sequencing: advances, challenges and opportunities. Nat Rev Genet 2011;12:87–98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang Z, Gerstein M, Snyder M. Rna-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009;10:57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Metzker ML. Sequencing technologies — the next generation. Nat Rev Genet 2010;11:31–46. 10.1038/nrg2626 [DOI] [PubMed] [Google Scholar]
- 29. Wu AR, Neff NF, Kalisky T, et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods 2014;11:41–6. 10.1038/nmeth.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen KH, Boettiger AN, Moffitt JR, et al. Rna imaging. spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015;348:aaa6090 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ståhl PL, Salmén F, Vickovic S, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016;353:78–82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 32. Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–8. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas R. Practical guide to ICP-MS : a tutorial for beginners. Boca Raton: CRC Press, 2008. [Google Scholar]
- 34. Gerdes MJ, Sevinsky CJ, Sood A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A 2013;110:11982–7. 10.1073/pnas.1300136110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Giesen C, Wang HAO, Schapiro D, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014;11:417–22. 10.1038/nmeth.2869 [DOI] [PubMed] [Google Scholar]
- 36. Angelo M, Bendall SC, Finck R, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med 2014;20:436–42. 10.1038/nm.3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chevrier S, Levine JH, Zanotelli VRT, et al. An immune atlas of clear cell renal cell carcinoma. Cell 2017;169:736–49. 10.1016/j.cell.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chew V, Lai L, Pan L, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci U S A 2017;114:E5900–9. 10.1073/pnas.1706559114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng C, Zheng L, Yoo J-K, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell 2017;169:1342–56. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 40. Azizi E, Carr AJ, Plitas G, et al. Single-Cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 2018;174:1293–308. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang L, Yu X, Zheng L, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- 42. Satpathy AT, Granja JM, Yost KE, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 2019;37:925–36. 10.1038/s41587-019-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keren L, Bosse M, Marquez D, et al. A structured Tumor-Immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 2018;174:1373–87. 10.1016/j.cell.2018.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lavin Y, Kobayashi S, Leader A, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell 2017;169:750–65. 10.1016/j.cell.2017.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chew V, Lee YH, Pan L, et al. Immune activation underlies a sustained clinical response to yttrium-90 radioembolisation in hepatocellular carcinoma. Gut 2019;68:335–46. 10.1136/gutjnl-2017-315485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Krieg C, Nowicka M, Guglietta S, et al. High-Dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 2018;24:144–53. 10.1038/nm.4466 [DOI] [PubMed] [Google Scholar]
- 47. Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jerby-Arnon L, Shah P, Cuoco MS, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 2018;175:984–97. 10.1016/j.cell.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Das R, Bar N, Ferreira M, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest 2018;128:715–20. 10.1172/JCI96798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schulz D, Zanotelli VRT, Fischer JR, et al. Simultaneous multiplexed imaging of mRNA and proteins with subcellular resolution in breast cancer tissue samples by mass cytometry. Cell Syst 2018;6:25–36. 10.1016/j.cels.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kokkat TJ, Patel MS, McGarvey D, et al. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank 2013;11:101–6. 10.1089/bio.2012.0052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moncada R, Chiodin M, Devlin JC, et al. Building a tumor atlas: integrating single-cell RNA-Seq data with spatial transcriptomics in pancreatic ductal adenocarcinoma. bioRxiv 2018;254375. [Google Scholar]
- 53. Baron M, Veres A, Wolock SL, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and Intra-cell population structure. Cell Syst 2016;3:346–60. 10.1016/j.cels.2016.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xia C, Fan J, Emanuel G, et al. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc Natl Acad Sci U S A 2019;116:19490–9. 10.1073/pnas.1912459116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature 2018;560:494–8. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moffitt JR, Bambah-Mukku D, Eichhorn SW, et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 2018;362:1–12. 10.1126/science.aau5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shahi P, Kim SC, Haliburton JR, et al. Abseq: Ultrahigh-throughput single cell protein profiling with droplet microfluidic barcoding. Sci Rep 2017;7:44447 10.1038/srep44447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14:865–8. 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saelens W, Cannoodt R, Todorov H, et al. A comparison of single-cell trajectory inference methods. Nat Biotechnol 2019;37:547–54. 10.1038/s41587-019-0071-9 [DOI] [PubMed] [Google Scholar]
- 60. Luecken MD, Theis FJ. Current best practices in single‐cell RNA‐seq analysis: a tutorial. Mol Syst Biol 2019;15:e8746 10.15252/msb.20188746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ringelhan M, Pfister D, O’Connor T, et al. The immunology of hepatocellular carcinoma. Nat Immunol 2018;19:222–32. 10.1038/s41590-018-0044-z [DOI] [PubMed] [Google Scholar]
- 62. Barnes TA, Amir E. Hype or hope: the prognostic value of infiltrating immune cells in cancer. Br J Cancer 2017;117:451–60. 10.1038/bjc.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]