Abstract
Alginate is a linear copolymer composed of 1→4 linked β-d-mannuronic acid (M) and its epimer α-l-guluronic acid (G). The polysaccharide is first produced as homopolymeric mannuronan and subsequently, at the polymer level, C-5 epimerases convert M residues to G residues. The bacterium Azotobacter vinelandii encodes a family of seven secreted and calcium ion-dependent mannuronan C-5 epimerases (AlgE1–AlgE7). These epimerases consist of two types of structural modules: the A-modules, which contain the catalytic site, and the R-modules, which influence activity through substrate and calcium binding. In this study, we rationally designed new hybrid mannuronan C-5 epimerases constituting the A-module from AlgE6 and the R-module from AlgE4. This led to a better understanding of the molecular mechanism determining differences in MG- and GG-block-forming properties of the enzymes. A long loop with either tyrosine or phenylalanine extruding from the β-helix of the enzyme proved essential in defining the final alginate block structure, probably by affecting substrate binding. Normal mode analysis of the A-module from AlgE6 supports the results.
Introduction
Alginate is a major polysaccharide constituent of brown algae1 and it is also synthesized by some bacteria of the Azotobacter(2) and Pseudomonas genera.3,4 Annual worldwide production of alginate is around 45,000 metric tons extracted from various types of brown seaweeds, with increasing applications and demand.5 Its numerous applications range from biomaterials in pharmaceutical utilization, as food additives (stabilizing, thickening, or gelling agent) to the use as technical materials in the textile printing industry.
Alginate is a linear copolymer of 1→4 linked β-d-mannuronic acid (M) and its epimer α-l-guluronic acid (G) differing only at C-5. It has no regular repeating structure but the residues occur in stretches of continuous M or G residues and sequences of alternating M and G, referred to as M-blocks, G-blocks, and MG-blocks, respectively.6 Alginate hydrogels are formed by ionic cross-linking of G-blocks with some contribution from the MG-blocks by certain divalent cations (e.g., Ca2+ and Ba2+).7,8 The relative amounts of M and G and the length of the different block structures therefore affect the gel-forming ability of alginates as well as other physicochemical properties.9 Extraction from different parts of the algae gives alginates with different properties. The stem of Laminaria hyperborea contains alginate with the highest levels of G (up to 70%) and is generally the most valuable and widely used in food and industrial applications because of excellent gelling properties.10−13 The leaf alginate has a lower G-content (less than 55%) and fewer applications, and thus a lower price in the market.
A unique feature for alginate is its synthesis as homopolymeric mannuronan, before G residues are introduced at the polymer level by enzymatic epimerization at C-5. Eight mannuronan C-5 epimerases have been identified in the alginate-producing bacterium Azotobacter vinelandii. One is a periplasmic epimerase (AlgG),14 which incorporates single G residues into the alginate during secretion of the polymer. Seven are extracellular (AlgE1–7)15,16 and convert M to G in different patterns. Additionally, one of the epimerases (AlgE7) displays alginate lyase activity.17
The AlgE epimerases all consist of two types of structural modules, designated A and R.15,16 Earlier studies demonstrated that only the A-modules are catalytically active,18 whereas the R-modules seem to modulate the catalytic rate by calcium and substrate binding.18−20 The seven extracellular A. vinelandii enzymes have one or two A-modules that can be classified as either MG-block formers or G-block formers.21 In addition, they consist of one to seven R-modules. Although the different A- and R-modules have high sequence similarities, the enzymes create quite different product patterns.16,18 The organism utilizes the differences in the enzymes to produce alginate suited for its various needs during its complex lifecycle. Similarly, the different C-5 epimerases can potentially be used to tailor specific alginate structures for commercial applications.22 However, the mode of action of these elusive enzymes and how it is related to their differences is still unclear.
AlgE6 (one A-module and three R-modules, AR1R2R3) and AlgE4 (one A-module and one R-module, AR) share a high sequence homology15 but produce alginate with different content and distribution of M and G residues. Both enzymes probably perform the same epimerization reaction, processively epimerizing every other M-residue into G-residues to form MG-blocks around 20 monomers long.23−25 This is supported by the fact that the residues in and around the active site are almost identical. However, whereas AlgE4 introduces only MG-blocks into the alginate chain, AlgE6 is able to epimerize MG-blocks and form G-blocks as well.26 As the main difference between the two enzymes is in their modular arrangements, individual modules of both enzymes have been studied extensively. The 3D structure of the A-module from AlgE427 and individual R-modules from AlgE4 and AlgE628,29 have been determined by X-ray crystallography and by nuclear magnetic resonance (NMR) spectroscopy, respectively. For the R-modules, the structural similarity is high, whereas their affinity for alginate is significantly different. The AlgE4 R-module shows interaction with dissociation constants in the μM range for alginate oligomers, with a clear preference for M-oligomers over MG-oligomers and no detected interaction with G-oligomers.20 In contrast, the AlgE6 R-modules do not display any binding to alginate oligomers when expressed individually. However, together (R1R2R3) they show weak interactions with long M-oligomers with dissociation constants in the mM range, but hardly any interaction with MG-oligomers.20
Tøndervik et al. 2013 have shown that switching the R-modules of AlgE4 and AlgE6 between mutant epimerases modulates the epimerization pattern.21 In 2014, Buchinger et al. made a hybrid enzyme AlgE64, consisting of AlgE6 A-module fused to the AlgE4 R-module.20 This enzyme led to ∼25% higher G-content compared to the native AlgE6 epimerase and it was hypothesized that increased G-formation was directly associated with the AlgE4 R-module’s stronger substrate interaction. The 3D structure of a full-length alginate epimerase has not been determined yet and the structure of the linker sequence connecting the A-modules to the R-modules is unknown. However, both small-angle X-ray scattering and NMR studies for the overall structure of AlgE4 suggest that the linker region between the A- and the R-modules is flexible and does not contain secondary structure elements.30
To understand why AlgE64 has increased G-block forming abilities, we focused on the regions close to the transition between the A- and R-modules. These regions are far away from the active site, but we found that they affected the product patterns of the enzymes. To study the molecular mechanisms underlying the MG- and G-block-forming properties of the mannuronan C-5 epimerases, we created new AlgE64 enzymes. This was done by rational design of the transition region based on the primary and tertiary structures of AlgE6 and AlgE4. The product profiles and mode of action of the new epimerases have been characterized with 1H NMR and time-resolved 13C NMR, respectively.
Materials and Methods
Cloning, Expression, and Purification of the Hybrid Epimerases
The hybrid mannuronan C-5 epimerase genes were all synthesized de novo (GenScript, Piscataway, USA). Genes coding for the hybrid enzymes were inserted into pMV23 vector31 as NdeI-NotI fragments. Epimerases used in this study are summarized in Table 1 and the gene sequences of AlgE64, AlgE64-A, AlgE64-B, AlgE64-B1, AlgE64-B2, and AlgE64-B3 are included in the Supporting Information.
Table 1. Epimerase Genes Used in This Study.
| insert | description of synthetic gene | refs |
|---|---|---|
| AlgE64 | encoding AlgE6 A-module (residues 1–386) combined with AlgE4 R-module (residues 387–530) | (20) |
| AlgE64-A | encoding AlgE6 A-module and first part of R-module (residues 1–409) combined with the last part of AlgE4 R-module (residues 410–531) | this study |
| AlgE64-B | encoding the first part of AlgE6 A-module (residues 1–300) combined with AlgE4 A- and R-modules (residues 301–534) | this study |
| AlgE64-B1 | encoding AlgE64-B with residues 305–322 from AlgE6 | this study |
| AlgE64-B2 | encoding AlgE64-B with residues 323–352 from AlgE6 | this study |
| AlgE64-B3 | encoding AlgE64-B with residues 353–375 from AlgE6 | this study |
| AlgE64-B F307Y | encoding AlgE64-B with point mutation F307Y | this study |
Standard recombinant DNA procedures were performed as described previously.32 Plasmids were purified by WizardPlus SV Minipreps DNA purification system (Promega). RbCl transformation protocol (New England BioLabs) was adopted for transformations of bacterial strains. Escherichia coli strain DH5α (Bethesda Research Laboratories) was used as a general cloning host, whereas E. coli RV308 (ATCC31608)33 was used for protein expression. Bacteria were grown at 37 °C in LB medium (yeast extract, 5 g/L; tryptone, 10 g/L; and NaCl, 10 g/L) or in LB agar (LB medium supplied with agar 20 g/L). For protein expression, strains were grown in a 3× LB medium (yeast extract, 15 g/L; tryptone, 30 g/L; and NaCl, 10 g/L) at 30 °C to A600nm ≈ 0.8–1.2. The cell cultures were successively incubated on ice for 5 min. Expression was induced by addition of m-toluate (final concentration 1 mM), and the cultures were incubated at 16 °C for 16–20 h. Media were supplemented with 200 μg/mL ampicillin when appropriate. The cells were harvested by centrifugation and the pellets were resuspended in 40 mM MOPS, pH 6.9, with 5 mM CaCl2 for sonication. After centrifugation, the crude protein extracts (supernatant) were filtered (0.22 μm) and loaded on a 5 mL HiTrap Q HP column (GE Healthcare). Fast protein liquid chromatography (ÄKTA FPLC system—GE Healthcare) was used for the purification and proteins were eluted using a stepwise NaCl gradient (0 to 1 M) in the 40 mM MOPS, pH 6.9, with 5 mM CaCl2 buffer. The purity of the protein-containing fractions was evaluated by SDS-PAGE, and the proteins were tested for epimerase activity by NMR.
Production of Alginate Substrates
High-molecular-weight mannuronan (poly-M) (FG = 0.00) was isolated from an epimerase-negative strain of Pseudomonas fluorescens NCIMB 10525.3413C-1 labeled mannuronan was produced by growing the mannuronan-producing P. fluorescens strain on a minimal medium with 99% d-13C-1 fructose as a carbon source. The obtained mannuronan was selectively enriched to 59% with 13C at carbon position C-1.35
Complete and Time-Resolved NMR Analysis of Epimerized Alginate Samples
Alginate epimerases can either epimerize an M-residue next to another M-residue or an M-residue next to a G-residue. As they are thought to epimerize every other residue in each processive event because of the orientation of monomers in the polymer chain, this will either create MG-blocks or G-blocks.20 In other words, their substrate specificities can explain product patterns and they can only create G-blocks if they are able to bind MG-blocks. As we can distinguish monomers based on their neighboring residues with 1H NMR,36,37 we can characterize the epimerization abilities of the enzymes extensively. Indirectly, we are then able to characterize their substrate specificities. After 48 h, the reaction has reached completion and the enzymes have reached their respective limits of how large a fraction of G-residues they can create. This will hereafter be denoted “complete epimerization”.
Samples of 2.5 mg/mL poly-M were epimerized with 25 μg/mL enzymes in 50 mM MOPS pH 6.9 with 75 mM NaCl and 4 mM CaCl2 buffer at 37 °C for 48 h. The epimerized samples were then subjected to two-step acid hydrolysis prior to complete NMR analysis. 3-(Trimethylsilyl)-propionic-2,2,3,3-d4 acid sodium salt (Aldrich, Milwaukee, WI) in D2O (2%, 5 μL) was added as the internal standard for the chemical shift, and triethylenetetra-amine hexa-acetate (Sigma-Aldrich) was added as a calcium chelator (0.3 M, 20 μL). 1H NMR spectra were recorded on Bruker AV III HD 600 or 800 MHz equipped with 5 mm with cryogenic CP-TCI, Bruker AV III HD 400 MHz equipped with 5 mm SmartProbe, Bruker Avance DPX 300 MHz equipped with 5 mm QNP (C/H) probe, or Bruker Avance DPX 400 MHz equipped with 5 mm z-gradient DUL (C/H) probe). Analysis of complete epimerized samples was recorded at 90 °C on the 300 or 400 MHz spectrometer, whereas time-resolved NMR recording the epimerization reaction was performed at 25 °C with 600 or 800 MHz spectrometer. For the time-resolved NMR analysis of epimerization reactions, a stock solution of 22 mg/mL 13C-1-enriched poly-M (average DPn ≈ 70) in 5 mM MOPS, pH 6.9, with 75 mM NaCl in 99.9% D2O was prepared. Purified enzyme fractions from ion exchange chromatography were subject to buffer exchange and concentrated (final concentration around 2.3 mg/mL) by spin columns (VivaSpin, Sartorius Stedim Biotech) with molecular cutoff of 10 kDa. Samples were washed with 5 mM MOPS, pH 6.9, with 75 mM NaCl and 27.5 mM CaCl2 in 99.9% D2O. Protein concentrations were determined with a Nanodrop ND-1000 to ensure similar enzyme concentration in the epimerization reaction. 13C-1-enriched poly-M stock solution (500 μL) was preheated in the NMR instrument and 1D proton and carbon spectra were recorded to ensure that the sample had not undergone any degradation or contamination prior to the time-resolved NMR experiment. Enzyme solution (50 μL) was added to the preheated substrate and mixed by inverting the sample two to three times. The sample was then immediately inserted into the preheated NMR instrument and the experiment was started. The recorded spectrum is a pseudo-2D-type experiment recording a 1D carbon NMR spectrum every 10 min with a total of 128 time points. The recorded 1D carbon spectrum (using inverse gated proton decoupling) contains 8K data points and has a spectral width of 80 ppm, 32 scans with a 30° flip angle, and relaxation delay of 1.1 s (total recording time of 60 s). The spectra were recorded using TopSpin 1.3, 2.1, 3.2 software (Bruker BioSpin) and processed and analyzed with TopSpin 3.0 software (Bruker BioSpin).
Normal Mode Analysis
The NMA was carried out using the WEBnm@-server38,39 and the elNémo server.40,41 A homology model structure for the A-module of AlgE6 was created with SWISS-MODEL42 using the crystal structure of AlgE4’s A-module (PDB ID 2PYH) as a template. The WEBnm@-server implements the elastic network model (ENM) with the C-α force field43 of Hinsen et al.44 The elNémo server employs the ENM model with the rotation translation-block (RTB) approximation. ENMs are known to sometimes exaggerate displacements, but still, the regions with the largest motions within a model can be identified.39
Results and Discussion
Hybrid Epimerases with Conserved Folding in the Transition between the A- and R-Modules
We have previously shown that swapping of the R-modules between AlgE6 and AlgE4 leads to changes in the epimerization pattern of the resulting hybrid epimerases. AlgE64 (A-module from AlgE6 and R-module from AlgE4) introduced a higher level of G-blocks when epimerizing poly-M than AlgE6.20 This is presumably because the AlgE4 R-module has a higher affinity for poly-M and poly-MG than the AlgE6 R-module. Increased contact time between enzyme and substrate could result in increased processivity (more sugar residues epimerized per interaction). Given the high similarity of the A- and R-modules of AlgE6 and AlgE4, we wanted to identify which residues in the epimerases contribute to the different epimerization patterns.
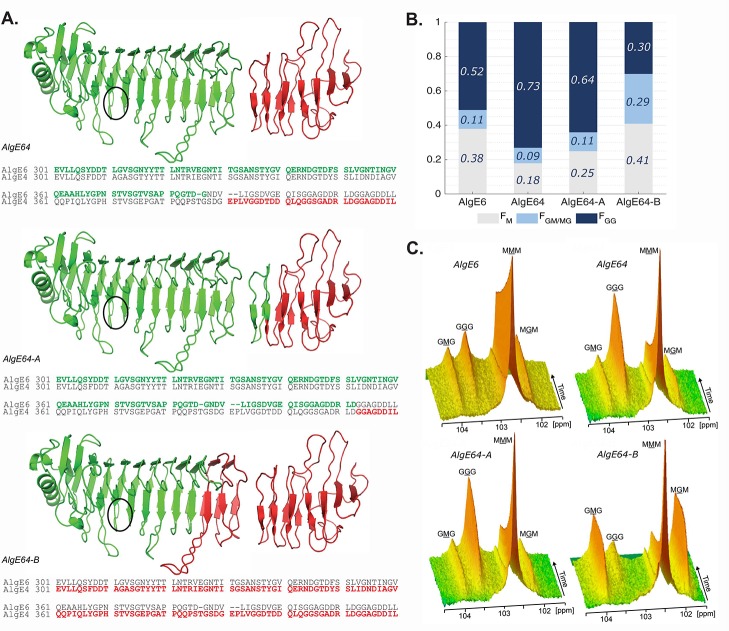
Based on AlgE64, two new hybrid epimerases were constructed: AlgE64-A and AlgE64-B. In these enzymes, we aimed at preserving the original folding of the transition region between the A- and the R-module to a larger extent than what was done in AlgE64. AlgE64 contains amino acid residues 1–386 from AlgE6 and the remaining residues from AlgE4, and no concern was taken in preserving the fold of the transition. As a crystal structure of a complete epimerase is lacking, the design of AlgE64-A and AlgE64-B (Figure 1A) was based on evaluations of conserved sequences and experimental27−29 and SWISS-MODEL42 homology model structures of individual modules. In AlgE64-A, residues 1–409 are from the AlgE6 A-module and the initial part of the AlgE6 R-module, whereas the remaining part of the R-module starting after the third β-string of the β-roll belongs to AlgE4 (residues 410–531 in AlgE64-A). In AlgE64-B, residues 1–300 are from the core part of the β-helix in the AlgE6 A-module, and the remainder residues 301–534 are from AlgE4. Thus, the transition region in AlgE64-A is based on AlgE6’s sequence and structure, whereas in AlgE64-B it is based on AlgE4. This preserves well-defined structure elements with conserved sequences and ensures correct folding of the modules and the transition between them. The epimerization patterns of AlgE64, AlgE64-A, AlgE64-B, and wild-type AlgE6 were tested on poly-M at complete epimerization (Figure 1B and Table S1). The relative intensities of peaks corresponding to specific monads, dyads, or triads give us fractions of G-blocks, M-residues, and MG-blocks in the alginate (denoted FGG, FM, and FGM/MG). We also get fractions of M-blocks and G-residues, FMM and FG (Table S1). As previously shown, AlgE64 is more effective at forming G-blocks than AlgE6.20 AlgE64 also displays better epimerization abilities than the two new hybrid epimerases both in terms of the total number of G residues and G-block content. AlgE64-A seems to conserve the nature of a G-block-forming epimerase like AlgE6, whereas AlgE64-B displays an epimerization pattern resembling an MG-block forming epimerase like AlgE4.
Figure 1.
(A) Structural model of the hybrid enzymes AlgE64, AlgE64-A, and AlgE64-B, based on the crystal structure of AlgE4 A-module (PDB code 2PYG) and the NMR-structure of AlgE4 R-module (PDB code 2AGM). Parts in green belong to AlgE6, whereas red parts correspond to AlgE4. The location of the active site is indicated with black circles. The structures are visualized with PyMOL.46 Sequence alignment of the transition region between the A- and the R-modules in AlgE6 and AlgE4 is shown for each hybrid enzyme at the bottom of their ribbon structure. Residues colored in green (AlgE6) and red (AlgE4) denote the amino acids present in the corresponding hybrid epimerases. (B) Product composition at complete epimerization with AlgE6, and hybrid enzymes AlgE64, AlgE64-A, and AlgE64-B, calculated from 1H NMR spectra. M residues are shown in gray bars, GM/MG dyads in light blue bars, and GG-dyads in dark blue bars. On the y-axis is the fraction of each monad and dyad, whereas the four different enzymes are shown along the x-axis. These values are also shown in Table S1. (C) Time-resolved NMR spectra showing epimerization of 13C1-labeled poly-M for the four enzymes from B. The position of the triads in the spectra is indicated, and the M or G moiety generating the signal is underlined.
To study the mode of action of the enzymes, we performed time-resolved NMR on epimerization of 13C1-labeled poly-M.35,45 With this method, the change in block composition in the substrate over time was inferred from the characteristic peaks of monomer triads (MMM, GGG, GMG, and MGM), that is, a decline in MMM and generation of G-containing triads (Figure 1C). These results confirmed the same trends as the complete epimerization. AlgE64-A produces G-blocks, shown as an increase in the peak marked GGG in the figure. Both AlgE64 and AlgE64-A give a simultaneous rapid decay of the M-blocks, shown as a decrease of the peak marked as MMM, as well as a slow increase of MG-blocks (peaks marked GMG and MGM). The GGM peak indicates the number of G-blocks and remains constant during the reaction, suggesting an introduction of G residues as elongation of pre-existing G-blocks (not apparent in Figure 1C but illustrated in Figure S1). AlgE64-B displays rapid incorporation of G residues in MG-blocks, shown as a fast increase of MGM and GMG peaks, and only at later stages a small amount of G residues is introduced as G-blocks.
From complete and time-resolved epimerization results, it is clear that the region differing in the two hybrid enzymes (defined by amino acids 300–410 in AlgE6) is important for the epimerization pattern. This is interesting, as it is located at least five to seven subsites away from the catalytic site (defined as residues Y149, D152, H154, E155, and D17827). We investigated this further by creating mutants differing in this region.
Identification of the Loop Protruding Out from the 10th Turn in the β-Helix as Important for Determining the Epimerization Pattern
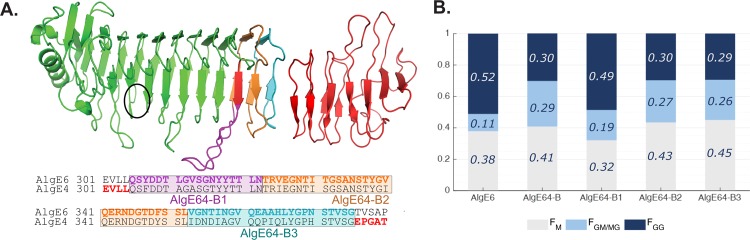
Based on AlgE64-B, three new hybrid epimerases were designed to identify which parts of the enzyme structure determine the epimerization pattern. Altered parts are shown in Figure 2A. Each hybrid enzyme contains a modified region where the amino acids in AlgE64-B originating from AlgE4 are converted to the ones in AlgE6. In AlgE64-B1, the amino acids from 305 to 322 were changed to correspond to the ones in AlgE6 (purple in Figure 2A and referring to AlgE6 numbering). In AlgE64-B2, the same was done with amino acids from 323 to 352 (orange), and in AlgE64-B3 these regions covered amino acids 353–375 (light blue).
Figure 2.
(A) Structural model overview of AlgE64-B with localization of three regions (indicated by purple (AlgE64-B1), orange (AlgE64-B2), and, light blue (AlgE64-B3) colored parts) in the last part of the A-module, which were mutated to create three new mutants. In the three new mutants, amino acids belonging to AlgE4 are substituted to amino acids present in AlgE6. Sequence alignment of AlgE6 and AlgE4 is shown at the bottom of the structural model. Colored bold residues show the three different modified parts, the same color scheme as in the model. A sequence alignment of the long loop and how it differs between these three mutants and AlgE64-B is shown in Figure S2B. The location of the active site is indicated with a black circle. (B) Product composition at complete epimerization for AlgE6 and the four different AlgE64-B mutants, calculated from 1H NMR spectra. M residues are represented as gray bars, GM/MG dyads as light blue bars, and GG-dyads as dark blue bars. The y-axis denotes the fraction of the three product types, whereas the four different enzymes are listed on the x-axis. These values are also shown in Table S1.
Data from NMR on poly-M epimerized with the new hybrid enzymes are shown in Figure 2B and Table S1. AlgE64-B2 and AlgE64-B3 have only minor differences in their epimerization patterns compared to AlgE64-B. On the other hand, the modified loop in AlgE64-B1 appears to have a considerable effect on the epimerization activity. AlgE64-B1 gave a higher FG fraction as well as increased GG sequences along with a decrease of M-content and MM sequences. Apparently, changes made in the loop are important for the G-block-forming properties of the epimerases as the hybrid enzyme AlgE64-B1 displays G-block formation similar to AlgE6.
Based on the results from the modeling, the loop mutated in AlgE64-B1 appears to be long enough to embrace the substrate-binding cleft. This is observed in other polysaccharide-active enzymes like alginate lyases (described further down).47 Glycosyl hydrolases also evolved long loops covering an open cleft, and this tunnel-like topology of the binding site creates the conditions for processivity.48−50 The tunnel allows the enzyme to remain firmly bound to the polysaccharide chain while it slides through the active site for several catalytic events.51,52 Cellobiohydrolase Cel6A from Trichoderma reeseii contains the active site located inside a tunnel-like cleft where a pair of loops forms a “roof” on the substrate-binding cleft. One of the loops undergoes conformational changes, which may be involved in catalysis by triggering the enzymatic process or threading the substrate through the active site. “Open and close” conformations of the loop could allow the enzyme to bind the polysaccharide throughout subsequent processive action.53 Likewise, we hypothesize that the long loop protruding out of the β-helix core of the A-module forms a tunnel-like cleft, which could embrace the alginate chain and affect catalysis. Indeed, mutations in the abovementioned loop of AlgE2–AlgE4 hybrids have previously been reported to influence the epimerization mechanism.54 However, these effects could be due to more than modification of the loop, as the hybrid enzymes were created by combining mutations in several regions.
Gatekeeper Residue for Determining the Epimerization Pattern of Alginate C-5 Epimerases
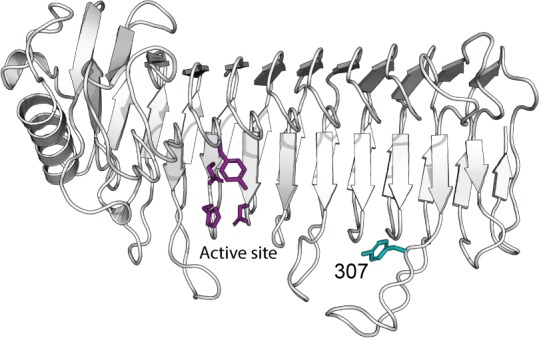
Tyrosine 307 (residue numbering for AlgE6) in the loop is conserved in all G-block-forming enzymes, whereas in MG-block-forming enzymes the same position is occupied by a phenylalanine. This was shown in an alignment of 14 AlgE wild-type and mutant A-module sequences.21 Tøndervik et al. isolated two mutant epimerases, which were able to introduce G-blocks more efficiently than the naturally occurring enzymes, and both have a tyrosine at position 307 like the native G-block-forming enzymes.
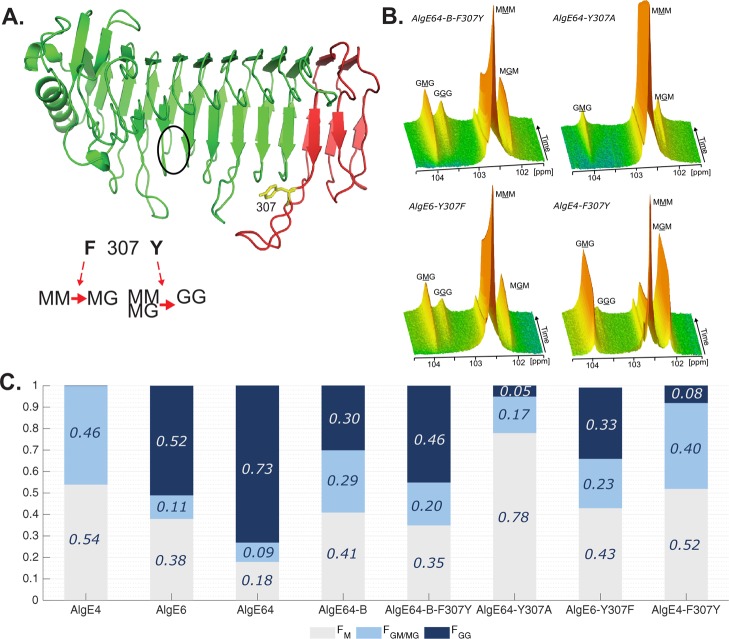
The point-mutant AlgE64-B-F307Y was therefore engineered to gain a better understanding of the effect of this particular loop residue in modulating the catalytic activity. AlgE64-B-F307Y only differs from AlgE64-B in the single residue 307, containing a tyrosine instead of a phenylalanine (Figure 3A). This mutant displays a significantly different epimerization profile than the parent enzyme AlgE64-B after epimerizing poly-M (Figure 3B and C; Table S1). In fact, AlgE64-B-F307Y gives similar values of monads and dyads to the ones obtained after complete epimerization of poly-M by the hybrid enzyme AlgE64-B1. Both enzymes are effective G-block formers, in contrast to AlgE64-B, which preferentially forms MG-blocks. The residue at position 307 thus appears to be important in determining the predominant epimerization pattern.
Figure 3.
(A) Ribbon structure of AlgE6 A-module represented in green (belonging to AlgE6) and red (belonging to AlgE4) colors as in point mutant AlgE64-B-F307Y. The location of the active site is indicated with a black circle. Tyrosine 307 in yellow is the point mutation of AlgE64-B-F307Y. Tyrosine 307 belongs to a loop in AlgE6 structure in proximity of the substrate binding groove. The model structure was obtained using SWISS-MODEL database.42 It is hypothesized that when residue 307 is a phenylalanine, the epimerases form an alternating (MG) block structure, whereas when it is a tyrosine they can form both MG and GG-blocks. (B) Time-resolved NMR spectra showing epimerization of 13C-labeled poly-M with AlgE64-B-F307Y, AlgE64-Y307A, AlgE6-Y307F, and AlgE4-F307Y. The position of the triads in the spectra is indicated, and the M or G moiety generating the signal is underlined. (C) Product composition at complete epimerization for the four enzymes from B and for AlgE4, AlgE6, AlgE64, and AlgE64-B, calculated from 1H NMR spectra. M residues are shown in gray bars, GM/MG dyads in light blue bars, and GG-dyads in dark blue bars. The y-axis denotes the fraction of each monad and dyad, whereas the seven different enzymes are listed on the x-axis. These epimerization patterns are also presented in Table S1.
To further confirm the role of the aromatic residue 307 in native epimerases, two additional mutants were generated. AlgE6-Y307F is the point mutant of AlgE6 (G-block former) where tyrosine 307 is mutated to phenylalanine, whereas AlgE4-F307Y is AlgE4 (a strict MG-block former unable to epimerize an M neighboring a G) with substitution of phenylalanine 307 to tyrosine. Figure 3B,C shows the results from poly-M epimerized by AlgE6-Y307F and AlgE4-F307Y. The epimerization activity, and particularly the G-block formation ability, of AlgE6 is considerably reduced when residue 307 is mutated (Figure 3C). AlgE4-F307Y gained G-block formation ability, albeit only to a minor extent (Figure 3C). Effects on the epimerization pattern are also found for a mutant of AlgE64 when substituting tyrosine 307 to alanine. AlgE64-Y307A showed a significant decrease of G-content and G-blocks of epimerized poly-M (Figure 3B,C, Table S1). In a predominantly MG-block-forming enzyme, an F307Y mutation increases the G-block-content, whereas a Y307F mutation in a G-block former results in the opposite. In addition, the large decrease in G-content created by AlgE64-Y307A compared to AlgE64 supports the hypothesis that residue 307 is directly involved in determining epimerization patterns.
Altogether, the data presented here strongly suggest that the loop and specifically the aromatic residue 307 play an important role in substrate-binding and affects the epimerization pattern of the epimerases. We speculate that the loop has a key role in G-block formation in AlgE64 and all the mutants in this study. As the loop is about five to six subsites away from the active site,21 we hypothesize an implication in substrate binding by enfolding the substrate-binding groove and clamping the substrate. Phenylalanine can only establish hydrophobic interactions with the alginate chain, via the CH-face of the alginate monomers. We believe that the loop in MG-block formers allows the alginate chain to be epimerized in a processive way every second M residue, sliding smoothly through the active site. For G-block-forming enzymes, we presume a tighter interaction where the tyrosine sidechain establishes both hydrogen bonds and hydrophobic interactions with the sugar rings. This strengthened interaction between the epimerase and alginate could allow epimerization of the initial product, poly-MG, creating G-blocks.
Flexible Loops Promote Processivity in AlgE6
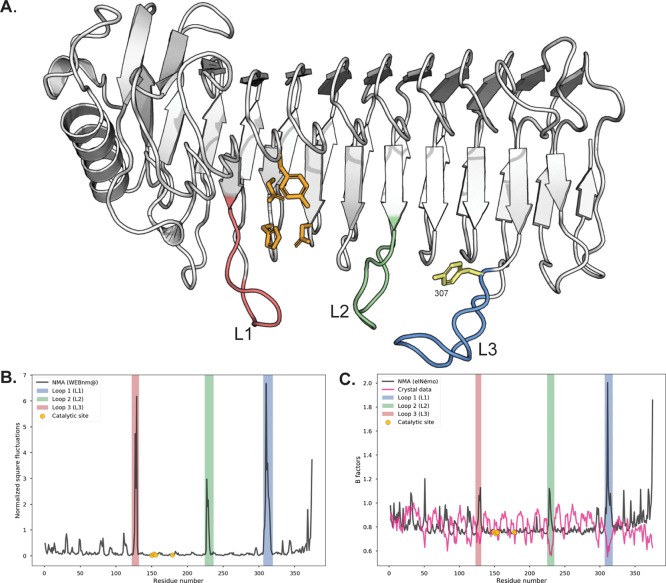
For this hypothesis to hold, the loop in question needs to be able to move quite flexibly. Recent findings on alginate lyases from the marine bacterium Zobellia galactanivorans reveal distinct topologies of the active sites involving several loops.47 In particular, the endolytic lyase AlyA1 presents an open cleft with three loops that possibly move to form a tunnel upon substrate binding. This results in “processive” catalysis where the lyase slides along the alginate chain while depolymerizing it. Apart from the loop harboring tyrosine 307, two other loops along the substrate binding groove are also present in the AlgE6 structure (see Figure 4A), similar to AlyA1. In the following, these loops will be labeled as L1 (loop 1, residues 123–131), L2 (loop 2, residues 225–235), and L3 (loop 3, residues 307–318 with tyrosine 307). To test if these loops are flexible enough to clamp the substrate in the groove in AlgE6, the flexibility of AlgE6 was investigated with normal mode analysis (NMA).38,39,55
Figure 4.
(A) Loops present along the substrate binding groove of the A-module of AlgE6. Loop 1 (L1, salmon) is defined by residues 123–131, loop 2 (L2, green) by residues 225–235, and loop 3 (L3, blue) by residues 307–318 (which includes tyrosine 307). The catalytic residues tyrosine 149, aspartate 152, histidine 154, and aspartate 178 are shown in orange sticks and the loop residue tyrosine 307 is shown in yellow sticks. Made with PyMOL. (B) Normalized square fluctuation for the C-α atoms of the residues, using the model structure. The fluctuations were obtained by the WEBnm@server as described in the text. The regions corresponding to the catalytic site and the investigated loops L1–L3 are highlighted in the figure. (C) Computed B factors by the elNémo server and the original B factors present in the model. The regions corresponding to the catalytic site and the investigated loops L1–L3 are highlighted in the figure.
In Figure 4B, we show the normalized square fluctuation for each C-α atom calculated using the 200 lowest (nontrivial) modes. The results in Figure 4B show that the largest fluctuations in the model structure are associated with residues in the loops. In particular, the residues in L1 and L3 demonstrate the potential flexibility of these loops. The lowest frequency modes found by an NMA indicate the most mobile parts and the direction of movements.56 Further visual inspection of the displacements associated with the lowest frequency modes show that these motions involve the movement of L3 (and L1) toward the substrate-binding groove, see Figures S3–S5 and Movies S1–S4 (available on-line). This supports the idea that these loops may be important for the binding of a substrate. The determined Debye–Waller factors, or B-factors, indicate how flexible different parts of a crystal structure are. The B-factors in the model do not indicate large fluctuations associated with the loops. However, this may be due to the crystallographic environment assumed in this case. To investigate this point further, we have recalculated the B-factors with an additional NMA and compare these B-factors to the B-factors assumed in the model in Figure 4C. Again, the NMA finds relatively large fluctuations associated with the loops. L3 has the largest fluctuations, indicating that it is flexible.
These findings are consistent with the hypothesis that the L3 loop containing tyrosine 307 has an important role in the enzyme function. A sequence alignment of the three loops in nine wild-type AlgE A-modules and four mutants shows that the amino acids in the loops are either highly conserved or bear low sequence identity. The only amino acid consistently different between G-block formers and MG-block formers is 307 (Figure S2). L1 and L2 could have similar roles of substrate clamping, but it is also possible that their flexibility promotes other, as yet unknown, functions.
Conclusions
As AlgE64 is an effective G-block forming epimerase, we made new hybrid AlgE64-descending epimerases differing in the transition regions between the A- and R-modules. The initial aim of explaining the improved G-block forming ability gave us insight into the molecular mechanism responsible for determining epimerization patterns. The last part of the A-module has a strong impact on the epimerization pattern, even though it is located far from the catalytic site. Loops protruding from the β-helix core of the A-module were suggested to be responsible for the variation in epimerization patterns, by forming a tunnel-like cleft embracing the alginate substrate. More specifically, amino acid 307 in loop L3 influenced the epimerization activity and the product profile. Finally, an NMA of the model A-module of AlgE6 supports the notion that flexible loops could interact with the substrate and influence the activity.
Acknowledgments
In memory of Professor Svein Valla, NTNU, who passed away during this work. Gerd Inger Sætrom and Randi Aune are gratefully acknowledged for technical assistance. MARPOL and FriPro Alginate Epimerase projects funded by the Research Council of Norway through grant 221576 and 250875.
Glossary
Abbreviations
- DPn
degree of polymerization
- ENM
elastic network model
- G
α-l-guluronic acid
- M
β-d-mannuronic acid
- NMA
normal mode analysis
- poly-M
polymannuronic acid, high-molecular-weight mannuronan
- RTB
rotation translation-block
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.9b04490.
NMR data of end-point epimerized alginate samples; NMR spectra at different time points; multiple sequence alignment of loops; traces of C-α′s from normal mode analysis and gene sequences of mutants (PDF)
Motions of displacements of loops associated with the lowest frequency modes of the normal mode analysis—mode7 (AVI)
Motions of displacements of loops associated with the lowest frequency modes of the normal mode analysis—mode7 (AVI)
Motions of displacements of loops associated with the lowest frequency modes of the normal mode analysis—mode8 (AVI)
Motions of displacements of loops associated with the lowest frequency modes of the normal mode analysis—mode9 (AVI)
Author Contributions
The paper was written through contributions of all the authors. All the authors have given approval to the final version of the paper. A.S., A.T., G.S.-B., H.S., and F.L.A. designed the research; A.S., A.T., A.L., and F.L.A. performed the research; A.S., A.T., M.G., A.L., G.S.-B., H.S. and F.L.A. analyzed the data; A.S., A.T., M.G., A.L., H.S., G.S.-B. and F.L.A. wrote the paper; and A.T., G.S.-B., H.S. and F.L.A. wrote the grant applications.
The authors declare no competing financial interest.
Supplementary Material
References
- Painter T. J.4 - Algal Polysaccharides; Aspinall G. O., Ed.; Academic Press: New York, 1983. [Google Scholar]
- Gorin P. A. J.; Spencer J. F. T. Exocellular Alginic Acid from Azotobacter Vinelandii. Can. J. Chem. 1966, 44, 993–998. 10.1139/v66-147. [DOI] [Google Scholar]
- Linker A.; Jones R. S. A New Polysaccharide Resembling Alginic Acid Isolated from Pseudomonads. J. Biol. Chem. 1966, 241, 3845–3851. [PubMed] [Google Scholar]
- Govan J. R. W.; Fyfe J. A. M.; Jarman T. R. Isolation of Alginate-Producing Mutants of Pseudomonas Fluorescens, Pseudomonas Putida and Pseudomonas Mendocina. Microbiology 1981, 125, 217–220. 10.1099/00221287-125-1-217. [DOI] [PubMed] [Google Scholar]
- Draget K. I.; Smidsrød O.; Skjåk-Bræk G.. Alginates from Algae. Biopolymers Online; Wiley-VCH Verlag GmbH & Co. KGaA, 2005; pp 215–224. [Google Scholar]
- Haug A.; Larsen B.; Smidsrød O.. A Study on the Constitution of Alginic Acid by Partial Acid Hydrolysis. Proceedings of the Fifth International Seaweed Symposium, Halifax, August 25–28, 1965, 1966; 5, pp 271–277.
- Mørch Ý. A.; Donati I.; Strand B. L.; Skjåk-Bræk G. Effect of Ca2+, Ba2+, and Sr2+on Alginate Microbeads. Biomacromolecules 2006, 7, 1471–1480. 10.1021/bm060010d. [DOI] [PubMed] [Google Scholar]
- Grant G. T.; Morris E. R.; Rees D. A.; Smith P. J. C.; Thom D. Biological Interactions between Polysaccharides and Divalent Cations: The Egg-Box Model. FEBS Lett. 1973, 32, 195–198. 10.1016/0014-5793(73)80770-7. [DOI] [Google Scholar]
- Smidsrød O.; Draget K. I. Alginates: Chemistry and Physical Properties. Carbohydrates Eur. 1996, 14, 6–13. [Google Scholar]
- Haug A.; Larsen B.; Smidsrød O. Uronic Acid Sequence in Alginate from Different Sources. Carbohydr. Res. 1974, 32, 217–225. 10.1016/s0008-6215(00)82100-x. [DOI] [Google Scholar]
- Andresen I.-L.; Skipnes O.; Smidsrød O.; Østgaard K.; Hemmer P. C.. Some Biological Functions of Matrix Components in Benthic Algae in Relation to Their Chemistry and the Composition of Seawater. Cellulose Chemistry and Technology; ACS Symposium Series; American Chemical Society, 1977; Vol. 48, pp 24–361. [Google Scholar]
- Skjåk-Bræk G.; Donati I.; Paoletti S.. Alginate Hydrogels: Properties and Applications. Polysaccharide Hydrogels: Characterization and Biomedical Applications; Pan Stanford Publishing, 2016; pp 449–498. [Google Scholar]
- Skjåk-Bræk G.; Martinsen A.. Applications of Some Algal Polysaccharides in Biotechnology. Seaweed resources in Europe: Uses and Potential, John Wiley and Sons, 1991; pp 219–257. [Google Scholar]
- Rehm B. H.; Ertesvåg H.; Valla S. A New Azotobacter Vinelandii Mannuronan C-5-Epimerase Gene (AlgG) Is Part of an Alg Gene Cluster Physically Organized in a Manner Similar to That in Pseudomonas Aeruginosa. J. Bacteriol. 1996, 178, 5884–5889. 10.1128/jb.178.20.5884-5889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svanem B. I. G.; Skjåk-Bræk G.; Ertesvåg H.; Valla S. Cloning and Expression of Three NewAzotobacter vinelandii Genes Closely Related to a Previously Described Gene Family Encoding Mannuronan C-5-Epimerases. J. Bacteriol. 1999, 181, 68–77. 10.1128/jb.181.1.68-77.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertesvåg H.; Høidal H. K.; Hals I. K.; Rian A.; Doseth B.; Valla S. A Family of Modular Type Mannuronan C-5-Epimerase Genes Controls Alginate Structure in Azotobacter Vinelandii. Mol. Microbiol. 1995, 16, 719–731. 10.1111/j.1365-2958.1995.tb02433.x. [DOI] [PubMed] [Google Scholar]
- Svanem B. I. G.; Strand W. I.; Ertesvåg H.; Skjåk-Bræk G.; Hartmann M.; Barbeyron T.; Valla S. The Catalytic Activities of the BifunctionalAzotobacter vinelandiiMannuronan C-5-Epimerase and Alginate Lyase AlgE7 Probably Originate from the Same Active Site in the Enzyme. J. Biol. Chem. 2001, 276, 31542–31550. 10.1074/jbc.m102562200. [DOI] [PubMed] [Google Scholar]
- Ertesvåg H.; Valla S. The A Modules of the Azotobacter Vinelandii Mannuronan-C-5-Epimerase AlgE1 Are Sufficient for Both Epimerization and Binding of Ca2+. J. Bacteriol. 1999, 181, 3033–3038. 10.1128/jb.181.10.3033-3038.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertesvåg H.; Doseth B.; Larsen B.; Skjåk-Braek G.; Valla S. Cloning and Expression of an Azotobacter Vinelandii Mannuronan C-5-Epimerase Gene. J. Bacteriol. 1994, 176, 2846–2853. 10.1128/jb.176.10.2846-2853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchinger E.; Knudsen D. H.; Behrens M. A.; Pedersen J. S.; Aarstad O. A.; Tøndervik A.; Valla S.; Skjåk-Bræk G.; Wimmer R.; Aachmann F. L. Structural and Functional Characterization of the R-modules in Alginate C-5 Epimerases AlgE4 and AlgE6 fromAzotobacter vinelandii. J. Biol. Chem. 2014, 289, 31382–31396. 10.1074/jbc.m114.567008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tøndervik A.; Klinkenberg G.; Aachmann F. L.; Svanem B. I. G.; Ertesvåg H.; Ellingsen T. E.; Valla S.; Skjåk-Bræk G.; Sletta H. Mannuronan C-5 Epimerases Suited for Tailoring of Specific Alginate Structures Obtained by High-Throughput Screening of an Epimerase Mutant Library. Biomacromolecules 2013, 14, 2657–2666. 10.1021/bm4005194. [DOI] [PubMed] [Google Scholar]
- Hoidal H. K.; Svanem B. I. G.; Gimmestad M.; Valla S. Mannuronan C-5 epimerases and cellular differentiation of Azotobacter vinelandii. Environ. Microbiol. 2000, 2, 27–38. 10.1046/j.1462-2920.2000.00074.x. [DOI] [PubMed] [Google Scholar]
- Høidal H. K.; Ertesvåg H.; Skjåk-Bræk G.; Stokke B. T.; Valla S. The RecombinantAzotobacter vinelandiiMannuronan C-5-Epimerase AlgE4 Epimerizes Alginate by a Nonrandom Attack Mechanism. J. Biol. Chem. 1999, 274, 12316–12322. 10.1074/jbc.274.18.12316. [DOI] [PubMed] [Google Scholar]
- Hartmann M.; Holm O. B.; Johansen G. A. B.; Skjåk-Braek G.; Stokke B. T. Mode of action of recombinantAzotobacter vinelandiimannuronan C-5 epimerases AlgE2 and AlgE4. Biopolymers 2002, 63, 77–88. 10.1002/bip.10017. [DOI] [PubMed] [Google Scholar]
- Campa C.; Holtan S.; Nilsen N.; Bjerkan T. M.; Stokke B. T.; Skjåk-Bræk G. Biochemical Analysis of the Processive Mechanism for Epimerization of Alginate by Mannuronan C-5 Epimerase AlgE4. Biochem. J. 2004, 381, 155–164. 10.1042/BJ20031265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarstad O. A.; Stanisci A.; Sætrom G. I.; Tøndervik A.; Sletta H.; Aachmann F. L.; Skjåk-Bræk G. Biosynthesis and Function of Long Guluronic Acid-Blocks in Alginate Produced byAzotobacter vinelandii. Biomacromolecules 2019, 20, 1613–1622. 10.1021/acs.biomac.8b01796. [DOI] [PubMed] [Google Scholar]
- Rozeboom H. J.; Bjerkan T. M.; Kalk K. H.; Ertesvåg H.; Holtan S.; Aachmann F. L.; Valla S.; Dijkstra B. W. Structural and Mutational Characterization of the Catalytic A-module of the Mannuronan C-5-epimerase AlgE4 fromAzotobacter vinelandii. J. Biol. Chem. 2008, 283, 23819–23828. 10.1074/jbc.m804119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aachmann F. L.; Svanem B. I. G. r.; Valla S.; Petersen S. B.; Wimmer R. NMR Assignment of the R-Module from the Azotobacter Vinelandii Mannuronan C5-Epimerase AlgE4. J. Biomol. NMR 2005, 31, 259. 10.1007/s10858-004-7911-7. [DOI] [PubMed] [Google Scholar]
- Aachmann F. L.; Svanem B. I. G.; Güntert P.; Petersen S. B.; Valla S.; Wimmer R. NMR Structure of the R-module. J. Biol. Chem. 2006, 281, 7350–7356. 10.1074/jbc.m510069200. [DOI] [PubMed] [Google Scholar]
- Buchinger E.; Aachmann F. L.; Aranko A. S.; Valla S.; Skjåk-BraeK G.; Iwaï H.; Wimmer R. Use of Protein Trans-splicing to Produce Active and Segmentally 2H, 15N Labeled Mannuronan C5-epimerase AlgE4. Protein Sci. 2010, 19, 1534–1543. 10.1002/pro.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakke I.; Berg L.; Aune T. E. V.; Brautaset T.; Sletta H.; Tondervik A.; Valla S. Random Mutagenesis of the PM Promoter as a Powerful Strategy for Improvement of Recombinant-Gene Expression. Appl. Environ. Microbiol. 2009, 75, 2002–2011. 10.1128/AEM.02315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J.; Russell D. W.; David W.. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press, 2001. [Google Scholar]
- Sletta H.; Tondervik A.; Hakvag S.; Aune T. E. V.; Nedal A.; Aune R.; Evensen G.; Valla S.; Ellingsen T. E.; Brautaset T. The Presence of N-Terminal Secretion Signal Sequences Leads to Strong Stimulation of the Total Expression Levels of Three Tested Medically Important Proteins during High-Cell-Density Cultivations of Escherichia Coli. Appl. Environ. Microbiol. 2007, 73, 906–912. 10.1128/AEM.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimmestad M.; Sletta H.; Ertesvag H.; Bakkevig K.; Jain S.; Suh S.-j.; Skjak-Braek G.; Ellingsen T. E.; Ohman D. E.; Valla S. The Pseudomonas Fluorescens AlgG Protein, but Not Its Mannuronan C-5-Epimerase Activity, Is Needed for Alginate Polymer Formation. J. Bacteriol. 2003, 185, 3515–3523. 10.1128/jb.185.12.3515-3523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M.; Duun A. S.; Markussen S.; Grasdalen H.; Valla S.; Skjåk-Bræk G. Time-Resolved 1H and 13C NMR Spectroscopy for Detailed Analyses of the Azotobacter Vinelandii Mannuronan C-5 Epimerase Reaction. Biochim. Biophys. Acta - Gen. Subj. 2002, 1570, 104–112. 10.1016/S0304-4165(02)00195-2. [DOI] [PubMed] [Google Scholar]
- Grasdalen H.; Larsen B.; Smidsrød O. A p.m.r. study of the composition and sequence of uronate residues in alginates. Carbohydr. Res. 1979, 68, 23–31. 10.1016/S0008-6215(00)84051-3. [DOI] [Google Scholar]
- Grasdalen H. High-Field, 1H-n.m.r. Spectroscopy of Alginate: Sequential Structure and Linkage Conformations. Carbohydr. Res. 1983, 118, 255–260. 10.1016/0008-6215(83)88053-7. [DOI] [Google Scholar]
- Hollup S.; Salensminde G.; Reuter N. WEBnm@: A Web Application for Normal Mode Analyses of Proteins. BMC Bioinformatics 2005, 6, 52. 10.1186/1471-2105-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S. P.; Fuglebakk E.; Hollup S. M.; Skjærven L.; Cragnolini T.; Grindhaug S. H.; Tekle K. M.; Reuter N. WEBnm@ v2.0: Web Server and Services for Comparing Protein Flexibility. BMC Bioinformatics 2014, 15, 427. 10.1186/s12859-014-0427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Sanejouand Y.-H. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004, 32, W610–W614. 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K.; Sanejouand Y.-H. On the Potential of Normal-Mode Analysis for Solving Difficult Molecular-Replacement Problems. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 796–799. 10.1107/S0907444904001982. [DOI] [PubMed] [Google Scholar]
- Biasini M.; Bienert S.; Waterhouse A.; Arnold K.; Studer G.; Schmidt T.; Kiefer F.; Cassarino T. G.; Bertoni M.; Bordoli L.; et al. SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 2014, 42, W252–W258. 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsen K.; Petrescu A.-J.; Dellerue S.; Bellissent-Funel M.-C.; Kneller G. R. Harmonicity in Slow Protein Dynamics. Chem. Phys. 2000, 261, 25–37. 10.1016/S0301-0104(00)00222-6. [DOI] [Google Scholar]
- Hinsen K. Analysis of Domain Motions by Approximate Normal Mode Calculations. Proteins Struct. Funct. Genet. 1998, 33, 417–429. . [DOI] [PubMed] [Google Scholar]
- Grasdalen H.; Larsen B.; Smisrod O. 13C-n.m.r. studies of monomeric composition and sequence in alginate. Carbohydr. Res. 1981, 89, 179–191. 10.1016/S0008-6215(00)85243-X. [DOI] [Google Scholar]
- Schrödinger L.The PyMOL Molecular Graphics System, Version 1.8, 2015.
- Thomas F.; Lundqvist L. C. E.; Jam M.; Jeudy A.; Barbeyron T.; Sandström C.; Michel G.; Czjzek M. Comparative Characterization of Two Marine Alginate Lyases fromZobellia galactanivoransReveals Distinct Modes of Action and Exquisite Adaptation to Their Natural Substrate. J. Biol. Chem. 2013, 288, 23021–23037. 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies G.; Henrissat B. Structures and Mechanisms of Glycosyl Hydrolases. Structure 1995, 3, 853–859. 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Varrot A.; Hastrup S.; Schülein M.; Davies G. J. Crystal structure of the catalytic core domain of the family 6 cellobiohydrolase II, Cel6A, from Humicola insolens, at 1.92 Å resolution. Biochem. J. 1999, 337, 297–304. 10.1042/BJ3370297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinen J.; Bergfors T.; Teeri T.; Knowles J. K. C.; Jones T. A. Three-Dimensional Structure of Cellobiohydrolase II from Trichoderma Reesei. Science 1990, 249, 1359. 10.1126/science.249.4975.1359-e. [DOI] [PubMed] [Google Scholar]
- Horn S. J.; Sorbotten A.; Synstad B.; Sikorski P.; Sorlie M.; Varum K. M.; Eijsink V. G. H. Endo/Exo Mechanism and Processivity of Family 18 Chitinases Produced by Serratia Marcescens. FEBS J. 2006, 273, 491–503. 10.1111/j.1742-4658.2005.05079.x. [DOI] [PubMed] [Google Scholar]
- Varrot A.; Frandsen T. P.; von Ossowski I.; Boyer V.; Cottaz S.; Driguez H.; Schülein M.; Davies G. J. Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens. Structure 2003, 11, 855–864. 10.1016/S0969-2126(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Zou J.-y.; Kleywegt G. J.; Ståhlberg J.; Driguez H.; Nerinckx W.; Claeyssens M.; Koivula A.; Teeri T. T.; Jones T. A. Crystallographic Evidence for Substrate Ring Distortion and Protein Conformational Changes during Catalysis in Cellobiohydrolase Ce16A from Trichoderma Reesei. Structure 1999, 7, 1035–1045. 10.1016/S0969-2126(99)80171-3. [DOI] [PubMed] [Google Scholar]
- Bjerkan T. M.; Lillehov B. E.; Strand W. I.; Skjåk-Bræk G.; Valla S.; Ertesvåg H. Construction and Analyses of Hybrid Azotobacter Vinelandii Mannuronan C-5 Epimerases with New Epimerization Pattern Characteristics. Biochem. J. 2004, 381, 813–821. 10.1042/bj20031580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykeman E. C.; Sankey O. F. Normal Mode Analysis and Applications in Biological Physics. J. Phys. Condens. Matter 2010, 22, 423202. 10.1088/0953-8984/22/42/423202. [DOI] [PubMed] [Google Scholar]
- Alexandrov V.; Lehnert U.; Echols N.; Milburn D.; Engelman D.; Gerstein M. Normal Modes for Predicting Protein Motions: A Comprehensive Database Assessment and Associated Web Tool. Protein Sci. 2005, 14, 633–643. 10.1110/ps.04882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.