Abstract
The long pre-cancerous state of colorectal cancer (CRC) provides an opportunity to prevent the occurrence and development of CRC. The detoxification of CRC food-borne carcinogenic heterocyclic amines is highly dependent on UDP glucuronosyltransferase 1A (UGT1A)-mediated glucuronidation. Sulforaphane (SFN), a phytochemical, possesses antioxidant, anti-inflammatory and anticarcinogenic effects on the prevention of CRC. Previous studies revealed that SFN upregulates the expression of UGT1A. The aim of the present study was to investigate the regulatory mechanism of SFN-induced UGT1A upregulation and provide novel understanding on the basic research and chemoprevention of CRC. In the present study, the viability and proliferation of CRC cells (HT-29 and SW480) treated with SFN were assessed by MTT, colony formation and EdU assays. Flow cytometry was used to detect the cell cycle arrest and apoptosis of cells treated with different concentrations of SFN. The motility of cells was determined by wound healing and Transwell assays. Nuclear factor, erythroid 2 like 2 (Nrf2) short hairpin RNA (shRNA) and negative control shRNA lentiviruses were used for cell transfection. Reverse transcription-quantitative polymerase chain reaction and western blotting were employed to verify the role of Nrf2 in SFN-induced UGT1A. HT-29 and SW480 cells were divided into a control, an SFN and a PD98059 [an extracellular signal-regulated kinase (ERK) inhibitor] + SFN group. Western blotting detected the protein levels of Nrf2 and UGT1A. Intracellular levels of reactive oxygen species (ROS) were detected using a reactive oxygen assay kit. The results revealed that SFN inhibits cell proliferation and colony formation, promotes apoptosis, and reduces the migratory ability of CRC cells. The phosphorylation of ERK induced by SFN promoted Nrf2 accumulation. Furthermore, a significant increase in the levels of UGT1A was observed, which coincided with SFN-induced upregulation of Nrf2 levels in nuclear fractions. Pretreatment with PD58059 reversed the SFN-induced subcellular translocation of Nrf2 and the expression of UGT1A. In addition, SFN-induced high levels of ROS in CRC cells may be associated with the ERK signaling pathway. Collectively, these results indicated that SFN inhibited the proliferation of CRC cells and upregulated the expression of UGT1A in CRC cells via the ERK/Nrf2 signaling pathway.
Keywords: sulforaphane, colorectal cancer, chemoprevention, ERK, Nrf2, UGT1A
Introduction
According to Global cancer statistics 2018 (1), colorectal cancer (CRC) ranks third in terms of incidence and second in terms of mortality among all cancers types in males and females. Among females, 9.2% of mortalities associated with cancer are due to CRC. Furthermore, the top three causes of mortality due to gastrointestinal diseases in the USA are CRC, followed by pancreatic cancer and liver cancer (2). Numerous cancer types are affected by lifestyle and environmental factors, and the incidence rates of the majority of cancers can be decreased by reducing potential risk factors. CRC is a potentially preventable cancer, and approximately 26.7% of CRC cases can be avoided by reducing the risk factors (3–5). The transformation from normal mucosa to occult adenoma to adenocarcinoma is a complicated multi-step and multi-stage process. This long-term pre-cancerous state provides opportunities to intervene in the development and progression of CRC. Among the numerous risk factors for CRC, diet factors account for ~80% (6), which is mainly associated with a high-fat, high-protein and low-fiber diet. A high intake of red meat and processed meat is an important factor in the pathogenesis of CRC (7–9). Heterocyclic amines (HAs), produced by high-temperature cooking of meat are one of the carcinogens of CRC (6,10,11). A study involving 407,270 participants with an overall median follow-up of 13.8 years demonstrated that the highest quintile of HAs was associated with increased risk of CRC (12). These HAs may cause chromosomal translocations, instability of cancer-associated gene microsatellites, chain mutations and oncogene activation, leading to the occurrence of CRC (13).
The metabolism of HAs is mainly catalyzed by metabolism II phase enzymes, and the polymorphic variations in the detoxifying enzymes may modulate the rate of conversion of toxic or carcinogenic compounds in the epithelium lining the lumen of the gastrointestinal tract (14). UDP glucuronosyltransferase 1A (UGT1A) is an important member of the family of metabolism II phase enzymes and is considered as an important system of detoxification. UGT1A metabolizes HAs-DNA adducts through glucuronidation, and serves a role in gene protection, which may be of great value in cancer prevention and therapy (15). Therefore, inducing the overexpression of metabolism phase II enzymes may aid to protect cells from the toxicity of carcinogens and DNA damage caused by the formation of adducts. Our previous study revealed that UGT1A expression was reduced in adenocarcinoma tissues compared with normal colonic mucosa tissues, indicating that the expression of UGT1A is altered in the early stage of colonic malignant transformation, and UGTlA may be of great significance in preventing tumor formation (16).
Sulforaphane (SFN), a phytochemical and derivative of isothiocyanate, is rich in cruciferous plants, and possesses antioxidant, anti-inflammatory, anticarcinogenic and preventive effects on CRC (17–19). Our previous study demonstrated that SFN could upregulate the expression of UGT1A in CRC cells, and promote cell-cycle arrest and apoptosis in human CRC cells (20). Based on the aforementioned detoxifying capacity of UGT1A, it was hypothesized that UGT1A may be an important molecular target for SFN in the prevention of CRC.
There is a cis-regulated structure in the UGT1A gene promoter region termed anti-oxidant responsive element (ARE). Nrf2, one of the multiple transcription factors that binds to ARE, can be induced by external factors and is considered to serve a key role in activating the transcription of various antioxidant and detoxifying enzymes, which protect cells from external stress (21,22). Our previous research revealed that SFN can increase the mRNA expression and nuclear translocation of Nrf2 in CRC cells, indicating that SFN may induce UGT1A expression through the activation of Nrf2 (20). However, the specific mechanism by which SFN induces the expression of Nrf2 and promotes its nuclear translocation remains unclear. Detailed knowledge concerning the pharmacological mechanism of action of SFN may aid to explore strategies for CRC prevention.
The present study provided evidence that intranuclear Nrf2 is upregulated and the extracellular signal-regulated kinase (ERK) signaling pathway is activated by SFN, which induces the expression of UGT1A.
Materials and methods
Cell culture and reagents
The human CRC cell lines HT-29 and SW480 (Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd.) were used in the present study and the cell lines were authenticated using STR profiles. HT-29 and SW480 cells were maintained in Dulbeccos modified Eagles medium (DMEM) (Gibco; Thermo Fisher Scientific, Inc.) and RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), respectively, supplemented with 10% fetal bovine serum (FBS) (Biological Industries) and antibiotics (100 U/ml penicillin and 0.1 mg/ml streptomycin) (EMD Millipore) under 5% CO2 in air at 37°C. The cell culture medium, including treatments, was changed every 48 h. All treatments and controls contained a final dimethyl sulfoxide (DMSO) concentration <0.1%. The anti-β-actin antibody (product no. 4970; 1:1,000), anti-phosphorylated-p38 (product no. 4511; 1:1,000) and anti-p38 (product no. 8690; 1:1,000) were obtained from Cell Signaling Technology, Inc., while the anti-Nrf2 (cat. no. ab62352; 1:1,000), anti-Lamin B1 (cat. no. ab133741; 1:1,000) and anti-phosphorylated-JNK (cat. no. ab124956; 1:1,000), anti-JNK (cat. no. ab179461; 1:1,000) anti-ERK (cat. no. ab184699; 1:10,000) antibodies were obtained from Abcam. The anti-phosphorylated-ERK (Thr202/Tyr204) antibody (clone S.812.9; cat. no. MA5-15173; 1:1,000) was obtained from Invitrogen (Thermo Fisher Scientific, Inc.). The anti-UGT1A antibody (cat. no. sc-271268) was obtained from Santa Cruz Biotechnology, Inc. The horseradish peroxidase (HRP)conjugated goat anti-rabbit immunoglobulin G (IgG) (cat. no. D110058; 1:5,000) and cyanine-3 (Cy3)-conjugated goat anti-rabbit IgG (cat. no. D111107; 1:100) antibodies were obtained from Sangon Biotech Shanghai, Co., Ltd. The peroxidaseconjugated goat anti-mouse IgG (cat. no. ZB2305; 1:2,500) was obtained from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. SFN (≥90% purity; product no. S4441; Fig. 1A) and the ERK inhibitor PD98059 (≥98% purity; product no. P215) were obtained from Sigma-Aldrich (Merck KGaA). All protocols of the manufacturers were followed.
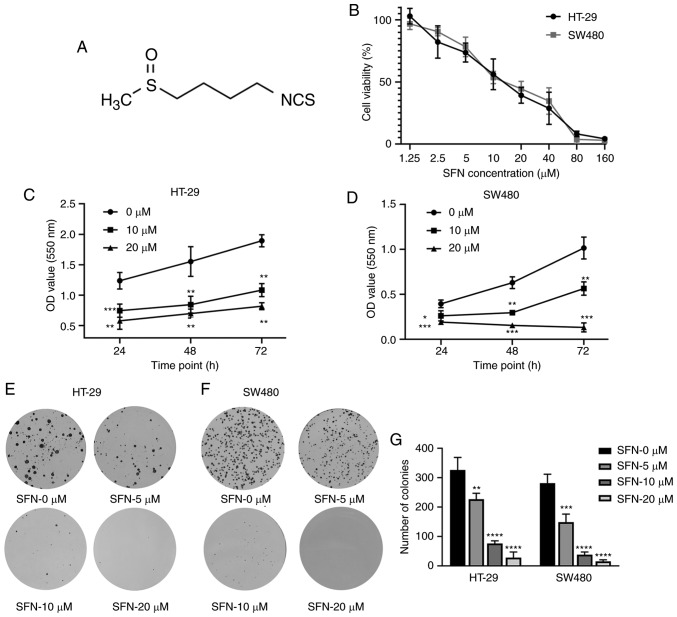
Figure 1.
SFN-induced cytotoxicity in colon cancer cells. (A) Structure of SFN, also known as 1-isothiocyanato-4-(methylsulfinyl)-butane (CAS, 4478-93-7; molecular formula, C6H11NOS2). (B) HT-29 and SW480 cells were incubated with 0, 1.25, 2.5, 5, 10, 20, 40, 80 and 160 µM SFN for 24 h. (C and D) An MTT assay was used to detect cell viability after treatment with various concentrations of SFN or for different time-points (24, 48 and 72 h). The statistical significance of the results was analyzed by two-way ANOVA. (E and F) Colon cancer cells (HT-29 and SW480) were cultured with various concentrations of SFN (0, 5, 10 and 20 µM). A representative image of colony formation from 3 independent experiments is presented. (G) The number of colonies of various groups presented in the graphs of parts E and F were quantified. The statistical significance of the results was analyzed by one-way ANOVA. The results are presented as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. SFN, sulforaphane; ANOVA, analysis of variance.
MTT assay
The effect of SFN on HT-29 and SW480 cell viability was determined by MTT assay. Briefly, cells were seeded at a density of 5×103 cells in 96-well plates and treated with various concentrations of SFN (1.25, 2.5, 5, 10, 20, 40, 80 and 160 µM) or blank medium for specific time-points (24, 48 and 72 h). Subsequently, 10 µl MTT reagent (5 mg/ml) was added to each well of the plates and the cells were incubated for 4 h at 37°C. Next, the medium was discarded and 100 µl DMSO was added. Upon agitation at room temperature for 10 min, the absorbance was measured at 550 nm using an Infinite M200 PRO Microplate Reader (Tecan Group Ltd.).
Colony formation assay
HT-29 and SW480 cells were inoculated into 6-well plates at a density of 700 cells/well and treated with various concentrations of SFN (0, 5, 10 and 20 µM) for 14 days. Finally, the cells were fixed with methanol at room temperature for 30 min, washed with PBS and stained with 0.1% crystal violet staining solution (Beyotime Institute of Biotechnology) at room temperature for 10 min. Upon washing the cells with PBS, the clusters (≥50 cells) were photographed by a NIKON camera and their number was counted.
EdU assay
HT-29 and SW480 cells were inoculated into 96-well plates at a density of 5×103 cells/well and treated with different concentrations of SFN (0, 10 and 20 µM) for 24 h. An EdU detection kit (cat. no. C10310-1; Guangzhou RiboBio Co., Ltd) was used to observe the groups. Cells were incubated with 100 µl medium and 50 µM EdU for 2 h, and then the medium was discarded. Cells were washed twice with PBS. The cells were fixed with methanol for 30 min at room temperature and then washed with PBS. Samples were incubated for 10 min with PBS containing 0.5% Triton X-100 (Beijing Solarbio Science & Technology Co., Ltd.) in a decolorization shaker and then washed with PBS. Cells were incubated with 100 µl 1X Apollo staining reaction solution for 30 min on a decolorization shaker in the dark, and then the staining reaction solution was discarded. Cells were washed twice with PBS containing 0.5% Triton X-100 for 10 min. The nucleuses were stained with 1X Hoechst 33342 reaction solution at room temperature for 30 min, prior to observation and capture of images with a fluorescence microscope (magnifications, ×100 and ×200).
Analysis of apoptosis and cell cycle arrest
HT-29 and SW480 cells were cultured in 60-mm dishes at a density of 60×104 cells/dish and treated with various concentrations (0, 10, 15 and 20 µM) of SFN for 24 h. An Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (cat. no. 556547; BD Biosciences) was used to observe the different states of the cells. Cells were washed twice with cold PBS and then resuspended in 1X Binding Buffer. Next, 5 µl Annexin V-FITC conjugate and 5 µl PI solution were added to the cell suspension, followed by incubation for 15 min at room temperature in the dark. Subsequently, 400 µl 1X Binding Buffer was added to each tube. The samples were analyzed by flow cytometry (BD Biosciences) within 1 h.
Cell cycle arrest was also detected by flow cytometry. The cells were collected and fixed with 75% ethanol at 4°C for 12 h, followed by flow cytometric analysis on a flow cytometer after PI staining at 4°C for 30 min.
Wound healing assay
Wound healing assays were carried out to evaluate the motility and metastatic potential of CRC cells. Cells were seeded in 6-well plates at a density of 6×105 cells/well. After 24 h, the cell monolayer was scratched with a 10-µl pipette tip. The wound healing assay was performed in monolayer culture, and then the cells were cultured in serum-free medium. Wounds were washed with PBS and incubated in serum-free medium in the presence of various concentrations of SFN (0, 10 and 20 µM). Images were captured at different time-points (0, 24 and 48 h). The wounded area was calculated using ImageJ software (V1.8.0.112; National Institutes of Health).
Transwell assay
Transwell assays were performed in Transwell chambers (pore size, 8.0 µm; Costar; Corning, Inc.). HT-29 and SW480 cells were cultured in medium containing various concentrations of SFN (0, 10 and 20 µM) for 2 days. Cells (2×105) were allowed to migrate from the upper chamber containing medium without FBS to the lower chambers containing medium with 30% FBS. The migrating cells were fixed with methanol at room temperature after 48 h of incubation and stained with 0.1% crystal violet at room temperature for 10 min. The cells that had migrated through the polycarbonate membrane were counted under a light microscope (5 random fields/well).
Lentivirus-mediated RNA interference
Lentiviral particles and Polybrene were purchased from Shanghai GenePharma Co., Ltd. The short hairpin RNA (shRNA) target sequence was 5′-GGGAGGAGCTATTATCCATTC-3′, and the negative control (NC) sequence was 5′-GTTCTCCGAACGTGTCACGT-3′. Nrf2 shRNA and NC shRNA were cloned into vectors containing the green fluorescent protein (GFP) gene. Then, the plasmid vectors were packed into lentiviral particles. Cells were seeded in 6-well plates at a density of 1×105 cells/well. Lentiviral particles with a multiplicity of infection of 80 were added to the cells. The GFP-expressing cells were detected using fluorescence microscopy, and puromycin was used for 7 days to screen stable-transfected cells with resistance to puromycin.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers protocol. RT reactions were carried out with 3 µg RNA, oligo-dT primer and M-MLV Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) in a volume of 20 µl with the Eppendorf Mastercycler nexus PCR instrument. The presence of Nrf2 and UGT1A transcripts was analyzed by qPCR, based on general fluorescence detection with SYBR-Green (Takara Bio, Inc.). β-actin was used as an internal control for normalization of the differences in RNA quantity and quality across samples. The qPCR conditions were 30 sec at 95°C followed by 40 cycles of 5 sec at 95°C, 10 sec at 60°C and 30 sec at 72°C. The data were quantified using the 2−ΔΔCq method (23), ΔΔCq=(Cq of target gene-Cq of β-actin) treatment-(Cq of target gene-Cq of β-actin)control. The gene-specific primers used were derived from PrimerBank and are summarized in Table I. All primers were synthesized by BioSune following sequence alignment with Basic Local Alignment Search Tool (National Center for Biotechnology Information).
Table I.
Primer sequences for RT-qPCR.
| Gene name | Primer sequence (5′→3′) |
|---|---|
| Nrf2 | Forward: TTCCCGGTCACATCGAGAG |
| Reverse: TCCTGTTGCATACCGTCTAAATC | |
| UGT1A | Forward: TCGAATCTTGCGAACAACACG |
| Reverse: ATGAAGGCCACTGTCAGCACG | |
| β-actin | Forward: CATGTACGTTGCTATCCAGGC |
| Reverse: CTCCTTAATGTCACGCACGAT |
UGT1A, UDP glucuronosyltransferase 1A; Nrf2, nuclear factor, erythroid 2 like 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Separation of the nucleus from cytoplasm
A Nucleoprotein extraction kit (BB-3102, BestBio Science) was used in the separation of the nucleus from cytoplasm. The cells were washed three times with cold PBS and collected into 1.5 ml-Eppendorf tubes using a cell scraper. Cells were centrifuged at 500 × g for 3 min at 4°C and then the supernatants were discarded. Cell lysates were added to the precipitates. The Eppendorf tubes were then vortexed for 15 sec every 5 min, three times in total. Subsequently, the suspensions were centrifuged for 5 min at 12,000 × g and the supernatants contained the cytoplasmic proteins. The nuclear lysates were added to the precipitates. After vortexing for 15 sec every 10 min, four times in total, the suspensions were centrifuged for 10 min at 12,000 × g and the supernatants contained the cytoplasmic proteins.
Immunocytochemistry
Cells were seeded into 24-well cell culture plates (6×104 cells/well) and divided into three groups: A control group, an SFN group and a PD98059 + SFN group. PD98059 was added to the medium 60 min prior to SFN addition, and cells were incubated for 24 h in the PD98059 + SFN group. Ultimately, the cells were fixed with methanol (chilled at −20°C) for 15 min and then washed three times with cold PBS. Samples were incubated for 30 min with PBS containing 0.2% Triton X-100 and then washed three times with cold PBS. Cells were next incubated in PBS containing 3% bovine serum albumin for 30 min at room temperature and washed three times with PBS. Then, cells were incubated with an anti-Nrf2 primary antibody (1:400) overnight at 4°C. The next day, the solution was removed and the cells were washed three times in PBS. Cells were then incubated with a Cy3-conjugated goat anti-rabbit IgG for 1 h at room temperature in the dark, prior to observation and capture of images with a fluorescence microscope (magnification, ×400).
Western blotting
Protein were isolated using the protein extraction kit (R0010; Beijing Solarbio Science & Technology Co., Ltd.). Protein extracts were heated at 99°C for 5 min and cooled in ice. The protein concentration was detected using a BCA assay kit (Tiangen Biotech Co., Ltd.). A total of 30 µg protein of each sample was loaded and separated on a 10% SDS polyacrylamide gel and electrophoretically transferred onto a polyvinylidene fluoride membrane. The membrane was blocked with 5% fat-free milk in TBS-Tween-20 (TBST) for 2 h and incubated with a specific primary antibody in Primary Antibody Diluent overnight at 4°C. The blots were washed three times for 5 min with TBST and then incubated with a secondary antibody for 1.5 h. After washing the membrane three times for 5 min with TBST, antibody binding was detected by ImageQuant LAS 4000 system (GE Healthcare Life Sciences). The optical density results were calculated using ImageJ 1.52 software.
Measurement of intracellular ROS
Intracellular ROS levels were detected using a Reactive Oxygen assay kit (Beyotime Institute of Biotechnology). Dichloro-dihydro-fluorescein diacetate was added to the medium to a final concentration of 10 µM and incubated for 20 min in a 37°C incubator. ROS levels were observed directly and images were obtained with a fluorescence microscope (magnification, ×100).
Statistical analysis
GraphPad Prism 8.0.1 software (GraphPad Software, Inc.) was used for statistical analysis. The results are presented as the mean ± standard deviation of ≥3 independent experiments. The statistical significance of the results was analyzed by Students t-test, one-way analysis of variance (ANOVA) with Dunnetts post hoc test and two-way ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
SFN inhibits the proliferation and clone formation of CRC cells
To evaluate the anticancer effect of SFN on CRC cells, the CRC cell lines HT-29 and SW480 were treated with different concentrations of SFN. An MTT assay was used to determine cell viability under various concentrations of SFN (0, 1.25, 2.5, 5, 10, 20, 40, 80 and 160 µM), as well as different treatment durations with SFN (24, 48 and 72 h). As presented in Fig. 1B, SFN significantly suppressed the cell viability relative to the control group in a dose-dependent and time-dependent manner (Fig. 1B-D). Next, the role of SFN in the colony-forming assay was assessed. Compared with the control group, the number of colonies in the SFN groups was significantly reduced (Fig. 1E and G). Furthermore, EdU staining was used to detect the effect of SFN on the proliferation of CRC cells (Fig. 2A and B). The results of EdU assay revealed that the cell proliferation activity in the SFN groups significantly decreased compared with the control group. All these results indicated that SFN may inhibit the proliferation of CRC cells.
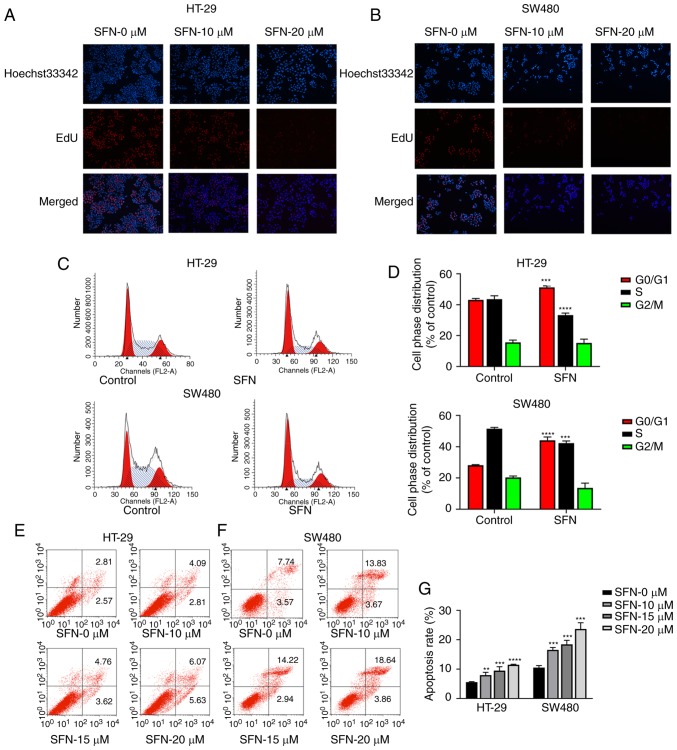
Figure 2.
SFN promotes the apoptosis of colon cancer cells and inhibits cell proliferation. (A and B) HT-29 and SW480 cells were treated with different concentrations of SFN in complete medium, and cell viability was assessed by EdU staining at 24 h. (C) HT-29 and SW480 cells were exposed to medium containing SFN for 24 h, and cell cycle arrest was detected by flow cytometry. (D) The cell cycle arrest of various groups was quantified. The statistical significance of the results was analyzed by Student's t-test. (E and F) HT-29 and SW480 cells were exposed to various conditions for 24 h, and cell apoptosis was detected by flow cytometry. (G) The cell apoptosis of various groups presented in the graphs of parts E and F was quantified. The statistical significance of the results was analyzed by two-way analysis of variance. The results are presented as the mean ± standard deviation of 3 independent experiments. **P<0.01, ***P<0.001 and ****P<0.0001. SFN, sulforaphane; EdU, 5-Ethynyl-2′- deoxyuridine.
SFN induces G0/M1-phase arrest and the apoptosis of CRC cells
To confirm the effect of SFN in cell cycle and apoptosis of HT-29 and SW480 cells, cells were treated with various concentrations (0, 10, 15 and 20 µM) of SFN for 24 h. Cells were analyzed by flow cytometry, and the results are presented in Fig. 2C-G. The results indicated that SFN could induce the G0/G1-phase arrest. Following treatment with SFN (Fig. 2C and D), there was a higher percentage of cells in the G0/G1 phase (HT-29: 51.20±0.96; SW480: 44.09±2.11%) compared with the control group (HT-29: 43.09±0.91; SW480: 28.14±0.37%). The accumulation of cells in the G0/G1 phase was accompanied by a decrease in the percentage of cells in the S phase (HT-29: 43.59±2.19 (control), 33.24±1.30 (SFN); SW480: 51.54±0.77 (control), 42.26±1.43% (SFN). The apoptosis rates (Fig. 2E and G) in HT-29 cells were 5.56±0.16, 7.97±0.93, 9.49±1.34 and 11.49±0.182% under treatment of SFN in 0, 10,15 20 µM, respectively. In addition, the apoptosis rates in SW480 cells were 10.50±0.70, 16.56±0.79, 18.43±1.38 and 23.67±2.12%. SFN could induce G0/G1 phase arrest and apoptosis in colorectal cancer.
SFN inhibits the motility of CRC cells
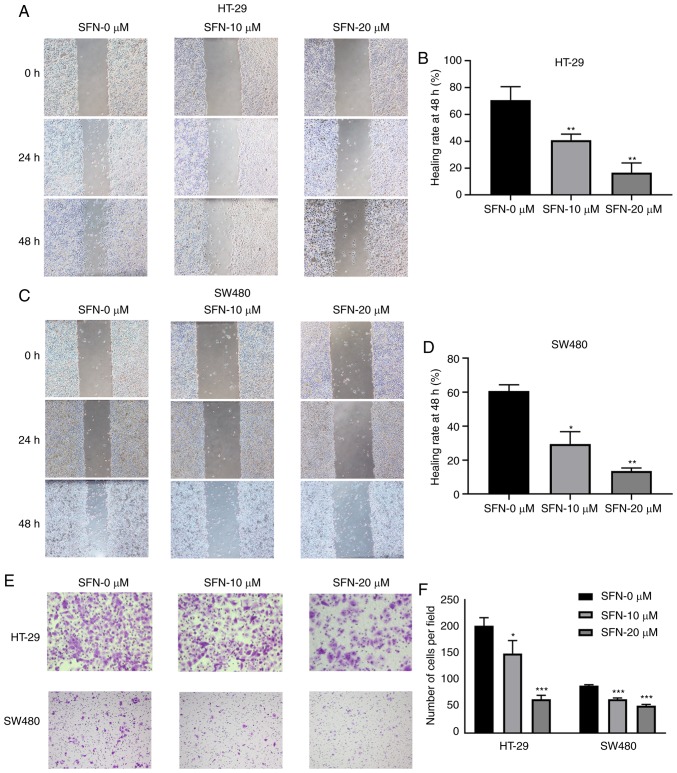
HT-29 and SW480 cells were treated in complete medium with or without various concentrations of SFN. Changes in the motility and migration capacity of the CRC cell lines HT-29 and SW480 were detected and analyzed by wound healing and Transwell assays. Decreased cell motility was observed in HT-29 and SW480 cell lines treated with SFN (Fig. 3A-D). The 48-h wound healing rates in HT-29 cells were 60.67±3.68, 29.48±7.30 and 13.59±1.76% (for 0, 10, 20 µM SFN, respectively), while in SW480 cells they were 70.77±9.90, 40.86±4.52 and 16.69±7.15% (for 0, 10, 20 µM SFN, respectively). The migrating capacity of cells was decreased in the SFN treatment group compared with the control group (Fig. 3E and F). The number of HT-29 cells migrating into the lower chamber were 201.33±15.04, 149.33±24.09 and 63.33±7.57 (for 0, 10, 20 µM SFN, respectively). The number of SW480 cells were 89.00±1.73, 63.33±2.52 and 51.33±2.52 (for 0, 10, 20 µM SFN, respectively). The results demonstrated that the metastasis of HT-29 and SW480 cells was significantly inhibited when SFN was added.
Figure 3.
SFN intervention decreases wound healing rates and cell migration. (A and C) HT-29 and SW480 cells were treated under different conditions, and their cell migratory ability was assessed by wound healing assay (magnification, ×40). (B and D) The wound healing rates of various groups presented in the graphs of parts A and C were quantified. The statistical significance of the results was analyzed by two-way ANOVA. (E) A total of 10×104 HT-29 or SW480 cells in 200 µl serum-free Dulbecco's modified Eagle's medium with different concentrations of SFN were seeded on the upper chamber, and cell migration was assessed by Transwell assay. Representative images are presented (magnification, ×100 and ×200). (F) The number of cells per field of various groups of part E was quantified. The statistical significance of the results was analyzed by two-way ANOVA. The results are presented as the mean ± standard deviation of 3 independent experiments. *P<0.05, **P<0.01 and ***P<0.001. SFN, sulforaphane; ANOVA, analysis of variance.
Effect of SFN on Nrf2 and UGT1A expression in CRC cells
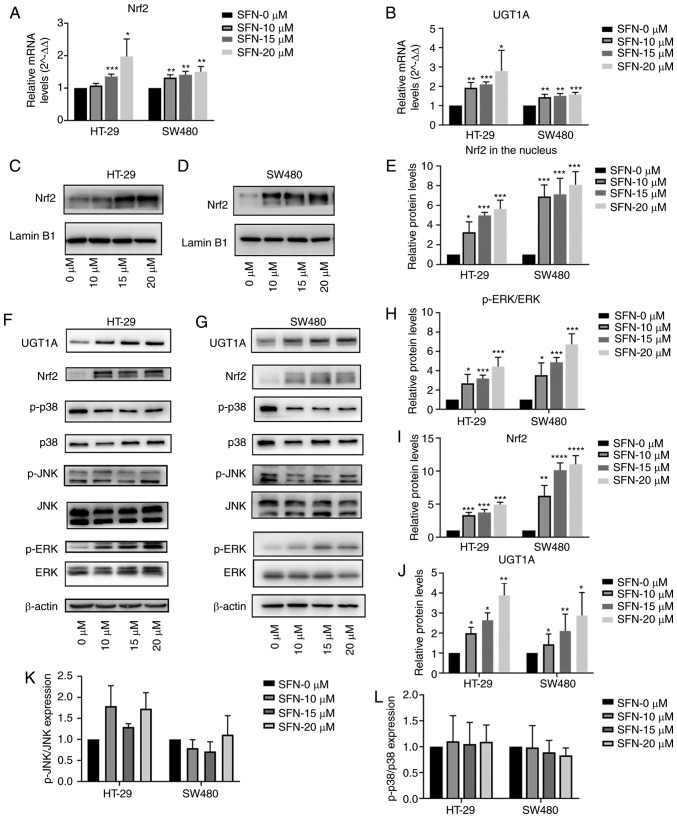
To assess the expression of Nrf2 regulated by SFN, CRC cells were treated with various concentrations (0, 10, 15 and 20 µM) of SFN for 48 h. As presented in Fig. 4A and B, treatment with SFN increased Nrf2 and UGT1A mRNA levels in a dose-dependent manner. Western blotting was used to detect the protein levels of Nrf2 and UGT1A with various concentrations of SFN. Fold-change values of Nrf2 expression in the nucleus (Fig. 4C and D) and total protein (Fig. 4F and G) are presented in Fig. 4E and I. In addition, significant increases in UGT1A expression (Fig. 4J) were observed in SFN treatment groups, which coincided with upregulation of Nrf2 levels in nuclear fractions.
Figure 4.
SFN upregulates Nrf2 and UGT1A in colon cancer cells. Cells were treated with different concentrations of SFN. (A and B) The transcriptional levels of Nrf2 and UGT1A were quantified by RT-qPCR and illustrated by a bar legend. (C-E) The Nrf2 expression level in the nucleus was assessed by immunoblotting and illustrated by a bar legend. (F-L) The immunoreactivity of p-ERK was normalized to that of total ERK. The immunoreactivity of Nrf2 and UGT1A, which was normalized to the expression of β-actin, and the immunoreactivity of p-JNK and p-p38 were normalized to that of total JNK and p38 was measured by immunoblotting and represented by respective bar graphs revealing the mean ± SD. Fold changes in optical density with the 0 h group normalized to 1. The statistical significance of the results was analyzed by two-way analysis of variance. The results are expressed as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. SFN, sulforaphane; Nrf2, nuclear factor, erythroid 2 like 2; UGT1A, UDP glucuronosyltransferase 1A; ERK, extracellular signal-regulated kinase; p-ERK, phosphorylated ERK; JNK, c-Jun NH2-terminal kinase; p-JNK, phosphorylated-JNK; p38, p38 kinase; SD, standard deviation; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
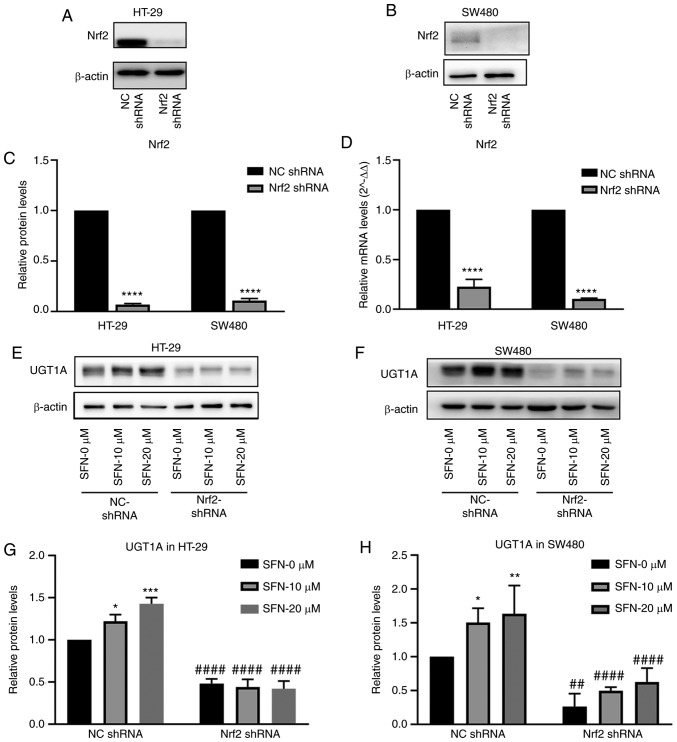
Nrf2 serves an important role in the SFN-induced upregulation of UGT1A expression
The expression of Nrf2 in the nucleus and UGT1A were increased upon treatment with SFN in the present study. Nrf2 is one of the multiple transcription factors that binds to ARE, which is a cis-regulated structure in the UGT1A gene promoter region (22). It was hypothesized that SFN upregulated UGT1A via Nrf2 binding to ARE. NC shRNA and Nrf2 shRNA were transfected into HT-29 and SW480 cells with lentiviruses, and stable-transfected cells were screened by puromycin and used for subsequent experiments. The levels of Nrf2 mRNA and protein expression (Fig. 5A-D) were detected by RT-qPCR and western blotting (P<0.0001). The NC shRNA and Nrf2 shRNA groups were cultured in complete medium containing 0, 10 and 20 µM SFN. Protein was collected after 48 h, and protein expression of UGT1A was detected by western blotting. The expression of UGT1A increased in the NC shRNA groups following treatment with SFN. However, the expression level of UGT1A decreased, and the induction effect of SFN on UGT1A expression disappeared upon inhibition of the expression of Nrf2 (Fig. 5E-H).
Figure 5.
Effect of Nrf2 on UGT1A expression induced by SFN. The Nrf2 gene was knocked down in SW480 and HT-29 cells. (A and B) The Nrf2 protein levels in HT-29 and SW480 cells were assessed by immunoblotting. (C) The expression levels of Nrf2 were quantified by ImageJ and illustrated by a bar legend. The statistical significance of the results was analyzed by Student's t-test. ****P<0.0001 vs. NC shRNA. (D) The transcriptional level of Nrf2 was quantified by RT-qPCR and illustrated by a bar legend. ****P<0.0001 vs. NC shRNA. The statistical significance of the results was analyzed by Student's t-test. (E and F) The expression levels of UGT1A in HT-29 and SW480 cells decreased in Nrf2-knockdown cells, and the induction effect of SFN on UGT1A expression decreased upon Nrf2-gene knockdown. (G and H) The UGT1A expression in various groups presented in the images of parts E and F was quantified by ImageJ. The statistical significance of the results was analyzed by two-way analysis of variance. *P<0.05, **P<0.01, ***P<0.001 vs. the SFN 0 µM group; ##P<0.01, #####P<0.0001 vs. the NC shRNA group. The results are expressed as the mean ± standard deviation of 3 independent experiments. SFN, sulforaphane; Nrf2, nuclear factor, erythroid 2 like 2; NC, negative control; shRNA, short hairpin RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
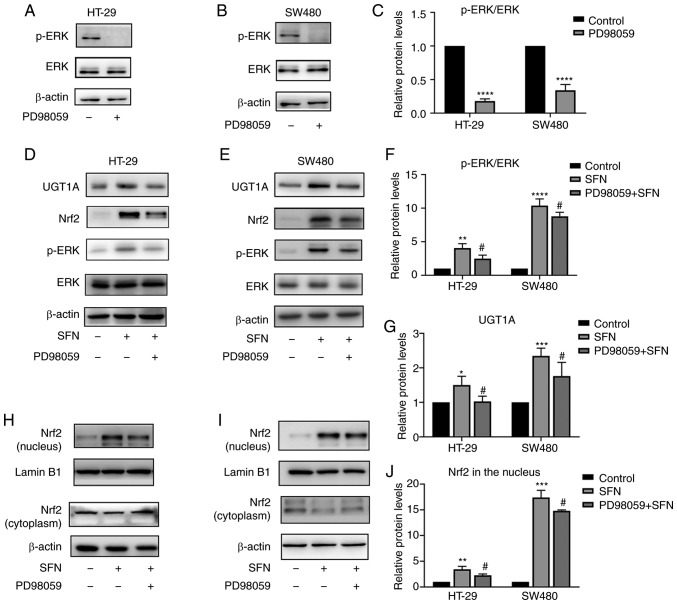
SFN promotes the expression of Nrf2 and UGT1A, and increases the levels of ROS through ERK
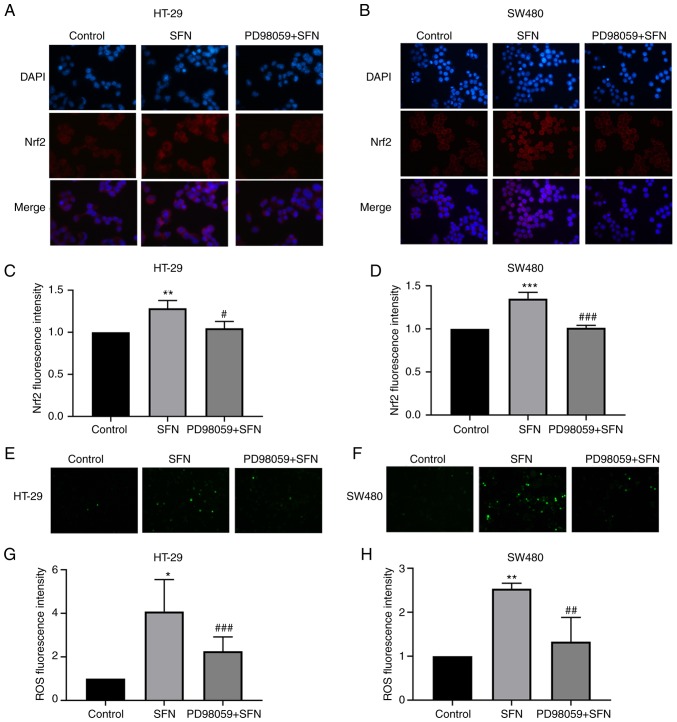
Notably, SFN treatment significantly increased the phosphorylation of ERK in a dose-dependent manner (Fig. 4F-H). In addition, the effect on JNK and p38 was not significantly altered (Fig. 4K and L). The results revealed that SFN may activate the ERK signaling pathway in a dose-dependent manner. PD98059 is a specific inhibitor of ERK and its inhibitory ability was verified in the present study (Fig. 6A-C). To confirm whether the SFN-modulated and Nrf2-induced UGT1A upregulation is associated with the ERK signaling pathway, cells were treated with PD98059 for 1 h to block the ERK pathway prior to treatment with SFN. Cells were divided into three groups: a control, an SFN and a PD98059 + SFN group. The control and SFN groups were treated with medium with and without SFN, respectively. The PD98059 + SFN group was pretreated with the ERK inhibitor PD98059 prior to being treated with SFN. Pretreatment with PD58059 reversed the nuclear accumulation of Nrf2 and the total protein levels of UGT1A (Figs. 6D-J and 7A-D). In addition to the aforementioned detection of the expression of Nrf2 and UGT1A, the levels of ROS were detected. The ERK inhibitor was able to inhibit the SFN-induced high levels of ROS in CRC cells (Fig. 7E-H).
Figure 6.
SFN induces UGT1A expression through ERK-dependent regulation of Nrf2. HT-29 and SW480 cells were pretreated with PD98059 (an ERK inhibitor) for 1 h. (A and B) p-ERK expression levels were assessed by immunoblotting. HT-29 and SW480 cells were pretreated with or without PD98059 for 1 h followed by 24 h of incubation with SFN. (C) The immunoreactivity of p-ERK was normalized to that of total ERK, and represented by a bar graph presenting the mean ± SD. Fold changes in optical density compared with the control group were normalized to 1. The statistical significance of the results was analyzed by Student's t-test. (D and E) UGT1A and Nrf2 expression levels of total protein were assessed by immunoblotting. (F and G) Bar graphs presenting the mean ± SD fold changes in OD (with the control group set as 1) of p-ERK normalized to ERK and UGT1A normalized to β-actin. The statistical significance of the results was analyzed by Student's t-test. (H and I) Nrf2 expression level in the nucleus and cytoplasm were assessed by immunoblotting. (J) Bar graph revealing the immunoreactivities (mean ± SD fold changes in OD, with control group set at 1) of Nrf2 normalized to Lamin B1 (marker for the nucleus). The statistical significance of the results was analyzed by Student's t-test. The results are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. the control; #P<0.05, vs. SFN. SFN, sulforaphane; Nrf2, nuclear factor, erythroid 2 like 2; UGT1A, UDP glucuronosyltransferase 1A; ERK, extracellular signal-regulated kinase; SD, standard deviation; OD, optical density; p-, phosphorylated.
Figure 7.
Nuclear translocation of Nrf2 and ROS levels observed by fluorescence microscopy. HT-29 and SW480 cells were pretreated with PD98059 (an ERK inhibitor) for 1 h. (A and B) Nuclear translocation of Nrf2 in HT-29 and SW480 cells. Fixed cells were incubated with anti-Nrf2 and fluorescein isothiocyanate-conjugated anti-rabbit immunoglobulin G antibodies (magnification, ×400). (C and D) Bar graphs revealing the fluorescence intensity (mean ± SD fold changes in OD, with control group set at 1) of Nrf2. (E and F) Intracellular ROS levels were observed by fluorescence microscopy (magnification, ×100). (G and H) Bar graphs revealing the fluorescence intensity (mean ± SD fold changes in OD, with control group set at 1) of ROS. The results are presented as the mean ± SD of 3 independent experiments. *P<0.05, **P<0.01, ***P<0.001 vs. the control; #P<0.05, ##P<0.01, ###P<0.001 vs. SFN. SFN, sulforaphane; Nrf2, nuclear factor, erythroid 2 like 2; SD, standard deviation.
Discussion
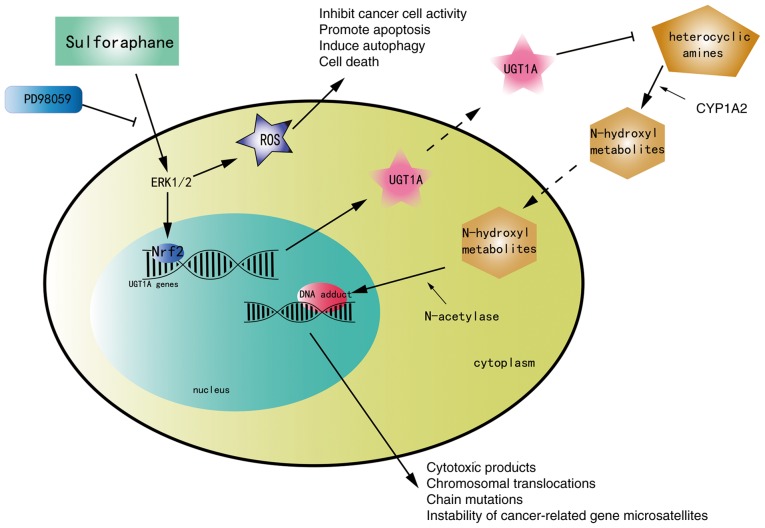
The roles of various phytochemicals in cancer prevention and treatment have recently attracted great attention. The present study investigated the antitumor effect and mechanism of SFN on the II phase metabolism phase enzyme UGT1A in colorectal cancer. UGT is one of the II phase metabolism phase enzymes. This supergene family has two subfamilies, UGT1A and UGT2, which can encode numerous isoenzymes. UGT1A encodes 9 isoenzymes, UGT1A1 and UGT1A3-UGT1A10, with different levels of expression in the intestinal tract, which are involved mainly in the metabolism of exogenous compounds. Upregulation of the human detoxifying enzyme UGT1A can metabolize HAs-DNA adducts through glucuronidation (24), which may aid to prevent the occurrence and development of CRC in advance. These HAs could be oxidized by cytochrome P450 family 1 subfamily A member 2 to N-hydroxyl metabolites along with the blood into the intestinal mucosa. Then, the N-hydroxyl metabolites could be acetylated by N-acetylase in intestinal epithelial cells and eventually bind to the DNA of intestinal epithelial cells to form DNA adducts. These adducts have been extensively studied and reported to serve a key role in chemically induced carcinogenesis (25), resulting in a series of cytotoxic products, including electrophilic groups and oxygen radicals. The aforementioned changes in DNA may cause chromosomal translocations, instability of cancer-associated gene microsatellites and chain mutations, leading to the occurrence of CRC (13). In phase II metabolism, with the introduction of polar groups, coupling enzymes usually add endogenous substituents, which greatly increase water solubility and facilitate excretion (25), thus increasing cell protection. Therefore, the rate of activation of HAs in tissues with reduced UGT1A activity is relatively high and tends to form DNA adducts compared with tissues with normal UGT1A activity. In the preliminary stages of cancer, the polymorphism of metabolic enzymes is key in determining the effects of environmental carcinogens. The high expression of UGT1A in the intestinal tract indicates that these enzymes may serve an important role in the metabolism of detoxification.
Our previous research revealed that the chemopreventive agent SFN upregulates the phase II metabolism enzyme UGT1A (20), which has a detoxifying effect on food-borne carcinogenic HAs. However, little is known concerning the signaling pathway that is involved in SFN-induced UGT1A expression. Consequently, the mechanism of action of SFN-induced UGT1A expression must be further explored. Several studies have revealed that the Nrf2 signaling pathway can be predictably induced by low concentrations of sulfhydryl-reactive molecules of various different chemical types, including SFN (26,27). In the present study, the expression of Nrf2 and UGT1A exhibited a synchronous increase following treatment of HT-29 and SW480 cells with SFN. It was observed that the trend was more obvious in HT-29 cells compared with in SW480 cells. Studies identified four consensus molecular subtypes (CMS) that expression subtypes have clinical relevance independent of the cancer stage. HT-29 belongs to CMS-3, and SW480 belongs to CMS-4. Previous studies have demonstrated that different subtypes of CRC cell lines have differences in RNA, DNA, and protein levels (28,29). Colon cell lines were either CMS2 or CMS3, expressed higher levels of gastro-intestinal marker genes, including some key transcription factors such as HNF4A and MYB. All CMS4 models were classified as undifferentiated, consistent with primary tumors. Concurrently, there are some differences in the sensitivity of different cell lines to the effects of drugs. This may explain the different expression levels of Nrf2 and UGT1A in the two cell lines examined in the present study. Next, the Nrf2 gene was knocked down to demonstrate that Nrf2 is crucial in the upregulation of UGT1A. The expression of UGT1A increased in the NC shRNA groups treated with SFN. However, the expression level of UGT1A decreased and the induction effect of SFN on UGT1A expression disappeared upon inhibition of the expression of the Nrf2 gene in the Nrf2 shRNA group. These results demonstrated that Nrf2 serves an integral role as a transcription factor in the transcriptional expression of UGT1A.
ERK is a member of the mitogen-activated protein kinase family. The ERK signaling pathway is known to be involved in numerous cell biological functions such as proliferation, migration, differentiation and death (30). A series of studies revealed that the regulation of Nrf2 may be associated with the activation of the ERK signaling pathway (31–33). In addition, a study observed that the expression of UGT1A was downregulated upon inhibition of the ERK signaling pathway with PD98059 (34). Subsequently, the present study verified the association between the ERK signaling pathway, Nrf2 and UGT1A through a series of experiments. Activation of ERK by phosphorylation was observed in the SFN group. To reveal the mechanism responsible for Nrf2 activation and to detect whether the activation of Nrf2 and the expression of UGT1A are associated with changes in the ERK signaling pathway, cells were pretreated with PD98059 to block the ERK signaling pathway, and then incubated in complete medium containing SFN. The present results revealed that SFN exhibited the potential to activate the ERK signaling pathway in a dose-dependent manner and to increase the protein expression of Nrf2 and UGT1A, while ERK1/2 inhibition impaired this trend. These results acknowledge that ERK serves an essential role in SFN-induced triggering of Nrf2-mediated UGT1A expression in HT-29 and SW480 cells, while inhibition of ERK attenuated the SFN-mediated upregulation of UGT1A transcription as well as the Nrf2 accumulation in nuclear compartments. The present study revealed for the first time that SFN upregulated the expression of UGT1A, which enhanced the metabolism of carcinogens via the ERK/Nrf2 signaling pathway in CRC cells (Fig. 8). However, blocking the ERK signaling pathway only partially reversed the upregulation of Nrf2 and UGT1A, and the pathway was not completely blocked. There may be other signaling pathways and multiple molecular regulatory mechanisms that may influence this process. It has been reported that SFN can activate Nrf2 via the PI3K/Akt pathway in arsenic-induced liver injury and nephrotoxicity (22,35). This effect may be associated with the high chemical electrophilicity of the central carbon of the isothiocyanate (-N=C=S) group. However, there is little research on this underlying mechanism for the activation of Nrf2 by SFN in colon cancer; therefore, further investigations are required.
Figure 8.
Mechanism of SFN on the II phase metabolism phase enzyme UGT1A. SFN, sulforaphane; UGT1A UDP glucuronosyltransferase 1A.
In addition to the aforementioned detection of Nrf2 and UGT1A expression, the present study detected the levels of ROS. A study on SFN and bladder cancer revealed that SFN induced a significant increase in ROS levels, which was necessary for SFN-induced mitochondrial dysfunction, serving an important role in SFN-induced apoptosis in cancer cells (36). In the present study, ROS levels were increased in CRC cells when these cells were cultured in SFN-containing medium. Notably, ERK inhibitors can inhibit SFN-induced high levels of ROS in CRC cells. This suggests that SFN-induced high levels of ROS in CRC cells may also be associated with the ERK signaling pathway. Oxidative stress is involved in three stages of cancer development: Initiation, promotion, and progression. Low-to-intermediate levels of ROS promote the occurrence and progression of tumors, but high levels of ROS can increase intrinsic oxidative stress of cancer cells, inhibit cancer cells activity, promote apoptosis, induce autophagy and even cause cell death (37,38). SFN induces apoptosis in glioblastoma cells via ROS-dependent inactivation of STAT3 phosphorylation (39) and has an anti-tumor effect on bladder cancer cells via the ROS-mediated intrinsic apoptosis pathway (36). Similar studies have also been performed in pancreatic cancer and hepatic cancer (40,41). These findings motivate further evaluation of SFN as a chemopreventive agent in cancer treatment. In addition, in studies investigating ROS-induced apoptosis, tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) can preferentially activate apoptosis of malignant cells; however, in a variety of human cancer cells the apoptosis induced by TRAIL is hard to control, resulting in TRAIL resistance. Notably, SFN induction of ROS can reduce the resistance and therefore SFN promotes TRAIL-induced cancer cell death (42). It was hypothesized that the inhibitory effect of SFN on colon cancer cells may be associated with the induction of high levels of ROS and may be regulated by the ERK signaling pathway. Collectively, these results indicated that the ERK signaling pathway and Nrf2 may represent strategic targets for the chemopreventive effects of SFN.
In a future experimental design, the mechanism may be studied and upstream and downstream channels may be further identified. In addition, given the opportunity, animal experiments may be added to the future experimental design and the mechanism may be studied thoroughly. Gene knockout mice would be designed and HA carcinogens could be used to establish mouse models with colorectal cancer. Colon tissues would be extracted for analysis at the gene, protein and cell levels to further explore the chemical preventive effect of SFN in colorectal cancer.
In addition, SFN is regarded as a sensitizer to anticancer drugs. A study has revealed that SFN analogs provide a novel approach to chemically sensitize CRC cells by modulating DNA damage/repair signaling pathways (43). In addition, SFN was revealed to inhibit the growth of C666 nasopharyngeal carcinoma cells and enhance the anti-tumor effect of cisplatin (44). Selectively preconditioning cancer cells with non-toxic amounts of a natural bioactive compound may safely enhance drug susceptibility. These compounds often activate drugs by upregulating the activity of drug metabolic enzymes; thus, they may significantly affect treatment outcomes despite low exposure (45–47).
The latest data in CRC revealed that the incidence rate of CRC has declined in recent years but remains high. CRC can be treated by surgery as well as radiotherapy and chemotherapy, but the 5-year survival rate is low, and the quality of life of patients is greatly reduced. As a multi-step and multi-stage process disease, CRC has a long precancerous stage. Thus, it is advisable to prevent it at its origin. Furthermore, the phytochemical SFN is readily available, and is widely present in cruciferous plants. Numerous studies have demonstrated the role of SFN in preventing tumors, inhibiting tumors and sensitizing anticancer drugs in colorectal tumors. If SFN could be used to prevent CRC and promote its early commercialization, patients with cancer and high-risk groups could be better protected. In summary, phytochemicals may aid to prevent CRC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science Foundation of China (grant no. 81372681).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors contributions
QH wrote the main manuscript and performed the experiments. MW and QH designed the study. MW, QH, CZ and WWZ performed data analysis. MW, YML, NXS, CL and FL contributed to the manuscript revisions and revised it critically for important intellectual content. All authors reviewed the manuscript, and read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Peery AF, Crockett SD, Barritt AS, Dellon ES, Eluri S, Gangarosa LM, Jensen ET, Lund JL, Pasricha S, Runge T, et al. Burden of gastrointestinal, liver, and pancreatic diseases in the United States. Gastroenterology. 2015;149:1731–1741.3. doi: 10.1053/j.gastro.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, McCullough ML, Patel AV, Ma J, Soerjomataram I, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 4.Wilson LF, Antonsson A, Green AC, Jordan SJ, Kendall BJ, Nagle CM, Neale RE, Olsen CM, Webb PM, Whiteman DC. How many cancer cases and deaths are potentially preventable? Estimates for Australia in 2013. Int J Cancer. 2018;142:691–701. doi: 10.1002/ijc.31088. [DOI] [PubMed] [Google Scholar]
- 5.Brown KF, Rumgay H, Dunlop C, Ryan M, Quartly F, Cox A, Deas A, Elliss-Brookes L, Gavin A, Hounsome L, et al. The fraction of cancer attributable to modifiable risk factors in England, Wales, Scotland, Northern Ireland, and the United Kingdom in 2015. Br J Cancer. 2018;118:1130–1141. doi: 10.1038/s41416-018-0029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Keefe SJ. Diet, microorganisms and their metabolites, and colon cancer. Nat Rev Gastroenterol Hepatol. 2016;13:691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner ND, Lloyd SK. Association between red meat consumption and colon cancer: A systematic review of experimental results. Exp Biol Med (Maywood) 2017;242:813–839. doi: 10.1177/1535370217693117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diallo A, Deschasaux M, Latino-Martel P, Hercberg S, Galan P, Fassier P, Allès B, Guéraud F, Pierre FH, Touvier M. Red and processed meat intake and cancer risk: Results from the prospective NutriNet-Sante cohort study. Int J Cancer. 2018;142:230–237. doi: 10.1002/ijc.31046. [DOI] [PubMed] [Google Scholar]
- 9.Klusek J, Nasierowska-Guttmejer A, Kowalik A, Wawrzycka I, Chrapek M, Lewitowicz P, Radowicz-Chil A, Klusek J, Głuszek S. The influence of red meat on colorectal cancer occurrence is dependent on the genetic polymorphisms of s-glutathione transferase genes. Nutrients. 2019;11(pii):E1682. doi: 10.3390/nu11071682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demeyer D, Mertens B, De Smet S, Ulens M. Mechanisms linking colorectal cancer to the consumption of (processed) red meat: A review. Crit Rev Food Sci Nutr. 2016;56:2747–2766. doi: 10.1080/10408398.2013.873886. [DOI] [PubMed] [Google Scholar]
- 11.Abid Z, Cross AJ, Sinha R. Meat, dairy, and cancer. Am J Clin Nutr. 2014;100(Suppl 1):386S–393S. doi: 10.3945/ajcn.113.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etemadi A, Abnet CC, Graubard BI, Beane-Freeman L, Freedman ND, Liao L, Dawsey SM, Sinha R. Anatomical subsite can modify the association between meat and meat compounds and risk of colorectal adenocarcinoma: Findings from three large US cohorts. Int J Cancer. 2018;143:2261–2270. doi: 10.1002/ijc.31612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai T, Yao L, Turesky RJ. Bioactivation of heterocyclic aromatic Amines by UDP glucuronosyltransferases. Chem Res Toxicol. 2016;29:879–891. doi: 10.1021/acs.chemrestox.6b00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Logt EM, Bergevoet SM, Roelofs HM, van Hooijdonk Z, te Morsche RH, Wobbes T, de Kok JB, Nagengast FM, Peters WH. Genetic polymorphisms in UDP-glucuronosyltransferases and glutathione S-transferases and colorectal cancer risk. Carcinogenesis. 2004;25:2407–2415. doi: 10.1093/carcin/bgh251. [DOI] [PubMed] [Google Scholar]
- 15.Liu M, Wang Q, Liu F, Cheng X, Wu X, Wang H, Wu M, Ma Y, Wang G, Hao H. UDP-glucuronosyltransferase 1A compromises intracellular accumulation and anti-cancer effect of tanshinone IIA in human colon cancer cells. PLoS One. 2013;8:e79172. doi: 10.1371/journal.pone.0079172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Qi YY, Chen S, Sun DF, Wang S, Chen J, Li YQ, Han W, Yang XY. Expression of UDP-glucuronosyltransferase 1A, nuclear factor erythroid-E2-related factor 2 and Kelch-like ECH-associated protein 1 in colonic mucosa, adenoma and adenocarcinoma tissue. Oncol Lett. 2012;4:925–930. doi: 10.3892/ol.2012.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin TF, Wang M, Qing Y, Lin YM, Wu D. Research progress on chemopreventive effects of phytochemicals on colorectal cancer and their mechanisms. World J Gastroenterol. 2016;22:7058–7068. doi: 10.3748/wjg.v22.i31.7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vuong LD, Nguyen QN, Truong VL. Anti-inflammatory and anti-oxidant effects of combination between sulforaphane and acetaminophen in LPS-stimulated RAW 264.7 macrophage cells. Immunopharmacol Immunotoxicol. 2019;41:413–419. doi: 10.1080/08923973.2019.1569049. [DOI] [PubMed] [Google Scholar]
- 19.Mazarakis N, Snibson K, Licciardi PV, Karagiannis TC. The potential use of l-sulforaphane for the treatment of chronic inflammatory diseases: A review of the clinical evidence. Clin Nutr. 2019 Mar 25; doi: 10.1016/j.clnu.2019.03.022. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 20.Wang M, Chen S, Wang S, Sun D, Chen J, Li Y, Han W, Yang X, Gao HQ. Effects of phytochemicals sulforaphane on uridine diphosphate-glucuronosyltransferase expression as well as cell-cycle arrest and apoptosis in human colon cancer Caco-2 cells. Chin J Physiol. 2012;55:134–144. doi: 10.4077/CJP.2012.BAA085. [DOI] [PubMed] [Google Scholar]
- 21.Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When NRF2 talks, who's listening? Antioxid Redox Signal. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thangapandiyan S, Ramesh M, Miltonprabu S, Hema T, Jothi GB, Nandhini V. Sulforaphane potentially attenuates arsenic-induced nephrotoxicity via the PI3K/Akt/Nrf2 pathway in albino Wistar rats. Environ Sci Pollut Res Int. 2019;26:12247–12263. doi: 10.1007/s11356-019-04502-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kalthoff S, Strassburg CP. Contribution of human UDP-glucuronosyltransferases to the antioxidant effects of propolis, artichoke and silymarin. Phytomedicine. 2018;56:35–39. doi: 10.1016/j.phymed.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Ewa B, Danuta MS. Polycyclic aromatic hydrocarbons and PAH-related DNA adducts. J Appl Genet. 2017;58:321–330. doi: 10.1007/s13353-016-0380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Palliyaguru DL, Kensler TW. Frugal chemoprevention: Targeting Nrf2 with foods rich in sulforaphane. Semin Oncol. 2016;43:146–153. doi: 10.1053/j.seminoncol.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lubelska K, Wiktorska K, Mielczarek L, Milczarek M, Zbroińska-Bregisz I, Chilmonczyk Z. Sulforaphane regulates NFE2L2/Nrf2-dependent xenobiotic metabolism phase ii and phase iii enzymes differently in human colorectal cancer and untransformed epithelial colon cells. Nutri Cancer. 2016;68:1338–1348. doi: 10.1080/01635581.2016.1224369. [DOI] [PubMed] [Google Scholar]
- 28.Vissenaekens H, Grootaert C, Rajkovic A, Van De Wiele T, Calatayud M. The response of five intestinal cell lines to anoxic conditions in vitro. Biol Cell. 2019;111:232–244. doi: 10.1111/boc.201800076. [DOI] [PubMed] [Google Scholar]
- 29.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, Bjørnslett M, Meza-Zepeda LA, Eknæs M, Lind GE, et al. Multi-omics of 34 colorectal cancer cell lines-a resource for biomedical studies. Mol Cancer. 2017;16:116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cagnol S, Chambard JC. ERK and cell death: Mechanisms of ERK-induced cell death-apoptosis, autophagy and senescence. FEBS J. 2010;277:2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 31.Jo C, Kim S, Cho SJ, Choi KJ, Yun SM, Koh YH, Johnson GV, Park SI. Sulforaphane induces autophagy through ERK activation in neuronal cells. FEBS Lett. 2014;588:3081–3088. doi: 10.1016/j.febslet.2014.06.036. [DOI] [PubMed] [Google Scholar]
- 32.Wong SY, Tan MG, Wong PT, Herr DR, Lai MK. Andrographolide induces Nrf2 and heme oxygenase 1 in astrocytes by activating p38 MAPK and ERK. J Neuroinflammation. 2016;13:251. doi: 10.1186/s12974-016-0723-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucolo C, Drago F, Maisto R, Romano GL, D'Agata V, Maugeri G, Giunta S. Curcumin prevents high glucose damage in retinal pigment epithelial cells through ERK1/2-mediated activation of the Nrf2/HO-1 pathway. J Cell Physiol. 2019;234:17295–17304. doi: 10.1002/jcp.28347. [DOI] [PubMed] [Google Scholar]
- 34.Svehlikova V, Wang S, Jakubikova J, Williamson G, Mithen R, Bao Y. Interactions between sulforaphane and apigenin in the induction of UGT1A1 and GSTA1 in CaCo-2 cells. Carcinogenesis. 2004;25:1629–1637. doi: 10.1093/carcin/bgh169. [DOI] [PubMed] [Google Scholar]
- 35.Thangapandiyan S, Ramesh M, Hema T, Miltonprabu S, Uddin MS, Nandhini V, Bavithra Jothi G. Sulforaphane potentially ameliorates arsenic induced hepatotoxicity in albino wistar rats: Implication of PI3K/Akt/Nrf2 signaling pathway. Cell Physiol Biochem. 2019;52:1203–1222. doi: 10.33594/000000082. [DOI] [PubMed] [Google Scholar]
- 36.Jo GH, Kim GY, Kim WJ, Park KY, Choi YH. Sulforaphane induces apoptosis in T24 human urinary bladder cancer cells through a reactive oxygen species-mediated mitochondrial pathway: The involvement of endoplasmic reticulum stress and the Nrf2 signaling pathway. Int J Oncol. 2014;45:1497–1506. doi: 10.3892/ijo.2014.2536. [DOI] [PubMed] [Google Scholar]
- 37.Xin Y, Bai Y, Jiang X, Zhou S, Wang Y, Wintergerst KA, Cui T, Ji H, Tan Y, Cai L. Sulforaphane prevents angiotensin II-induced cardiomyopathy by activation of Nrf2 via stimulating the Akt/GSK-3ß/Fyn pathway. Redox Biol. 2018;15:405–417. doi: 10.1016/j.redox.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chikara S, Nagaprashantha LD, Singhal J, Horne D, Awasthi S, Singhal SS. Oxidative stress and dietary phytochemicals: Role in cancer chemoprevention and treatment. Cancer Lett. 2018;413:122–134. doi: 10.1016/j.canlet.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 39.Miao Z, Yu F, Ren Y, Yang J. D,L-Sulforaphane induces ROS-dependent apoptosis in human gliomablastoma cells by inactivating STAT3 signaling pathway. Int J Mol Sci. 2017;18(pii):E72. doi: 10.3390/ijms18010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramani R, Gonzalez E, Arumugam A, Nandy S, Gonzalez V, Medel J, Camacho F, Ortega A, Bonkoungou S, Narayan M, et al. Nimbolide inhibits pancreatic cancer growth and metastasis through ROS-mediated apoptosis and inhibition of epithelial-to-mesenchymal transition. Sci Rep. 2016;6:19819. doi: 10.1038/srep19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pocasap P, Weerapreeyakul N, Thumanu K. Alyssin and iberin in cruciferous vegetables exert anticancer activity in HepG2 by increasing intracellular reactive oxygen species and tubulin depolymerization. Biomol Ther (Seoul) 2019;27:540–552. doi: 10.4062/biomolther.2019.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin CY, Molagoda IMN, Karunarathne WAHM, Kang SH, Park C, Kim GY, Choi YH. TRAIL attenuates sulforaphane-mediated Nrf2 and sustains ROS generation, leading to apoptosis of TRAIL-resistant human bladder cancer cells. Toxicol Appl Pharmacol. 2018;352:132–141. doi: 10.1016/j.taap.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Okonkwo A, Mitra J, Johnson GS, Li L, Dashwood WM, Hegde ML, Yue C, Dashwood RH, Rajendran P. Heterocyclic analogs of sulforaphane trigger DNA damage and impede DNA repair in colon cancer cells: Interplay of HATs and HDACs. Mol Nutr Food Res. 2018;62:e1800228. doi: 10.1002/mnfr.201800228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L, Chan LS, Lung HL, Yip TTC, Ngan RKC, Wong JWC, Lo KW, Ng WT, Lee AWM, Tsao GSW, et al. Crucifera sulforaphane (SFN) inhibits the growth of nasopharyngeal carcinoma through DNA methyltransferase 1 (DNMT1)/Wnt inhibitory factor 1 (WIF1) axis. Phytomedicine. 2019;63:153058. doi: 10.1016/j.phymed.2019.153058. [DOI] [PubMed] [Google Scholar]
- 45.Erzinger MM, Bovet C, Hecht KM, Senger S, Winiker P, Sobotzki N, Cristea S, Beerenwinkel N, Shay JW, Marra G, et al. Sulforaphane Preconditioning Sensitizes Human Colon Cancer Cells towards the Bioreductive Anticancer Prodrug PR-104A. PLoS One. 2016;11:e0150219. doi: 10.1371/journal.pone.0150219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Erzinger MM, Sturla SJ. Bioreduction-mediated food-drug interactions: Opportunities for oncology nutrition. Chimia (Aarau) 2011;65:411–415. doi: 10.2533/chimia.2011.411. [DOI] [PubMed] [Google Scholar]
- 47.Milczarek M, Mielczarek L, Lubelska K, Dąbrowska A, Chilmonczyk Z, Matosiuk D, Wiktorska K. In vitro evaluation of sulforaphane and a natural analog as potent inducers of 5-fluorouracil anticancer activity. Molecules. 2018;23(pii):E3040. doi: 10.3390/molecules23113040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.