Abstract
Colorectal cancer (CRC) is one of the most common digestive tract tumors worldwide. Catalpol exerts inhibitory effects on the progression of several cancer types by regulating microRNAs (miRs). However, the precise role and carcinostatic mechanism of catalpol on CRC cells are poorly understood which limits the application of catalpol treatment. In the present study, miR-34a and sirtuin 1 (SIRT1) expression levels were detected in CRC tissues and CRC cell lines by RT-qPCR. Computational software analysis, luciferase assays and western blotting were used to demonstrate the downstream target of miR-34a in CRC cells. Effects of catalpol on cell viability, apoptosis, autophagic flux and the miR-34a/SIRT1 axis in the CRC cells were assessed by CCK-8 assay, flow cytometry, electron microscopy and western blotting, respectively. Whether the miR-34a/SIRT1 axis participated in catalpol-mediated autophagy and apoptosis was investigated. The effects of catalpol on the miR-34a/SIRT1 axis and malignant behavior were evaluated in a rat model of azoxymethane (AOM)-induced CRC. It was revealed that miR-34a expression levels were significantly decreased while SIRT1 was overexpressed in most of the CRC tissues and all the CRC cell lines. Clinically, a low level of miR-34a was correlated with poor clinicopathological characteristics in CRC patients. Catalpol reduced cell viability, suppressed autophagy, promoted apoptosis, and regulated the expression of SIRT1 by inducing miR-34a in vitro and in vivo. The autophagy-inhibiting effect of catalpol may be a mechanism to promote apoptosis of CRC cells. miR-34a mimic transfection resulted in autophagy-suppressive activity similar to that of catalpol, while the miR-34a inhibitor attenuated the antiautophagic effects of catalpol. In conclusion, miR-34a is involved in regulating catalpol-mediated autophagy and malignant behavior by directly inhibiting SIRT1 in CRC.
Keywords: colorectal cancer, catalpol, autophagy, miR-34a, SIRT1
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer-related deaths worldwide (1). CRC has become a critical health problem with an increase in its incidence and mortality over the past decade (2). Despite advances in surgical resection combined with chemotherapy and radiotherapy, to the best of our knowledge, progress in treatment of CRC remains sluggish in recent years (3). Therefore, numerous studies have attempted to illuminate the underlying mechanisms on the development of CRC and develop novel methods to improve therapeutic effects.
Catalpol, a bioactive component extracted from traditional Chinese medicine Rehmannia glutinosa (Di Huang), has various pharmacological activities (4). Several studies indicated that catalpol has inhibitory effects on proliferation and induced apoptosis in several cancer types including human CRC (3–5). Our previous study also focused on the therapeutic effect of catalpol against acute liver injury via alleviation of inflammatory response (6). However, the underlying mechanisms of catalpol in CRC treatment are still unclear, which limits the application of catalpol in CRC treatment.
microRNAs (miRs) are small noncoding RNAs which are involved in numerous biological processes. Accumulating evidence has demonstrated that miRs are aberrantly expressed in multiple human cancers and stimulate inappropriate cellular programs (7). The antitumor and antiproliferative activities of catalpol are closely associated with aberrant versions of miRs (8–10), which provides insights on the interaction between catalpol and the expression of miRs. Our previous studies indicated that miR-34a has a tumor-suppressive role in human CRC and cholangiocarcinoma (11,12); thus, we hypothesized that there may be a correlation between catalpol and miR-34a in the progression of CRC.
Recently, the roles of autophagy regulated by miRs in the progression of human cancers have been of concern (13,14). Our previous studies demonstrated that the resistance of oxaliplatin in CRC cells was connected with miR-34a-mediated autophagy (11). In addition, it was reported that inhibition of autophagy attenuated the anti-inflammatory effect regulated by catalpol on liver fibrosis (15). Moreover, catalpol was revealed to suppress autophagy to prevent denervated muscular atrophy (16). Considering the association between autophagy, miRNAs and catalpol, it was surmised that the autophagy regulated by miRNAs may be involved in mediating the functional role of catalpol. However, the exact role of miR-34a mediated by catalpol in CRC cell autophagy and apoptosis awaits to be elucidated.
Materials and methods
Patients and tissue samples
CRC samples and adjacent normal colon tissues were obtained from 60 CRC patients (median age, 62 years; range, 47–76 years; 26 mmale and 34 mfemale patients) between January 2018 and December 2018 at the Second Affiliated Hospital of Harbin Medical University. All patients provided written informed consent according to our institutional guidelines, and the study protocol was approved by the Institutional Review Board of Harbin Medical University (KY2018-208). The samples were trimmed and snap frozen in liquid nitrogen immediately after surgical resection. The tumor stage was classified according to the 7th tumor-node-metastasis (TNM) classification of the International Union against Cancer (UICC) (17). Data of clinical characteristics were collected from medical records.
Animal model of azoxymethane (AOM)-induced CRC
Three-week-old male Wistar rats (40–60 g) obtained from the Animal Center of the Second Affiliated Hospital of Harbin Medical University were housed with free access to sterile food and water under a standard 12-h light/dark cycle and a controlled temperature (22±2°C) and humidity (55±5%). Animal experiments were carried out in strict accordance with the Harbin Medical University Institutional Animal Care guidelines and was approved by the Animal Research Ethics Use Committee of Harbin Medical University (KY2018-208). Thirty-six rats were randomly divided into 3 groups of 12 rats each. Animals in the experimental groups (AOM and catalpol) received AOM (15 mg/kg i.p.) and the control group received an equal volume of normal saline, once weekly for 2 weeks. Catalpol dissolved in phosphate-buffered saline (PBS; 10 mg/kg) was administered intragastrically daily from week 5 to 25. The AOM group was administered the same volume of PBS orally. Rats health and behavior were monitored every 2 days. At the end of the experiment, euthanasia by anesthesia overdose with intraperitoneal injection of sodium pentobarbital (Nembutal; 200 mg/kg body weight) was perfomed. Then colon tissue samples were obtained from each rat after decapitation was performed to confirm euthanasia (6,18).
Cell culture and treatment
The human colon epithelial cell line FHC obtained from the American Type Culture Collection (ATCC) was cultured in DMEM/F12 medium (GIBCO; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere. The human CRC cell lines HCT116, HT29, SW620 and SW480 were purchased from the Cell Bank of the Chinese Academy of Sciences and were routinely cultured using RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS. The miR-34a mimics or miR-34a inhibitor purchased from Shanghai GenePharma Co., Ltd., were transfected into the CRC cells for 48 h using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) in indicated concentrations according to the supplier's instructions. Rapamycin (10 nM; Cell Signaling Technology, Inc.) and 3-methyladenine (3-MA; 5 mM; Sigma-Aldrich; Merck KGaA) were added to further activate or inhibit the flux of autophagy in CRC cells as previously described (12).
Bioinformatics analysis and luciferase assay
Since miRNAs function by base-pairing with sequences in the 3′-UTR of their target mRNA, we used TargetScan (http://www.targetscan.org/) to search for potential targets of miR-34a. For luciferase reporter assay, the CRC cells were co-transfected with 80 ng mutant reporter plasmids, 10 ng pRL-TK-Renilla-luciferase plasmid, and sirtuin 1 (SIRT1) RNAs (20 nmol/l). Luciferase activity was assessed by quantifying the ratio of firefly luciferase activity to Renilla luciferase activity using the Dual-Luciferase Reporter Assay system (Promega Corporation) as previously described (12) at 24 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA or miRs were isolated from cultured cells and fresh surgical tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) or mirVana miRNA Isolation kit (Ambion; Thermo Fisher Scientific, Inc.). They were reverse-transcribed to cDNA by High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Inc.). RT-qPCR was performed in triplicate with the Power SYBR Green (Thermo Fisher Scientific, Inc.) using the ABI StepOne system. The thermocycling parameters were as follows: 10 min at 95°C for polymerase activation followed by 40 cycles consisting of 95°C for 15 sec and 60°C for 60 sec. The expression levels of SIRT1 were normalized to β-actin, and U6 small nuclear RNA (U6) was used as the internal control for miR-34a. The 2−∆∆Cq method was used to normalize the relative levels of the target genes. The details of all the primers for RT-qPCR used in this study are presented in Table I (19).
Table I.
Primers used for RT-qPCR.
| Primer name | Primer sequence: 5′-3′ |
|---|---|
| SIRT1 | F: CCCAGAACATAGACACGCTGGA |
| R: ATCAGCTGGGCACCTAGGACA | |
| β-actin | F: ATGTTGAGACCTTCAACACC |
| R: AGGTAGTCAGTCAGGTCCCGGCC | |
| miR-34a | F: TGGTGTCGTGGAGTCG |
| R: GGCATCTCTCGCTTCATCTT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT |
SIRT1, sirtuin 1; miR, microRNA; F, forward; R, reverse.
Western blotting
Cell extracts were collected and proteins were extracted using RIPA buffer (Beyotime Institute of Biotechnology). The concentration of total protein was measured using a BCA Protein Assay Kit (Beyotime Institute of Biotechnology). Thirty µg protein lysates was electrophoresed by 12% SDS-PAGE and then transferred to nitrocellulose membranes (Bio-Rad Laboratories, Inc.). After blocking with 5% fat-free milk for 2 h, the membranes were probed with the relevant primary antibodies (SIRT1, LC3, Beclin 1, Bcl-2, Bax, cytochrome c and β-actin; Table II) at 4°C overnight. Finally an enhanced chemiluminescence detection reagents Alexa Fluor 680 donkey anti-mouse IgG or Alexa Fluor 680 donkey anti-rabbit IgG (Table II) secondary antibody was incubated with the membranes for 12 h at 4°C and visualized with an Odyssey™ Infrared Imaging system (LI-COR Biosciences). Densitometry was performed using Alpha Imager 2200 (Alpha Innotech Corp.). β-actin was used as a control for whole-cell lysates.
Table II.
Antibodies used for western blotting.
| Antibody | Manufacturer | Species | Dilution |
|---|---|---|---|
| SIRT1 | cat. no. sc-135792 (Santa Cruz Biotechnology, Inc.) | Mouse | 1:500 |
| LC3-II (MAPLC3β) | cat. no. sc-28266 (Santa Cruz Biotechnology, Inc.) | Rabbit | 1:500 |
| Beclin 1 | cat. no. sc-11427 (Santa Cruz Biotechnology, Inc.) | Rabbit | 1:500 |
| Bcl-2 | cat. no. sc-7382 (Santa Cruz Biotechnology, Inc.) | Mouse | 1:200 |
| Bax | cat. no. sc-526 (Santa Cruz Biotechnology, Inc.) | Mouse | 1:200 |
| Cytochrome c | cat. no. sc-13156 (Santa Cruz Biotechnology, Inc.) | Mouse | 1:200 |
| β-actin | cat. no. sc-69879 (Santa Cruz Biotechnology, Inc.) | Mouse | 1:500 |
| Alexa Fluor 680 donkey anti-mouse IgG anti-mouse IgG | A10038 (Thermo Fisher Scientific, Inc.) | Donkey | 1:5,000 |
| Alexa Fluor 680 donkey anti-rabbit IgG | A10043 (Thermo Fisher Scientific, Inc.) | Donkey | 1:5,000 |
SIRT1, sirtuin 1.
Cell viability and cytotoxicity assay
Cell Counting Kit-8 (CCK-8; Boster Biological Technology) was used to assess cell viability as previously described (11). The CRC cells were seeded at a density of 3×103/well in 96-well plates and maintained at 37°C overnight. CCK-8 solution (10 µl) was added into each well and incubated for another 1 h. The absorbance at 450 nm was measured using a Microplate Reader (Bio-Rad Laboratories, Inc.). For the cytotoxicity assay, lactate dehydrogenase (LDH) activity in culture medium was determined using an LDH release assay kit (Boster Biological Technology).
Evaluation of autophagy and apoptosis by flow cytometry
Cell autophagy was assessed by detecting the mono-dansylcadaverine (MDC; Sigma-Aldrich; Merck KGaA) positively stained cells using flow cytometry. Cells (104) were washed with PBS and then incubated with 0.05 mmol/l MDC in PBS at 37°C for 45 min. After washing three times with PBS, the cell suspension was analyzed by flow cytometry immediately. For cell apoptosis detection, 104 cells were collected in 200 ml RPMI-1640 medium without FBS. Following resuspension, approximately 10 ml Annexin V solution (BD Biosciences) was added. Fifteen min later, 300 ml medium buffer and 5 ml propidium iodide (PI; BD Biosciences) were added, and the cell suspension was analyzed by flow cytometry (BD Biosciences) immediately (11).
Electron microscopy
Cells from each group were harvested and fixed with 2.5% glutaraldehyde at 4°C for 2 h. The samples were suspended in PBS with 1% osmic acid. After dehydration and embedding, the ultrathin sections were prepared on uncoated copper grids with an Ultrotome (Reichert/Leica Ultracut S) and stained with 3% uranyl acetate and lead citrate for 5 min at room temperature. Images were acquired using a JEM 1230 transmission electron microscope (JEOL, Ltd.).
Statistical analysis
Statistical analysis was carried out by SPSS version 21 (IBM Corp.) or GraphPad Prism version 5.0 (GraphPad Software, Inc.). All the data from at least three independent experiments were expressed as the mean ± standard deviation. Comparisons of quantitative data between two groups were performed using paired two-tailed Student's t-tests. Comparisons among three or more groups were performed by one-way ANOVA test followed by Tukey's test. Correlation analysis between miR-34a and SIRT1 mRNA expression levels was performed by Pearson's rank correlation coefficient analysis. The impact of clinical parameters was estimated by multivariate logistic regression analysis and Cox regression analysis (Table III). P<0.05 was considered to indicate a statistically significant difference.
Table III.
Relationship between miR-34a expression and clinicopathological features in CRC patients.
| miR-34a | ||||
|---|---|---|---|---|
| Characteristics | n | Low | High | P-value |
| Age (years) | 0.603 | |||
| <60 | 27 | 14 | 13 | |
| ≥60 | 33 | 16 | 17 | |
| Sex | 0.397 | |||
| Male | 26 | 14 | 12 | |
| Female | 34 | 16 | 18 | |
| Tumor size (cm) | 0.094 | |||
| <5 | 36 | 19 | 17 | |
| ≥5 | 24 | 11 | 13 | |
| Differentiation | <0.001 | |||
| Well/moderate | 20 | 2 | 18 | |
| Poor | 40 | 28 | 12 | |
| Lymphatic node metastasis | <0.001 | |||
| Present | 43 | 28 | 15 | |
| Absent | 17 | 2 | 15 | |
| Clinical stages | <0.001 | |||
| I/II | 17 | 2 | 15 | |
| III/IV | 43 | 28 | 15 | |
| CA199 level (U/ml) | 0.500 | |||
| <37 | 31 | 15 | 16 | |
| ≥37 | 29 | 15 | 14 | |
| CEA level (ng/ml) | <0.001 | |||
| <5 | 16 | 2 | 14 | |
| ≥5 | 44 | 28 | 16 | |
The median value of miR-34a expression level in CRC patients was 0.6725. P<0.05 is considered to indicate a statistically significant difference. CRC, colorectal cancer; CA, carbohydrate antigen; CEA, carcinoembryonic antigen.
Results
miR-34a is downregulated and SIRT1 is upregulated in CRC tissues and cell lines
The expression of miR-34a in CRC tissues was significantly decreased compared with the adjacent non-tumor tissues (Fig. 1A). SIRT1 mRNA expression was upregulated in CRC tissues compared to the adjacent normal colon tissues (Fig. 1B). Notably, the expression levels of miR-34a and SIRT1 had a significant inverse correlation (Fig. 1C) as revealed by Spearman correlation analysis. The 60 CRC patients were divided into two groups according to the median value (0.6725) of the miR-34a expression level (0.6725) in CRC tissues to further evaluate the association between the expression levels of miR-34a and the clinicopathological characteristics of CRC (Table III). The data indicated that miR-34a expression was decreased in samples with poor cell differentiation, lymphatic metastasis, high expression of carcinoembryonic antigen (CEA) and advanced clinical stages. However, age, sex, tumor size, expression of carbohydrate antigen (CA)19-9 had no association with miR-34a expression (P>0.05). miR-34a expression was further detected in FHC and HT29, HCT116, SW480 and SW620 CRC cell lines. As revealed in Fig. 1D, the expression levels of miR-34a were significantly decreased in all the CRC cell lines compared to FHC cells. The expression levels of SIRT1 in CRC cell lines were significantly higher than that in FHC cells (Fig. 1E). HT29 and HCT116 exhibited moderate expression levels of miR-34a and SIRT1, thus these two cell lines were selected to further explore the role of the miR-34a/SIRT1 axis in the progression of CRC.
Figure 1.
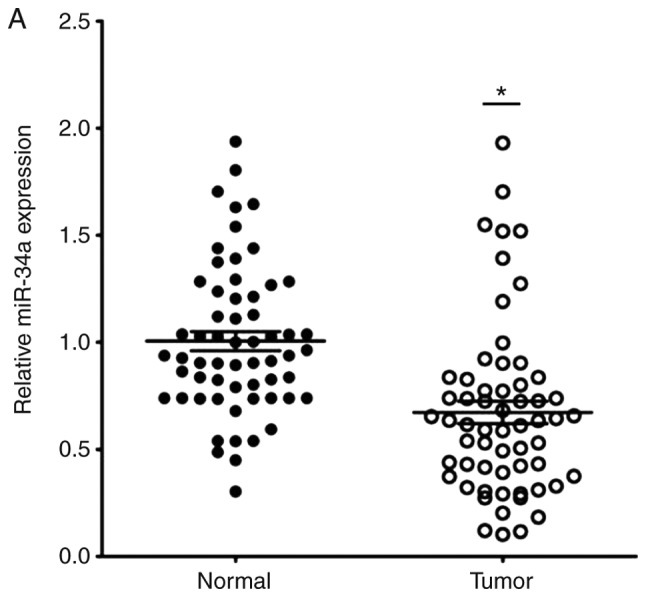
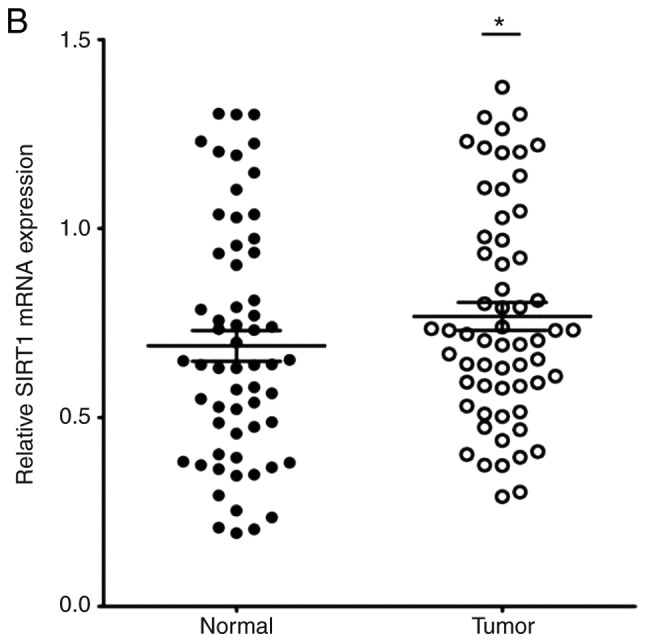
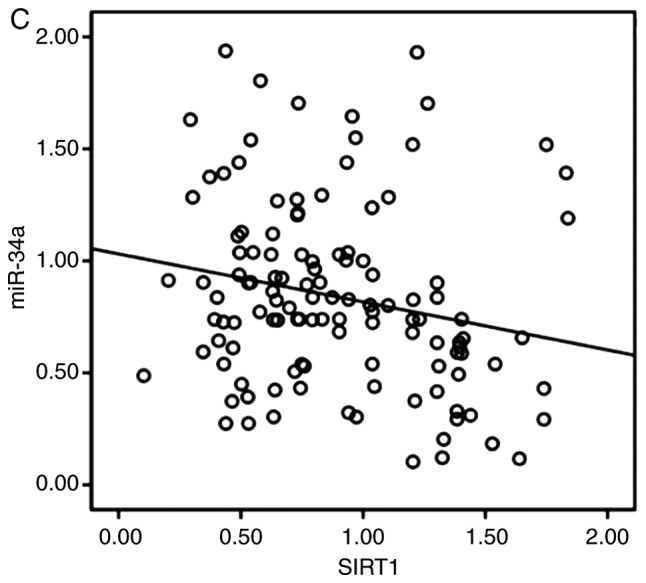
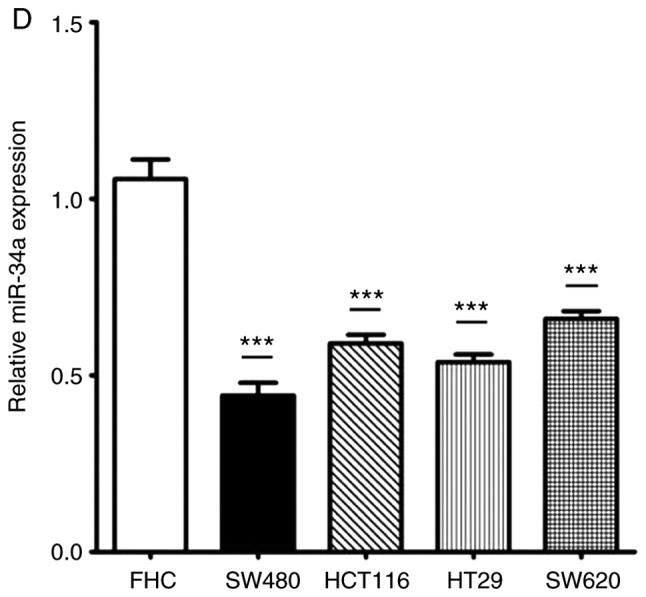
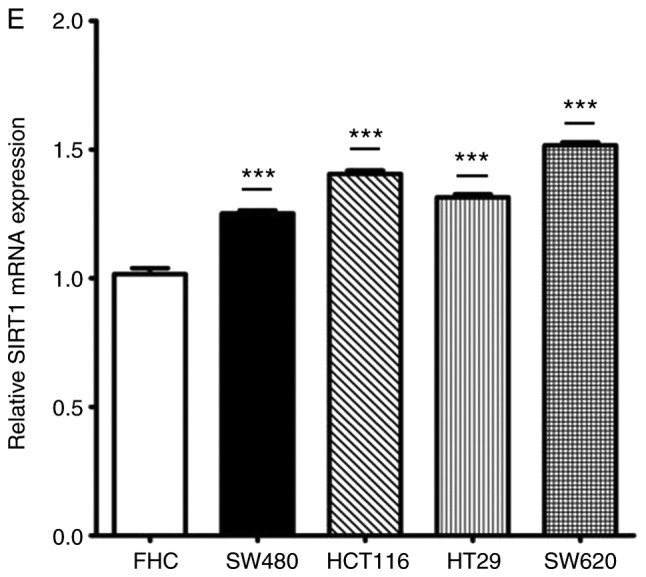
miR-34a is downregulated and SIRT1 is upregulated in CRC tissues and cell lines. (A) RT-qPCR was performed to examine downregulated miR-34a expression in 60 CRC samples (*P<0.05). (B) RT-qPCR was performed to examine upregulated SIRT1 mRNA expression in CRC specimens compared to the adjacent non-tumor tissues (*P<0.05). (C) Inverse correlation between miR-34a and SIRT1 expression was determined by Spearman's rank correlation coefficient analysis (R=−0.241, P=0.008). (D) RT-qPCR revealed downregulated expression of miR-34a in CRC cell lines (SW480, HCT116, HT29 and SW620) compared to FHC cells (***P<0.001). (E) RT-qPCR revealed overexpression of SIRT1 mRNA in CRC cell lines compared to FHC cells (***P<0.001). *P<0.05 indicated a significant difference vs. the control group; ***P<0.001 indicated a significant difference vs. the FHC cells. miR, microRNA; SIRT1, sirtuin 1; CRC, colorectal cancer.
miR-34a directly targets SIRT1 in CRC cells
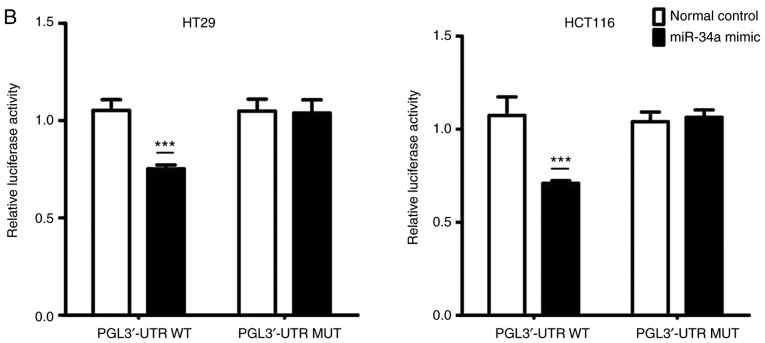
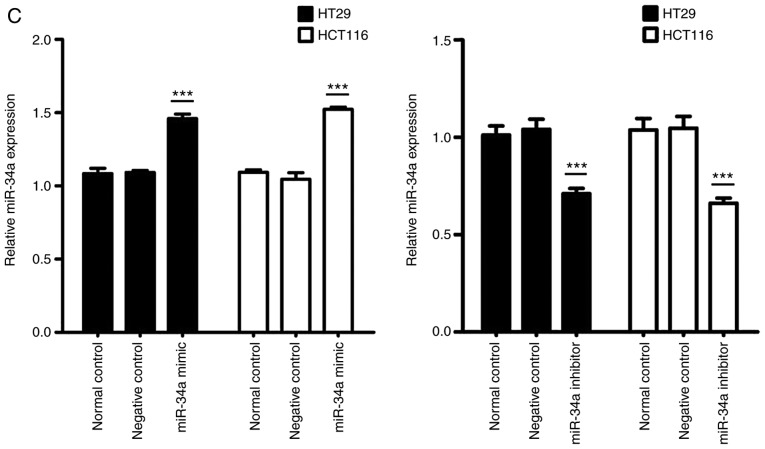
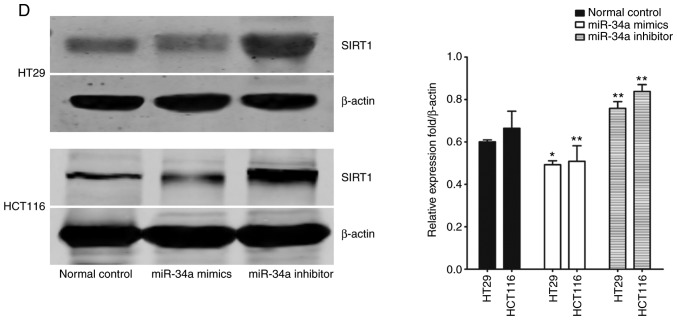
Data from TargetScan (http://www.targetscan.org/) revealed that the 3′-UTR of SIRT1 contained the complementary site which may play a role in translational repression for the seed region of miR-34a (Fig. 2A). A dual luciferase assay was conducted in HT29 and HCT116 cells. As revealed in Fig. 2B, miR-34a mimic transfection (PGL3′-UTR WT) decreased luciferase activity compared to the control group. Additionally, transfection with the mutated 3′-UTR (PGL3′-UTR MUT) of the SIRT1 gene, revealed no significant difference in luciferase activities between the miR-34a mimic transfection and the control groups. Moreover, compared to the normal control group, miR-34a mimic transfection and miR-34a inhibitor successfully increased and decreased, respectively, the endogenous miR-34a expression levels in CRC cells (Fig. 2C). Results of western blotting revealed that the transfection with the miR-34a mimics decreased SIRT1 expression while transfection with the miR-34a inhibitor promoted SIRT1 expression at the protein level in CRC cells (Fig. 2D). These data indicated that SIRT1 was one of the direct targets of miR-34a in CRC cells.
Figure 2.

SIRT1 is a direct miR-34a target in CRC cell lines. (A) Data from TargetScan revealed the miR-34a binding site in the 3′-UTR of SIRT1. (B) Luciferase reporter assay revealed a reduction in luciferase activity of WT SIRT1 3′-UTR by transfection with miR-34a mimic in HT29 and HCT116 cells (***P<0.001). (C) RT-qPCR analysis of the expression of miR-34a in CRC cells by miR-34a mimic or inhibitor transfection (***P<0.001). (D) SIRT1 protein expression was examined by western blotting (*P<0.05 and **P<0.01 indicates a significant difference vs. the control group). SIRT1, sirtuin 1; miR, microRNA; CRC, colorectal cancer; WT, wild-type.
Catalpol suppresses autophagy and induces apoptosis through the miR-34a/SIRT1 axis in CRC cell lines
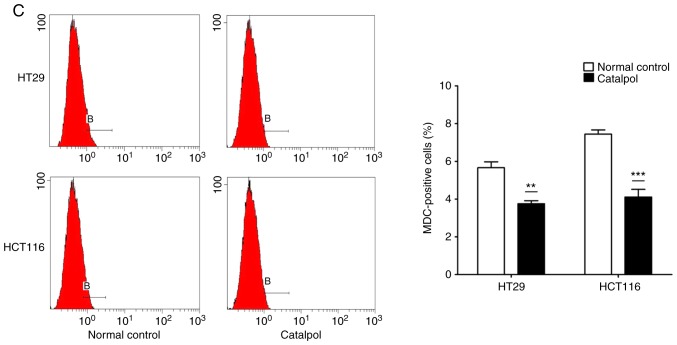
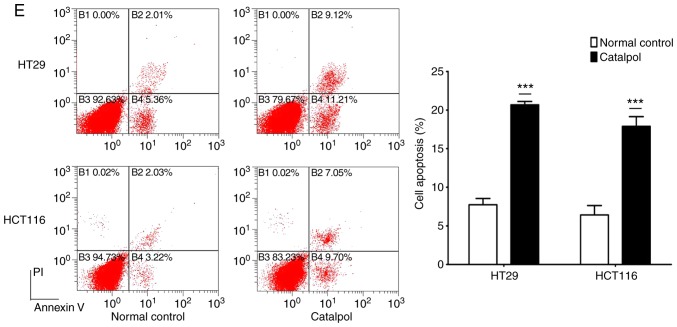
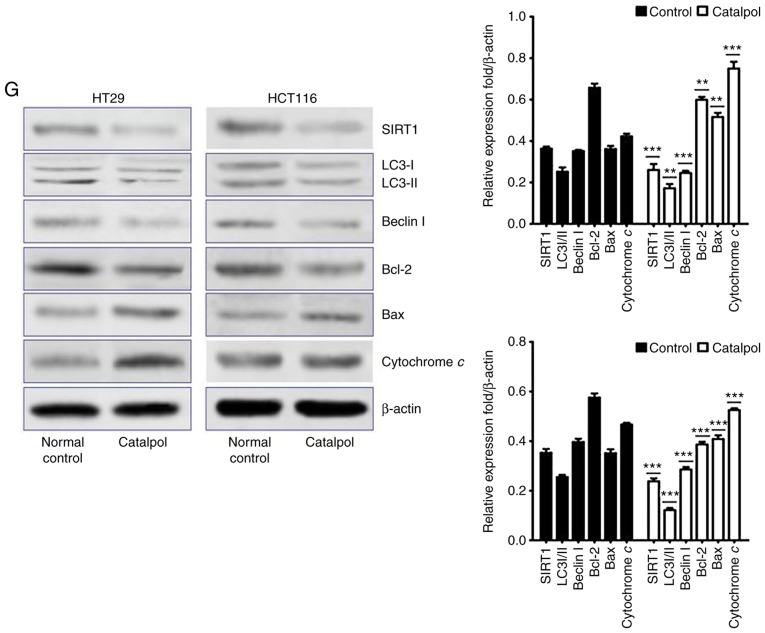
The effects of catalpol on CRC cell viability and cytotoxicity were initially evaluated in vitro. CRC cell viability was significantly decreased by 30–50 µM catalpol treatment compared to the control group (Fig. 3A). Cytotoxicity was revealed at a concentration of 40 µM (Fig. 3B). Thus, 30 µM was used in the following experiments. Whether catalpol significantly inhibited autophagy in CRC cells was next investigated. MDC-positively stained CRC cells were significantly decreased by catalpol treatment (Fig. 3C). Consistent with the flow cytometric results, Fig. 3D reveals the decreased autophagosomes associated with catalpol treatment in CRC cells. Compared with the normal controls, expression of autophagy-related protein Beclin 1 and LC3-II (also referred as MAPLC3β) was significantly downregulated by catalpol (Fig. 3G). Induction of apoptosis of CRC cells by catalpol was demonstrated by flow cytometry (Fig. 3E) and Bax, cytochrome c expression levels were increased, while Bcl-2 protein expression levels were reduced (Fig. 3G). Consistent with previous studies (8–10), expression of miR-34a in CRC cells was markedly upregulated by catalpol treatment compared to control cells (Fig. 3F). As anticipated, expression of SIRT1 in both HT29 and HCT116 cells was downregulated by catalpol (Fig. 3G). These results indicated that suppression of autophagy and induction of apoptosis of CRC cells by catalpol may be partly achieved through the miR-34a/SIRT1 axis.
Figure 3.
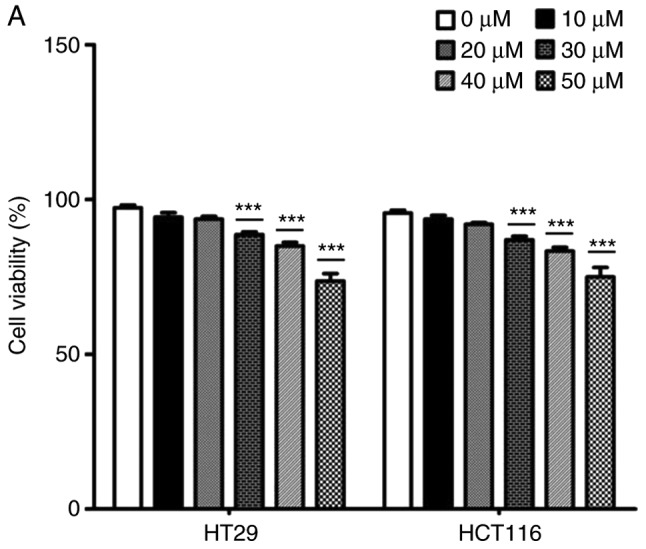
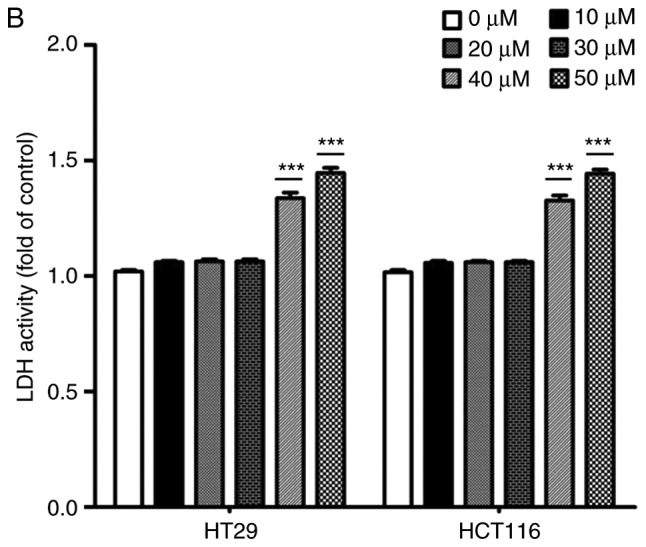
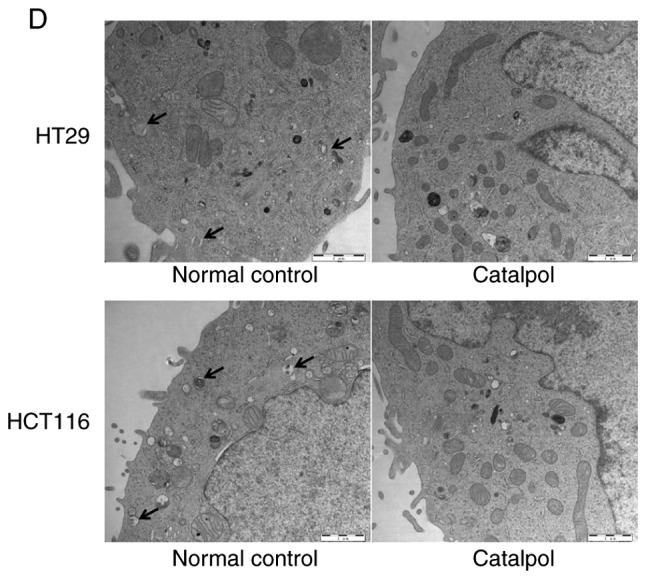
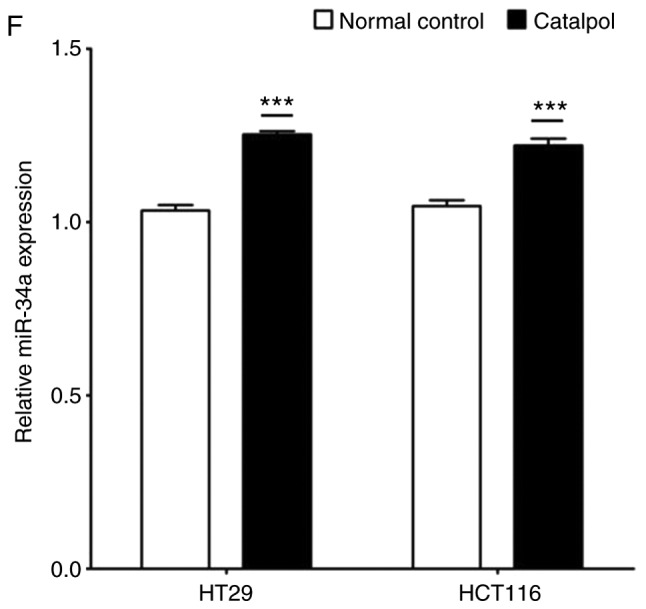
Catalpol reduces cell viability, suppresses autophagy and induces apoptosis through the miR-34a/SIRT1 axis in vitro. (A) The inhibited effect of catalpol on CRC cell viability determined by CCK-8 analysis (***P<0.001). (B) LDH activity assay was used to examined the toxicity of catalpol on HT29 and HCT116 cells (***P<0.001). (C) Autophagy of CRC cells was determined using flow cytometry (**P<0.01 and ***P<0.001). (D) Autophagic vacuole formation was revealed by representative electron micrographs in CRC cell lines. The arrows indicate the autophagosomes. Catalpol reduces cell viability, suppresses autophagy and induces apoptosis through the miR-34a/SIRT1 axis in vitro. (E) Analysis of apoptosis by flow cytometry in HT29 and HCT116 cells (***P<0.001). (F) Expression of miR-34a was determined by RT-qPCR (***P<0.001). (G) Western blotting was performed to detect SIRT1 and autophagy, and apoptosis-related proteins in CRC cells (**P<0.01 and ***P<0.001 indicates a significant difference vs. the normal control group). miR, microRNA; SIRT1, sirtuin 1; CRC, colorectal cancer; LDH, lactate dehydrogenase; CCK-8, Cell Counting Kit-8.
Catalpol-regulated autophagy is involved in mediating apoptosis in CRC cells
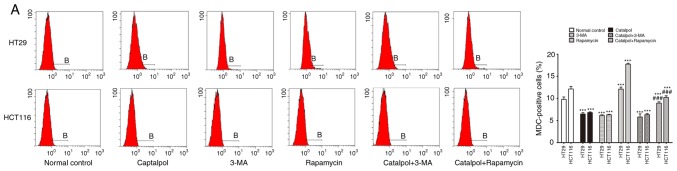
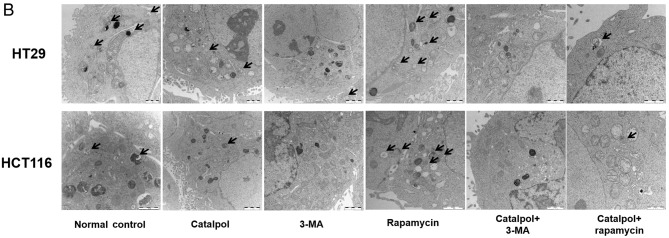
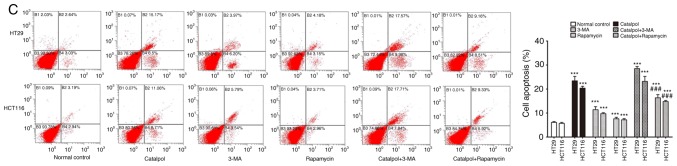
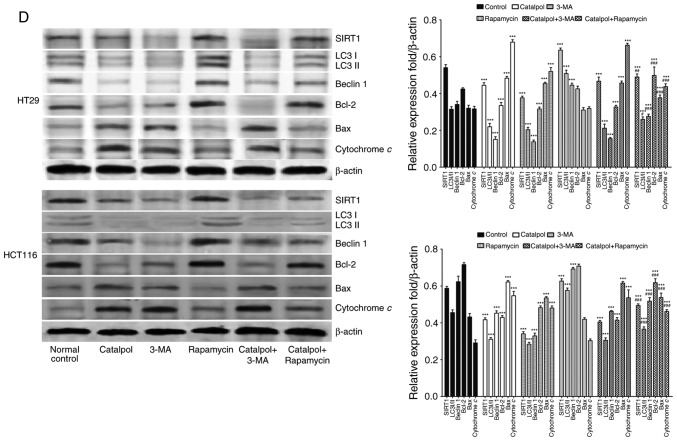
To address whether catalpol regulated autophagy-modulated apoptosis of CRC cells, the CRC cells were subjected to catalpol with or without 3-MA and rapamycin treatment. The results revealed that catalpol inhibited autophagy and induced apoptosis of CRC cells, as indicated by decreased MDC-positive cells (Fig. 4A) and autophagosomes (Fig. 4B); decreased expression levels of LC3-II, Beclin 1 and Bcl-2; and higher expression levels of Bax and cytochrome c (Fig. 4D) as well as the apoptotic rate (Fig. 4C). 3-MA significantly suppressed autophagy and induced apoptosis of CRC cells while rapamycin activated autophagy but had no effect on apoptosis in CRC cells, as revealed by increased MDC-positive cells (Fig. 4A) and autophagosomes (Fig. 4B) and higher expression of autophagy-related proteins (Fig. 4D). In addition, 3-MA combined with catalpol did not affect either autophagy activity or apoptosis of CRC cells as compared with treatment with catalpol alone. However, compared with treatment with catalpol alone, rapamycin in combination with catalpol attenuated the inhibitory effect on autophagy and induced apoptosis of CRC cells (Fig. 4). These results strongly indicated that inhibition of autophagy by catalpol may exert an accelerated effect on apoptosis of CRC cells.
Figure 4.
Inhibition of autophagy regulated by catalpol contributes to apoptosis in CRC cells. (A) CRC cells were exposed to 30 µM catalpol for 24 h and treated with or without 3-MA and rapamycin, and then flow cytometry was used to detect the autophagic levels (***P<0.001 and ###P<0.001). (B) Autophagic vacuole formation was revealed by representative electron micrographs in CRC cell lines. The arrows indicate the autophagosomes. (C) Apoptosis was analyzed by flow cytometry in CRC cells after various treatments (***P<0.001 and ###P<0.001). (D) Western blotting revealed the expression of SIRT1, LC3II, Beclin 1, Bcl-2, Bax, and cytochrome c in HT29 and HCT116 cells. β-Actin was used as a loading control. Quantitation of the results was performed (***P<0.001 indicates a significant difference vs. the normal control group; #P<0.05, ##P<0.01 and ###P<0.001 indicates a significant difference vs. the group treated with catalpol). CRC, colorectal cancer; SIRT1, sirtuin 1; 3-MA, 3-methyladenine.
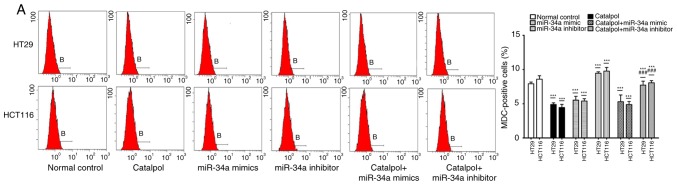
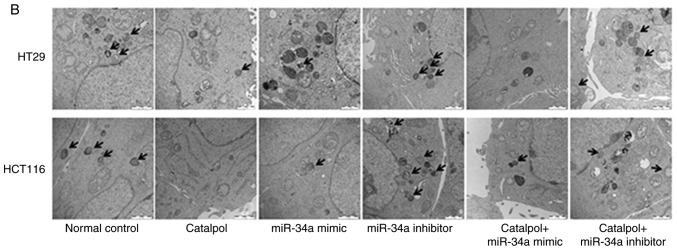
The miR-34a/SIRT1 pathway participates in regulating catalpol-suppressed CRC cell autophagy and induces apoptosis
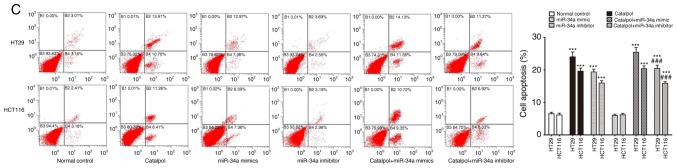
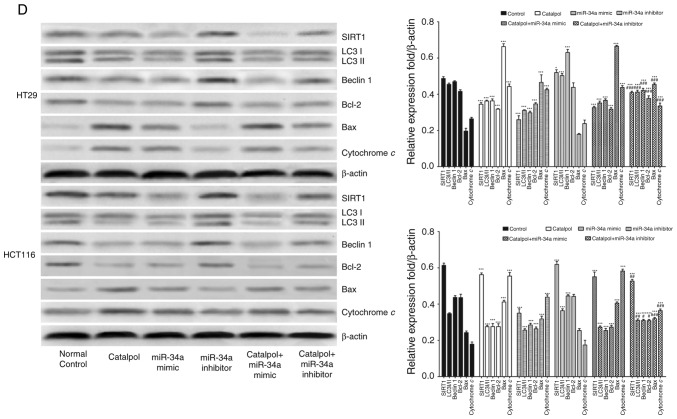
miR-34a and catalpol have been demonstrated as key regulators of autophagy (11,15); thus, it was investigated whether miR-34a was involved in mediating catalpol-regulated autophagy and apoptosis of CRC cells. Both the cell lines were treated by catalpol and transfected with miR-34a mimics or inhibitors. The number of MDC-positive cells detected by flow cytometry (Fig. 5A) and autophagosomes detected by electron microscopy (Fig. 5B) were significantly decreased compared to the controls. Western blotting revealed decreased expression of SIRT1, LC3-II, Beclin 1 and Bcl-2, and higher expression of Bax and cytochrome c (Fig. 5D) in CRC cells in the catalpol treatment and miR-34a mimics transfection groups compared to the control group. Similarly, the apoptotic rate was significantly increased in both cell lines by treatment with catalpol or miR-34a mimics compared to the control group (Fig. 5C). The level of autophagy was enhanced but apoptosis was not weakened in both HT29 and HCT116 cell lines by the miR-34a inhibitor, since the results revealed increased MDC-positive cells (Fig. 5A) and autophagosomes (Fig. 5B), along with increased SIRT1, LC3-II and Beclin 1 protein expression levels (Fig. 5D). The effect on CRC cell apoptosis was not significant compared to the control group (Fig. 5C and D). Treatment with catalpol in combination with miR-34a mimic had no significant effect on autophagy flux or apoptosis compared with catalpol alone in HT29 and HCT116 cells (Fig. 5). Compared with catalpol alone, the inhibitory effect of autophagy and induction of apoptosis were impaired by catalpol in combination with miR-34a inhibitor in CRC cells, as indicated by increased MDC-positive cells (Fig. 5A) and autophagosomes (Fig. 5B), increased expression of SIRT1, LC3-II, Beclin 1 and Bcl-2, and decreased expression of Bax and cytochrome c (Fig. 5D) in CRC cells. Moreover, the inhibitory effect on apoptosis was significantly reduced in both HT29 and HCT116 cell lines compared to catalpol treatment alone (Fig. 5C). These data indicated that the miR-34a/SIRT1 axis at least partly participated in mediating catalpol-suppressed autophagy and catalpol-induced apoptosis of CRC cells.
Figure 5.
The miR-34a/SIRT1 axis participates in regulating catalpol-suppressed CRC cell autophagy and induces apoptosis. (A) HT29 and HCT116 cells were treated with 30 µM catalpol for 24 h and in combination with miR-34a mimic or inhibitor, and then flow cytometry was used to examine autophagic levels (***P<0.001 and ###P<0.001). (B) Autophagic vacuole formation was revealed by representative electron micrographs in CRC cell lines. The arrows indicate the autophagosomes. (C) The CRC cell apoptotic rate was analyzed by flow cytometry (***P<0.001 and ###P<0.001). (D) Western blotting revealed the expression of SIRT1, LC3, Beclin 1, Bcl-2, Bax, and cytochrome c in HT29 and HCT116 cells (*P<0.05 and ***P<0.001 indicates a significant difference vs. the normal control group; #P<0.05, ##P<0.01 and ###P<0.001 indicates a significant difference vs. the group treated with catalpol). miR, microRNA; SIRT1, sirtuin 1; CRC, colorectal cancer.
Catalpol inhibits tumorigenesis through regulation of the miR-34a/SIRT1 pathway in a rat model
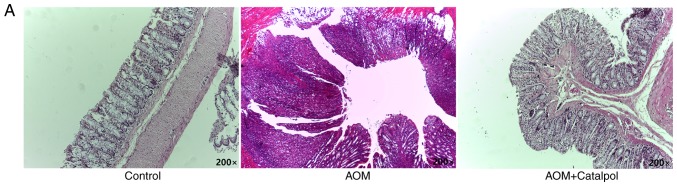
The AOM-induced CRC rat model was used to evaluate the chemopreventive effect and toxicity of catalpol in vivo. First, percentage weight gain among each group was observed, however the differences were not significant (data not shown). As revealed in Fig. 6A, catalpol intervention markedly prevented the formation of colon tumors. All rats treated with AOM developed preneoplastic aberrant crypt foci (ACF) (Fig. 6B). The catalpol group had a significantly decreased number of ACFs with ≤4 crypts as well as total ACF compared to the AOM group (Fig. 6B). No significant difference was revealed in the number of ACFs containing >5 crypts among the groups (Fig. 6B). To determine the potential mechanism of catalpol in vivo, miR-34a and SIRT1 mRNA expression levels in colon tissues from each group were examined by RT-qPCR. Consistent with the results in vitro, expression of miR-34a was significantly decreased in the AOM-treated group compared to the control group, and catalpol treatment promoted the expression of miR-34a compared to AOM treatment (Fig. 6C). The expression of SIRT1 mRNA was significantly upregulated by treatment with AOM compared to the control while treatment with catalpol in combination with AOM inhibited the promoting effect on the expression of SIRT1 compared to AOM treatment alone (Fig. 6D). These results provided evidence that the catalpol-mediated miR-34a/SIRT1 pathway was involved in inhibition of malignant behavior in vivo.
Figure 6.
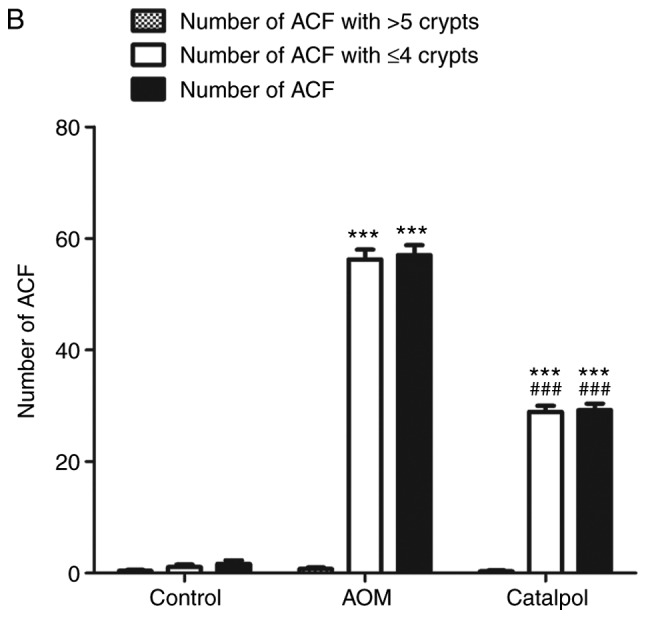
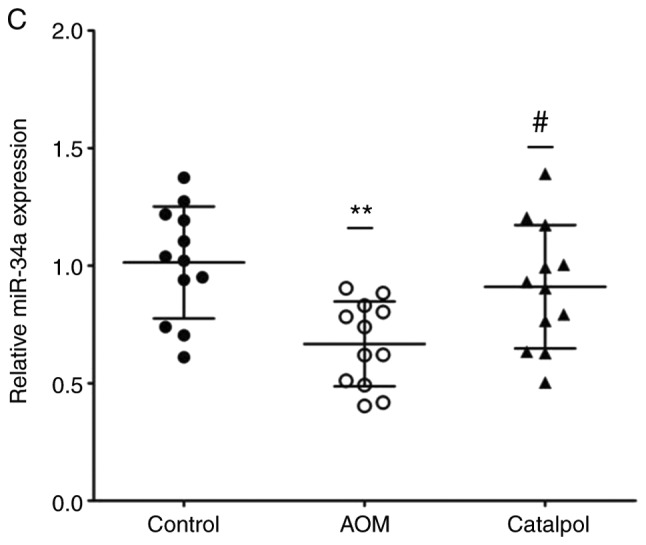
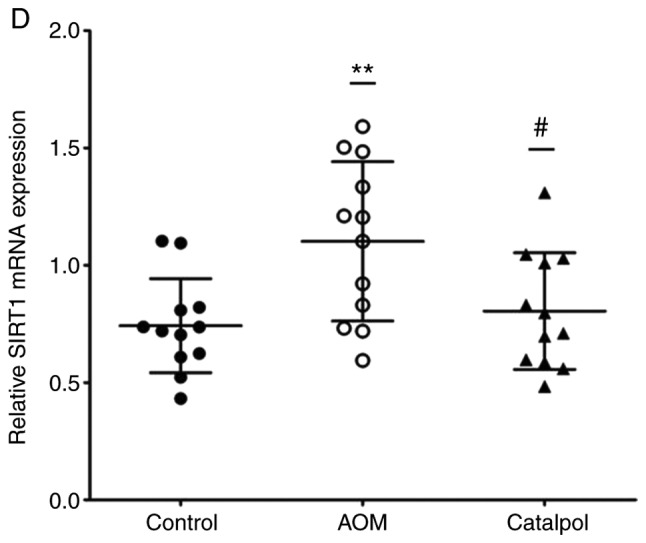
Catalpol inhibits tumorigenesis in a rat model of AOM-induced CRC by regulating the miR-34a/SIRT1 axis. (A) Catalpol treatment suppressed the formation of colon tumors. (B) Number of ACFs with ≤4 crypts from the colon of rats (***P<0.001 and ###P<0.001). Number of ACFs with >5 crypts in the entire colon (P>0.05). Total number of ACFs in the entire colon (***P<0.001 and ###P<0.001). (C) Expression of miR-34a in colon samples determined by RT-qPCR (**P<0.01 and #P<0.05). (D) SIRT1 mRNA expression levels in the colon samples determined by RT-qPCR (**P<0.01, #P<0.05) (**P<0.01 and ***P<0.001 indicates a significant difference vs. the control group; #P<0.05 and ###P<0.001 indicates a significant difference vs. the group treated with AOM). AOM, azoxymethane; CRC, colorectal cancer; miR, microRNA; ACF, aberrant crypt foci; SIRT1, sirtuin 1.
Discussion
Catalpol is one of the main active ingredients of R. glutinosa (Di Huang) which has been revealed to have antitumor effects on various of human cancers (5,8–10). Recently, the inhibited effect on inflammation and tumor angiogenesis of catalpol in CRC cells was revealed and catalpol was considered as a potential candidate compound for treating CRC (3,10,20). Significant changes in the miRNA expression profiles were observed in cancer cells after catalpol treatment (8–10,21). Moreover, the present findings indicated that miR-34a played a tumor-suppressive role during progression of human CRC and cholangiocarcinoma (11,12). Thus, miR-34a was selected as a candidate gene for catalpol in the present study. The present research revealed that miR-34a was aberrantly downregulated in most of human CRC tissues and all the CRC cell lines. Since miRNAs exert physiological functions by directly modulating their target mRNA expression, bioinformatics analysis was performed to predict if SIRT1 may be one of the potential targets of miR-34a. SIRT1 is a class III nuclear deacetylase that can mediate genetic programs by modifying histones and transcription factors (22). SIRT1 is reported to inactivate p53 by deacetylating a critical lysine residue, and blocking SIRT1 may lead to the activation of the p53 pathway and suppress the progression of human cancers including CRC (23). Moreover, miR-34a could bind the site within the 3′-UTR of SIRT1 to silence the mRNA as revealed in a previous study (24). Consistent with previous research, ectopic expression of miR-34a interposed SIRT1 protein expression in CRC cell lines, and miR-34a may be a tumor-suppressor and function by directly inhibiting SIRT1 expression in the progression of CRC.
Mounting evidence has revealed that catalpol has inhibitory effects on proliferation and inducing effects on apoptosis in several types of human malignancies (8–10,21). Moreover, catalpol was revealed to inhibit autophagy and decrease apoptosis via the mammalian target of rapamycin pathway in denervated muscular atrophy (16). In addition, it was reported that resistance of autophagy attenuated the catalpol-induced anti-inflammatory effect on liver fibrosis (15). In the present study, it was revealed that 30–50 µM catalpol suppressed cell viability and 40 µM catalpol resulted in toxicity in CRC cells. Thus, 30 µM was used in the subsequent experiments. As documented previously, modest autophagy may protect cells from apoptosis under different conditions (11,25). The present data indicated that CRC cell autophagy was significantly inhibited and apoptosis was induced by catalpol treatment. Moreover, it was revealed that catalpol upregulated miR-34a expression and suppressed SIRT1 expression in CRC cells. Catalpol has been revealed to target multiple signaling pathways, including Bcl-2/Bax, JNK, NF-κB to exert its pharmacological effects (26,27). Thus, the antitumor effects and the potential mechanisms underlying the catalpol-mediatied miR-34a/SIRT1 axis may be regulated by complex signaling pathways. The present study indicated that the miR-34a/SIRT1 axis was likely involved in regulating catalpol suppression of autophagy and induction of apoptosis in CRC cells, however, more studies are required in the future.
To date, there is wide recognition that autophagy plays a dual role in regulating the fate of cancer cells (28). Although increasing evidence has focused on the association between autophagy and progression of malignant tumors, it is far from being clarified. The present study indicated that catalpol and 3-MA attenuated activation of autophagy and induced apoptosis of CRC cells. In addition, 3-MA in combination with catalpol did not show any effect on autophagy or apoptosis of CRC cells. Notably, rapamycin impaired the promoting effect of catalpol on CRC cell apoptosis. Therefore, it was suggested that catalpol may induce CRC cell apoptosis by suppressing autophagy.
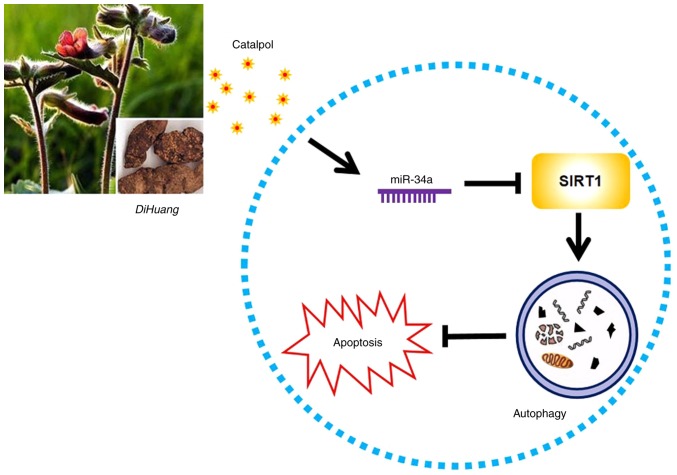
To determine whether miR-34a participated in mediating catalpol-regulated autophagy and apoptosis, CRC cells were treated by catalpol and concurrent promotion or silencing of miR-34a expression. It was confirmed that both catalpol and miR-34a overexpression antagonized cell autophagy and promoted apoptosis of CRC cells. The autophagy was aggravated but apoptosis was not restrained when miR-34a was exogenously inhibited. Combination treatment with catalpol and miR-34a mimic revealed no effect on autophagy or apoptosis in CRC cells. Moreover, the antitumor effects of catalpol were neutralized by concurrent silencing of miR-34a expression. Previous studies indicated that catalpol administration significantly alleviated symptoms in a rat colitis model (21,29). The present results indicated that there was no significant weight gain in any of the experimental groups, which may be because the experiment period was not long enough. However, all rats treated with AOM developed preneoplastic ACF in the colon. Catalpol treatment impaired the carcinogenic effects of AOM. Moreover, as was anticipated, catalpol promoted the expression of miR-34a and inhibited the expression of SIRT1 in the colon of rats. Conversely, Xiong et al reported that activation of SIRT1 by catalpol attenuated infiltration of inflammatory cells, cytokine profiles, oxidative responses, and epithelial cell apoptosis in colitis rats (21). The appearance of this discrepancy in SIRT1 expression by catalpol treatment may be due to the totally different diseases and AOM-induced stages as well as the different cell lines used in in vitro investigations. Thus, it was speculated that catalpol may induce apoptosis and inhibit autophagy at least partly by upregulating miR-34a, along with suppression of SIRT1 expression (Fig. 7).
Figure 7.
Catalpol-inhibited autophagy is involved in inducing CRC cell apoptosis by mediating the miR-34a/SIRT1 axis. CRC, colorectal cancer; miR, microRNA; ACF, aberrant crypt foci; SIRT1, sirtuin 1.
In summary, it was demonstrated that miR-34a functioned as an antioncogene in human CRC, which was at least in part through the suppression of SIRT1 expression. Aberrantly downregulated expression of miR-34a in CRC patients was correlated with adverse clinical characteristics. In addition, cell autophagy was suppressed while apoptosis was induced by catalpol treatment in human CRC cells. Catalpol-regulated autophagy was demonstrated to participate in mediating apoptosis in CRC cells. Furthermore, miR-34a was upregulated by catalpol treatment and catalpol may function by modulating the miR-34a/SIRT1 axis, which at least partly provides an insight into the mechanisms underlying catalpol treatment and may be considered as a novel therapeutic target for CRC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CRC
colorectal cancer
- SIRT1
sirtuin 1
- miRs
miRNAs/microRNAs
- UTR
untranslated region
- AOM
azoxymethane
- PBS
phosphate-buffered saline
- MDC
mono-dansylcadaverine
- NC
negative control
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- CA
carbohydrate antigen
- CEA
carcinoembryonic antigen
Funding
The present study was supported by Heilongjiang Postdoctoral Funds for Scientific Research Initiation (LBH-Q17129) and the Science Foundation of the Health Commission of Heilongjiang Province (2018346).
Availability of data and materials
The data used in the present study are available from the corresponding author upon reasonable request.
Authors' contributions
PFQ designed the experiments. PFQ and ZLZ performed the experiments. LY analyzed the data and wrote the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All patients provided written informed consent according to our institutional guidelines and the study protocol was approved by the Institutional Review Board of Harbin Medical University. The animal experiments were carried out in strict accordance with the Harbin Medical University Institutional Animal Care and Use Committee (KY2018-208).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, up-regulating expression of microRNA-21. Gastroenterology. 2017;152:851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu P, Wu Y, Yang A, Fu X, Mao M, Liu Z. Catalpol suppressed proliferation, growth and invasion of CT26 colon cancer by inhibiting inflammation and tumor angiogenesis. Biomed Pharmacother. 2017;95:68–76. doi: 10.1016/j.biopha.2017.08.049. [DOI] [PubMed] [Google Scholar]
- 4.Xiu LZ, Bo J, Zhi BL, Hao S, An LJ. Catalpol ameliorates cognition deficits and attenuates oxidative damage in the brain of senescent mice induced by D-galactose. Pharmacol Biochem Behav. 2007;88:64–72. doi: 10.1016/j.pbb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZH, Zhansheng H. Catalpol inhibits migration and induces apoptosis in gastric cancer cells and in athymic nude mice. Biomed Pharmacother. 2018;103:1708–1719. doi: 10.1016/j.biopha.2018.03.094. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Jia R, Wang F, Qiu G, Qiao P, Xu X, Wu D. Catalpol protects mice against Lipopolysaccharide/D-galactosamine-induced acute liver injury through inhibiting inflammatory and oxidative response. Oncotarget. 2018;9:3887–3894. doi: 10.18632/oncotarget.23242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian MicroRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 8.Gao N, Tian JX, Shang YH, Zhao DY, Wu T. Catalpol suppresses proliferation and facilitates apoptosis of OVCAR-3 ovarian cancer cells through upregulating microRNA-200 and downregulating MMP-2 expression. Int J Mol Sci. 2014;15:19394–19405. doi: 10.3390/ijms151119394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C, Wu F, Liu Y, Meng C. Catalpol suppresses proliferation and facilitates apoptosis of MCF-7 breast cancer cells through upregulating microRNA-146a and downregulating matrix metalloproteinase-16 expression. Mol Med Rep. 2015;12:7609–7614. doi: 10.3892/mmr.2015.4361. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Gao H, Wang H, Zhang Y, Xu W, Lin S, Wang H, Wu Q, Guo J. Catalpol promotes cellular apoptosis in human HCT116 colorectal cancer cells via microRNA-200 and the downregulation of PI3K-Akt signaling pathway. Oncol Lett. 2017;14:3741–3747. doi: 10.3892/ol.2017.6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23:1816–1827. doi: 10.3748/wjg.v23.i10.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao P, Li G, Bi W, Yang L, Yao L, Wu D. microRNA-34a inhibits epithelial mesenchymal transition in human cholangiocarcinoma by targeting Smad4 through transforming growth factor-beta/Smad pathway. BMC Cancer. 2015;15:469. doi: 10.1186/s12885-015-1359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Hu L, Cogdell D, Lu L, Gao C, Tian W, Zhang Z, Kang Y, Fleming JB, Zhang W. MIR506 induces autophagy-related cell death in pancreatic cancer cells by targeting the STAT3 pathway. Autophagy. 2017;13:703–714. doi: 10.1080/15548627.2017.1280217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Zhu P, Zhang L, Xiong B, Tao J, Guan W, Li C, Chen C, Gu J, Duanmu J, Zhang W. Autophagy inhibition attenuates the induction of anti-inflammatory effect of catalpol in liver fibrosis. Biomed Pharmacother. 2018;103:1262–1271. doi: 10.1016/j.biopha.2018.04.156. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Shao Y, Gao Y, Wan G, Wan D, Zhu H, Qiu Y, Ye X. Catalpol prevents denervated muscular atrophy related to the inhibition of autophagy and reduces BAX/BCL2 ratio via mTOR pathway. Drug Des Devel Ther. 2018;13:243–253. doi: 10.2147/DDDT.S188968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobin LH, Gospodarowicz KM, Wittekind CH, editors. UICC International Union Against Cancer. 7th. Wiley-Blackwell; 2009. TNM classification of malignant tumors. [Google Scholar]
- 18.Xu Y, Yang X, Mei S, Sun Y, Li J. Acquisition of temozolomide resistance by the rat C6 glioma cell line increases cell migration and side population phenotype. Oncol Rep. 2019;42:2355–2362. doi: 10.3892/or.2019.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Dong J, Liang W, Wang T, Sui J, Wang J, Deng Z, Chen D. Saponins regulate intestinal inflammation in colon cancer and IBD. Pharmacol Res. 2019;144:66–72. doi: 10.1016/j.phrs.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Xiong Y, Shi L, Wang L, Zhou Z, Wang C, Lin Y, Luo D, Qiu J, Chen D. Activation of sirtuin 1 by catalpol-induced down-regulation of microRNA-132 attenuates endoplasmic reticulum stress in colitis. Pharmacol Res. 2017;123:73–82. doi: 10.1016/j.phrs.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 22.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, Wu X, Yan D. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett. 2017;410:50–62. doi: 10.1016/j.canlet.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Lai M, Du G, Shi R, Yao J, Yang G, Wei Y, Zhang D, Xu Z, Zhang R, Li Y, et al. miR-34a inhibits migration and invasion by regulating the SIRT1/p53 pathway in human SW480 cells. Mol Med Rep. 2015;11:3301–3307. doi: 10.3892/mmr.2015.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao PF, Yao L, Zhang XC, Li GD, Wu DQ. Heat shock pretreatment improves stem cell repair following ischemia-reperfusion injury via autophagy. World J Gastroenterol. 2015;21:12822–12834. doi: 10.3748/wjg.v21.i45.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Xu G, Ma S, Li F, Yuan M, Xu H, Huang K. Catalpol ameliorates high-fat diet-induced insulin resistance and adipose tissue inflammation by suppressing the JNK and NF-κB pathways. Biochem Biophys Res Commun. 2015;467:853–858. doi: 10.1016/j.bbrc.2015.10.054. [DOI] [PubMed] [Google Scholar]
- 27.Li DQ, Bao YM, Li Y, Wang CF, Liu Y, An LJ. Catalpol modulates the expressions of Bcl-2 and Bax and attenuates apoptosis in gerbils after ischemic injury. Brain Res. 2006;1115:179–185. doi: 10.1016/j.brainres.2006.07.063. [DOI] [PubMed] [Google Scholar]
- 28.Jin S, White E. Role of autophagy in cancer: Management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crespo I, San-Miguel B, Prause C, Marroni N, Cuevas MJ, González-Gallego J, Tuñón MJ. Glutamine treatment attenuates endoplasmic reticulum stress and apoptosis in TNBS-induced colitis. PLoS One. 2012;7:e50407. doi: 10.1371/journal.pone.0050407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in the present study are available from the corresponding author upon reasonable request.