Abstract
Human cytomegalovirus (HCMV) infection remains an important cause of neurodevelopmental sequelae in infants infected in utero. Unique to the natural history of perinatal HCMV infections is the occurrence of congenital HCMV infections (cCMV) in women with existing immunity to HCMV, infections that have been designated as nonprimary maternal infection. In maternal populations with a high HCMV seroprevalence, cCMV that follows nonprimary maternal infections accounts for 75%–90% of all cases of cCMV infections as well as a large proportion of infected infants with neurodevelopmental sequelae. Although considerable effort has been directed toward understanding immune correlates that can modify maternal infections and intrauterine transmission, the source of virus leading to nonprimary maternal infections and intrauterine transmission is not well defined. Previous paradigms that included reactivation of latent virus as the source of infection in immune women have been challenged by studies demonstrating acquisition and transmission of antigenically distinct viruses, a finding suggesting that reinfection through exposure to an exogenous virus is responsible for some cases of nonprimary maternal infection. Additional understanding of the source(s) of virus that leads to nonprimary maternal infection will be of considerable value in the development and testing of interventions such as vaccines designed to limit the incidence of cCMV in populations with high HCMV seroprevalence.
Keywords: congenital CMV infection, maternal non-primary infection, recurrent infection, CMV infection in pregnancy
Human cytomegalovirus (HCMV) infections are ubiquitous in all populations and in most regions of the world are acquired during infancy and childhood. Although HCMV infection results in lifelong persistence, there are limited definitive data linking HCMV infection to definable clinical syndromes in the vast majority of individuals with intact adaptive and innate immune systems. However, there is an extensive literature describing associations between HCMV infections and human cancers, cardiovascular disease, and immunosenescence [1–6]. Although far from definitive, several lines of evidence are often presented that convincingly argue for a role of HCMV in either the development and/or phenotypic expression of these diseases. Ongoing studies will hopefully clarify the role of HCMV in these diseases. In contrast, HCMV infections in individuals with compromised immune systems, secondary to untreated HIV infection, treatment with immunosuppressive agents to limit allograft rejection, or following treatments for autoinflammatory or autoimmune diseases, often result in disease in multiple organ systems that can be directly attributed to HCMV replication. In addition, severe multisystem disease can be observed in newborn infants who are infected in utero and in infants with severe immunodeficiencies, particularly those with deficits in T-lymphocyte function [7, 8].
The source of HCMV infections in most normal hosts in community settings can be traced to close contact with individuals shedding virus. Because young infants infected with HCMV commonly shed large amounts of virus for prolonged periods of time, observational studies have frequently identified exposure to young children as a major risk factor for HCMV infection [9–11]. Other studies have demonstrated the transmission of HCMV through sexual contact with increased HCMV seroprevalence being observed in couples discordant for HCMV infection, women attending sexually transmitted infection (STI) clinics, and in early studies of homosexual men [12–14]. Perhaps the most common route for HCMV transmission in many populations in the world is through ingestion of breast milk from a previously infected mother, with reported rates of transmission being as high as 50%–70% [15–18].
In contrast to community sources of HCMV, infections identified in hospital settings such as those in transplant recipients can frequently be traced to specific sources such as the transplanted organ, blood products from donors with HCMV infections, or from reactivation of an existing latent infection in the immunocompromised host [19–24]. Notably, infections acquired from the infected allograft and/or blood products can occur in individuals with preexisting immunity to HCMV, although these new infections are established in the presence of deficits in adaptive and innate immune responses. Similarly, observations made in decade-old studies of HCMV infection in homosexual men reported that these individuals were not infrequently infected with genotypically diverse strains of HCMV, a finding that suggested reinfection in this population [14, 25]. However, the underlying immunostatus of these populations of homosexual men was not well defined, so it could be argued that reinfections in this population also resulted from deficits in adaptive immunity. Yet it is important to note that infection (reinfection) of the previously immunocompetent host, including pregnant women, also appears to take place [26, 27]. Studies in solid organ allograft recipients have provided definitive evidence that reinfections with HCMV occur and contribute to HCMV-associated disease in these individuals, yet mechanisms leading to reinfection in the presence of existing adaptive immunity to HCMV remained incompletely defined in allograft recipients and even less well understood in previously infected, immunocompetent women. In contrast to allograft recipients, reinfections that presumably follow community acquisition of HCMV are similar to those occurring in nonimmune individuals in that they are rarely, if ever associated with clinical symptomatology or documented laboratory abnormalities. Yet, reinfections in pregnant seroimmune women are believed to represent a major source of infection leading to intrauterine transmission and congenital HCMV (cCMV) infection [28–30]. Thus, understanding the source(s) of reinfection in pregnant seroimmune women and mechanisms responsible for acquisition of a new virus are of considerable importance in the design and potential testing of prophylactic vaccines as well as other interventions that could limit the incidence of cCMV, particularly in populations with high HCMV seroprevalence [31].
REINFECTION IN SEROIMMUNE WOMEN: INFECTIONS FOLLOWING EXPOSURE TO NEW VIRAL VARIANTS
Early studies in women attending STI clinics provided evidence that women could be infected with multiple genotypes of HCMV as defined by comparison of viral DNA fragment size following restriction endonuclease digestion [32]. Because individual women shed different viral genotypes at different times, the authors concluded that reinfection from an exogenous source of HCMV was relatively frequent [32]. Similarly, studies in children in group care facilities demonstrated shedding of different HCMV genotypes over time in individual children [33]. Interestingly, in this study about 16% (6/37) of the children who shed virus in longitudinal samples were found to have different viral genotypes in sequential specimens collected during the study [33]. These findings were interpreted as evidence that some children were reinfected multiple times within the time frame of this study [33]. Last, a number of investigators documented infections with multiple strains of HCMV in homosexual men utilizing similar methodologies, thus providing additional evidence of reinfection with genotypic variants of HCMV [14, 25].
Reinfection of seroimmune pregnant women with new genotypic variants of HCMV as a source of nonprimary maternal infections and resultant cCMV infection was described nearly 20 years ago [26]. Results from these studies were based on the presence of polymorphisms in a major antigenic site of the virion envelop glycoprotein H (gH) that allowed serological detection of new infections in previously HCMV seroimmune women [34]. Using antigens derived from this region of gH, acquisition of new viruses during pregnancy were described as a seroconversion, that is, the detection of new antibody reactivities to a specific antigenic site on gH [26]. In this study, the authors documented that women transmitted newly acquired viruses to their fetuses using sequence analysis of the UL75 viral gene (gH) from viruses recovered from infants with cCMV [26]. Subsequently, combinations of polymorphisms in antigenic sites on both envelope proteins gB and gH of HCMV have been used to further increase the sensitivity of serological detection of reinfection in pregnant seroimmune women [35, 36]. Using this assay system, the incidence of serologically defined reinfections in normal women during an intrapartum period was estimated to be about 30% over a period of 36 months [27]. Thus, reinfection of immunocompetent women as defined by the development of antibody reactivity to new antigenic determinants appears to be relatively frequent in some populations.
REINFECTION IN SEROIMMUNE WOMEN: PERSISTENT INFECTION VERSUS INFECTION WITH EXOGENOUS VIRUS
Two sources of virus that can result in infection of HCMV immune women have been proposed: (1) reactivation of a latent virus infection leading to recurrent infection and (2) infection following exposure to an exogenous source of virus. Although either mechanism could lead to intrauterine transmission and cCMV, distinguishing between these two sources of virus is of considerable importance in the design of strategies to limit the incidence of cCMV following nonprimary maternal infections, regardless if such strategies will rely on vaccine-induced immunity or behavioral interventions to limit exposures.
Reactivation of latent HCMV has long been argued to be the source of recurrent or nonprimary maternal infections leading to cCMV [37]. This paradigm is based on early studies that relied entirely on restriction enzyme digestions and comparison of restriction fragment length polymorphisms (RFLPs) as a methodology to genotype viral isolates, including studies designed to establish genetic relatedness of viruses recovered following transmission from an index case [38]. This approach represented a major advance in epidemiological studies of HCMV infection because early attempts to develop serological grouping of HCMV to aid epidemiological investigations of HCMV infections were unsuccessful, even though serological differences could be demonstrated [39, 40]. However, results from many of the published studies that utilized RFLPs to genotype viral isolates can no longer be considered as definitive evidence of genetic relatedness of HCMV isolates as the use of limited combinations of restriction enzymes that were often included in these studies sampled very limited amounts of the genetic diversity of HCMV. Furthermore, in these early studies virus isolates were often extensively passaged in vitro. More recent studies have shown that even brief passaging in vitro can introduce a variety of mutations into viral isolates, including deletions of regions of the genome. Such mutations could further confound a definitive assignment of genetic relatedness between viral isolates. Thus, even though the reactivation of latent virus leading to nonprimary infections in pregnant women remains consistent with an established paradigm in the biology of HCMV in the immunocompromised transplant recipient, this paradigm has not been rigorously validated in pregnant women in studies using contemporary technologies. Although definitive data are lacking, it is important to note that the frequency of reactivation of latent HCMV infections during pregnancy has not been defined but could be significant if intermittent reactivations of latent infections account for virus shedding in seroimmune women during pregnancy. However, available data that would argue that virus shedding during pregnancy reflects frequent reactivations of latent infections are confounded because most studies of virus shedding in pregnant seroimmune women have included women with increased risk for exposure to HCMV from known sources such as young children.
More recent findings from population studies have provided epidemiological data that are inconsistent with reactivation of latent virus as a sole source of nonprimary infections in pregnant seroimmune women. An epidemiological feature of cCMV infection that was noted decades ago is that as the seroprevalence increases in a maternal population, the rate of cCMV infections also increases, such that populations with the highest seroprevalence also have the highest prevalence of cCMV infections [31, 41, 42]. This observation is consistent with reactivation of latent maternal infections and resulting intrauterine transmission being a stochastic event such that as more women in the population are persistently infected, the prevalence of cCMV could be expected to increase. However, studies in some maternal populations argue against such a direct relationship between seroprevalence, reactivation, and cCMV infection. This is illustrated by the results from a recent study in Finland in which the prevalence of cCMV was calculated as 2 per 1000 based on results from screening of 20 000 newborn infants for HCMV shedding [45]. This result was somewhat surprising as the overall maternal seroprevalence in Finland has been reported to be about 70%, a rate that has been associated with a much higher prevalence of cCMV in many countries, including urban populations in the United States [46] (Table 1). Second, comparison of the seroprevalence in women of childbearing age and the prevalence of cCMV in 2 different ethnic groups in the United States again revealed a population-specific discrepancy between maternal seroprevalence and the prevalence of cCMV, such that the prevalence of cCMV is about 3-fold higher in black maternal populations as compared to Hispanic women [47, 48] (Table 2). Thus, a simple explanation based on a stochastic reactivation of latent HCMV in pregnant women as a source of infection is not entirely consistent with existing epidemiological data and suggests that other risk factors specific to maternal populations are associated with the delivery of an infant with cCMV in women with HCMV seroimmunity prior to conception.
Table 1.
Human Cytomegalovirus (HCMV) Seroreactivity and Congenital HCMV Prevalence in Maternal Populations
| Location | Maternal Seroreactivity | cCMV Prevalence |
|---|---|---|
| Brazil (Mussi-Pinhata, 2018) [29] | 98% | 6.1 |
| China (Wang, 2011) [28] | 96% | 7 |
| Japan (Tanimura, 2017) [43] | 71% | 6.4 |
| France (Leruez-Ville, 2017) [44] | 61% | 3.7 |
| Finland (Puhakka, 2018) [43]a | 71% | 2.0 |
Abbreviation: cCMV, congenital human cytomegalovirus.
aMaternal seroprevalence in Finland estimated based on data reported by Puhakka et al, 2016.
Table 2.
Maternal Seroreactivity and Congenital Human Cytomegalovirus Prevalence in 2 Maternal Populations
| Maternal Population | Maternal Seroimmunitya | cCMV Prevalenceb |
|---|---|---|
| Black | 75%–80% | 9.5/1000 |
| Hispanic | 75%–80% | 3.0/1000 |
In contrast to limited definitive data supporting reactivation of latent HCMV infection as a source of infection in immune pregnant women, reinfection of immunocompetent pregnant women with a new variant of HCMV defined by detection of new antibody reactivities has been described in several maternal populations and represents a plausible mechanism of nonprimary maternal infections leading to intrauterine transmission and cCMV [26, 35, 36]. However, it is important to stress that these studies remain incomplete, as the source of virus leading to reinfections in the pregnant women has not been definitively identified in available studies. Notably, studies that have described reinfection of pregnant women have been carried out in populations with high HCMV seroprevalence and in one study, women experiencing nonprimary infections leading to intrauterine transmission were more likely to be exposed to children <3 years of age, a well-described risk factor for virus exposure and infection (Figure 1) [35]. Interestingly, this risk factor for reinfection for pregnant women aligned with results from studies of virus infection in seronegative maternal populations in which exposure to young children has been the most consistently reported risk factor for HCMV infection and delivery of an infant with cCMV [49–54]. Thus, it appears that an established risk factor for HCMV infections during pregnancy—that is, exposure to young children—is common to both nonimmune and immune women undergoing HCMV infection during pregnancy. These observations would argue that reinfections (nonprimary) infections in seroimmune women during pregnancy can be explained by exposure to new viruses, presumably from household contacts and not solely by reactivations/recurrences of existing infections. Last, an alternative explanation for reinfections detected by development of new antibody reactivity has also been proposed. In this mechanism, the production of new serotype specific antibody reactivity that have been used to define reinfections in immune women is proposed to be generated following de novo expansion of minor populations of resident viruses in persistently infected women. This mechanism requires the establishment of persistent infection by multiple genotypic viral variants following initial infection and then, at a later time, expansion of a minor population to a sufficient level that can induce a measurable antibody response. Although current data cannot exclude this potential explanation, recent studies from next-generation sequencing of longitudinal specimens from immunocompromised children and congenitally infected infants have shown remarkable stability of virus populations within a host (intrahost) without periodic expansion of previously undetectable populations [55, 56]. Interestingly, the genetic complexity of virus populations in these individuals were markedly altered when new viral populations were introduced into the host, presumably as the result of reinfection with a new virus population [55, 56].
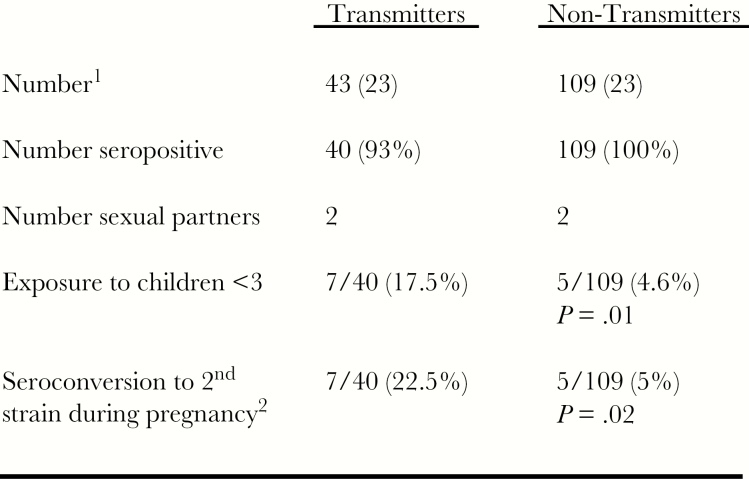
Figure 1.
Increased rate of intrauterine transmission of human cytomegalovirus (HCMV) in seroimmune women with seroconversion to new antigenic variants. Sequential serum samples from women who transmitted virus to their fetuses (transmitters) and matched controls who did not transmit virus (nontransmitters) from a highly HCMV-seroimmune population were assayed for polymorphic antigenic determinants on glycoprotein B and glycoprotein H to determine reinfection with new HCMV variants during pregnancy. aApproximately 4000 women were enrolled in this prospective study of HCMV infection during pregnancy. Demographic features of the population including number (median age at enrollment), HCMV seroreactivity, number of sexual partners, and exposure to children <3 years of age are shown. bThe rate of seroconversion to a new antigenic variant of HCMV is shown (22.5% in transmitters and 5% in nontransmitters). Note that exposure to children <3 years of age was associated with seroconversion to a new variant of HCMV. Adapted from Yamamoto et al [35].
MECHANISMS OF REINFECTIONS: VIRUS ESCAPE FROM HCMV-SPECIFIC ADAPTIVE IMMUNITY
Regardless if a serologically defined reinfection in a previously immune woman can be attributed to exposure to a new virus from a close contact or alternatively from expansion of a minor variant from a site of persistence, it can be inferred that new viral variants escape control by existing adaptive immunity, presumably virus-specific antibodies. Proposed mechanisms that facilitate evasion of HCMV from existing antiviral antibodies range from variations in primary sequence of virion proteins that are targets of protective antibodies to virus-encoded immune evasion functions, findings that suggest that HCMV acquires polymorphisms in virus-encoded targets of protective immune responses to allow escape and persistence in individuals and in populations. Sequence variations in several major HCMV envelope glycoproteins including gB, gH, gO, gN, and UL128 could limit recognition by potentially protective antiviral antibodies as each of these proteins has been shown to either be a target of functional antibodies or a component of protein complex recognized by functional antibodies, findings consistent with the critical role of these envelope proteins in the replication and infectivity of HCMV [57]. Examples of these include gH and gN in which infection with a new virus that results in seroconversion to a new gH or gN genotype results in the development of functional antibody responses reactive with the new antigenic variants (Figure 2) [26, 58, 59]. Other mechanisms that could limit the activity of existing antiviral antibodies include the presence of extensive carbohydrate modifications in several abundant envelope glycoproteins, including gB, gO, and gN in which carbohydrate modifications comprise approximately 40%, 50%, and 75%, respectively, of the mass of the virion protein [57]. Evidence of the importance of the carbohydrate modifications in the recognition of HCMV by antiviral antibodies against several glycoproteins including, gB, gH, and gN has been demonstrated by generation of recombinant viruses lacking a portion of the carbohydrate modifications of gN [57, 60]. Although variations in linear antibody binding sites could limit recognition of variant viruses, it is perhaps more likely that variations in the potential multitude of conformation-dependent antibody binding sites present on gB, gH, gN, and components of the pentamer (gH, gL, UL128-131) and trimer (gH, gL, gO) complexes could provide additional modes of escape from antiviral antibodies in the immune host. Findings from an early study demonstrating that anti-gB virus-neutralizing antibodies were directed at strain-specific sites in clinical HCMV isolates are consistent with variations in conformation-dependent antibody binding sites on gB [61]. Similarly, potent anti-gH virus neutralizing monoclonal antibodies have been shown to exhibit considerable differences in neutralization activity when assayed on different strains of HCMV, suggesting that viral strain–specific conformation-dependent epitopes could impact functional antibody activity in the host with existing immunity to HCMV [62, 63]. Last, investigators analyzing responses to a subunit gB vaccine have suggested that initial response of the immune system to nonprotective epitope of gB limited the subsequent development of functional antibody and potentially protective responses to virus infection, that is, a concept referred to as original antigenic sin in older literature [64].
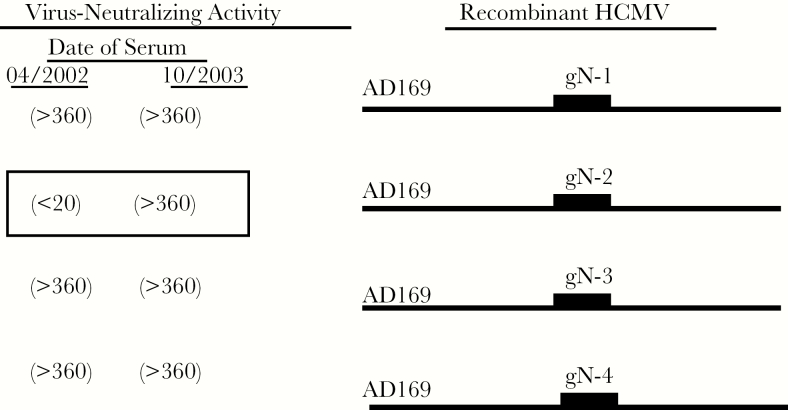
Figure 2.
Seroconversion to new glycoprotein N (gN) genotype. Congenic human cytomegalovirus (HCMV) was derived from the HB-5 HCMV BAC clone with replacement of UL73 with UL73 encoding gN genotypes 1–4 (Burkhardt, 2009) [56]. These viruses differed only in the amino acid sequence encoded by UL73 (gN). These viruses were used in a microneutralization assay to define neutralizing activity of sequential sera from an immunocompetent woman during the intrapartum period. This is unit of virus neutralizing activity the reciprocal of the serum dilution resulting in 50% reduction in infectivity is shown. Note the increase in neutralizing capacity of sera from the later date for AD169 gN2 indicating the development of new antibody reactivity for this gN genotype.
A large number of virus-encoded immune evasion functions have been shown to target both adaptive and innate immune responses to HCMV. In general, these immune evasion functions have been shown to target cellular effector functions of the immune response and less so humoral responses to HCMV. However, it is of interest that studies in a rhesus macaque model of CMV reinfection demonstrated that immune evasion functions encoded by rhesus CMV (RhCMV) that target CD8+ T-cell responses played an important role in reinfection of immune animals [65]. Deletion of these viral genes limited the capacity of a mutant RhCMV to reinfect immune animals, a finding that argues for a contribution of T-cell immunity in prevention of infection [65]. Similarly, the expression of a virus-encoded interleukin 10 functional homolog in RhCMV has also been shown to play a role in the early events of infection [66]. The importance of this immune modulating function of RhCMV in reinfection of animals with existing immunity remains to be determined.
Much of the preceding discussion is based on in vitro assay functional antibody activities that are projected to be protective in vivo. Yet there are few, if any, well-studied in vitro measures of protective antibody activity that can be assigned to a specific function(s) of antibody. Perhaps the most obvious example of the redundancy of in vitro antiviral antibody activities is virus neutralization. HCMV neutralizing antibodies have been shown to target gB, gH, gN, the trimer complex, and the pentamer complex [58, 67–70]. In addition, virus-neutralizing antibodies reactive with these virion envelope proteins or protein complexes can bind virus and in many cases, also bind to cells infected with HCMV and thus potentially lead to antibody dependent cellular cytotoxicity (ADCC)–mediated destruction of infected cells and potentially antibody-dependent phagocytosis (ADP) of virus. Of note, antiviral antibodies that participate in ADCC or ADP can be directed at epitopes present on viral envelope proteins that do not lead to in vitro virus neutralization and potentially, nonenvelope viral proteins (ADCC). In agreement with these possibilities, informative animal models and studies in recipients of a gB subunit vaccine have clearly demonstrated that in vitro virus-neutralizing activities do not correlate with in vivo protection and that in some cases antibodies that are nonneutralizing in vitro can have substantial protective activity in vivo [71, 72]. Finally, in assays quantifying the capacity of anti-envelope antibodies to limit cell-to-cell virus spread, a significant discrepancy was reported between the relative potency of antibodies that limit cell-to-cell spread as compared to their capacity to neutralize cell-free virus [73]. Thus, it appears that there are a number of mechanisms through which HCMV can potentially escape preexisting adaptive humoral immunity when antibody activity is measured by conventional assays, and it is an almost certainty that other mechanisms of escape from control by existing antiviral antibodies will be defined as additional mechanisms of functional antiviral antibody activity are defined.
CONCLUSIONS
The frequent occurrence of nonprimary maternal infections leading to cCMV infections and the similar long-term outcomes in infants with cCMV born to women undergoing primary and nonprimary infections presents one of the most vexing questions in the design, testing, and deployment of prophylactic HCMV vaccines [31, 74]. Current strategies in HCMV vaccine development have been forced to rely on the benchmark of adaptive immune responses that follow community-acquired primary infections of immunocompetent women. Even if these responses could be protective in the face of limited exposures to HCMV, it is possible that these responses will provide incomplete protection in maternal populations in which exposures to HCMV are more frequent and potentially of greater magnitude. In these populations, protective responses could require the induction of a greater magnitude and breadth of antiviral antibody reactivity to limit reinfection by new viruses of differing antigenic content. Thus, the performance of prophylactic vaccines that induce adaptive immune responses similar to those following naturally acquired infections may be quite different in highly seroimmune populations such as are present in South America, Asia, Africa, and in some urban populations in the United States when compared to maternal populations with low to intermediate seroprevalence and presumably less frequent exposures to HCMV. Additional understanding of the source(s) of nonprimary HCMV infections in seroimmune women should be considered a critical parameter in the design and eventual testing of candidate vaccines in seroimmune populations.
Notes
Acknowledgments. The author thanks Dr Suresh Boppana for critical reading of this manuscript; Dr Michael Mach for providing the reagents that were essential for many of the studies described in this manuscript; and Drs Marisa Mussi-Pinhata, Aparecida Yamamoto, and Geraldo Duarte for the collaborative studies carried out in Brazil.
Financial support. This work was supported by grants from the National Institute of Allergy and Infectious Diseases (grant numbers AI035602 and AI089956) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant number HD061959) of the US National Institutes of Health.
Supplement sponsorship. This supplement was sponsored by NIAID and NICHD.
Potential conflicts of interest. The author has received compensation from Sanofi Pasteur as a consultant.
The author has submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Cobbs CS. Cytomegalovirus and brain tumor: epidemiology, biology and therapeutic aspects. Curr Opin Oncol 2013; 25:682–8. [DOI] [PubMed] [Google Scholar]
- 2. Lawler SE. Cytomegalovirus and glioblastoma; controversies and opportunities. J Neurooncol 2015; 123:465–71. [DOI] [PubMed] [Google Scholar]
- 3. Joseph GP, McDermott R, Baryshnikova MA, Cobbs CS, Ulasov IV. Cytomegalovirus as an oncomodulatory agent in the progression of glioma. Cancer Lett 2017; 384:79–85. [DOI] [PubMed] [Google Scholar]
- 4. Nikolich-Žugich J, van Lier RAW. Cytomegalovirus (CMV) research in immune senescence comes of age: overview of the 6th International Workshop on CMV and Immunosenescence. Geroscience 2017; 39:245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nikitskaya E, Lebedeva A, Ivanova O, et al. Cytomegalovirus-productive infection is associated with acute coronary syndrome. J am Heart Assoc 2016; 5. doi:10.1161/JAHA.116.003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johansson I, Andersson R, Friman V, et al. Cytomegalovirus infection and disease reduce 10-year cardiac allograft vasculopathy-free survival in heart transplant recipients. BMC Infect Dis 2015; 15:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vora SB, Englund JA. Cytomegalovirus in immunocompromised children. Curr Opin Infect Dis 2015; 28:323–9. [DOI] [PubMed] [Google Scholar]
- 8. Vicetti Miguel CP, Mejias A, Ramilo O, Ardura MI, Sánchez PJ. Cytomegalovirus meningitis in an infant with severe combined immunodeficiency. J Pediatr 2016; 173:235–7. [DOI] [PubMed] [Google Scholar]
- 9. Cannon MJ, Stowell JD, Clark R, et al. Repeated measures study of weekly and daily cytomegalovirus shedding patterns in saliva and urine of healthy cytomegalovirus-seropositive children. BMC Infect Dis 2014; 14:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Murph JR, Souza IE, Dawson JD, et al. Epidemiology of congenital cytomegalovirus infection: maternal risk factors and molecular analysis of cytomegalovirus strains. Am J Epidemiol 1998; 147:940–7. [DOI] [PubMed] [Google Scholar]
- 11. Fowler KB, Stagno S, Pass RF. Maternal age and congenital cytomegalovirus infection: screening of two diverse newborn populations, 1980-1990. J Infect Dis 1993; 168:552–6. [DOI] [PubMed] [Google Scholar]
- 12. Chandler SH, Handsfield HH, McDougall JK. Isolation of multiple strains of cytomegalovirus from women attending a clinic for sexually transmitted disease. J Infect Dis 1987; 155:655–60. [DOI] [PubMed] [Google Scholar]
- 13. Handsfield HH, Chandler SH, Caine VA, et al. Cytomegalovirus infection in sex partners: evidence for sexual transmission. J Infect Dis 1985; 151:344–8. [DOI] [PubMed] [Google Scholar]
- 14. Collier AC, Chandler SH, Handsfield HH, Corey L, McDougall JK. Identification of multiple strains of cytomegalovirus in homosexual men. J Infect Dis 1989; 159:123–6. [DOI] [PubMed] [Google Scholar]
- 15. Martins-Celini FP, Yamamoto AY, Passos DM, et al. Incidence, risk factors, and morbidity of acquired postnatal cytomegalovirus infection among preterm infants fed maternal milk in a highly seropositive population. Clin Infect Dis 2016; 63:929–36. [DOI] [PubMed] [Google Scholar]
- 16. Hamprecht K, Goelz R. Postnatal cytomegalovirus infection through human milk in preterm infants: transmission, clinical presentation, and prevention. Clin Perinatol 2017; 44:121–30. [DOI] [PubMed] [Google Scholar]
- 17. Jobe AH. CMV transmission in human milk. J Pediatr 2009; 154:A1. [DOI] [PubMed] [Google Scholar]
- 18. Stagno S, Reynolds DW, Pass RF, Alford CA. Breast milk and the risk of cytomegalovirus infection. N Engl J Med 1980; 302:1073–6. [DOI] [PubMed] [Google Scholar]
- 19. Koval CE. Prevention and treatment of cytomegalovirus infections in solid organ transplant recipients. Infect Dis Clin North Am 2018; 32:581–97. [DOI] [PubMed] [Google Scholar]
- 20. Haidar G, Singh N. Viral infections in solid organ transplant recipients: novel updates and a review of the classics. Curr Opin Infect Dis 2017; 30:579–88. [DOI] [PubMed] [Google Scholar]
- 21. Ariza-Heredia EJ, Nesher L, Chemaly RF. Cytomegalovirus diseases after hematopoietic stem cell transplantation: a mini-review. Cancer Lett 2014; 342:1–8. [DOI] [PubMed] [Google Scholar]
- 22. Beam E, Razonable RR. Cytomegalovirus in solid organ transplantation: epidemiology, prevention, and treatment. Curr Infect Dis Rep 2012; 14:633–41. [DOI] [PubMed] [Google Scholar]
- 23. Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med 2007; 357:2601–14. [DOI] [PubMed] [Google Scholar]
- 24. Ljungman P, Hakki M, Boeckh M. Cytomegalovirus in hematopoietic stem cell transplant recipients. Hematol Oncol Clin North Am 2011; 25:151–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drew WL, Sweet ES, Miner RC, Mocarski ES. Multiple infections by cytomegalovirus in patients with acquired immunodeficiency syndrome: documentation by Southern blot hybridization. J Infect Dis 1984; 150:952–3. [DOI] [PubMed] [Google Scholar]
- 26. Boppana SB, Rivera LB, Fowler KB, Mach M, Britt WJ. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med 2001; 344:1366–71. [DOI] [PubMed] [Google Scholar]
- 27. Ross SA, Arora N, Novak Z, Fowler KB, Britt WJ, Boppana SB. Cytomegalovirus reinfections in healthy seroimmune women. J Infect Dis 2010; 201:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang C, Zhang X, Bialek S, Cannon MJ. Attribution of congenital cytomegalovirus infection to primary versus non-primary maternal infection. Clin Infect Dis 2011; 52:e11–3. [DOI] [PubMed] [Google Scholar]
- 29. Mussi-Pinhata MM, Yamamoto AY, Aragon DC, et al. Seroconversion for cytomegalovirus infection during pregnancy and fetal infection in a highly seropositive population: “the BraCHS study.” J Infect Dis 2018; 218:1200–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adachi K, Xu J, Ank B, et al. NICHD HPTN 040 Study Team Cytomegalovirus urinary shedding in HIV-infected pregnant women and congenital cytomegalovirus infection. Clin Infect Dis 2017; 65:405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Britt WJ. Maternal immunity and the natural history of congenital human cytomegalovirus infection. Viruses 2018; 10. doi:10.3390/v10080405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chandler SH, Handsfield HH, McDougall JK. Isolation of multiple strains of cytomegalovirus from women attending a clinic for sexually transmitted disease. J Infect Dis 1987; 155:655–60. [DOI] [PubMed] [Google Scholar]
- 33. Bale JF Jr, Petheram SJ, Souza IE, Murph JR. Cytomegalovirus reinfection in young children. J Pediatr 1996; 128:347–52. [DOI] [PubMed] [Google Scholar]
- 34. Urban M, Klein M, Britt WJ, Hassfurther E, Mach M. Glycoprotein H of human cytomegalovirus is a major antigen for the neutralizing humoral immune response. J Gen Virol 1996; 77(Pt 7):1537–47. [DOI] [PubMed] [Google Scholar]
- 35. Yamamoto AY, Mussi-Pinhata MM, Boppana SB, et al. Human cytomegalovirus reinfection is associated with intrauterine transmission in a highly cytomegalovirus-immune maternal population. Am J Obstet Gynecol 2010; 202:297.e1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ikuta K, Minematsu T, Inoue N, et al. Cytomegalovirus (CMV) glycoprotein H-based serological analysis in Japanese healthy pregnant women, and in neonates with congenital CMV infection and their mothers. J Clin Virol 2013; 58:474–8. [DOI] [PubMed] [Google Scholar]
- 37. Alford CA, Stagno S, Pass RF. Natural history of perinatal cytomegaloviral infection. Ciba Found Symp 1979; 77:125–47. [DOI] [PubMed] [Google Scholar]
- 38. Huang ES, Alford CA, Reynolds DW, Stagno S, Pass RF. Molecular epidemiology of cytomegalovirus infections in women and their infants. N Engl J Med 1980; 303:958–62. [DOI] [PubMed] [Google Scholar]
- 39. Faix RG. Cytomegalovirus antigenic heterogeneity can cause false-negative results in indirect hemagglutination and complement fixation antibody assays. J Clin Microbiol 1985; 22:768–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stagno S, Reynolds DW, Lakeman A, Charamella LJ, Alford CA. Congenital cytomegalovirus infection: consecutive occurrence due to viruses with similar antigenic compositions. Pediatrics 1973; 52:788–94. [PubMed] [Google Scholar]
- 41. Stagno S, Pass RF, Dworsky ME, Alford CA. Congenital and perinatal cytomegalovirus infections. Semin Perinatol 1983; 7:31–42. [PubMed] [Google Scholar]
- 42. Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 2007; 17:253–76. [DOI] [PubMed] [Google Scholar]
- 43.Tanimura K, Tairaku S, Morioka I, et al. Universal screening with use of immunoglobulin G avidity for congenital cytomegalovirus infection. Clin Infect Dis 2017; 65:1652–1658. [DOI] [PubMed] [Google Scholar]
- 44.Leruez-Ville M, Magny JF, Couderc S, et al. Risk factors for congenital cytomegalovirus infection following primary and nonprimary maternal infection: a prospective neonatal screening study using polymerase chain reaction in saliva. Clin Infect Dis 2017;65:398–404. [DOI] [PubMed] [Google Scholar]
- 45. Puhakka L, Lappalainen M, Lonnqvist T, et al. The burden of congenital cytomegalovirus infection: a prospective cohort study of 20 000 infants in Finland. J Pediatric Infect Dis Soc 2018. doi: 10.1093/jpids/piy027. [DOI] [PubMed] [Google Scholar]
- 46. Puhakka L, Sarvikivi E, Lappalainen M, Surcel HM, Saxen H. Decrease in seroprevalence for herpesviruses among pregnant women in Finland: cross-sectional study of three time points 1992, 2002 and 2012. Infect Dis (Lond) 2016; 48:406–10. [DOI] [PubMed] [Google Scholar]
- 47. Fowler KB, Ross SA, Shimamura M, et al. Racial and ethnic differences in the prevalence of congenital cytomegalovirus infection. J Pediatr 2018; 200:196–201.e1. [DOI] [PubMed] [Google Scholar]
- 48. Colugnati FA, Staras SA, Dollard SC, Cannon MJ. Incidence of cytomegalovirus infection among the general population and pregnant women in the United States. BMC Infect Dis 2007; 7:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stagno S, Cloud G, Pass RF, Britt WJ, Alford CA. Factors associated with primary cytomegalovirus infection during pregnancy. J Med Virol 1984; 13:347–53. [DOI] [PubMed] [Google Scholar]
- 50. Pass RF, Hutto C, Ricks R, Cloud GA. Increased rate of cytomegalovirus infection among parents of children attending day-care centers. N Engl J Med 1986; 314:1414–8. [DOI] [PubMed] [Google Scholar]
- 51. Adler SP. Cytomegalovirus transmission among children in day care, their mothers and caretakers. Pediatr Infect Dis J 1988; 7:279–85. [DOI] [PubMed] [Google Scholar]
- 52. Zheng QY, Huynh KT, van Zuylen WJ, Craig ME, Rawlinson WD. Cytomegalovirus infection in day care centres: a systematic review and meta-analysis of prevalence of infection in children. Rev Med Virol 2019; 29:e2011. [DOI] [PubMed] [Google Scholar]
- 53. Marshall BC, Adler SP. The frequency of pregnancy and exposure to cytomegalovirus infections among women with a young child in day care. Am J Obstet Gynecol 2009; 200:163.e1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hyde TB, Schmid DS, Cannon MJ. Cytomegalovirus seroconversion rates and risk factors: implications for congenital CMV. Rev Med Virol 2010; 20:311–26. [DOI] [PubMed] [Google Scholar]
- 55. Cudini J, Roy S, Houldcroft CJ, et al. Human cytomegalovirus haplotype reconstruction reveals high diversity due to superinfection and evidence of within-host recombination. Proc Natl Acad Sci U S A 2019; 116:5693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pokalyuk C, Renzette N, Irwin KK, et al. Characterizing human cytomegalovirus reinfection in congenitally infected infants: an evolutionary perspective. Mol Ecol 2017; 26:1980–90. [DOI] [PubMed] [Google Scholar]
- 57. Gardner TJ, Tortorella D. Virion glycoprotein-mediated immune evasion by human cytomegalovirus: a sticky virus makes a slick getaway. Microbiol Mol Biol Rev 2016; 80:663–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Burkhardt C, Himmelein S, Britt W, Winkler T, Mach M. Glycoprotein N subtypes of human cytomegalovirus induce a strain-specific antibody response during natural infection. J Gen Virol 2009; 90:1951–61. [DOI] [PubMed] [Google Scholar]
- 59. Pati SK, Novak Z, Purser M, et al. Strain-specific neutralizing antibody responses against human cytomegalovirus envelope glycoprotein N. Clin Vaccine Immunol 2012; 19:909–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kropff B, Burkhardt C, Schott J, et al. Glycoprotein N of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS Pathog 2012; 8:e1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Britt WJ. Recent advances in the identification of significant human cytomegalovirus-encoded proteins. Transplant Proc 1991; 23:64–9, discussion 69. [PubMed] [Google Scholar]
- 62. Simpson JA, Chow JC, Baker J, et al. Neutralizing monoclonal antibodies that distinguish three antigenic sites on human cytomegalovirus glycoprotein H have conformationally distinct binding sites. J Virol 1993; 67:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gardner TJ, Stein KR, Duty JA, et al. Functional screening for anti-CMV biologics identifies a broadly neutralizing epitope of an essential envelope protein. Nat Commun 2016; 7:13627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Baraniak I, Kern F, Holenya P, Griffiths P, Reeves M. Original antigenic sin shapes the immunological repertoire evoked by human cytomegalovirus glycoprotein B/MF59 vaccine in seropositive recipients. J Infect Dis 2019; 220:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hansen SG, Powers CJ, Richards R, et al. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 2010; 328:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deere JD, Chang WLW, Villalobos A, et al. Neutralization of rhesus cytomegalovirus IL-10 reduces horizontal transmission and alters long-term immunity. Proc Natl Acad Sci U S A 2019; 116:13036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Britt WJ, Vugler L, Butfiloski EJ, Stephens EB. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol 1990; 64:1079–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shimamura M, Mach M, Britt WJ. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol 2006; 80:4591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vanarsdall AL, Chin AL, Liu J, et al. HCMV trimer- and pentamer-specific antibodies synergize for virus neutralization but do not correlate with congenital transmission. Proc Natl Acad Sci U S A 2019; 116:3728–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Macagno A, Bernasconi NL, Vanzetta F, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol 2010; 84:1005–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bootz A, Karbach A, Spindler J, et al. Protective capacity of neutralizing and non-neutralizing antibodies against glycoprotein B of cytomegalovirus. PLoS Pathog 2017; 13:e1006601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Baraniak I, Kropff B, Ambrose L, et al. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A 2018; 115:6273–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Murrell I, Bedford C, Ladell K, et al. The pentameric complex drives immunologically covert cell-cell transmission of wild-type human cytomegalovirus. Proc Natl Acad Sci U S A 2017; 114:6104–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Britt WJ. Congenital human cytomegalovirus infection and the enigma of maternal immunity. J Virol 2017; 91. doi:10.1128/JVI.02392-16. [DOI] [PMC free article] [PubMed] [Google Scholar]