Abstract
Motor-neuron specific microRNA-218 (miR-218) has recently received attention because of its roles in mouse development. However, miR-218 relevance to human motor neuron disease was not yet explored. Here, we demonstrate by neuropathology that miR-218 is abundant in healthy human motor neurons. However, in amyotrophic lateral sclerosis (ALS) motor neurons miR-218 is downregulated and its mRNA targets are reciprocally upregulated (de-repressed). We further identify the potassium channel Kv10.1 as a new miR-218 direct target that controls neuronal activity. In addition, we screened thousands of ALS genomes and identified six rare variants in the human miR-218-2 sequence. miR-218 gene variants fail to regulate neuron activity, suggesting the importance of this small endogenous RNA for neuronal robustness. The underlying mechanisms involve inhibition of miR-218 biogenesis and reduced processing by DICER. Therefore, miR-218 activity in motor neurons may be susceptible to failure in human ALS suggesting that miR-218 may be a potential therapeutic target in motor neuron disease.
One Sentence Summary:
Genetics, pathology and molecular studies demonstrate that miR-218 is modulated and might play a role in amyotrophic lateral sclerosis.
Introduction
microRNA-218 (miR-218) is an endogenous small RNA that is enriched in motor neurons. Its relevance to motor neuron diseases was recently suggested by showing that miR-218 is essential for perinatal neuromuscular survival (1, 2), it is decreased in human amyotrophic lateral sclerosis (ALS) post-mortem spinal cord (3, 4), that cell-free miR-218 can serve as marker for motor neuron loss in a rodent model of ALS (4) and as a neuron-to-astrocyte signal (5). However, miR-218 was not yet studied in human motor neurons and relevance to human ALS is still missing.
ALS is a fatal disease of the human motor neuron system, characterized by the selective degeneration of cortical and ventral spinal motor neurons.
More than two dozen different genes have been associated with ALS in families or via genome-wide association studies. Mutations in these genes explain only a small fraction of the cases (6-9). Thus, ALS genetic variants in SOD1, NEK1, TARDBP or FUS are observed in <1-3% of cases and the fraction of disease explained by the hexanucleotide repeat at the first exon of C9orf72 is <10% (6, 10). ALS-associated genes are ubiquitously expressed and therefore provide limited insight as to why ALS shows motor neuron-selective vulnerability (8, 11).
Differential susceptibilities could be explained by the dysregulated activity of cell-type specific genes, including miRNAs. Indeed, we and others have shown that miRNA dysregulation is involved in ALS (3, 12-18).
In this study, we demonstrate (i) that miR-218 is specifically enriched in human spinal motor neurons and is downregulated in ALS, (ii) that miR-218 orchestrates neuronal activity in a new pathway upstream of Kv10.1 (Kcnh1) voltage-gated potassium channel and (iii) that rare genetic miR-218 variants, identified in patients with ALS, are detrimental to its biogenesis and function, providing a connection from human genetics to motor neuron-specific functions.
Results
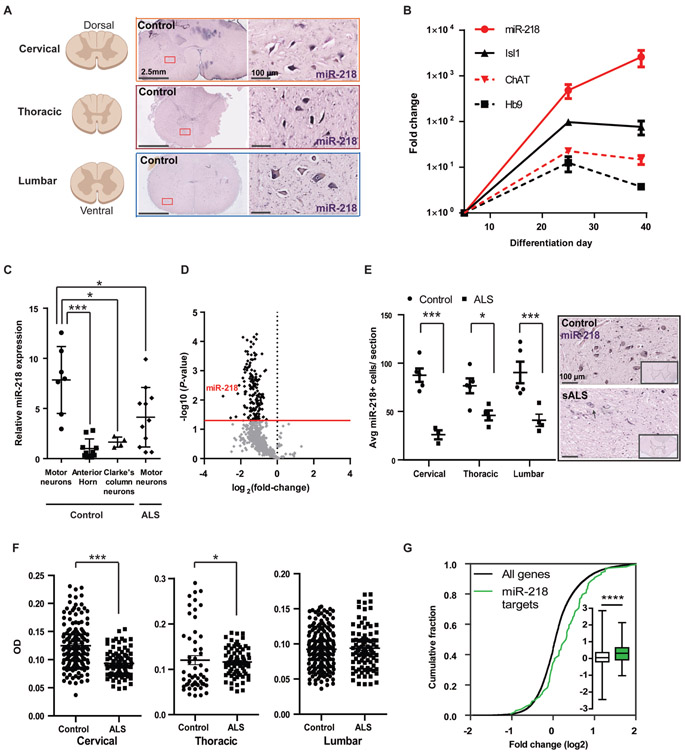
miR-218 is highly and specifically expressed in mature human and murine motor neurons We sought to evaluate the relevance of miR-218 to human motor neuron and its relevance to ALS. First, miRNA in situ hybridization in human tissues depicted motor neuron specific expression pattern of miR-218 in ventral motor neurons throughout the human spinal cord (Fig. 1A). In parallel, we differentiated human inducible pluripotent stem cells (iPSCs) into motor neurons, following a protocol developed by Kiskinis et al. (19). Accordingly, several mRNA markers of motor neuron differentiation were upregulated, namely, Isl1, Hb9 and ChAT. miR-218 expression was upregulated >2000 fold from undifferentiated pluripotent state to human motor neurons (Fig. 1B). We then assessed miR-218 expression in laser capture microdissection motor neurons from lumbar spinal cords of samples where neurological disease was not reported, by revisiting data that was generated in our previous work (3). miR-218 is specifically enriched in control motor neurons relative to surrounding non motor neuron tissue at the ventral horn of the human lumbar spinal cord or relative to proprioceptive neurons at Clarke’s column. Furthermore, assessing miR-218 expression in laser capture microdissection-enriched surviving lumbar motor neurons of patients with ALS that suffered from bulbar onset disease, revealed ~2 fold repression relative to control lumbar motor neurons (Fig. 1C and Datafile S1). We further tested another independent set of postmortem tissues with an orthogonal nanoString nCounter miRNA profiler. This RNA study revealed that miR-218 was the most downregulated miRNA in lumbar ventral horns of sporadic ALS (sALS) nervous systems, relative to non-neurodegeneration controls (Fig. 1D and Datafile S2). Reduced miR-218 in ALS may be explained by loss of motor neurons and / or by molecular downregulation in motor neurons that are still present in the ventral horn. Accordingly, we have performed miR-218 in situ hybridization that revealed reduced numbers of miR-218+ cells in ALS patient tissue relative to non-neurodegeneration controls (Fig. 1E and Datafile S1) and a reduction in the densitometric miR-218 in situ hybridization signal in ALS motor neurons (Fig. 1F). Finally, we demonstrated that there is a global upregulation (de-repression) of miR-218-5p targets in human ALS spinal motor neurons by comparing the expression of top 100 predicted miR-218-5p mRNA targets (TargetScan (20)), in laser capture microdissection-enriched surviving motor neurons from lumbar spinal cords of patients with sALS relative to all expressed mRNAs and to the expression in non-neurodegeneration controls (Fig. 1G (21)). Taken together, our results show that miR-218 is a highly sensitive marker of human spinal motor neurons, whose expression rises high in developing human motor neuron and is maintained in the adult. miR-218 expression is reduced in motor neuron disease because of both molecular downregulation and of motor neuron loss and the mRNA targets of miR-218 are reciprocally upregulated. Therefore, miR-218 might serve as marker of motor neuron mass in the human ventral horn in ALS.
Fig. 1. miR-218 is expressed in the human spinal motor neurons and is downregulated in human ALS.
(A-F) Three orthogonal miRNA quantification studies in human motor neurons from 20 ALS cases and 14 non-neurodegeneration controls: (A) miR-218 chromogenic in situ hybridization depicting broad expression along the cervical, thoracic and lumbar regions of the adult human spinal cord. (B) qPCR analysis of miR-218, Hb9, Isl1 and ChAT in human iPSCs and differentiated motor neurons. miR-218 normalized to U6 expression. mRNAs normalized to average of HPRT and β–actin expression, presented on a log scale; n=3 independent wells per time point. (C) miR-218 expression in laser-capture micro-dissected human lumbar motor neurons. miR-218 expression in non-neurodegeneration motor neurons (n=7 human spinal cords), relative to surrounding non-motor neuron anterior horn tissue (n=10), to Clarke’s column proprioceptive neurons (n=4), or to ALS motor neurons (n=9 sporadic and 2 familial nervous systems carrying the SOD1 A4V mutation). TaqMan qPCR analysis of miR-218 normalized to the average of RNU48/SNORD48, RNU44/SNORD44 and U6 in the same sample, and to the average miR-218 expression in the anterior horn. One-way ANOVA followed by Newman-Keuls multiple comparison test performed on log-transformed data, Means ± SD. (D) Volcano plot of relative miRNA expression in ALS lumbar ventral horns (n=5), versus non-neurodegeneration controls (n=2; x-axis log2 scale), screened by nanoString nCounter platform. y-axis depicts the differential expression p-values (−log10 scale). Black dots indicate P < 0.05; light gray dots are non-significant. miR-218 is the most downregulated miRNA in ALS nervous systems. Data normalized to the average of five control mRNAs (ACTB, B2M, GAPDH, RPL19, RPLP0). (E) Reduced miR-218+ cell numbers in sALS patient anterior horns (n=4), relative to non-neurodegeneration controls (n=5) and representative miRNA in situ hybridization micrographs. Two way ANOVA followed by Bonferroni’s multiple comparison test, Means ± SEM, and (F) chromogenic miR-218 in situ hybridization signal densitometry in motor neurons at different spinal cord levels (non-neurodegeneration control/ALS cases: Cervical n=151/85 cells; Thoracic n=54/75; Lumbar 189/92). One-tailed Mann-Whitney test, Means ± SEM. (G) Cumulative distribution function (CDF) plot of top 100 predicted miR-218-5p targets (TargetScan (20)), or all expressed mRNAs, in laser capture microdissection-enriched surviving motor neurons from lumbar spinal cords of patients with sALS with rostral onset and caudal progression (n=13) relative to non-neurodegeneration controls (n=6; (21)) and Box-Plot (inset) depicting median, upper and lower quartiles and extreme points. P-value calculated using Kolmogorov–Smirnov test comparing miR-218-5p targets subset distribution to all genes. * P<0.05; *** P<0.001; **** P<0.0001.
miR-218 regulates motor neuron network activity
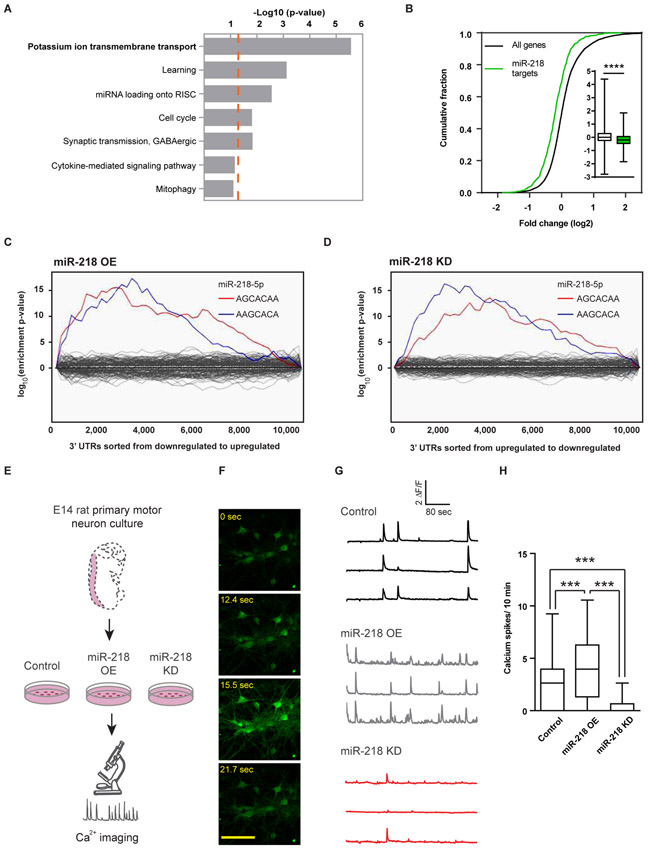
To study miR-218 function, we moved to rodent models, whereby miR-218 is specifically expressed in mouse motor neurons, without any preference to motor neuron subtypes (Fig. S1 and (1, 2)). We first performed ontology analysis (22) of predicted miR-218 targets (20). This study identified enrichment in biological processes related to potassium ion transmembrane transport (Fig. 2A). Therefore, we tested the hypothesis that miR-218 regulates primary motor neuron gene expression and activity. Dissociated embryonic mouse spinal cords, were enriched for motor neurons via optiprep gradient sedimentation (23) and transduced with lentiviruses encoding miR-218 overexpression (OE) or miR-218 knockdown (KD). Next generation sequencing (NGS) of RNA revealed that predicted miR-218 targets (TargetScan (20)), were significantly down-regulated following OE of miR-218 (Fig. 2B, p<0.0001). Accordingly, enrichment for two miR-218-5p seed-matches was depicted among mRNAs that were down / up regulated following miR-218 OE / KD, respectively (Sylamer study (24), Fig. 2C,D). No signatures were identified for the target set of any other miRNA. Therefore, the vectors used were specifically affecting miR-218 expression or silencing functions. Expression data are available at gene expression Omnibus (GSE136409).
Fig. 2. miR-218 controls motor neuron network activity.
(A) Seven most enriched gene ontology terms (22) of predicted miR-218 targets (20). p-value of term enrichment (−log10, dashed orange line indicates P = 0.05). (B) CDF plot of miR-218 predicted targets, relative to all expressed mRNAs, following OE of miR-218 and box-plot (insets), depicting median, upper and lower quartiles and extreme points. P-value calculated using Kolmogorov–Smirnov test comparing miR-218-5p subset distribution to all genes. **** P<0.0001. Binding site enrichment of all known miRNAs, in ~10,000 expressed mRNAs, was tested after (C) miR-218 OE, or (D) miR-218 KD, relative to control virus. Significant enrichment for two miR-218-5p seed-matches (blue, red) and lack of enrichment for any other miRNA (gray) via a Sylamer study (24). (E) Diagram of calcium transient imaging in embryonic rat spinal motor neurons, transduced with lentiviruses encoding control vector, miR-218 OE or a miR-218 KD. Neuron time lapse micrographs (F), representative traces (G) and (H) quantification of spontaneous calcium spike frequencies (ΔF/F >0.5) from Fluo2 HighAff AM study after 12 days in vitro. Recorded from 58 / 76 / 41 control / OE /KD cells, respectively. Box-Plot depicting the median, upper and lower quartiles and extreme points. Kruskal-Wallis test followed by Dunn’s multiple comparison test, *** P<0.001. This experiment was repeated 3 independent times with similar results.
We then monitored intracellular calcium transients in primary rat motor neurons that overexpressed (~8-fold), or knocked-down (~50%) miR-218. Calcium dynamics were monitored on days 12-13 in vitro, using the Ca2+-sensitive dye, Fluo2 HighAff AM, setting the spike threshold for activity as delta F / F > 2 over baseline (Fig. 2E,F). miR-218 OE increased the frequency of spontaneous calcium bursts by ~70%, compared to cells that were transduced with control viruses, whereas miR-218 KD attenuated neuronal Ca2+ transient by ~80%, relative to control (Fig. 2G,H). Changes in miR-218 expression did not alter motor neuron viability or morphology (Fig. S2). Therefore, miR-218 regulates neuronal activity.
miR-218 regulates neuronal intrinsic excitability
To test if miR-218 is involved in the regulation of active or passive conductance in neurons, we further employed patch-clamp. However, since primary motor neurons displayed an elevated resting membrane potential of >−50mV in our hands, consistent with a previous study (25), we were forced to use primary rat hippocampal neurons as alternative, a well-established cell type for patch clamp studies, which expresses miR-218, though less than spinal motor neurons (26-28). Current clamp electrophysiological experiments were performed with CNQX (6-cyano-7-nitroquinoxaline-2,3-dione, AMPA/Kainate blocker) and APV (2-amino-5-phosphonopentanoate, NMDA receptor blocker), on culture days 15-21. In response to current injection (300 pA, 500ms) neuronal firing frequency was ~twofold higher with miR-218 OE, relative to miR-218 KD (17.9 Hz ± 1.3 vs. 9.4 ± 1.9 Hz, p<0.01, Fig. S3A,B) and rheobase, the current input required to generate an action potential (500ms −100 to +500 pA steps in 20 pA increments), was ~35% lower in miR-218 OE, relative miE-218 KD (157 ± 12 pA vs. 242 ± 19 pA, p<0.001, Fig. S3C,D). Mean voltage threshold for triggering the first spike was unchanged between the different conditions (Fig. S3E) and resting membrane potential (RMP) correlated in a bidirectional manner with miR-218 expression (miR-218 OE −58.8± 0.7 mV, n=65; control, −60.9±0.7 mV, n=44; miR-218 KD −63.7 ± 0.7 mV, n=30, p-value<0.001, Fig. S3F). Taken together, network and intrinsic activity studies support the hypothesis that miR-218 regulates neuronal excitability, at least in rat hippocampal neurons.
Kv10.1 functions downstream of miR-218 in motor neurons
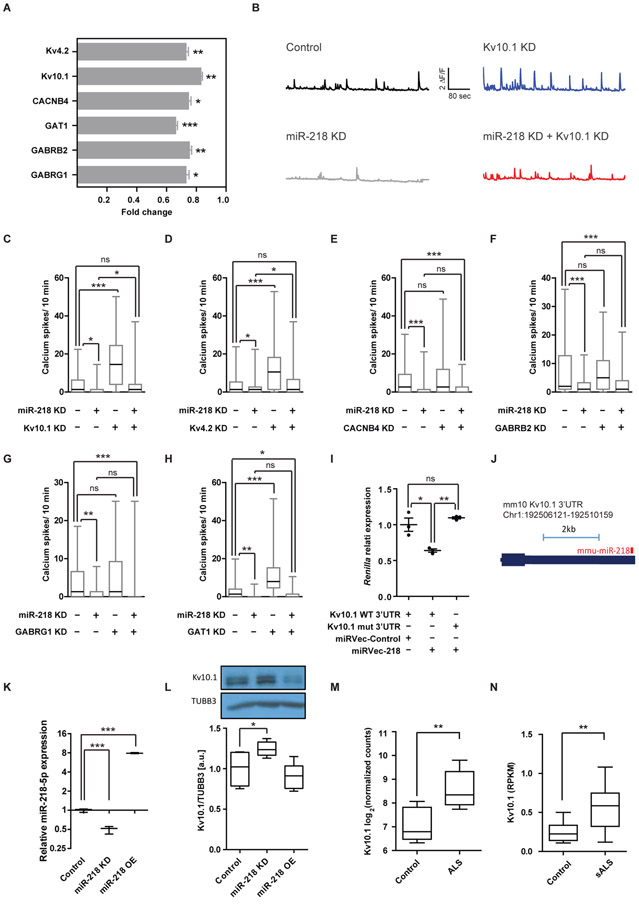
To gain molecular insight into the mechanisms by which miR-218 regulates network activity, we next focused on a selected set of relevant targets in the context of neuronal activity. This group includes the potassium channels Kv4.2 (Kcnd2) and Kv10.1 (Kcnh1), GABA receptor subunits Gabrb2 and Gabrg1, GABA transporter GAT1 (Slc6a1) and the calcium channel beta subunit Cacnb4. The changes in the expression of the above six targets, in response to miR-218 overexpression, were validated in an independent set of experiments using qPCR on RNA extracted from rat primary motor neurons (Fig. 3A).
Fig. 3. The potassium channel Kv10.1 acts downstream of miR-218.
(A) qPCR measuring the expression of mRNAs targets, following miR-218 OE (n=15). Data normalized to control virus (n=12) and to average expression of HPRT and β–actin, two technical duplicates, two-sided student’s t-test, Means ± SEM. (B) Representative traces of individual motor neurons and (C-H) quantification of spontaneous calcium spike frequencies (ΔF/F >0.5) of embryonic rat spinal motor neurons, transduced with lentiviruses encoding a control vector or miR-218 KD and further transfected with siRNA for specific target KD or a non-targeting siRNA control (minus sign). ≥55 cells recorded per each experimental condition; N≥2 independent experimental repeats with similar results. Kruskal-Wallis test followed by Dunn’s multiple comparison test. (I) Relative Renilla luminescence upstream of a wild-type Kv10.1 3′UTR or a mutated 3′UTR that is insensitive to miR-218, normalized to co-expressed firefly luciferase and to a negative control miRNA vector. n=3 independent wells per experimental condition. One-way ANOVA followed by Bonferronie’s multiple comparison test. Means ± SEM. (J) miR-218:Kv10.1 3’UTR chimera from an AGO2 CLEAR-CLIP experiment in mouse cortex (29). (K) miR-218 expression (qPCR n=3, normalized to U6) and (L) Kv10.1 protein expression (Western blot n =5), upon miR-218 lentiviral KD or OE, in primary rat motor neurons and a representative blot detected with anti Kv10.1 and anti Tubulin Beta-III (TUBB3) antibodies. Box-Plots depict median, upper / lower quartiles & extreme points, one-way ANOVA followed by Newman-Keuls multiple comparison test. (M) Kv10.1 mRNA expression, as log2 normalized counts, from NGS study of induced ALS motor neurons (n=4 different donors in duplicates) or non-neurodegeneration controls (n=3 different donors in duplicates; (30)). Box-Plots depict median, upper / lower quartiles & extreme points, DESeq analysis. (N) Kv10.1 mRNA expression, as Reads Per Kilobase Million (RPKM) from NGS study of laser capture microdissection-enriched surviving motor neurons from lumbar spinal cords of patients with sALS with rostral onset and caudal progression (n=12) and non-neurodegeneration controls (n=8; (21), GSE76220). Box-Plots depict median, upper / lower quartiles & extreme points, two-sided student’s t-test. * P<0.05; ** P<0.01; *** P<0.001; ns – non-significant.
Because miR-218 enhances neuronal activity, we hypothesized that relevant mRNA targets potentially encode for proteins acting downstream of miR-218 in inhibiting neuronal activity. Thus, their KD should increase bursting, reminiscent of miR-218 OE, and concomitant KD of both miR-218 and its target may rescue neuronal activity.
We therefore analyzed the frequency of spontaneous calcium transients in primary motor neurons following candidate target KD, with siRNA nanoparticles that exhibited 20%-80% target mRNA KD (Fig. S4). Non-targeting siRNAs were used as control. Knockdown of either Kv10.1 (Kcnh1) or Kv4.2 (Kcnd2) enhanced the frequency of spontaneous calcium transients and was sufficient to rescue neuronal excitation upon miR-218 inhibition (Fig. 3B-D). In addition, we tested the calcium channel Cacnb4 and GABA pathway components Gabrb2, Gabrg1 and GAT1, which did not obey the requirements to be considered as epistatic downstream effectors of miR-218 in the motor neuron system, under our experimental conditions (Fig. 3E-H).
To substantiate the evidence for the relevance of voltage-gated potassium channels, we performed a series of additional studies that collectively increased our confidence in the relevance of Kv10.1 and were not sufficiently supportive of Kv4.2 in this context.
We demonstrated that both Kv10.1 (Kcnh1) or Kv4.2 (Kcnd2) mRNAs can be directly targeted by miR-218, by measuring the luminescence of a Renilla reporter, harboring the 3′ untranslated region (3′UTR) of either Kv10.1 (Kcnh1) or Kv4.2 (Kcnd2). miR-218 silencing was abrogated by mutated miRNA recognition sequences (Fig. 3I and Fig. S5A). We also mined miRNA-mRNA chimera data from AGO2 cross linking and immunoprecipitation study in the mouse cortex (29). This study revealed miR-218 binding to the 3’UTR of Kv10.1, in vivo in the unmanipulated cortex (Fig. 3J). We next transduced primary rat motor neurons with viral vectors that either overexpress or knockdown miR-218 (Fig. 3K). miR-218 expression reciprocally correlated with Kv10.1 protein under miR-218 KD (Fig. 3L, Fig. S6 and Datafile S3), as could be expected from a genuine target. miR-218 OE did not affect Kv10.1 expression, which might be because of the high basal miR-218 expression in motor neurons. Finally, to test if Kv10.1 is upregulated in ALS, along with miR-218 downregulation, we mined human NGS data, which revealed higher Kv10.1 mRNA expression in ALS, in both induced human motor neurons of patients with ALS ((30) Fig. 3M) and in laser capture microdissection-enriched surviving motor neurons from lumbar spinal cords of patients with sporadic ALS (sALS) with rostral onset and caudal progression ((21) Fig. 3N). A parallel analysis of Kv4.2 expression and regulation was not equally supportive and it is therefore less likely to be regulated by miR-218 in motor neurons and in ALS (Fig. S5B-F and Datafile S4). Therefore, Kv10.1 appear as a relevant miR-218 target in vitro and in vivo and might be relevant also in human ALS.
Rare miR-218 genetic variants are detected in human patients with ALS
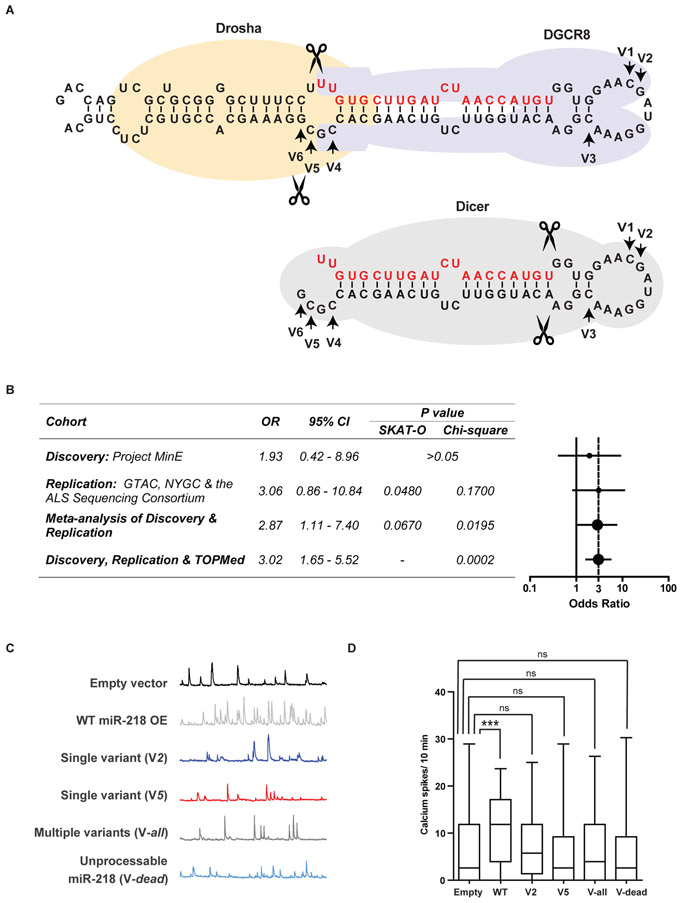
To examine the relevance of miR-218 to human disease we screened for rare genetic variations (minor allele frequency <0.01) in the human miR-218-1 (Chr. 4) and miR-218-2 (Chr. 5) genes in ALS and controls cohorts of Project MinE ALS sequencing consortium data (31). We observed 6 unique rare variants in the precursor miR-218-2 (pre-miR-218-2) gene and a single variant (rs371622197) in pre-miR-218-1 (Fig. 4A and table S1) in multi-national cohorts, which were matched geographically and for ancestry (see methods). None of these variants were harbored within the ~22 nucleotides of mature miR-218-5p [miRBase v20 (32)]. Region-based rare variant association testing by the Optimized Sequence Kernel Association Test (SKAT-O) (33) was non-significant (adjusted p value >0.05). However, odds ratio (OR) was 1.93 with 95% confident interval (CI): 0.42-8.96 (Fig. 4B). We then performed an independent replication study on additional cohorts of Genomic Translation for ALS Care (GTAC), the ALS Sequencing Consortium and the New York Genome Center (NYGC) ALS Consortium for rare miR-218-2 variant association.
Fig. 4. Rare genetic miR-218 variants disrupt its ability to regulate neuronal activity.
(A) Diagrams of miR-218-2 pri-miRNA (upper) and the pre-miRNA hairpin (lower), with demarcation of DROSHA, DGCR8 and DICER binding and arrows, revealing variant nucleotides (V1-V6). Guide RNA in red. (B) Table and forest plot depicting odds ratio (OR) estimates with 95% confidence intervals (CI), across study cohorts and p-values, calculated with SKAT-O or Chi-squared test with Yate’s correction. Vertical dotted line denotes OR=3. (C) Representative motor neuron traces and (D) quantification of spontaneous calcium spike frequencies (ΔF/F >0.5) in embryonic rat spinal motor neurons, transduced with lentiviruses encoding WT or mutated human miR-218-2. Number of cells recorded in a single experiment: Control, n=131; WT miR-218-2, n=114; single variant V2, n=137; single variant V5, n=119; multiple variant Vall, n=118; Unprocessable miR-218-2 Vdead, n=111. N=4 independent times with similar results. Kruskal-Wallis test followed by Dunn’s multiple comparison test. *** P<0.001; ns – non-significant.
Rare miR-218-2 variants were enriched in cases (p = 0.048 by SKAT-O; OR=3.06, 95% CI: 0.86 - 10.84). Meta-analysis of both discovery and replication cohorts p value was 0.067 by SKAT-O, (34) and a joint analysis p value was 0.0195 (Chi squared with Yate’s correction; OR=2.87, 95% CI: 1.11-7.40; Fig. 4B). Therefore, the burden of variants showed nominal association to the trait (p < 0.05), although it did not reach genome-wide significance (p = 5.0 × 10−8) with ALS in our study. Finally, we assessed an independent large cohort of 62,784 non-ALS genomes from NHLBI's Trans-Omics for Precision Medicine (TOPMed). This validation effort yielded a joint p value of 0.0002 by Chi-Square test with Yate’s correction with OR=3.02 (95% CI: 1.65 - 5.52), which confirmed the robustness of the findings (Fig. 4B). This modest excess of rare pre-miR-218-2 variants in ALS did not survive genome wide statistical correction. Taken together, individuals harboring miR-218-2 sequence variants have a risk that is almost three time as high to suffer from ALS, relative to the general population.
Rare miR-218 genetic variants disrupt its ability to regulate neuronal excitability
miRNA genes exhibit high evolutionary conservation and sequence mutations may be detrimental to their function. We sought to test the impact of mutated miR-218 on neuronal activity by intracellular calcium transient recording. The variants were aggregated in two main domains, namely in the loop region, that is supposed to bind DGCR8 (35) and in the miRNA 3' terminal, which is cleaved by Drosha (35, 36) and then becomes an important element of recognition by Dicer (37, 38). To test these variants functionally, we created vectors that represent loop and 3' terminal variants. Then, we transduced primary rat motor neurons with the following miR-218-2 vectors: (i) Control vector, (ii) wild-type human miR-218-2 (WT); (iii) the predominant pre-miRNA loop variant (Chr5:168,195,207, V2); (iv) the most abundant patient variant at the miRNA 3' terminal (Chr5:168,195,174, V5); (V) a miR-218-2 version, harboring a collection of variants, superimposed from cases (Vall); or (vi) a miR-218-2 sequence that we designed to be resistant to Drosha activity, which yields no mature miR-218 (Vdead). Wild-type miR-218 increased spontaneous calcium burst frequency as expected, whereas miR-218 with variant sequences failed to upregulate neuronal Ca2+ transient frequency (Fig. 4C,D).
Rare miR-218 genetic variants inhibit its biogenesis
We tested the hypothesis that miR-218 variants impair neuronal bursting, through inhibition of biogenesis or creation of abnormal forms of the mature miRNA. We used HEPG2 cells, which do not express the endogenous miR-218 gene, to over express wild-type or mutated forms of primary miR-218 (pri-miR-218). In addition, we co-transfected miR-214-3p mimics, which served as spike-in control for downstream normalization.
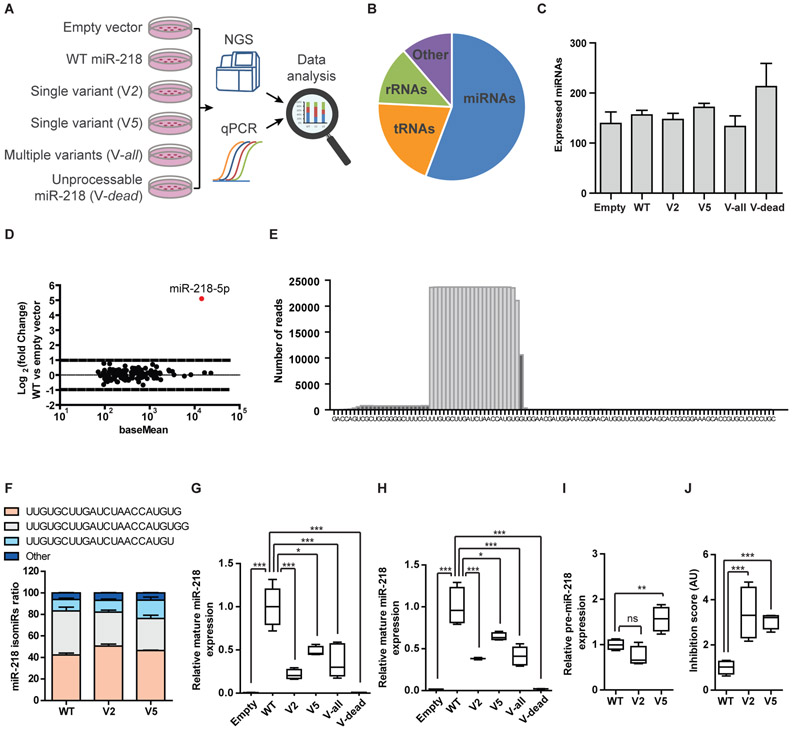
We performed small RNA NGS, on RNA extracted from transfected HEPG2 cells (Fig. 5A). miRNAs were the dominant RNAs in the libraries (56%, Fig. 5B), at approximately a million miRNA reads / library and complexity of ~160 different miRNA species (Fig. 5C). The expression of mature miR-218 following transfection was comparable with the most abundant endogenous miRNAs in HepG2 cells (Fig. 5D). miR-218-5p dominated the expression profile, whereas sequences aligned to the loop or to miR-218-3p were less prevalent, as expected (Fig. 5E). Furthermore, the isomiR-218 profile was comparable across different variants (Fig. 5F). The expression of mature miR-218, derived from mutated forms of pri-miR-218, was lower compared to the wild-type form (Fig. 5G). We validated the drop in mature miR-218 expression, when harboring variants, with quantitative real time PCR (Fig. 5H). We also detected the accumulation of pre-miR-218 forms, following transfection with a vector harboring the most abundant variant (V5; Fig. 5I), a hallmark of failed biogenesis. The inhibition score (3), describing the ratio of DICER substrate (pre-miR-218) to product (mature miR-218), was increased by 3.4 fold for the predominant pre-miRNA loop variant (V2) and by 3.1 fold for the most abundant variant (V5), relative to wild-type miR-218, demonstrating inhibition of miR-218 biogenesis (Fig. 5J). Taken together, mutated miR-218 exhibits impaired biogenesis, providing a conceivable mechanism for insufficient regulation of neuronal activity.
Fig. 5. Rare genetic variants in miR-218 inhibit biogenesis.
(A) Diagram of experimental design. HEPG2 cells transfected with WT miR-218-2 or miR-218-2 genetic variants and processing of RNA for NGS and qPCR studies. (B) Pie chart of relative representation of different RNA families in NGS data (percentage of reads aligned to miRNA- 56%; tRNA −20%; rRNA – 13%; other RNA types – 11%). (C) The number of expressed miRNAs was comparable across samples. Means ± SEM. (D) MA plot of miRNA expression in HEPG2 cells transfected with wild-type miR-218-2, relative to control vector. Abundance (x-axis; presented on a log scale) against ratio of miRNA in cells overexpressing WT miR-218 vs a control vector (log 2 fold change). (E) Histogram of number of reads-per-base for WT miR-218-2 sequences, aligned over the genomic sequence. (F) Bar graph of miR-218-2-5p isotypes (isomiR-218-2-5p, sequence denoted) in HEPG2 transfected with WT miR-218-2, or V2 / V5 variants. Relative expression of mature miR-218-2 from (G) NGS or (H) TaqMan qPCR studies, normalized to miR-214-3p spike-in mimics. (I) Pre-miR-218-2 expression from NGS. (J) The ratio of pre-miR-218-2 (substrate) to mature miR-218 (product), defined as “inhibition score”. Inhibition score approximates a value of 1 in the WT condition, whereas a value > 1, reflects reduced DICER activity. Control, n=3; WT miR-218-2, n=5; single variant V2, n=4; single variant V5, n=4; multiple variant Vall, n=5; Unprocessable miR-218-2; Vdead, n=3. Box-Plots depict median, upper / lower quartiles & extreme points, One-way ANOVA followed by Bonferroni’s multiple comparison test performed on data (I) or log-transformed data (G, H, J), * P<0.05; ** P<0.01; *** P<0.001; ns – non-significant.
Discussion
miR-218 was recently put in the spotlight for its roles in motor neuron development (1, 2). The link between perinatal death of mice deficient of miR-218 and a potential deleterious effect in adult humans requires further investigations. In the current work, we demonstrated miR-218 relevance to human motor neurons in a systematic effort that explains how miR-218 contributes to a previously unappreciated facet of motor neuron specificity and disease susceptibility. ALS neuropathology establishes miR-218 as marker of human motor neuron mass and well-being that is downregulated in ALS. Accordingly, mRNA targets of miR-218 are upregulated / de-repressed.
We identified rare sequence variants in the miR-218-2 gene that impair miR-218 biogenesis and its ability to regulate motor neuron activity. These sequence variants are relevant for the understanding of motor neuron health and disease. We suggest that miR-218-2 variants are sub-optimal for a Dicer-dependent step of biogenesis, thus reducing mature miR-218 expression and contributing to selective motor neuron vulnerability. Subtle miR-218 downregulation in humans, plausibly contributes to failed homeostasis in adults, potentially because of broad upregulation (de-repression) of dozens of miR-218 targets in human motor neurons. Furthermore, because miR-218 expression is downregulated in motor neurons of sporadic and familial patients with ALS, individuals harboring miR-218 variants suffer two sequential hits to miR-218 expression and function. Therefore, miR-218 is a relevant candidate for genetic screening in additional ALS genetics cohort.
A previously unrecognized pathway downstream of miR-218 controls neuronal activity by regulating the voltage-gated potassium channel, Kv10.1. Altered motor neuron excitability and ion channel dysfunction have been reported in patients, rodent and ALS iPSC models (39-51) and drugs such as ezogabine (retigabine) (52), or riluzole, which control potassium and sodium channels, respectively, elute to the relevance of therapeutically altering neuronal activity in ALS.
Additionally, increased expression of voltage-gated potassium channel subtypes have been reported in iPSC-derived ALS motor neurons with FUS and SOD1 mutations and targeting potassium currents with 4-Aminopyridine, a potassium channel blocker, recovered neuronal activity patterns in culture (53). These observations resonate with miR-218 activity upstream of voltage gated potassium channel and suggest that aberrant neuronal activity is an important contributing factor at the ALS milieu.
Our study does not rule out that additional targets may play parallel roles in controlling neuron activity downstream of miR-218. miR-218 is a member of an expanding class of miRNAs that regulate neuronal activity in flies (54, 55) and mammals (56-59), including miR-128 (57), miR-101 (28) and miR-324-5p (60). The emerging regulation of neuronal activity by miRNAs depends on their capacity to fine-tune the expression of dosage-sensitive proteins locally, at dendrites, axons and synapses.
miR-218 regulates a myriad of targets designated Target218 (1). Our work, along with reported specific targets in astrocytes and neuronal progenitors (2, 5), contribute to deconvoluting the Target218 network. Interestingly, Amin et al. recently showed by a patch clamp study in lumbar spinal slices that miR-218 contributes to inhibiting neuronal activity (1). Reconciling this observation with ours requires new conditional miR-218 alleles that will allow uncoupling miR-218 roles in interneuron differentiation (2) and plausibly in establishing interneuron-motor neuron circuitry, from miR-218 roles in adult motor neurons. Furthermore, developmental loss of miR-218 causes motor neuron death, further complicating the comparison to the moderate KD in the post-mitotic motor neurons.
In summary, motor-neuron enriched miR-218 might serve as a marker of motor neuron mass in the human ventral horn in ALS and miR-218 functions uncovers previously unappreciated facets of motor neuron specificity that may be particularly susceptible to failure in human patients with ALS. Currently, it is not clear if the global miR-218 downregulation in human neuropathology is a consequence of Dicer inhibition (3) and how such a downregulation might impact non-cell autonomous effects of miR-218 (5). Mouse modelling can be beneficial for exploring miR-218 allele genetic interactions with other ALS-associated mutations and the functional implications of the discovered variants in the miR-218-2 gene sequence. Therefore, the study contributes to an emerging view of ALS as a disease with a prominent RNA component and suggests that miR-218 is a potential therapeutic target for motor neuron disease (graphically summarized in Fig. S7).
Materials and Methods
Study design
The overall objective of our study was to investigate the relevance of motor-neuron specific miR-218 to human motor neuron specificity and disease (summary in Fig. S7), by employing molecular, neurogenetics and neuropathology approaches. First, we performed four orthogonal miRNA quantification studies in human motor neurons: (1) chromogenic miR-218 in situ hybridization in human spinal cord, (2) nanoString nCounter, (3) miR-218 qPCR, and (4) analysis of mRNA expression of miR-218 targets from laser capture microdissection-enriched surviving motor neurons from lumbar spinal cords of patients with sALS. These experiments established miR-218 as marker of human motor neuron mass and well-being. To test whether miR-218 regulates motor neuron activity we transduced primary motor neuron with lentiviruses encoding miR-218 OE or KD and monitored intracellular calcium transients and intrinsic activity by patch-clamp electrophysiological experiments. A series of bioinformatics and experimental steps collectively directed us to conclude that Kv10.1 is a direct target of miR-218 in this system. Using statistical genetics and burden studies of rare variants, we identified miR-218 genetic variants in large ALS cohorts. The variants were shown to inhibit biogenesis and to impair miR-218 function. Experimentalists were blinded while analyzing data. Outliers were excluded if deviated ±2 SDs away from mean. The number of samples that were taken for case-control cohort in neuropathology (Human motor neuron systems: 20 ALS cases, 14 non-neurodegeneration controls) and neurogenetics (Human genomes: 7,738 ALS, 71,656 controls). These numbers reflect the maximal availability at the time of the study.
Statistical analysis
Statistics performed with Prism Origin (GraphPad Software Inc.). Shapiro-Wilk test was used to assess normality of the data. Pair-wise comparisons passing normality test were analyzed with Student’s t-test whereas the Mann-Whitney test was used for pairwise comparison of nonparametric data. Multiple group comparisons passing normality test were analyzed using ANOVA with post hoc tests, whereas nonparametric multiple group comparisons were analyzed using the Kruskal-Wallis test with Dunn’s post hoc testing, when ANOVA assumptions were not met. Statistical P values <0.05 were considered significant. Data presented as specified in the figure legends. Data are shown as means ± SEM or SD or graphed using boxplots, as noted in the text. Individual subject level data are reported in Datafile S1.
Supplementary Material
Fig. S1. miR-218 is highly and specifically expressed in human and murine spinal motor neurons.
Fig. S2. High content analysis (HCA) of neuronal morphology following miR-218 perturbation.
Fig. S3. miR-218 regulates intrinsic excitability.
Fig. S4. qPCR validation of miR-218 target knockdown.
Fig. S5. Evaluation of miR-218 upstream of the mRNA encoding for the potassium channel Kv4.2 (Kcnd2).
Fig. S6. Kv10.1 (Kcnh1) protein quantification by western blot, following miR-218 KD.
Fig. S7. A summary diagram of key observations.
Table S1. Identified hsa-miR-218-2 variants.
Table S2. DsiRNA sequences employed in the study.
Table S3. Synthetic miR-218 sequences used for cloning into pMA-T vectors.
Table S4. Primers used for quantitative real-time PCR.
Acknowledgments:
We gratefully acknowledge the contributions of all participants and the investigators who provided biological samples and data for Project Mine ALS sequencing consortium, the Genomic Translation for ALS Care (GTAC), the ALS Sequencing Consortium, the New York Genome Center (NYGC) ALS Consortium and TOPMed (Trans-Omics for Precision Medicine) of the National Heart, Lung, and Blood Institute (NHLBI, https://www.nhlbiwgs.org/topmed-banner-authorship). Samples used in this research were in part obtained from the UK National DNA Bank for MND Research, funded by the MND Association and the Wellcome Trust. We would like to thank people with MND and their families for their participation in this project. We acknowledge sample management undertaken by Biobanking Solutions funded by the Medical Research Council at the Centre for Integrated Genomic Medical Research, University of Manchester. The authors would like to thank Tzofar Hajbi and Osnat Amram for veterinary services and husbandry. We thank Andres Mauricio, Jaramillo Flautero, Igor Ulitsky, Menahem Segal, Bernard Atalli, Avraham Yaron, Uri Asheri, Rotem Tal and Giordano Lippi for insightful comments on the manuscript. We are grateful for Noa Sadeh and Menahem Segal for training in calcium imaging, to Haim Barr, Noga Kozer and Alexander Plotnikov for assistance with automated imaging, Tali Shalit and Zeev Melamed for assistance with bioinformatics, Katayun Cohen-Kashi Malina for assistance with patch clamp and Maria J. Rodriguez for histology. Hornstein lab is supported by friends of Dr. Sydney Brenner. EH is Head of Nella and Leon Benoziyo Center for Neurological Diseases and incumbent of Ira & Gail Mondry Professorial chair.
Funding: The work is funded by Target ALS (118945 To E.H., J.M.R. and S.L.P.), Legacy Heritage Fund, Bruno and Ilse Frick Foundation for Research on ALS, Teva Pharmaceutical Industries Ltd as part of the Israeli National Network of Excellence in Neuroscience (NNE) and Minna-James-Heineman Stiftung through Minerva. The research leading to these results has received funding to E.H. from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 617351. Israel Science Foundation, the ALS-Therapy Alliance, AFM Telethon (20576 to E.H.), Motor Neuron Disease Association (UK), The Thierry Latran Foundation for ALS research, ERA-Net for Research Programmes on Rare Diseases (FP7), A. Alfred Taubman through IsrALS, Yeda-Sela, Yeda-CEO, Israel Ministry of Trade and Industry, Y. Leon Benoziyo Institute for Molecular Medicine, Kekst Family Institute for Medical Genetics, David and Fela Shapell Family Center for Genetic Disorders Research, Crown Human Genome Center, Nathan, Shirley, Philip and Charlene Vener New Scientist Fund, Julius and Ray Charlestein Foundation, Fraida Foundation, Wolfson Family Charitable Trust, Adelis Foundation, MERCK (UK), Maria Halphen, Estates of Fannie Sherr, Lola Asseof, Lilly Fulop. S.M.K.F. is supported by the ALS Canada Tim E. Noël Postdoctoral Fellowship. C.E. and I.M. were supported by scholarship from Teva Pharmaceutical Industries Ltd as part of the Israeli National Network of Excellence in Neuroscience (NNE). Work at C. Gross lab was supported by NIH grant (R01NS092705to C.G.). Work at T.M Miller lab was supported by grants from Project5 for ALS, Target ALS, the National Institute of Neurological Disorders and Stroke (R01NS078398 to T.M.M.; F31NS092340 to M.L.H. and T.M.M.), the Robert Packard Center for ALS Research, University of Missouri Spinal Cord Injury/Disease Research Program and the Hope Center for Neurological Disorders. Work at M. B. Harms lab is funded by grants from ALS Association and form Biogen. R. H. Brown Jr. was funded by ALS Association, ALS Finding a Cure, Angel Fund, ALS-One, Cellucci Fund and NIH grants (R01 NS104022, R01 NS073873 and NS111990-01 to R.H.B.J.). A.A.C. was supported through the following funding organizations under the aegis of JPND - www.jpnd.eu (United Kingdom, Medical Research Council (MR/L501529/1; MR/R024804/1)) and through the Motor Neurone Disease Association. This study represents independent research part funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. A.N.B. was supported by Suna and Inan Kirac Foundation. J.E.L. is supported by the National Institute of Health/NINDS (R01 NS073873). H.P. was supported by a grant from the ALS Association.
Footnotes
Competing interests: M.A.B., N.M.B. and K.A.L. are employed by Integrated DNA Technologies, Inc (IDT), which manufactures reagents similar to some described in the manuscript; M.A.B. owns equity in DHR, the parent company of IDT; T.M.M. holds licensing agreement with Ionis Pharmaceuticals and with C2N, he is on the Advisory Board of Ionis Pharmaceuticals and Biogen and is consulting to Cytokinetics; T.M.M. and M.L.H. are inventors on patent/patent application (PCT/US2016/019602, now U.S. patent application number 15/553,922) submitted by Washington University that covers methods to detect motor neuron disease using miRNAs and to target motor neuron disease miRNAs; T.M.M. is an inventor on patent/patent application (PCT/US2015/053283, now U.S. patent application number 15/515,909) submitted by Washington University that covers Tau Kinetic Measurements; T.M.M. is an inventor on patent/patent application (PCT/US2013/031500, now U.S. patent application number 16/298,607with corresponding national stage applications or issued patents in Australia, Canada, Europe and Japan) that is jointly owned with Ionis Pharmaceuticals that covers Methods for modulating tau expression for reducing neurodegenerative syndromes; T.M.M. is an inventor on patent/patent application (Issue #10,273,474) that is jointly owned with Ionis Pharmaceuticals that covers Methods for Modulating Tau Expression for Reducing Seizure and Modifying a Neurodegenerative Syndrome; T.M.M. is an inventor on patent/patent application (61/547,890) submitted by Washington University that covers Metabolism of SOD1 in the CSF; I.R. and E.H. are inventors on pending patent family PCT/IL2016/050328 entitled “Methods of treating motor neuron diseases”. All other authors declare that they have no competing interests.
Data and materials availability: Gene Expression Omnibus accession number: GSE136409. miR-218-2-5p isotypes counting code: https://github.com/TsviyaOlender/mir-218. Human miR-218 precursors (miRBase v20 (32)), are at Chr5:168195173-168195236; Chr4: 20529922-20529986 of human genome build 19 (hg19). Human genetics data is publically available from the sequencing consortia: Project MinE, Genomic Translation for ALS Care (GTAC), ALS Sequencing Consortium, New York Genome Center (NYGC) and NHLBI's Trans-Omics for Precision Medicine (TOPMed). All Other data used for this manuscript are available in the manuscript.
References and Notes:
- 1.Amin ND, Bai G, Klug JR, Bonanomi D, Pankratz MT, Gifford WD, Hinckley CA, Sternfeld MJ, Driscoll SP, Dominguez B, Lee KF, Jin X, Pfaff SL, Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 350, 1525–1529 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiebes KP, Nam H, Cambronne XA, Shen R, Glasgow SM, Cho HH, Kwon JS, Goodman RH, Lee JW, Lee S, Lee SK, miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat Commun 6, 7718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emde A, Eitan C, Liou LL, Libby RT, Rivkin N, Magen I, Reichenstein I, Oppenheim H, Eilam R, Silvestroni A, Alajajian B, Ben-Dov IZ, Aebischer J, Savidor A, Levin Y, Sons R, Hammond SM, Ravits JM, Moller T, Hornstein E, Dysregulated miRNA biogenesis downstream of cellular stress and ALS-causing mutations: a new mechanism for ALS. The EMBO journal 34, 2633–2651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoye ML, Koval ED, Wegener AJ, Hyman TS, Yang C, O'Brien DR, Miller RL, Cole T, Schoch KM, Shen T, Kunikata T, Richard JP, Gutmann DH, Maragakis NJ, Kordasiewicz HB, Dougherty JD, Miller TM, MicroRNA Profiling Reveals Marker of Motor Neuron Disease in ALS Models. J Neurosci 37, 5574–5586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoye ML, Regan MR, Jensen LA, Lake AM, Reddy LV, Vidensky S, Richard JP, Maragakis NJ, Rothstein JD, Dougherty JD, Miller TM, Motor neuron-derived microRNAs cause astrocyte dysfunction in amyotrophic lateral sclerosis. Brain, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renton AE, Chio A, Traynor BJ, State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 17, 17–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, Mitra RD, Ravits J, Harms MB, Baloh RH, Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol 77, 100–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown RH, Al-Chalabi A, Amyotrophic Lateral Sclerosis. N Engl J Med 377, 162–172 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Al-Chalabi A, van den Berg LH, Veldink J, Gene discovery in amyotrophic lateral sclerosis: implications for clinical management. Nat Rev Neurol 13, 96–104 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Kenna KP, van Doormaal PT, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, van Rheenen W, van Eijk KR, Jones AR, Keagle P, Shatunov A, Sproviero W, Smith BN, van Es MA, Topp SD, Kenna A, Miller JW, Fallini C, Tiloca C, McLaughlin RL, Vance C, Troakes C, Colombrita C, Mora G, Calvo A, Verde F, Al-Sarraj S, King A, Calini D, de Belleroche J, Baas F, van der Kooi AJ, de Visser M, Ten Asbroek AL, Sapp PC, McKenna-Yasek D, Polak M, Asress S, Munoz-Blanco JL, Strom TM, Meitinger T, Morrison KE, Consortium S, Lauria G, Williams KL, Leigh PN, Nicholson GA, Blair IP, Leblond CS, Dion PA, Rouleau GA, Pall H, Shaw PJ, Turner MR, Talbot K, Taroni F, Boylan KB, Van Blitterswijk M, Rademakers R, Esteban-Perez J, Garcia-Redondo A, Van Damme P, Robberecht W, Chio A, Gellera C, Drepper C, Sendtner M, Ratti A, Glass JD, Mora JS, Basak NA, Hardiman O, Ludolph AC, Andersen PM, Weishaupt JH, Brown RH Jr., Al-Chalabi A, Silani V, Shaw CE, van den Berg LH, Veldink JH, Landers JE, NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet 48, 1037–1042 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor JP, Brown RH Jr., Cleveland DW, Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haramati S, Chapnik E, Sztainberg Y, Eilam R, Zwang R, Gershoni N, McGlinn E, Heiser PW, Wills AM, Wirguin I, Rubin LL, Misawa H, Tabin CJ, Brown R Jr., Chen A, Hornstein E, miRNA malfunction causes spinal motor neuron disease. Proc Natl Acad Sci U S A 107, 13111–13116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koval ED, Shaner C, Zhang P, du Maine X, Fischer K, Tay J, Chau BN, Wu GF, Miller TM, Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum Mol Genet 22, 4127–4135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ, Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci 7, 178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos-Melo D, Droppelmann CA, He Z, Volkening K, Strong MJ, Altered microRNA expression profile in Amyotrophic Lateral Sclerosis: a role in the regulation of NFL mRNA levels. Mol Brain 6, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eitan C, Hornstein E, Vulnerability of microRNA biogenesis in FTD-ALS. Brain Res, (2016). [DOI] [PubMed] [Google Scholar]
- 17.Figueroa-Romero C, Hur J, Lunn JS, Paez-Colasante X, Bender DE, Yung R, Sakowski SA, Feldman EL, Expression of microRNAs in human postmortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol Cell Neurosci 71, 34–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung YT, Peng KC, Chen YC, Yen YP, Chang M, Thams S, Chen JA, Mir-17 approximately 92 Confers Motor Neuron Subtype Differential Resistance to ALS-Associated Degeneration. Cell Stem Cell, (2019). [DOI] [PubMed]
- 19.Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Di Costanzo S, Atwater N, Maeder ML, Goodwin MJ, Nemesh J, Handsaker RE, Paull D, Noggle S, McCarroll SA, Joung JK, Woolf CJ, Brown RH, Eggan K, Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 14, 781–795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V, Bell GW, Nam JW, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs. Elife 4, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krach F, Batra R, Wheeler EC, Vu AQ, Wang R, Hutt K, Rabin SJ, Baughn MW, Libby RT, Diaz-Garcia S, Stauffer J, Pirie E, Saberi S, Rodriguez M, Madrigal AA, Kohl Z, Winner B, Yeo GW, Ravits J, Transcriptome-pathology correlation identifies interplay between TDP-43 and the expression of its kinase CK1E in sporadic ALS. Acta Neuropathol 136, 405–423 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milligan C, Gifondorwa D, Isolation and culture of postnatal spinal motoneurons. Methods Mol Biol 793, 77–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dongen S, Abreu-Goodger C, Enright AJ, Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods 5, 1023–1025 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das M, Molnar P, Devaraj H, Poeta M, Hickman JJ, Electrophysiological and morphological characterization of rat embryonic motoneurons in a defined system. Biotechnol Prog 19, 1756–1761 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Kaalund SS, Veno MT, Bak M, Moller RS, Laursen H, Madsen F, Broholm H, Quistorff B, Uldall P, Tommerup N, Kauppinen S, Sabers A, Fluiter K, Moller LB, Nossent AY, Silahtaroglu A, Kjems J, Aronica E, Tumer Z, Aberrant expression of miR-218 and miR-204 in human mesial temporal lobe epilepsy and hippocampal sclerosis-convergence on axonal guidance. Epilepsia 55, 2017–2027 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM, A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 11, 705–716 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lippi G, Fernandes CC, Ewell LA, John D, Romoli B, Curia G, Taylor SR, Frady EP, Jensen AB, Liu JC, Chaabane MM, Belal C, Nathanson JL, Zoli M, Leutgeb JK, Biagini G, Yeo GW, Berg DK, MicroRNA-101 Regulates Multiple Developmental Programs to Constrain Excitation in Adult Neural Networks. Neuron 92, 1337–1351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, Darnell RB, miRNA-target chimeras reveal miRNA 3'-end pairing as a major determinant of Argonaute target specificity. Nat Commun 6, 8864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson L, iMN (Exp 2) - ALS, SMA and Control (unaffected) iMN cell lines differentiated from iPS cell lines using a long differentiation protocol - RNA-seq. (2017).
- 31.E. A. L. S. S. C. Project Min, Project MinE: study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ, miRBase: tools for microRNA genomics. Nucleic Acids Res 36, D154–158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, N. G. E. S. P.-E. L. P. Team, Christiani DC, Wurfel MM, Lin X, Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. Am J Hum Genet 91, 224–237 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S, Teslovich TM, Boehnke M, Lin X, General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet 93, 42–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS, Functional Anatomy of the Human Microprocessor. Cell 161, 1374–1387 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN, The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Elbashir SM, Lendeckel W, Tuschl T, RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15, 188–200 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nykanen A, Haley B, Zamore PD, ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Kanai K, Kuwabara S, Misawa S, Tamura N, Ogawara K, Nakata M, Sawai S, Hattori T, Bostock H, Altered axonal excitability properties in amyotrophic lateral sclerosis: impaired potassium channel function related to disease stage. Brain 129, 953–962 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Vucic S, Nicholson GA, Kiernan MC, Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain 131, 1540–1550 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Kuo JJ, Siddique T, Fu R, Heckman CJ, Increased persistent Na(+) current and its effect on excitability in motoneurones cultured from mutant SOD1 mice. J Physiol 563, 843–854 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zona C, Pieri M, Carunchio I, Voltage-dependent sodium channels in spinal cord motor neurons display rapid recovery from fast inactivation in a mouse model of amyotrophic lateral sclerosis. J Neurophysiol 96, 3314–3322 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Bories C, Amendola J, Lamotte d'Incamps B, Durand J, Early electrophysiological abnormalities in lumbar motoneurons in a transgenic mouse model of amyotrophic lateral sclerosis. Eur J Neurosci 25, 451–459 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Pieri M, Albo F, Gaetti C, Spalloni A, Bengtson CP, Longone P, Cavalcanti S, Zona C, Altered excitability of motor neurons in a transgenic mouse model of familial amyotrophic lateral sclerosis. Neurosci Lett 351, 153–156 (2003). [DOI] [PubMed] [Google Scholar]
- 45.van Zundert B, Peuscher MH, Hynynen M, Chen A, Neve RL, Brown RH Jr., Constantine-Paton M, Bellingham MC, Neonatal neuronal circuitry shows hyperexcitable disturbance in a mouse model of the adult-onset neurodegenerative disease amyotrophic lateral sclerosis. J Neurosci 28, 10864–10874 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan KA, Schuster JE, Fu R, Siddique T, Heckman CJ, Altered postnatal maturation of electrical properties in spinal motoneurons in a mouse model of amyotrophic lateral sclerosis. J Physiol 589, 2245–2260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs A, Kutterer S, Muhling T, Duda J, Schutz B, Liss B, Keller BU, Roeper J, Selective mitochondrial Ca2+ uptake deficit in disease endstage vulnerable motoneurons of the SOD1G93A mouse model of amyotrophic lateral sclerosis. J Physiol 591, 2723–2745 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxena S, Roselli F, Singh K, Leptien K, Julien JP, Gros-Louis F, Caroni P, Neuroprotection through excitability and mTOR required in ALS motoneurons to delay disease and extend survival. Neuron 80, 80–96 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Devlin AC, Burr K, Borooah S, Foster JD, Cleary EM, Geti I, Vallier L, Shaw CE Chandran S, Miles GB, Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun 6, 5999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Ni W, Horwich AL, Kaczmarek LK, An ALS-Associated Mutant SOD1 Rapidly Suppresses KCNT1 (Slack) Na+-Activated K+ Channels in Aplysia Neurons. J Neurosci 37, 2258–2265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang YM, Yamamoto M, Kobayashi Y, Yoshihara T, Liang Y, Terao S, Takeuchi H, Ishigaki S, Katsuno M, Adachi H, Niwa J, Tanaka F, Doyu M, Yoshida M, Hashizume Y, Sobue G, Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann Neurol 57, 236–251 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH Jr., Cudkowicz ME, Bean BP, Eggan K, Woolf CJ, Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep 7, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Naujock M, Stanslowsky N, Bufler S, Naumann M, Reinhardt P, Sterneckert J, Kefalakes E, Kassebaum C, Bursch F, Lojewski X, Storch A, Frickenhaus M, Boeckers TM, Putz S, Demestre M, Liebau S, Klingenstein M, Ludolph AC, Dengler R, Kim KS, Hermann A, Wegner F, Petri S, 4-Aminopyridine Induced Activity Rescues Hypoexcitable Motor Neurons from Amyotrophic Lateral Sclerosis Patient-Derived Induced Pluripotent Stem Cells. Stem Cells 34, 1563–1575 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Verma P, Augustine GJ, Ammar MR, Tashiro A, Cohen SM, A neuroprotective role for microRNA miR-1000 mediated by limiting glutamate excitotoxicity. Nat Neurosci 18, 379–385 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Guven-Ozkan T, Busto GU, Schutte SS, Cervantes-Sandoval I, O'Dowd DK, Davis RL, MiR-980 Is a Memory Suppressor MicroRNA that Regulates the Autism-Susceptibility Gene A2bp1. Cell Rep 14, 1698–1709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harraz MM, Eacker SM, Wang X, Dawson TM, Dawson VL, MicroRNA-223 is neuroprotective by targeting glutamate receptors. Proc Natl Acad Sci U S A 109, 18962–18967 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan CL, Plotkin JL, Veno MT, von Schimmelmann M, Feinberg P, Mann S, Handler A, Kjems J, Surmeier DJ, O'Carroll D, Greengard P, Schaefer A, MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science 342, 1254–1258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sakai A, Saitow F, Miyake N, Miyake K, Shimada T, Suzuki H, miR-7a alleviates the maintenance of neuropathic pain through regulation of neuronal excitability. Brain 136, 2738–2750 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Fiore R, Rajman M, Schwale C, Bicker S, Antoniou A, Bruehl C, Draguhn A, Schratt G, MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. The EMBO journal 33, 2231–2246 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gross C, Yao X, Engel T, Tiwari D, Xing L, Rowley S, Danielson SW, Thomas KT, Jimenez-Mateos EM, Schroeder LM, Pun RY, Danzer SC, Henshall DC, Bassell GJ, MicroRNA-Mediated Downregulation of the Potassium Channel Kv4.2 Contributes to Seizure Onset. Cell Rep 17, 37–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, Chuang HY, Kallberg M, Kumar SA, Liao A, Little KM, Stromberg MP, Tanner SW, Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 29, 2041–2043 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, Genomes Project Analysis G, The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM, Robust relationship inference in genome-wide association studies. Bioinformatics 26, 2867–2873 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rungta RL, Choi HB, Lin PJ, Ko RW, Ashby D, Nair J, Manoharan M, Cullis PR, Macvicar BA, Lipid Nanoparticle Delivery of siRNA to Silence Neuronal Gene Expression in the Brain. Mol Ther Nucleic Acids 2, e136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gage GJ, Kipke DR, Shain W, Whole animal perfusion fixation for rodents. Journal of visualized experiments : JoVE, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nuovo GJ, In situ detection of precursor and mature microRNAs in paraffin embedded, formalin fixed tissues and cell preparations. Methods 44, 39–46 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T, miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods 6, 139–141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obernosterer G, Martinez J, Alenius M, Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc 2, 1508–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 69.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I, Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anders S, Pyl PT, Huber W, HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. miR-218 is highly and specifically expressed in human and murine spinal motor neurons.
Fig. S2. High content analysis (HCA) of neuronal morphology following miR-218 perturbation.
Fig. S3. miR-218 regulates intrinsic excitability.
Fig. S4. qPCR validation of miR-218 target knockdown.
Fig. S5. Evaluation of miR-218 upstream of the mRNA encoding for the potassium channel Kv4.2 (Kcnd2).
Fig. S6. Kv10.1 (Kcnh1) protein quantification by western blot, following miR-218 KD.
Fig. S7. A summary diagram of key observations.
Table S1. Identified hsa-miR-218-2 variants.
Table S2. DsiRNA sequences employed in the study.
Table S3. Synthetic miR-218 sequences used for cloning into pMA-T vectors.
Table S4. Primers used for quantitative real-time PCR.