Abstract
Comprehensive inhibition of RUNX1, RUNX2, and RUNX3 led to marked cell suppression compared with inhibition of RUNX1 alone, clarifying that the RUNX family members are important for proliferation and maintenance of diverse cancers, and “cluster regulation of RUNX (CROX)” is a very effective strategy to suppress cancer cells. Recent studies reported by us and other groups suggested that wild-type RUNX1 is needed for survival and proliferation of certain types of leukemia, lung cancer, gastric cancer, etc. and for their one of metastatic target sites such as born marrow endothelial niche, suggesting that RUNX1 often functions oncogenic manners in cancer cells. In this review, we describe the significance and paradoxical requirement of RUNX1 tumor suppressor in leukemia and even solid cancers based on recent our findings such as “genetic compensation of RUNX family transcription factors (the compensation mechanism for the total level of RUNX family protein expression)”, “RUNX1 inhibition-induced inhibitory effects on leukemia cells and on solid cancers through p53 activation”, and “autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells”. Taken together, these findings identify a crucial role for the RUNX cluster in the maintenance and progression of cancers and suggest that modulation of the RUNX cluster using the pyrrole-imidazole polyamide gene-switch technology is a potential novel therapeutic approach to control cancers.
Keywords: CROX, gene-switch technology
INTRODUCTION
RUNX1 is one of the RUNX family proteins (RUNX1, RUNX2, RUNX3), and each forms a heterodimer with core-binding factor β (CBFB: PEBP2B) (Liu et al., 1993; Sood et al., 2017). RUNX regulates transcription of target genes by recognizing and binding to the core consensus sequence, 5’-TGTGGT-3’, and in rare cases, to 5’-TGCGGT-3’, through the RUNT domain (Warren et al., 2000; Yan et al., 2004). which is homologous to the p53 family (Tahirov et al., 2001), but functional crosstalk between RUNX1 and p53 in carcinogenicity, proliferation, and maintenance has not been elucidated at the physiological level. CBFB is non-DNA-binding subunit that enhances the transcription activity of RUNX by potentiating the DNA binding ability and stability of the RUNX family (Warren et al., 2000; Yan et al., 2004). However, the compensation mechanism for the total level of RUNX family protein expression and CBFB is still unknown. The carcinogenic properties of RUNX1 in the developmental mechanism of cancers, but the carcinogenetic molecular basis of RUNX1, including that of other RUNX family members, for proliferation, maintenance and metastasis of cancer is unknown (Ito et al., 2015; Kamikubo, 2018).
Pyrrole-imidazole (PI) polyamides (PI-Polyamides) enter the minor groove of the DNA double-helix structure and recognize a specific nucleotide sequence (Best et al., 2003; Trauger et al., 1996). Using an advanced technique to modify PI-Polyamides with an alkylating agent, chlorambucil (Chb), developed by our collaborator, Professor Hiroshi Sugiyama, at the Department of Chemistry of Kyoto University Graduate School of Science (Bando and Sugiyama, 2006), we are newly developing Chb-M’ and Chb-50, which recognize and bind to the core consensus sequences (consensus RUNX-binding sequences) common to the RUNX family: 5’-TGTGGT-3’ and 5’-TGCGGT-3’. Chb-M’ and Chb-50 can comprehensively inhibit the target genes of the RUNX family by antagonistically inhibiting recruitment of the RUNX family members to the RUNX-binding consensus sequences (Morita et al., 2017a).
OUR NOVEL FINDINGS
Inhibition of RUNX1 induces cell cycle arrest and apoptosis in cancer cells depending on stabilization and activation of p53 protein
Antileukemic effect mediated by RUNX1 depletion is highly dependent on a functional p53-mediated cell death pathway. BCL11A and TRIM24, factors that degrade and inhibit p53, are directly transcriptionally regulated by the RUNX family. Disappearance of both factors by specific inhibition of RUNX1 results in stabilization of the p53 protein, demonstrating that the RUNX1 inhibition-induced inhibitory effects on cancer cells are dependent on stabilization and activation of wild-type p53 protein (Morita et al., 2017a). It was suggested that high expression of RUNX1 in cancer cells induces high expression of p53-degrading BCL11A and TRIM24, destabilizing p53 protein especially in leukemia cells and some kinds of p53 wild-type solid cancers. This indicates a previously unknown role of RUNX1 in cancers (Kamikubo, 2018; Morita et al., 2017a).
The cancer inhibition effects by CROX: cluster regulation of RUNX
We investigated the transcriptional regulatory mechanism of RUNX family members. The consensus RUNX-binding sequences are present in the promotor region and control expression of each other through inhibitory transcriptional regulation, suggesting that regulation by mutual inhibition maintains the protein level of the RUNX family at a constant level (Morita et al., 2017a). The total protein level of the RUNX family (RUNX1 + RUNX2 + RUNX3) was positively correlated with the CBFB protein level. Moreover, the outcome was significantly poorer in the high CBFB expression group for almost all cancer types, suggesting that the total RUNX family expression level is correlated with CBFB expression, and the RUNX family cluster is a common antitumor target among cancers (Morita et al., 2017a). We name the mechanism as “genetic compensation of RUNX family transcription factors” and the comprehensive inhibition of RUNX1, RUNX2, and RUNX3 led to marked cell suppression compared with inhibition of RUNX1 alone, clarifying that the RUNX family members are important for proliferation and maintenance of cancer cells. Our novel theory “cluster regulation of RUNX (CROX)” to overcome the mechanism “genetic compensation of RUNX family transcription factors” maybe a very effective strategy to suppress diverse kinds of cancers (Kamikubo, 2018; Morita et al., 2017a).
Paradoxical enhancement of leukemogenesis in acute myeloid leukemia with moderately attenuated RUNX1 expressions
Inv16 leukemia (AML M4Eo) is caused by the CBFβ-SMMHC fusion protein (Hyde et al., 2010; Liu et al., 1993; 1995; Lukasik et al., 2002). The RUNX1 high-affinity binding domain (HABD) is present in the fusion protein and it markedly enhances the binding ability between the fusion protein and RUNX1 compared with that between CBFβ (wild-type) and RUNX1. The fusion protein captures RUNX1 and transfers it to the cytoplasm from the nucleus, and RUNX1 is dominantly inhibited by the fusion protein. We reported that in D43 mice with the HABD deletion, the RUNX1-transferring effect was inhibited and RUNX1 remained in the nucleus, which moderately improved the dominant RUNX1 inhibition. However, D43 chimeric mice developed Inv16 leukemia at around one week after birth even though no additional gene abnormality was present, suggesting that dominant RUNX1 inhibition is not essential for Inv16 leukemia (Kamikubo et al., 2010). And RUNX1 promotes survival of acute myeloid leukemia cells (Goyama et al., 2013; Hyde et al., 2015), suggesting that RUNX1 is not only a tumor suppressor, but also an oncogenic factor (Goyama et al., 2013; Hyde et al., 2015; Ito et al., 2015; Kamikubo et al., 2010; 2013).
Analysis of clinical data revealed that intermediate level gene expression of RUNX1 marked the poorest-prognostic cohort in relation to acute myelogenous leukemia (AML) patients with highor low-level RUNX1 expressions (Morita et al., 2017b). Through our series of RUNX1 knockdown experiments with various RUNX1 attenuation potentials, we found that moderate attenuation of RUNX1 contributed to the enhanced propagation of AML cells through accelerated cell-cycle progression, whereas profound RUNX1 depletion led to cell-cycle arrest and apoptosis (Morita et al., 2017b). In these RUNX1-silenced tumors, amounts of compensative upregulation of RUNX2 and RUNX3 expressions were roughly equivalent and created an absolute elevation of total RUNX (RUNX1 + RUNX2 + RUNX3) expression levels in RUNX1 moderately attenuated AML cells through “genetic compensation of RUNX family transcription factors” theory (Morita et al., 2017a; 2017b). This elevation resulted in enhanced transactivation of GSTA2 expression, a vital enzyme handling the catabolization of intracellular reactive oxygen species as well as advancing the cell-cycle progressions, and thus ultimately led to the acquisition of proliferative advantage in RUNX1 moderately attenuated AML cells. Our ndings indicate that moderately attenuated RUNX1 expressions paradoxically enhance leukemogenesis in AML cells through intracellular environmental change via GSTA2, suggesting that the inhibition of GSTA2 through “cluster regulation of RUNX (CROX)” is a very effective strategy to suppress AML (Morita et al., 2017b).
Development and proposal of a RUNX family cluster regulation method using PI-Polyamides
PI-Polyamide is a superior medium-sized compound with selective binding ability to the specific nucleotide sequence in the DNA double-helix structure (Best et al., 2003; Trauger et al., 1996). We designed original molecules, PI-Polyamide: Chb-M’ and Chb-50s, which specifically bind to the RUNX-binding consensus sequences, and evaluated the gene expression pattern of the comprehensively regulated RUNX family (Bando and Sugiyama, 2006; Morita et al., 2017a). The RUNX family pathway was negatively controlled and strong cell suppression effects were observed in vitro and in vivo and no toxicity was noted in several preliminary toxicity tests (Morita et al., 2017a). As all RUNX family members share the RUNX-binding consensus sequences: 5’-TGTGGT-3’ or 5’-TGCGGT-3’, cluster regulation of the RUNX family was possible. This was not possible with the RUNX1 inhibitor alone, and the compensatory mechanism of the RUNX family was successfully overcome (Morita et al., 2017a).
Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells
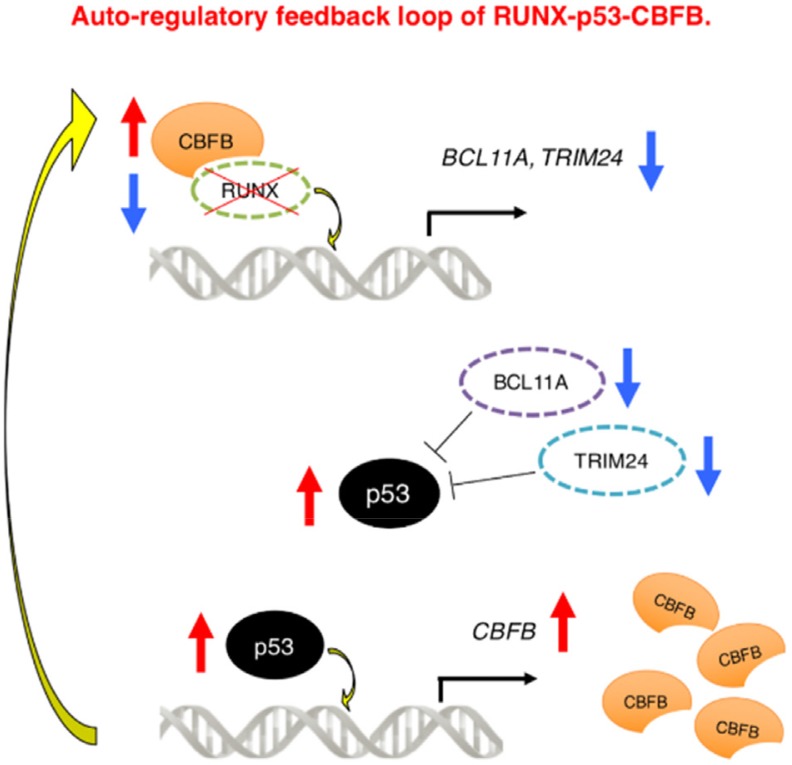
We have found for the first time that CBFB transcription is modulated by RUNX family members, p53 directly transactivates CBFB promoter through its p53-responsive element-like sequences, it is conceivable that RUNX1 regulates the expression of CBFB not directly but indirectly via p53 (Morita et al., 2017c). In addition to the transcriptional regulatory mechanism of CBFB, we have also demonstrated that RUNX1-p53-CBFB regulatory circuit contributes to the acquisition of treatment resistance of AML cells strongly suggesting that an autonomous RUNX1-p53-CBFB regulatory triangle plays a vital role in the maintenance and the acquisition of chemo-resistance of AML cells (Morita et al., 2017c) (Fig. 1).
Fig. 1. Auto-regulatory feedback loop of RUNX1-p53-CBFB.
RUNX1-inhibition treatment induces p53. Induced p53 directly binds to CBFB promoter and stimulates its transcription and translation, which in turn acts as a platform for the stabilization of RUNX1, thereby creating the compensative RUNX1-p53-CBFB feedback loop. Modified from the article of Morita et al. (2017c) (Sci. Rep. 7, 16604) in accordance with the Creative Commons Attribution 4.0 International (CC BY 4.0) license.
RUNX1 positively regulates the ErbB2/HER2 signaling pathway through modulating SOS1 expression in gastric cancer cells
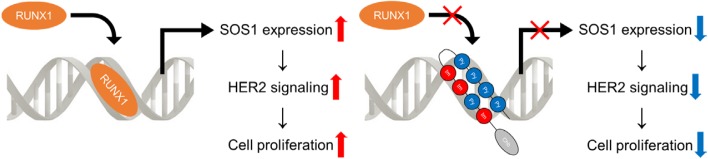
Up-regulation of the ErbB2/HER2 signaling pathway is frequently-encountered in gastric cancer in addition to Her2 breast cancer. This signaling cascade is partly mediated by son of sevenless homolog (SOS) family, which function as adaptor proteins in the RTK cascades (Mitsuda et al., 2018). We have found for the first time that RUNX1 regulates the ErbB2/HER2 signaling pathway in gastric cancer cells through transactivating SOS1 expression, rendering itself an ideal target in anti-tumor strategy toward this cancer (Mitsuda et al., 2018). Mechanistically, RUNX1 interacts with the RUNX1 binding DNA sequence located in SOS1 promoter and positively regulates it. Knockdown of RUNX1 led to the decreased expression of SOS1 as well as dephosphorylation of ErbB2/HER2, subsequently suppressed the proliferation of gastric cancer cells (Mitsuda et al., 2018). Chb-M’ consistently led to the deactivation of the ErbB2/HER2 signaling pathway and was effective against several gastric cancer cell lines (Mitsuda et al., 2018). We identified a novel interaction of RUNX1 and the ErbB2/HER2 signaling pathway in gastric cancer, which can potentially be exploited in the management of this malignancy (Fig. 2).
Fig. 2. The mechanism of Her2 signaling activation by RUNX1, and the inhibition of cell proliferation by RUNX inhibitor (Chb-M').
Schematic abstract showing that Left: RUNX1 interacts with the RUNX1 binding DNA sequence located in SOS1 promoter and positively regulates it. Upon stimulation of Her2, SOS proteins act as adaptors to augment the HER2 signaling, thereby thought to significantly contribute to the proliferation of the cells. Right: Novel RUNX inhibitor (Chb-M’) led to the decreased expression of SOS1 as well as dephosphorylation of HER2, subsequently suppressed the proliferation of Her2 gastric cancer.
RUNX transcription factors potentially control E-selectin expression in the bone marrow vascular niche in mice
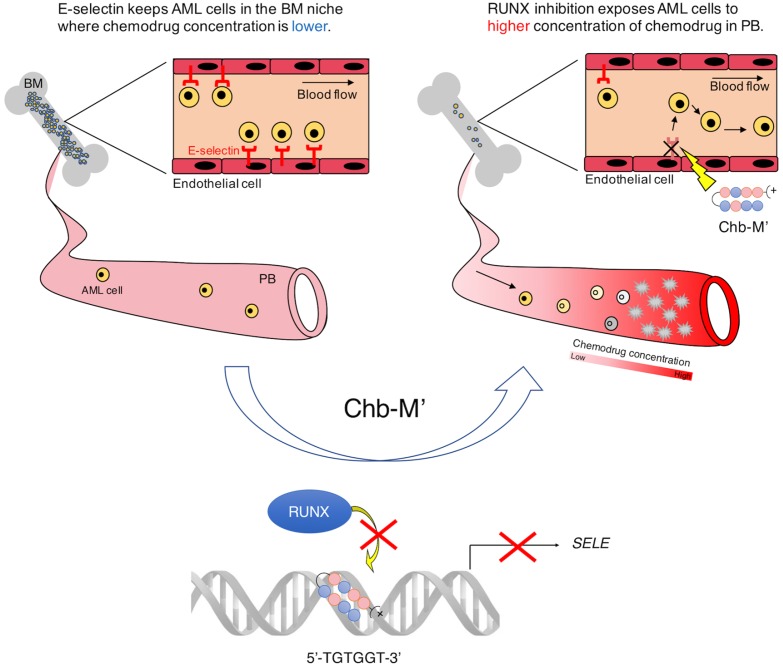
We reported that the evidence suggesting the possible involvement of RUNX transcription factors in the transactivation of E-selectin, a member of selectin family of cell adhesion molecules, on the vascular endothelial cells of the mice bone marrow niche (Morita et al., 2018). The gene switch mediated silencing of RUNX downregulated E-selectin expression in the vascular niche and negatively controlled the engraftment of AML cells in the bone marrow, extending the overall survival of leukemic mice (Morita et al., 2018). This work identi ed the novel role of RUNX family genes in the vascular niche and showed that the vascular niche, a home for AML cells, could be strategically targeted with RUNX-silencing antileukemia therapies. Considering the excellent efficacy of RUNX-inhibition therapy through CROX on AML cells themselves, this strategy potentially targets AML cells both directly and indirectly, thus providing a better chance of cure for poor-prognostic AML patients (Fig. 3).
Fig. 3. Schematic abstract showing that RUNX1 enhances leukemia cell engraftment in the vascular niche possibly through modulating E-selectin expressions.
Left: E-Selectin keeps AML cells in the bone marrow (BM) niche where chemodrug concentration is lower. Right: RUNX1 inhibition esposes AML cells to higher concenyrationof chemodrug in periferal blood (PB). Modified from the article of Morita et al. (2018) (Blood Adv. 2, 509-515) with original copyright holder's permission.
CONCLUSION AND DISCUSSION
Dominant inhibition of the RUNX1 was previously considered to be the main cause of the disease-developing proliferation maintenance mechanism, mainly in CBF leukemia (Sood et al., 2017). However, our studies clarified that wild-type RUNX1 is needed for survival and proliferation of leukemia cells, suggesting that a treatment strategy targeting RUNX1 is more effective (Morita et al., 2017a). As the RUNX family has a mechanism to compensate for loss among the family members, moderate inhibition of RUNX1 increases the total RUNX family protein level through the mechanism “Genetic compensation of RUNX family transcription factors” (Morita et al., 2017a), and the RUNX family transcriptionally regulates the p53 inhibitors such as BCL11A and TRIM24. Therefore, several groups have tried to regulate RUNX1 alone, but suppression of leukemia cells was incomplete. To overcome this, we propose comprehensive RUNX family cluster regulation “cluster regulation of RUNX (CROX)” as a leukemia treatment strategy because it is difficult to individually inhibit RUNX family proteins (Morita et al., 2017a). As PI-Polyamide: Chb-M’ and Chb-50, currently in development, target the common consensus sequences of the RUNX family, individual inhibition of the members is not necessary (Morita et al., 2017a). The RUNX family pathway was negatively controlled and strong cell suppression effects were observed in vitro and in vivo through Chb-M’ treatments. Actually, Chb-M’ consistently led to the deactivation of the ErbB2/HER2 signaling pathway and was effective against Her2 gastric cancer (Mitsuda et al., 2018) and downregulated E-selectin expression in the vascular niche and negatively controlled the engraftment of AML cells in the bone marrow (Morita et al., 2018). Although the comprehensive RUNX family cluster regulation has yet to be applied in the clinical fields, we suggest the RUNX gene switch method using PI-Polyamides as a new modality that may contribute to the next generation of cancer treatment strategies.
ACKNOWLEDGMENTS
This review depends on the results of researches supported by Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]), and Basic Science and Platform Technology Program for Innovative Biological Medicine from Japan Agency for Medical Research and Development (AMED): grant No. 15am0301005h0002, Grant-in-Aid for Scientific Research (KAKENHI): grant No. 17H03597 and 16K14632.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Bando T., Sugiyama H. Synthesis and biological properties of sequence-specific DNA-alkylating pyrrole-imidazole polyamides. Acc. Chem. Res. 2006;39:935–944. doi: 10.1021/ar030287f. [DOI] [PubMed] [Google Scholar]
- Best T.P., Edelson B.S., Nickols N.G., Dervan P.B. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K.A., et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Invest. 2013;123:3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R.K., Kamikubo Y., Anderson S., Kirby M., Alemu L., Zhao L., Liu PP. Cbfb/Runx1 repression-independent blockage of differentiation and accumulation of Csf2rb-expressing cells by Cbfb-MYH11. Blood. 2010;115:1433–1443. doi: 10.1182/blood-2009-06-227413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R.K., Zhao L., Alemu L., Liu P.P. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia. 2015;29:1771–1778. doi: 10.1038/leu.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Kamikubo Y. Genetic compensation of RUNX family transcription factors in leukemia. Cancer Sci. 2018;109:2358–2363. doi: 10.1111/cas.13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikubo Y., Hyde R.K., Zhao L., Alemu L., Rivas C., Garrett L.J., Liu P.P. The C-terminus of CBFβ-SMMHC is required to induce embryonic hematopoietic defects and leukemogenesis. Blood. 2013;121:638–642. doi: 10.1182/blood-2012-06-434688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikubo Y., Zhao L., Wunderlich M., Corpora T., Hyde R.K., Paul T.A., Kundu M., Garrett L., Compton S., Huang G., et al. Accelerated leukemogenesis by truncated CBF beta-SMMHC defective in high-affinity binding with RUNX1. Cancer Cell. 2010;17:455–468. doi: 10.1016/j.ccr.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Tarle S.A., Hajra A., Claxton D.F., Marlton P., Freedman M., Siciliano M.J., Collins F.S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- Liu P.P., Hajra A., Wijmenga C., Collins F.S. Molecular pathogenesis of the chromosome 16 inversion in the M4Eo subtype of acute myeloid leukemia. Blood. 1995;85:2289–2302. doi: 10.1182/blood.V85.9.2289.bloodjournal8592289. [DOI] [PubMed] [Google Scholar]
- Lukasik S.M., Zhang L., Corpora T., Tomanicek S., Li Y., Kundu M., Hartman K., Liu P.P., Laue T.M., Biltonen R.L., et al. Altered affinity of CBF beta-SMMHC for Runx1 explains its role in leukemogenesis. Nat. Struct. Biol. 2002;9:674–679. doi: 10.1038/nsb831. [DOI] [PubMed] [Google Scholar]
- Mitsuda Y., Morita K., Maeda S., Kashiwazaki G., Taniguchi J., Bando T., Hirata M., Kataoka T.R., Muto M., Kaneda Y., et al. RUNX1 positively regulates ErbB2/HER2 signaling pathway through modulating SOS1 expression in gastric cancer cells. Sci. Rep. 2018;8:6423. doi: 10.1038/s41598-018-24969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Maeda S., Suzuki K., Kiyose H., Taniguchi J., Liu P.P., Sugiyama H., Adachi S., Kamikubo Y. Paradoxical enhancement of leukemogenesis in acute myeloid leukemia with moderately-attenuated RUNX1 expressions. Blood Adv. 2017b;1:1440–1451. doi: 10.1182/bloodadvances.2017007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Noura M., Tokushige C., Maeda S., Kiyose H., Kashiwazaki G., Taniguchi J., Bando T., Yoshida K., Ozaki T., et al. Autonomous feedback loop of RUNX1-p53-CBFB in acute myeloid leukemia cells. Sci. Rep. 2017c;7:16604. doi: 10.1038/s41598-017-16799-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Suzuki K., Maeda S., Matsuo A., Mitsuda Y., Tokushige C., Kashiwazaki G., Taniguchi J., Maeda R., Noura M., et al. Genetic regulation of the RUNX transcription factor family has antitumor effects. J. Clin. Invest. 2017a;127:2815–2828. doi: 10.1172/JCI91788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Tokushige C., Maeda S., Kiyose H., Noura M., Iwai A., Yamada M., Kashiwazaki G., Taniguchi J., Bando T., et al. RUNX transcription factors potentially control E-selectin expressions in the vascular niche of mice bone marrow. Blood Adv. 2018;2:509–515. doi: 10.1182/bloodadvances.2017009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R., Kamikubo Y., Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirov T.H., Inoue-Bungo T., Morii H., Fujikawa A., Sasaki M., Kimura K., Shiina M., Sato K., Kumasaka T., Yamamoto M., et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/S0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- Trauger J.W., Baird E.E., Dervan P.B. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382:559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- Warren A.J., Bravo J., Williams R.L., Rabbitts T.H. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFbeta. EMBO J. 2000;19:3004–3015. doi: 10.1093/emboj/19.12.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Liu Y., Lukasik S.M., Speck N.A., Bushweller J.H. CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat. Struct. Mol. Biol. 2004;11:901–906. doi: 10.1038/nsmb819. [DOI] [PubMed] [Google Scholar]