Abstract
The transcription factor RUNX1 first came to prominence due to its involvement in the t(8;21) translocation in acute myeloid leukemia (AML). Since this discovery, RUNX1 has been shown to play important roles not only in leukemia but also in the ontogeny of the normal hematopoietic system. Although it is currently still challenging to fully assess the different parameters regulating RUNX1 dosage, it has become clear that the dose of RUNX1 can greatly affect both leukemia and normal hematopoietic development. It is also becoming evident that varying levels of RUNX1 expression can be used as markers of tumor progression not only in the hematopoietic system, but also in non-hematopoietic cancers. Here, we provide an overview of the current knowledge of the effects of RUNX1 dosage in normal development of both hematopoietic and epithelial tissues and their associated cancers.
Keywords: development, dosage, hematopoiesis, runx1, tumorigenesis
INTRODUCTION
RUNX1 is the founding member of the mammalian core-binding transcription factor family which also consists of RUNX2, RUNX3 and their non-DNA binding co-factor core-binding factor beta (CBFβ) (Ito et al., 2015; Mevel et al., 2019). In humans, RUNX1 is localized on chromosome 21 and was first identified by Miyoshi et al. (1991) as the acute myeloid leukemia gene 1 (AML1) due to its involvement in the t(8;21) translocation in acute myeloid leukemia (AML). Shortly after this discovery, the murine version of Runx1 was identified (Bae et al., 1993; Ogawa et al., 1993b; Wang et al., 1993) which paved the way for the development of Runx1 knockout mouse models. These models revealed that RUNX1 plays a crucial role in the establishment of the hematopoietic system during embryogenesis (North et al., 1999; Okuda et al., 1996; Wang et al., 1996a). Both in ontogeny and disease, there are indications that the dose of wild-type (WT) RUNX1 can have profound effects on cell survival and differentiation. Although arguably best studied in hematopoiesis and leukemia, RUNX1 has also been found to play important roles in the development and tumorigenesis of epithelial tissues (Hong et al., 2019; Mevel et al., 2019; Taniuchi et al., 2012). Here, we aim to provide an overview of the current knowledge of the effects of RUNX1 dosage, in mouse and human, during normal development and homeostasis of hematopoietic and epithelial tissues as well as the known requirements for endogenous WT RUNX1 in cancers.
Dosage reflects both the amount of protein as well as its activation status. Indeed, RUNX1 protein levels can be regulated by the rate of transcription, translation and stability. RUNX1 activity (either as activator or repressor) is also further modulated through protein conformation, intracellular localization, post-translational modifications (PTMs) and interactions with additional proteins. RUNX1 has many interacting partners and their availability depends on cell type, differentiation status and cell cycle. Describing these interactors fully lies outside the scope of this brief review and has been covered in great details in excellent recent reviews (Chuang et al., 2013; Goyama et al., 2015; Ito et al., 2015). In vertebrates, there is a high degree of homology between the different RUNX proteins both within and across species (Rennert et al., 2003). However this high degree of inter-gene similarity does not necessarily mean that mechanisms of action and regulation of RUNX1 can be extrapolated to other RUNX family members (Bruno et al., 2019). This review focuses specifically on what is known about RUNX1 in human and mouse.
RUNX1 PROTEIN LEVELS AND RUNX1 ACTIVITY
Two promoters control RUNX1 transcription, the P1 (distal) and the P2 (proximal) promoter, whose major generated transcripts are respectively the distal RUNX1c and the proximal RUNX1b isoforms (Ghozi et al., 1996; Miyoshi et al., 1995). The two promoters are differentially active depending on the cell context and developmental stage (Bee et al., 2009; Draper et al., 2016; Sroczynska et al., 2009). P1 transcripts are longer than P2 transcripts due to the presence of a 150 kb intron suggesting that the former takes longer to produce (Levanon et al., 2001; Pozner et al., 2000). Furthermore, both isoforms possess different 5' and 3' untranslated regions containing motifs known to affect post-transcriptional events like RNA stability and the rate of translation initiation (Levanon et al., 2001; Levanon and Groner, 2004; Pozner et al., 2000). At the protein level, the two isoforms only differ in their most N-terminal amino acid sequence (Fig. 1A). The unique N-terminus of RUNX1b has been implicated in protein stability (Nieke et al., 2017), while the unique N-terminal sequence of RUNX1c has been shown to have higher binding capacity on certain genes (Telfer and Rothenberg, 2001). The common regions of both isoforms consist of a N-terminal region, which potentially plays a role in transcriptional activation (Liu et al., 2006), followed by the DNA binding Runt homology domain which also forms the interaction domain for the RUNX family co-factor CBFβ. CBFβ is the heterodimeric binding partner of all RUNX proteins (Nagata et al., 1999; Ogawa et al., 1993a; 1993b). CBFβ enhances RUNX DNA-binding affinity and protects it from degradation (Bravo et al., 2001; Huang et al., 2001; Tahirov et al., 2001; Yan et al., 2004). Interestingly, two different isoforms of CBFβ have been described which, at least in the case of RUNX2, have been shown to differentially affect DNA binding (Jiang et al., 2016). The C-terminal half of RUNX1 harbors a transactivation domain, flanked by inhibitory regions (Aronson et al., 1997; Kanno et al., 1998; Levanon et al., 1998). In primates there is a third commonly expressed RUNX1 isoform, RUNX1a, transcribed from the P2 promoter (Miyoshi et al., 1995). This isoform lacks most of the C-terminus including the transactivation domain. In mice, it is thought that an exon 6 skipping variant of Runx1b is fulfilling a similar role (Komeno et al., 2014).
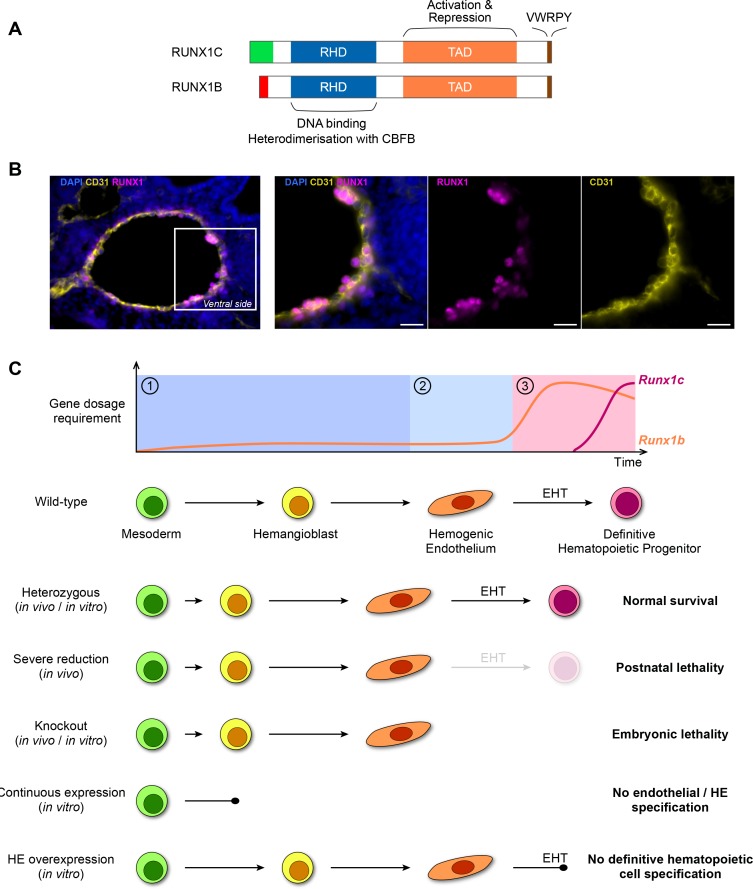
Fig. 1. RUNX1 dosage in hematopoietic development.
(A) Schematic representation of the two most abundant RUNX1 isoforms. Except for the most N-terminal sequence (RUNX1C N-terminal in green, RUNX1B N-terminal in red) the proteins are identical and they both contain the highly conserved Runt homology domain (RHD, blue) followed by a transactivation domain (TAD, orange) which is flanked by inhibitory regions. The C-terminal inhibitory region contains a highly conserved VWRPY motif (brown). (B) Immunofluorescence on the AGM of a E10.5 mouse embryo. The dorsal aortic endothelial cells are marked by the endothelial marker CD31 (yellow). The majority of the cells on the ventral side of the dorsal aorta (constituting both endothelial and rare HE cells) are positive for the RUNX1 protein (magenta). Scale bars = 20 μm. (C) Current model of RUNX1 dosage in hematopoietic development. Top: RUNX1 dosage requirement can be divided in three phases. Phase ①: early in differentiation RUNX1 is not required but its (low) dose influences the timing and dynamics of HE cells appearance. Phase ②: although RUNX1 levels are still low in HE cells, its presence is required for the initiation of the EHT. Phase ③: an increased dose of RUNX1 is required for the completion of EHT and the generation of the first mature hematopoietic cells. The whole differentiation process is predominantly controlled by the RUNX1b isoform. Bottom: schematic overview of the currently available phenotypic data on RUNX1 dosage during the establishment of the hematopoietic system in the embryo.
Finally, RUNX1 activity and stability can be modulated by various PTMs including phosphorylation, methylation, acetylation, ubiquitination, sumoylation and prolyl isomerisation (Blumenthal et al., 2017; Goyama et al., 2015; Ito et al., 2015). In Table 1, we have listed the residues in RUNX1 that have been shown to be the target of PTM and their effect on RUNX1. Few of these PTM have been extensively studied in vivo neither in development nor in cancer models. In general multiple residues have to be mutated to see clear phenotypes in vivo suggesting, perhaps not unexpectedly, that there is a high degree of redundancy and/or compensation in place (Goyama et al., 2004; Huang et al., 2012; Tachibana et al., 2008; Yoshimi et al., 2012).
Table 1.
Post translational modifications of RUNX1
| Post translational modification | Effect | Modifier | Target domain | Target residues (Runx1b) |
|---|---|---|---|---|
| Serine/threonine phosphorylation (1) | Increased transactivation, decreased stability | ERK | Predominatly C-term transactivaton domain | S249, S266, S276, S435, T273 |
| Serine/threonine phosphorylation (1) | Increased transactivation, decreased stability | Hip2k | Predominatly C-term transactivaton domain | S249, S276, T273 |
| Serine/threonine phosphorylation (1, 2) | Increased transactivation, decreased stability | CDK | Predominatly C-term transactivaton domain | S21, S249, S266, S276, S397, T273 |
| Tyrosine phosphorylation (3) | Increased transactivation, increased stability, reduced HDAC interaction, increased DNA binding | Src kinase | Predominatly C-term inhibitory domain | Y260, Y375, Y378, Y379, Y386 |
| Methylation (4) | Reduced SIN3a interaction, increased transactivation activity | PRMT1 | C-term inhibitory domain | R2016 and R210 |
| Methylation (5) | Reduced transactivation via increased co-repressor DPF2 binding | PRMT4 | C-term transactivation domain | R223 |
| Acetylation (6) | Reduced DNA binding, reduced transactivation | p300/CBP | N-terminus | K24, K43 |
| Ubiquitination (7) | Increased degradation | STUB1 E3 ubiquitin ligase | Predominantly runt domain | K24, K43, K83, 90, 125, 144, 167, 182, 188 (potential targets) |
| SUMOylation (8) | Unknown (reduced transactivation shown for RUNX3) | PIAS1 | Runt domain | K144 |
| Prolyl isomerization (9) | Increased acetylation, stability and transactivation activity | PIN1 | Not defined | Not defined |
Currently described post translational modifications of RUNX1 and their effect on RUNX1. All amino acid residues are numbered based on RUNX1b. The number between brackets (#) refers to the following citations: 1, (Aikawa et al., 2006; Biggs et al., 2006; Imai et al., 2004; Tanaka et al., 1996; Wee et al., 2008; Zhang et al., 2004); 2, (Guo and Friedman, 2011); 3, (Huang et al., 2012; Leong et al., 2016); 4, (Zhao et al., 2008); 5, (Vu et al., 2013); 6, (Yamaguchi et al., 2004); 7, (Shang et al., 2009; Yonezawa et al., 2017); 8, (Kim et al., 2014); 9, (Islam et al., 2014).
RUNX1 IN HEMATOPOIESIS AND LEUKEMIA
RUNX1 dosage in hematopoietic development
In mammalian embryogenesis, the hematopoietic system is established via several consecutive waves of blood cell generation (Dzierzak and Bigas, 2018). In mice, the first wave generates primitive erythrocytes at embryonic day 7.25 (E7.25). It is followed by the emergence of erythroid myeloid progenitors at E8.25, and lymphoid myeloid progenitors at E9.5. The final wave of hematopoiesis at E10.5 takes place in the aorta-gonad-mesonephros (AGM) region of the embryo proper and generates the first hematopoietic stem cells (HSCs). The HSCs then migrate to the fetal liver (E12.5) where they multiply and mature before colonizing the bone marrow (E16.5). Except for the first wave, RUNX1 is absolutely required for blood cell formation (Chen et al., 2009; Lancrin et al., 2009; North et al., 1999; Okuda et al., 1996; Wang et al., 1996a; Yokomizo et al., 2008). At all sites of de-novo blood cell generation in the embryo, the hematopoietic cells have been found to arise from a specialized endothelium (hemogenic endothelium or HE), via a process termed the endothelial-to-hematopoietic transition (EHT) (Boisset et al., 2010; Chen et al., 2009; Eilken et al., 2009; Lancrin et al., 2009; Ottersbach, 2019; Zovein et al., 2008). RUNX1 is required for EHT (Chen et al., 2009; Lancrin et al., 2009; Liakhovitskaia et al., 2009; Menegatti et al., 2019) and there are indications that RUNX1 dosage is important for the progression and timing of this process.
Detailed studies using reporter mice and mouse embryonic stem cell lines (mESCs) demonstrated that the P2 promoter (Runx1b isoform) is activated first during ontogeny (Bee et al., 2009; Sroczynska et al., 2009). The mESCs system further revealed that the P2 promoter is active from the hemangioblasts (the mesodermal precursor to HE) stage onwards (Lie-A-Ling et al., 2018; Sroczynska et al., 2009). Both in vivo and in the mESCs system, it is clear that the P2 promoter is dominant in the HE, while afterwards, as the first hematopoietic stem and progenitor cells (HSPCs) emerge, the P1 promoter becomes active (Bee et al., 2009; Sroczynska et al., 2009). In vivo, upon migration of the HSPC to the fetal liver, P2 activity decreases and P1 becomes the dominant promoter (Bee et al., 2009; Sroczynska et al., 2009). Quantification of Runx1 RNA levels in bulk sorted populations derived from mESCs suggest Runx1 expression is higher in hematopoietic progenitors than in the preceding differentiation stages including HE (Goode et al., 2016; Lie-A-Ling et al., 2018). Similar observations have been made by single cell polymerase chain reaction analyses (Swiers et al., 2013) and RNA-seq (Baron et al., 2018) of cells isolated from mouse AGM, with the frequency of Runx1 expressing cells increasing according to differentiation stage. Despite potentially lower levels of Runx1 expression in HE, immunofluorescence analyses of the AGM in mice has demonstrated that the majority of the cells on the ventral side of the dorsal aorta (constituting both endothelial and rare HE cells) are positive for the presence of the RUNX1 protein (Fig. 1B) (North et al., 1999).
In human, the picture is less clear. Early publications indicate that during human ESCs (hESCs) differentiation the expression of RUNX1 isoforms is similar to that of the mESCs system whereby RUNX1b precedes RUNX1c (Challen and Goodell, 2010; Ditadi et al., 2015; Ng et al., 2016), whereas recent papers report that RUNX1c is expressed first (Angelos et al., 2018; Navarro-Montero et al., 2017). Similar to data obtained in mice, RNA-seq on single cells from human embryos demonstrated that RUNX1 expression can be detected in cells with arterial endothelial gene expression profiles (likely constituting both endothelium and HE), and as the cells differentiate to HSPCs, the proportion of RUNX1 expressing cells increases (Zeng et al., 2019).
Modulation of gene dosage has been extensively used to assess the effect of RUNX1 dosage changes in ontogeny. Although total Runx1 KO is embryonic lethal, heterozygous mice appear unaffected (North et al., 1999; Okuda et al., 1996; Wang et al., 1996a). However, closer inspection revealed profound effects on the window of HSC emergence which is expedited by approximately half a day (Cai et al., 2000; Mukouyama et al., 2000; Wang et al., 1996a; 1996b). In contrast, a more severe reduction of Runx1 levels, by homozygous disruption of the P2 promoter, leads to postnatal death (Bee et al., 2010; Pozner et al., 2007). Potential dosage effects are also observed when the RUNX1 non-DNA binding partner Cbfβ is deleted (Niki et al., 1997; Sasaki et al., 1996; Wang et al., 1996b). Indeed, although Cbfβ knockout mice appear to phenocopy the Runx1 KO models, generation of hypomorphic Cbfβ alleles resulted in a slight delay in the window of mortality when compared to the Runx1 KO animals and the presence of a few hematopoietic progenitors in these embryos (Wang et al., 1996b). Evidence from the mESCs system, closely modeling yolk sac hematopoiesis, is in line with the data obtained in vivo and demonstrated that the reduction of RUNX1 through haploinsufficiency expedites blood development by 12 h (Lacaud et al., 2002; Lacaud et al., 2004). Conversely, overexpression of RUNX1 in both human and mESCs blocks hematopoiesis. In hESCs, RUNX1 overexpression from the ESC stage onwards has no effect on mesoderm commitment but disrupts subsequent endothelial and HE specification (Chen et al., 2017). Overexpression in mESCs derived HE appears to induce an accelerated EHT without the emergence of mature hematopoietic cells, while low levels of Runx1 can induce a productive EHT (Lie-A-Ling et al., 2018). Furthermore, it was also demonstrated that RUNX1 is required for both the initiation and completion of EHT and that both events may require a different dose of RUNX1.
Taken together, the current data indicate that the initial establishment of the hematopoietic system relies on a low dose of RUNX1 and that careful modulation of this low dose controls the dynamic and progression of blood formation (Fig. 1C).
RUNX1 mutations and requirement in leukemia
Considering its importance in the ontogeny of the hematopoietic system, it is not surprising that RUNX1 has been found to be a recurrent target of genomic alterations in hematological disorders (reviewed in Bellissimo and Speck, 2017; Sood et al., 2017). RUNX1 is implicated in more than 50 chromosomal translocations leading to pediatric acute lymphoblastic leukemia (ALL), AML and myelodysplastic syndrome (MDS). In addition to translocations, mono or bi-allelic somatic mutations of RUNX1 have been documented in MDS, AML, ALL and chronic myelomonocytic leukemia (CMML). Finally, germline mono-allelic mutations of RUNX1 are associated with familial platelet disorder with predisposition to AML (FPD/ AML).
In terms of dosage, high levels of RUNX1 mRNA are frequently observed in AML, T cell-ALL (T-ALL) and B cell-ALL (B-ALL) (Sun et al., 2019). Increased RUNX1 transcription is in particular observed in B-ALLs and is associated with the fusion of ETV6 to RUNX1 (TEL/AML1) (Gandemer et al., 2007; Robinson et al., 2003; Soulier et al., 2003). In this context, increased RUNX1 mRNA is a positive prognostic marker although its precise role is unclear. In T-ALL, the non-mutated WT RUNX1 allele is important for leukemogenesis and tumor survival (Choi et al., 2017). Here, RUNX1 is required for the expression of a subset of TAL-1 and Notch regulated genes, including MYB and MYC, which are required for maintenance of the leukemia. Consequently, the deletion of WT Runx1 in a mouse T-ALL model or small molecule mediated inhibition of RUNX1 in patient samples can impair leukemic growth. Interestingly, RUNX1 inhibition did not affect normal hematopoietic cells, indicating a specific requirement for WT RUNX1 in T-ALL cells (Choi et al., 2017).
In AMLs, increased RUNX1 transcript levels have been associated with both, de-novo AMLs and AMLs harboring the FLT3-ITD (internal tandem duplication) (Behrens et al., 2017; Salarpour et al., 2017). In the latter case, RUNX1 cooperates with FLT3 to induce leukemia. Also, it is striking that RUNX1 mutations appear to be absent in patients with leukemogenic fusion protein leukemias (Patel et al., 2012; Schnittger et al., 2011; Tang et al., 2009). In this context, dependency on WT RUNX1 has been shown for AML1-ETO (t8;21), CBFB-SMMHC (inv16), MLL-AF9, and CBFB-MYH11 (inv16) translocation leukemias (Ben-Ami et al., 2013; Goyama et al., 2013; Hyde et al., 2015). In the case of AML1-ETO, WT RUNX1 and the RUNX1-ETO fusions both target many identical sites in the genome. However, binding is mutually exclusive and it is the balance between the two proteins that is driving the transcriptional networks maintaining leukemia (Ptasinska et al., 2014). Investigation of CBFB-MYH11 (inv16) has shown that leukemia containing fusion protein variants with reduced WT RUNX1 binding/inhibition are more leukemogenic than their stronger RUNX1 inhibitory counterparts (Hyde et al., 2015; Kamikubo et al., 2010). The need for the right balance between oncogenic mutation/fusion and WT RUNX1 is further highlighted by the finding that patient samples with intermediate WT RUNX1 levels tend to have a poor prognosis (Morita et al., 2017a). Additionally, depletion of RUNX1 has been shown to lead to compensation by the other RUNX family members RUNX2 and RUNX3 (Morita et al., 2017a; 2017b). The addiction of leukemia to WT RUNX1 extends to AML expressing mutated forms of RUNX1, with its knockdown negatively affecting leukemic cells (Mill et al., 2019). Finally, patient studies demonstrated allelic imbalances in the transcriptional activity of mutant and WT alleles, further highlighting the potential importance of the dosage of WT RUNX1 in a leukemic context (Batcha et al., 2019).
RUNX1 IN EPITHELIAL TISSUES AND CANCERS
The role of RUNX1 dosage during the development and homeostasis of epithelial tissues remains less documented than in the hematopoietic setting. However, increasing evidence suggests a role for RUNX1 in various non-hematopoietic tissues of epithelial origin (reviewed in Mevel et al., 2019). Indeed, high throughput next-generation sequencing has revealed relatively high frequencies of genomic alterations of RUNX1, and CBFβ in solid cancers (Blyth et al., 2005; Ito et al., 2015), albeit to lower levels than in leukemia (Figs. 2A and 2B). Interestingly, while it is yet to be fully determined to what extent these alterations contribute to tumor biology, mutations of RUNX1 have been associated with loss of function (van Bragt et al., 2014). Beyond the presence of these mutations, earlier studies identified RUNX1 mRNA as part of a 17-gene signature associated with metastasis in a panel of adenocarcinomas, including breast and prostate cancers, with its expression inversely correlating with tumor aggressiveness (Ramaswamy et al., 2003). Overall, under- and over-expression of endogenous RUNX1 has been found in several solid tumors, reinforcing the idea that it is broadly implicated in the biology and pathology of epithelial tissues (Blyth et al., 2005; Ito et al., 2015; Scheitz et al., 2012).
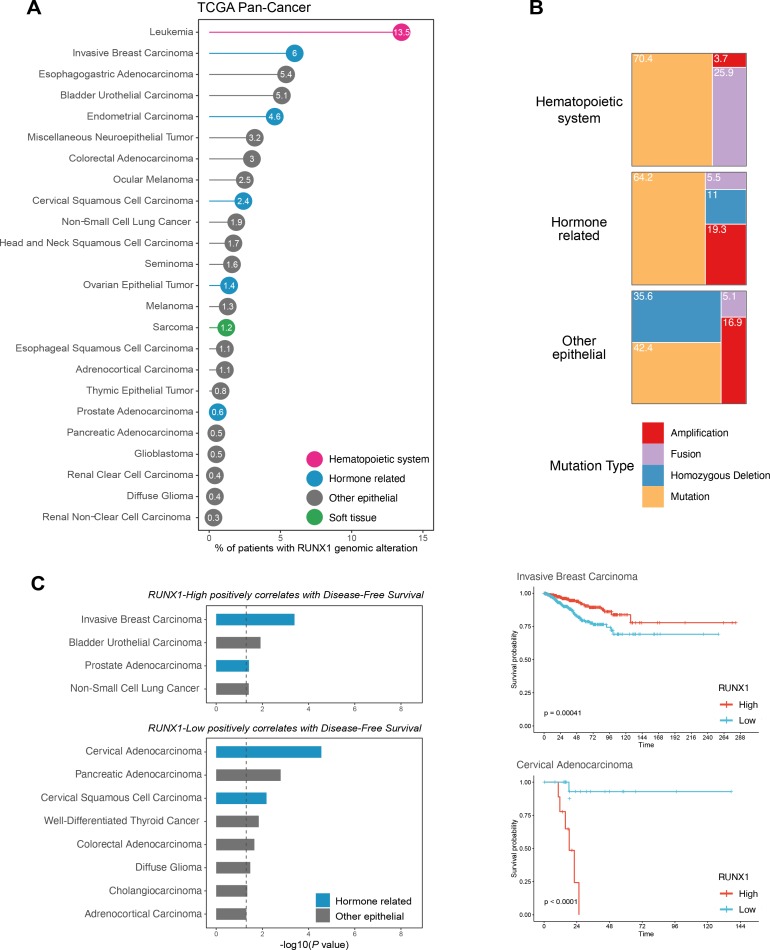
Fig. 2. Meta-analysis of RUNX1 alterations and prognostic value in the TCGA PanCancer atlas.
(A) Frequency of RUNX1 genomic alterations across the TCGA PanCancer atlas. Cancers with no alterations were excluded. Cancers affecting the hematopoietic system are colored in pink, hormone related cancers in blue, cancers of soft tissues in green, and other epithelial cancers in grey. (B) Proportion of RUNX1 amplification, homozygous deletion, fusion and mutation in cancers affecting the hematopoietic system, hormone related cancers, and additional epithelial cancers. Soft tissue cancers were excluded from these analyses due to the small number of patients affected. (C) Prognostic value of RUNX1 mRNA expression using the TCGA PanCancer Atlas expression data, in terms of Disease-Free Survival. Datasets of the TCGA PanCancer Atlas were downloaded from cBioPortal (https://www.cbioportal.org/). Briefly, patients were split in RUNX1-High and RUNX1-Low groups using the “surv_cutpoint” function of the “survminer” R package (“minprop” argument set to 0.1). Cancers were then separated into two groups, depending on whether RUNX1-High and RUNX1-Low groups are significantly associated with a better prognosis (P value < 0.05 using the univariate log-rank test). Representative examples of the corresponding Kaplan–Meier curves are shown for the Invasive Breast Carcinoma and Cervical Adenocarcinoma datasets (defined by the “Cancer Type” column of the TCGA PanCancer Atlas clinical data).
RUNX1 in hormone-related cancers
Hormone-related cancers constitute some of the most common cancers in women and men, and RUNX1 alterations have been reported in all of these malignancies. To date, the role of RUNX1 in solid tumorigenesis has been best studied in mammary tissue (Riggio and Blyth, 2017). The normal breast epithelium is one of the few epithelial tissues for which changes in RUNX1 dosage have been reported during normal physiology/homeostasis. In addition to differential expression levels of RUNX1 in the basal and luminal compartments of the mammary ducts, RUNX1 levels have also been shown to fluctuate during pregnancy and lactation (Blyth et al., 2010; McDonald et al., 2014; van Bragt et al., 2014). In mice, Runx1 was demonstrated to be a crucial regulator of the ER+ mammary luminal lineage. Deletion of Runx1 led to a reduction of ER+ mature luminal cells, which could be rescued by the loss of either Trp53 or Rb1 (van Bragt et al., 2014). With regards to cancer, several studies involving large patient cohorts have identified recurrent CBFβ and RUNX1 mutations (Banerji et al., 2012; Cancer Genome Atlas Network, 2012; Ellis et al., 2012; Kas et al., 2017; Nik-Zainal et al., 2016; Pereira et al., 2016). At the protein level, high-grade primary breast tumors also displayed in general reduced levels of RUNX1 compared to low/mid-grade tumors (Kadota et al., 2010). These observations have led to the hypothesis that RUNX1 could have a tumor suppressor function. The proliferation of ER+ breast cancer cells was increased upon RUNX1 knockdown, which led to estrogen-mediated AXIN1 suppression and enhanced β-catenin activation (Chimge et al., 2016). In agreement with a tumor suppressor role, a link has emerged between RUNX1 and suppression of the epithelial-to-mesenchymal transition (EMT) process. Indeed, downregulation of RUNX1 in the normal mammary epithelial cell line MCF10A was sufficient to induce hyperproliferation and abnormal morphogenesis (Wang et al., 2011). The morphological changes observed upon RUNX1 knockdown were characteristic of an EMT, and associated with the activation of transforming growth factor β (TGFβ) and WNT signaling pathways (Hong et al., 2017). Both RUNX1 and RUNX3 were also shown to prevent the induction of YAP-mediated EMT in this same cell line (Kulkarni et al., 2018). Likewise, the RUNX1-CBFβ complex was able to prevent the migration potential of the ER+ breast cancer cell line MCF7 in an ER-dependent manner (Pegg et al., 2019). The emerging role of RUNX1 in EMT is not unexpected considering its well documented role in EHT, a process often referred to as ‘EMT-like’ (Hamidi and Sheng, 2018; Monteiro et al., 2016). However, while RUNX1 is critical for the induction of EHT during hematopoietic development, it appears to act as a gatekeeper of EMT in breast cancer cells.
In contrast to its putative tumor suppressive functions, RUNX1 is also believed to be associated with oncogenic roles. Indeed, higher RUNX1 mRNA levels were found in the triple-negative breast cancer subgroup (Karn et al., 2011; Rody et al., 2011). This was later corroborated by a strong correlation between high RUNX1 protein levels and poor prognosis in triple-negative and ER-negative breast cancers (Ferrari et al., 2014). Increased expression of RUNX1 was also associated with disease progression in patient samples and in the MMTV‐PyMT mouse model. Interestingly, the invasiveness of the cells isolated from this mouse model could be repressed by knocking-down Runx1 expression (Browne et al., 2015), suggesting that its role in EMT may be context-dependent.
Beyond breast cancer, overexpression of RUNX1 was correlated with overexpression of p21WAF1/CIP1 in invasive endometrioid carcinoma, where it was suggested to play a role in promoting myometrial infiltration (Planaguma et al., 2004; 2006). In this respect, Doll and colleagues found that ectopic overexpression of RUNX1 in the endometrial cancer cell line HEC1A was associated with the establishment of distant metastasis (Doll et al., 2009). High levels of RUNX1 were also reported in human epithelial ovarian tumors, and its knockdown in the SKOV-3 cell line led to a decrease in proliferation, migration, and invasion (Keita et al., 2013).
Although less substantial than in female-related cancers, there is accumulating evidence for a potential role of RUNX1 in prostate cancer. Single-nucleotide polymorphisms within the RUNX1 gene—such as the rs2253319 polymorphism— were associated with an increased risk of prostate cancer progression and metastasis (Huang et al., 2011). RUNX1 was also found amplified in a significant proportion of neuroendocrine castration-resistant prostate cancer (Beltran et al., 2016). However, the biological relevance of these alterations, if any, remains unknown. Contrasting studies looking at RUNX1 expression in prostate cancer have reported that RUNX1 mRNA increases with pathological stage (Yeh et al., 2009), while protein levels have been reported to be decreased in advanced forms of the disease (Takayama et al., 2015). Interestingly, the links between RUNX1 and hormones reported in breast cancer (Riggio and Blyth, 2017) seem to extend to the prostate gland which is particularly rich in androgens. In Nkx3.1/Pten mutant mice, prolonged exposure to reduced androgens levels resulted in prostate tumors with up-regulated Runx1 (Banach-Petrosky et al., 2007). RUNX1 has also been shown to be a downstream target of androgen receptor signaling, and is thought to play divergent roles in AR-dependent and castration-resistant prostate cancer cell lines (Takayama et al., 2015). With regards to the growing importance of stroma-cancer interactions, downregulation of RUNX1 expression in mesenchymal stem cells was shown to reduce their proliferative potential in response to TGFβ, before their differentiation into prostate cancer-associated myofibroblasts (Kim et al., 2014).
RUNX1 in skin cancers
In keeping with its role in hematopoiesis, Runx1 dosage has been found to be important for hair follicle stem cells. During homeostasis, reduced levels of Runx1 favors self-renewal of bulge stem cells (Hoi et al., 2010), while high Runx1 expression promotes differentiation into early progenitor hair germ cells (Lee et al., 2014). RUNX1 has been linked to skin cancer in mice, where its activated expression during chemically induced skin carcinogenesis was proposed to be oncogenic (Hoi et al., 2010). In line with this, loss of RUNX1 impaired the proliferation of human oral and skin squamous cell carcinoma cell lines (Scheitz et al., 2012). Runx1 was also found essential for the survival and proliferation of cultured keratinocytes (Hoi et al., 2010), notably by regulating fatty acid production (Jain et al., 2018).
Other tissues
RUNX1 has also been linked with tumors of the gastrointestinal tract, where it was found to be frequently downregulated (Miyagawa et al., 2006; Sakakura et al., 2005). In conditional mouse models, Runx1 deletion is sufficient to induce intestinal tumorigenesis (Fijneman et al., 2012). In gastric cancer cell lines, both the knockdown of RUNX1 and its therapeutic inhibition resulted in reduced tumorigenic potential via suppression of the ErbB2/HER2 signaling pathway (Mitsuda et al., 2018). Finally, the previously noted emerging link between RUNX1 and EMT has also been documented in colorectal cancer (Li et al., 2019), and renal fibrosis (Zhou et al., 2018) in which RUNX1 acts as an inducer of EMT. Increased expression of RUNX1 was also predictive of poor prognosis in patients diagnosed with clear cell renal cell carcinoma (Fu et al., 2019).
CONCLUSIONS
It is now well established that RUNX1 dosage is important during normal development and homeostasis of hematopoietic tissues, and there is a growing body of evidence indicating that it is important in epithelial tissues as well. These studies highlight the multifaceted characteristics of RUNX1, in particular in non-hematopoietic tissues, where it was not originally thought to be involved. Alterations of RUNX1 dosage in these tissues were initially revealed by large scale genomic studies and these results are reinforced by growing experimental evidence implicating RUNX1 in crucial hallmarks of cancer progression such as cell proliferation, EMT or DNA repair (Tay et al., 2018). It has now become clear that RUNX1 can act both as an oncogenic or a tumor-suppressive factor (Blyth et al., 2005; Ito et al., 2015; Neil et al., 2017). Intriguingly, the implication of RUNX1 in both female and male related cancers has revealed a close relationship with ER and AR, which warrants further investigations. While the functional evidence between RUNX1 dosage and cancer development is often still lacking and requires further work, it has become evident that varying levels of RUNX1 expression can be used as markers of tumor progression in specific clinical cohorts (Fig. 2C).
Finally, although systems modifying RUNX1 dosage via (conditional) knock-out alleles as well as controlled transcriptional regulation provide valuable information on how RUNX1 dosage can affect normal physiology and cancer, detailed stage and cell type-specific information on physiological RUNX1 dosage levels would drive our understanding even further. In this context, it should be emphasized that when evaluating RUNX1 dosage, both the amount of protein as well as its activation status should be taken into consideration. Currently, it is still very challenging to fully assess the different parameters regulating RUNX1 dosages. However, the continuous improvement of single-cell technologies might soon allow us to interrogate, at a single-cell level, the quantity and ratios of RUNX1 isoforms, as well as their PTMs. Such data would provide valuable insights on RUNX1 dosage at the single cell level and would allow us to better investigate their functions.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Aikawa Y., Nguyen L.A., Isono K., Takakura N., Tagata Y., Schmitz M.L., Koseki H., Kitabayashi I. Roles of HIPK1 and HIPK2 in AML1- and p300-dependent transcription, hematopoiesis and blood vessel formation. EMBO J. 2006;25:3955–3965. doi: 10.1038/sj.emboj.7601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelos M.G., Abrahante J.E., Blum R.H., Kaufman D.S. Single cell resolution of human hematoendothelial cells defines transcriptional signatures of hemogenic endothelium. Stem Cells. 2018;36:206–217. doi: 10.1002/stem.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson B.D., Fisher A.L., Blechman K., Caudy M., Gergen J.P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol. 1997;17:5581–5587. doi: 10.1128/MCB.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.C., Yamaguchi-Iwai Y., Ogawa E., Maruyama M., Inuzuka M., Kagoshima H., Shigesada K., Satake M., Ito Y. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993;8:809–81. [PubMed] [Google Scholar]
- Banach-Petrosky W., Jessen W.J., Ouyang X., Gao H., Rao J., Quinn J., Aronow B.J., Abate-Shen C. Prolonged exposure to reduced levels of androgen accelerates prostate cancer progression in Nkx3.1; Pten mutant mice. Cancer Res. 2007;67:9089–9096. doi: 10.1158/0008-5472.CAN-07-2887. [DOI] [PubMed] [Google Scholar]
- Banerji S., Cibulskis K., Rangel-Escareno C., Brown K.K., Carter S.L., Frederick A.M., Lawrence M.S., Sivachenko A.Y., Sougnez C., Zou L., et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron C.S., Kester L., Klaus A., Boisset J.C., Thambyrajah R., Yvernogeau L., Kouskoff V., Lacaud G., van Oudenaarden A., Robin C. Single-cell transcriptomics reveal the dynamic of haematopoietic stem cell production in the aorta. Nat. Commun. 2018;9:251. doi: 10.1038/s41467-018-04893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batcha A.M.N., Bamopoulos S.A., Kerbs P., Kumar A., Jurinovic V., Rothenberg-Thurley M., Ksienzyk B., Philippou-Massier J., Krebs S., Blum H., et al. Allelic imbalance of recurrently mutated genes in acute myeloid leukaemia. Sci. Rep. 2019;9:11796. doi: 10.1038/s41598-019-48167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bee T., Liddiard K., Swiers G., Bickley S.R., Vink C.S., Jarratt A., Hughes J.R., Medvinsky A., de Bruijn M.F. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol. Dis. 2009;43:35–42. doi: 10.1016/j.bcmd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Bee T., Swiers G., Muroi S., Pozner A., Nottingham W., Santos A.C., Li P.S., Taniuchi I., de Bruijn M.F. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- Behrens K., Maul K., Tekin N., Kriebitzsch N., Indenbirken D., Prassolov V., Muller U., Serve H., Cammenga J., Stocking C. RUNX1 cooperates with FLT3-ITD to induce leukemia. J. Exp. Med. 2017;214:737–752. doi: 10.1084/jem.20160927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellissimo D.C., Speck N.A. RUNX1 mutations in inherited and sporadic leukemia. Front. Cell Dev. Biol. 2017;5:111. doi: 10.3389/fcell.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H., Prandi D., Mosquera J.M., Benelli M., Puca L., Cyrta J., Marotz C., Giannopoulou E., Chakravarthi B.V., Varambally S., et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami O., Friedman D., Leshkowitz D., Goldenberg D., Orlovsky K., Pencovich N., Lotem J., Tanay A., Groner Y. Addiction of t(8;21) and inv(16) acute myeloid leukemia to native RUNX1. Cell Rep. 2013;4:1131–1143. doi: 10.1016/j.celrep.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Biggs J.R., Peterson L.F., Zhang Y., Kraft A.S., Zhang D.E. AML1/RUNX1 phosphorylation by cyclin-dependent kinases regulates the degradation of AML1/RUNX1 by the anaphase-promoting complex. Mol. Cell. Biol. 2006;26:7420–7429. doi: 10.1128/MCB.00597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal E., Greenblatt S., Huang G., Ando K., Xu Y., Nimer S.D. Covalent modifications of RUNX proteins: structure affects function. Adv. Exp. Med. Biol. 2017;962:33–44. doi: 10.1007/978-981-10-3233-2_3. [DOI] [PubMed] [Google Scholar]
- Blyth K., Cameron E.R., Neil J.C. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Blyth K., Vaillant F., Jenkins A., McDonald L., Pringle M.A., Huser C., Stein T., Neil J., Cameron E.R. Runx2 in normal tissues and cancer cells: a developing story. Blood Cells Mol. Dis. 2010;45:117–123. doi: 10.1016/j.bcmd.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Boisset J.C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Bravo J., Li Z., Speck N.A., Warren A.J. The leukemia-associated AML1 (Runx1)--CBF beta complex functions as a DNA-induced molecular clamp. Nat. Struct. Biol. 2001;8:371–378. doi: 10.1038/86264. [DOI] [PubMed] [Google Scholar]
- Browne G., Taipaleenmaki H., Bishop N.M., Madasu S.C., Shaw L.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. J. Cell. Physiol. 2015;230:2522–2532. doi: 10.1002/jcp.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L., Ramlall V., Studer R.A., Sauer S., Bradley D., Dharmalingam G., Carroll T., Ghoneim M., Chopin M., Nutt S.L., et al. Selective deployment of transcription factor paralogs with submaximal strength facilitates gene regulation in the immune system. Nat. Immunol. 2019;20:1372–1380. doi: 10.1038/s41590-019-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., de Bruijn M., Ma X., Dortland B., Luteijn T., Downing R.J., Dzierzak E. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity. 2000;13:423–431. doi: 10.1016/S1074-7613(00)00042-X. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challen G.A., Goodell M.A. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp. Hematol. 2010;38:403–416. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Teng J., Liu H., Pan X., Zhou Y., Huang S., Lai M., Bian G., Mao B., Sun W., et al. Inducible overexpression of RUNX1b/c in human embryonic stem cells blocks early hematopoiesis from mesoderm. J. Mol. Cell Biol. 2017;9:262–273. doi: 10.1093/jmcb/mjx032. [DOI] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., Speck N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge N.O., Little G.H., Baniwal S.K., Adisetiyo H., Xie Y., Zhang T., O'Laughlin A., Liu Z.Y., Ulrich P., Martin A., et al. RUNX1 prevents oestrogen-mediated AXIN1 suppression and beta-catenin activation in ER-positive breast cancer. Nat. Commun. 2016;7:10751. doi: 10.1038/ncomms10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A., Illendula A., Pulikkan J.A., Roderick J.E., Tesell J., Yu J., Hermance N., Zhu L.J., Castilla L.H., Bushweller J.H., et al. RUNX1 is required for oncogenic Myb and Myc enhancer activity in T-cell acute lymphoblastic leukemia. Blood. 2017;130:1722–1733. doi: 10.1182/blood-2017-03-775536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang L.S., Ito K., Ito Y. RUNX family: regulation and diversification of roles through interacting proteins. Int. J. Cancer. 2013;132:1260–1271. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- Ditadi A., Sturgeon C.M., Tober J., Awong G., Kennedy M., Yzaguirre A.D., Azzola L., Ng E.S., Stanley E.G., French D.L., et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll A., Gonzalez M., Abal M., Llaurado M., Rigau M., Colas E., Monge M., Xercavins J., Capella G., Diaz B., et al. An orthotopic endometrial cancer mouse model demonstrates a role for RUNX1 in distant metastasis. Int. J. Cancer. 2009;125:257–263. doi: 10.1002/ijc.24330. [DOI] [PubMed] [Google Scholar]
- Draper J.E., Sroczynska P., Tsoulaki O., Leong H.S., Fadlullah M.Z., Miller C., Kouskoff V., Lacaud G. RUNX1B expression is highly heterogeneous and distinguishes megakaryocytic and erythroid lineage fate in adult mouse hematopoiesis. PLoS Genet. 2016;12:e1005814. doi: 10.1371/journal.pgen.1005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E., Bigas A. Blood development: hematopoietic stem cell dependence and independence. Cell Stem Cell. 2018;22:639–651. doi: 10.1016/j.stem.2018.04.015. [DOI] [PubMed] [Google Scholar]
- Eilken H.M., Nishikawa S., Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Ellis M.J., Ding L., Shen D., Luo J., Suman V.J., Wallis J.W., Van Tine B.A., Hoog J., Goiffon R.J., Goldstein T.C., et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari N., Mohammed Z.M., Nixon C., Mason S.M., Mallon E., McMillan D.C., Morris J.S., Cameron E.R., Edwards J., Blyth K. Expression of RUNX1 correlates with poor patient prognosis in triple negative breast cancer. PLoS One. 2014;9:e100759. doi: 10.1371/journal.pone.0100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijneman R.J., Anderson R.A., Richards E., Liu J., Tijssen M., Meijer G.A., Anderson J., Rod A., O'Sullivan M.G., Scott P.M., et al. Runx1 is a tumor suppressor gene in the mouse gastrointestinal tract. Cancer Sci. 2012;103:593–599. doi: 10.1111/j.1349-7006.2011.02189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sun S., Man X., Kong C. Increased expression of RUNX1 in clear cell renal cell carcinoma predicts poor prognosis. PeerJ. 2019;7:e7854. doi: 10.7717/peerj.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandemer V., Rio A.G., de Tayrac M., Sibut V., Mottier S., Ly Sunnaram B., Henry C., Monnier A., Berthou C., Le Gall E., et al. Five distinct biological processes and 14 differentially expressed genes characterize TEL/AML1-positive leukemia. BMC Genomics. 2007;8:385. doi: 10.1186/1471-2164-8-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghozi M.C., Bernstein Y., Negreanu V., Levanon D., Groner Y. Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proc. Natl. Acad. Sci. U. S. A. 1996;93:1935–1940. doi: 10.1073/pnas.93.5.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode D.K., Obier N., Vijayabaskar M.S., Lie A.L.M., Lilly A.J., Hannah R., Lichtinger M., Batta K., Florkowska M., Patel R., et al. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev. Cell. 2016;36:572–587. doi: 10.1016/j.devcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S., Huang G., Kurokawa M., Mulloy J.C. Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene. 2015;34:3483–3492. doi: 10.1038/onc.2014.305. [DOI] [PubMed] [Google Scholar]
- Goyama S., Schibler J., Cunningham L., Zhang Y., Rao Y., Nishimoto N., Nakagawa M., Olsson A., Wunderlich M., Link K.A., et al. Transcription factor RUNX1 promotes survival of acute myeloid leukemia cells. J. Clin. Invest. 2013;123:3876–3888. doi: 10.1172/JCI68557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S., Yamaguchi Y., Imai Y., Kawazu M., Nakagawa M., Asai T., Kumano K., Mitani K., Ogawa S., Chiba S., et al. The transcriptionally active form of AML1 is required for hematopoietic rescue of the AML1-deficient embryonic para-aortic splanchnopleural (P-Sp) region. Blood. 2004;104:3558–3564. doi: 10.1182/blood-2004-04-1535. [DOI] [PubMed] [Google Scholar]
- Guo H., Friedman A.D. Phosphorylation of RUNX1 by cyclin-dependent kinase reduces direct interaction with HDAC1 and HDAC3. J. Biol. Chem. 2011;286:208–215. doi: 10.1074/jbc.M110.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidi S., Sheng G. Epithelial-mesenchymal transition in haematopoietic stem cell development and homeostasis. J. Biochem. 2018;164:265–275. doi: 10.1093/jb/mvy063. [DOI] [PubMed] [Google Scholar]
- Hoi C.S., Lee S.E., Lu S.Y., McDermitt D.J., Osorio K.M., Piskun C.M., Peters R.M., Paus R., Tumbar T. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol. Cell. Biol. 2010;30:2518–2536. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D., Fritz A.J., Gordon J.A., Tye C.E., Boyd J.R., Tracy K.M., Frietze S.E., Carr F.E., Nickerson J.A., Van Wijnen A.J., et al. RUNX1-dependent mechanisms in biological control and dysregulation in cancer. J. Cell. Physiol. 2019;234:8597–8609. doi: 10.1002/jcp.27841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D., Messier T.L., Tye C.E., Dobson J.R., Fritz A.J., Sikora K.R., Browne G., Stein J.L., Lian J.B., Stein G.S. Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget. 2017;8:17610–17627. doi: 10.18632/oncotarget.15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Shigesada K., Ito K., Wee H.J., Yokomizo T., Ito Y. Dimerization with PEBP2beta protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 2001;20:723–733. doi: 10.1093/emboj/20.4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Woo A.J., Waldon Z., Schindler Y., Moran T.B., Zhu H.H., Feng G.S., Steen H., Cantor A.B. A Src family kinase-Shp2 axis controls RUNX1 activity in megakaryocyte and T-lymphocyte differentiation. Genes Dev. 2012;26:1587–1601. doi: 10.1101/gad.192054.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.P., Lan Y.H., Lu T.L., Pao J.B., Chang T.Y., Lee H.Z., Yang W.H., Hsieh C.J., Chen L.M., Huang L.C., et al. Clinical significance of runt-related transcription factor 1 polymorphism in prostate cancer. BJU Int. 2011;107:486–492. doi: 10.1111/j.1464-410X.2010.09512.x. [DOI] [PubMed] [Google Scholar]
- Hyde R.K., Zhao L., Alemu L., Liu P. P. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia. 2015;29:1771–1778. doi: 10.1038/leu.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kurokawa M., Yamaguchi Y., Izutsu K., Nitta E., Mitani K., Satake M., Noda T., Ito Y., Hirai H. The corepressor mSin3A regulates phosphorylation-induced activation, intranuclear location, and stability of AML1. Mol. Cell. Biol. 2004;24:1033–1043. doi: 10.1128/MCB.24.3.1033-1043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Yoon W.J., Woo K.M., Baek J.H., Ryoo H.M. Pin1-mediated prolyl isomerization of Runx1 affects PU.1 expression in pre-monocytes. J. Cell. Physiol. 2014;229:443–452. doi: 10.1002/jcp.24462. [DOI] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Jain P., Nattakom M., Holowka D., Wang D.H., Thomas Brenna J., Ku A.T., Nguyen H., Ibrahim S.F., Tumbar T. Runx1 role in epithelial and cancer cell proliferation implicates lipid metabolism and Scd1 and Soat1 activity. Stem Cells. 2018;36:1603–1616. doi: 10.1002/stem.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Qin X., Kawane T., Komori H., Matsuo Y., Taniuchi I., Ito K., Izumi S., Komori T. Cbfb2 isoform dominates more potent Cbfb1 and is required for skeletal development. J. Bone Miner. Res. 2016;31:1391–1404. doi: 10.1002/jbmr.2814. [DOI] [PubMed] [Google Scholar]
- Kadota M., Yang H.H., Gomez B., Sato M., Clifford R.J., Meerzaman D., Dunn B.K., Wakefield L.M., Lee M.P. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLoS One. 2010;5:e9201. doi: 10.1371/journal.pone.0009201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikubo Y., Zhao L., Wunderlich M., Corpora T., Hyde R.K., Paul T.A., Kundu M., Garrett L., Compton S., Huang G., et al. Accelerated leukemogenesis by truncated CBF beta-SMMHC defective in high-affinity binding with RUNX1. Cancer Cell. 2010;17:455–468. doi: 10.1016/j.ccr.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Kanno Y., Chen L.F., Ogawa E., Kim W.Y., Ito Y. Intrinsic transcriptional activation-inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor alpha subunit revealed in the presence of the beta subunit. Mol. Cell. Biol. 1998;18:2444–2454. doi: 10.1128/MCB.18.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karn T., Pusztai L., Holtrich U., Iwamoto T., Shiang C.Y., Schmidt M., Muller V., Solbach C., Gaetje R., Hanker L., et al. Homogeneous datasets of triple negative breast cancers enable the identification of novel prognostic and predictive signatures. PLoS One. 2011;6:e28403. doi: 10.1371/journal.pone.0028403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas S.M., de Ruiter J.R., Schipper K., Annunziato S., Schut E., Klarenbeek S., Drenth A.P., van der Burg E., Klijn C., Ten Hoeve J.J., et al. Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat. Genet. 2017;49:1219–1230. doi: 10.1038/ng.3905. [DOI] [PubMed] [Google Scholar]
- Keita M., Bachvarova M., Morin C., Plante M., Gregoire J., Renaud M.C., Sebastianelli A., Trinh X.B., Bachvarov D. The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle. 2013;12:972–986. doi: 10.4161/cc.23963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Barron D.A., San Martin R., Chan K.S., Tran L.L., Yang F., Ressler S.J., Rowley D.R. RUNX1 is essential for mesenchymal stem cell proliferation and myofibroblast differentiation. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16389–16394. doi: 10.1073/pnas.1407097111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeno Y., Yan M., Matsuura S., Lam K., Lo M.C., Huang Y.J., Tenen D.G., Downing J.R., Zhang D.E. Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood. 2014;123:3760–3769. doi: 10.1182/blood-2013-08-521252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni M., Tan T.Z., Syed Sulaiman N.B., Lamar J.M., Bansal P., Cui J., Qiao Y., Ito Y. RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget. 2018;9:14175–14192. doi: 10.18632/oncotarget.24419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud G., Gore L., Kennedy M., Kouskoff V., Kingsley P., Hogan C., Carlsson L., Speck N., Palis J., Keller G. Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood. 2002;100:458–466. doi: 10.1182/blood-2001-12-0321. [DOI] [PubMed] [Google Scholar]
- Lacaud G., Kouskoff V., Trumble A., Schwantz S., Keller G. Haploinsufficiency of Runx1 results in the acceleration of mesodermal development and hemangioblast specification upon in vitro differentiation of ES cells. Blood. 2004;103:886–889. doi: 10.1182/blood-2003-06-2149. [DOI] [PubMed] [Google Scholar]
- Lancrin C., Sroczynska P., Stephenson C., Allen T., Kouskoff V., Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Sada A., Zhang M., McDermitt D.J., Lu S.Y., Kemphues K.J., Tumbar T. High Runx1 levels promote a reversible, more-differentiated cell state in hair-follicle stem cells during quiescence. Cell Rep. 2014;6:499–513. doi: 10.1016/j.celrep.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong W.Y., Guo H., Ma O., Huang H., Cantor A.B., Friedman A.D. Runx1 phosphorylation by Src increases trans-activation via augmented stability, reduced histone deacetylase (HDAC) binding, and increased DNA affinity, and activated Runx1 favors granulopoiesis. J. Biol. Chem. 2016;291:826–836. doi: 10.1074/jbc.M115.674234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Glusman G., Bangsow T., Ben-Asher E., Male D.A., Avidan N., Bangsow C., Hattori M., Taylor T.D., Taudien S., et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene. 2001;262:23–33. doi: 10.1016/S0378-1119(00)00532-1. [DOI] [PubMed] [Google Scholar]
- Levanon D., Goldstein R.E., Bernstein Y., Tang H., Goldenberg D., Stifani S., Paroush Z., Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon D., Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- Li Q., Lai Q., He C., Fang Y., Yan Q., Zhang Y., Wang X., Gu C., Wang Y., Ye L., et al. RUNX1 promotes tumour metastasis by activating the Wnt/beta-catenin signalling pathway and EMT in colorectal cancer. J. Exp. Clin. Cancer Res. 2019;38:334. doi: 10.1186/s13046-019-1330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakhovitskaia A., Gribi R., Stamateris E., Villain G., Jaffredo T., Wilkie R., Gilchrist D., Yang J., Ure J., Medvinsky A. Restoration of Runx1 expression in the Tie2 cell compartment rescues definitive hematopoietic stem cells and extends life of Runx1 knockout animals until birth. Stem Cells. 2009;27:1616–1624. doi: 10.1002/stem.71. [DOI] [PubMed] [Google Scholar]
- Lie-A-Ling M., Marinopoulou E., Lilly A.J., Challinor M., Patel R., Lancrin C., Kouskoff V., Lacaud G. Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development. 2018;145:dev149419. doi: 10.1242/dev.149419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Carlsson L., Grundstrom T. Identification of an N-terminal transactivation domain of Runx1 that separates molecular function from global differentiation function. J. Biol. Chem. 2006;281:25659–25669. doi: 10.1074/jbc.M603249200. [DOI] [PubMed] [Google Scholar]
- McDonald L., Ferrari N., Terry A., Bell M., Mohammed Z.M., Orange C., Jenkins A., Muller W.J., Gusterson B.A., Neil J.C., et al. RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray, and ectopic expression of Runx2 perturbs differentiation in the mouse mammary gland. Dis. Model. Mech. 2014;7:525–534. doi: 10.1242/dmm.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegatti S., de Kruijf M., Garcia-Alegria E., Lacaud G., Kouskoff V. Transcriptional control of blood cell emergence. FEBS Lett. 2019;593:3304–3315. doi: 10.1002/1873-3468.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel R., Draper J.E., Lie-A-Ling M., Kouskoff V., Lacaud G. RUNX transcription factors: orchestrators of development. Development. 2019;146:dev148296. doi: 10.1242/dev.148296. [DOI] [PubMed] [Google Scholar]
- Mill C.P., Fiskus W., DiNardo C.D., Qian Y., Raina K., Rajapakshe K., Perera D., Coarfa C., Kadia T.M., Khoury J.D., et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood. 2019;134:59–73. doi: 10.1182/blood.2018893982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda Y., Morita K., Kashiwazaki G., Taniguchi J., Bando T., Obara M., Hirata M., Kataoka T.R., Muto M., Kaneda Y., et al. RUNX1 positively regulates the ErbB2/HER2 signaling pathway through modulating SOS1 expression in gastric cancer cells. Sci. Rep. 2018;8:6423. doi: 10.1038/s41598-018-24969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa K., Sakakura C., Nakashima S., Yoshikawa T., Kin S., Nakase Y., Ito K., Yamagishi H., Ida H., Yazumi S., et al. Down-regulation of RUNX1, RUNX3 and CBFbeta in hepatocellular carcinomas in an early stage of hepatocarcinogenesis. Anticancer Res. 2006;26:3633–3643. doi: 10.1038/sj.onc.1210403. [DOI] [PubMed] [Google Scholar]
- Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995;23:2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R., Pinheiro P., Joseph N., Peterkin T., Koth J., Repapi E., Bonkhofer F., Kirmizitas A., Patient R. Transforming growth factor beta drives hemogenic endothelium programming and the transition to hematopoietic stem cells. Dev. Cell. 2016;38:358–370. doi: 10.1016/j.devcel.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Maeda S., Suzuki K., Kiyose H., Taniguchi J., Liu P.P., Sugiyama H., Adachi S., Kamikubo Y. Paradoxical enhancement of leukemogenesis in acute myeloid leukemia with moderately attenuated RUNX1 expressions. Blood Adv. 2017a;1:1440–1451. doi: 10.1182/bloodadvances.2017007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K., Suzuki K., Maeda S., Matsuo A., Mitsuda Y., Tokushige C., Kashiwazaki G., Taniguchi J., Maeda R., Noura M., et al. Genetic regulation of the RUNX transcription factor family has antitumor effects. J. Clin. Invest. 2017b;127:2815–2828. doi: 10.1172/JCI91788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama Y., Chiba N., Hara T., Okada H., Ito Y., Kanamaru R., Miyajima A., Satake M., Watanabe T. The AML1 transcription factor functions to develop and maintain hematogenic precursor cells in the embryonic aorta-gonad-mesonephros region. Dev. Biol. 2000;220:27–36. doi: 10.1006/dbio.2000.9617. [DOI] [PubMed] [Google Scholar]
- Nagata T., Gupta V., Sorce D., Kim W.Y., Sali A., Chait B.T., Shigesada K., Ito Y., Werner M.H. Immunoglobulin motif DNA recognition and heterodimerization of the PEBP2/CBF Runt domain. Nat. Struct. Biol. 1999;6:615–619. doi: 10.1038/10658. [DOI] [PubMed] [Google Scholar]
- Navarro-Montero O., Ayllon V., Lamolda M., Lopez-Onieva L., Montes R., Bueno C., Ng E., Guerrero-Carreno X., Romero T., Romero-Moya D., et al. RUNX1c regulates hematopoietic differentiation of human pluripotent stem cells possibly in cooperation with proinflammatory signaling. Stem Cells. 2017;35:2253–2266. doi: 10.1002/stem.2700. [DOI] [PubMed] [Google Scholar]
- Neil J.C., Gilroy K., Borland G., Hay J., Terry A., Kilbey A. The RUNX genes as conditional oncogenes: insights from retroviral targeting and mouse models. Adv. Exp. Med. Biol. 2017;962:247–264. doi: 10.1007/978-981-10-3233-2_16. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Azzola L., Bruveris F.F., Calvanese V., Phipson B., Vlahos K., Hirst C., Jokubaitis V.J., Yu Q.C., Maksimovic J., et al. Differentiation of human embryonic stem cells to HOXA(+) hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat. Biotechnol. 2016;34:1168–1179. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- Nieke S., Yasmin N., Kakugawa K., Yokomizo T., Muroi S., Taniuchi I. Unique N-terminal sequences in two Runx1 isoforms are dispensable for Runx1 function. BMC Dev. Biol. 2017;17:14. doi: 10.1186/s12861-017-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki M., Okada H., Takano H., Kuno J., Tani K., Hibino H., Asano S., Ito Y., Satake M., Noda T. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5697–5702. doi: 10.1073/pnas.94.11.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North T., Gu T.L., Stacy T., Wang Q., Howard L., Binder M., Marin-Padilla M., Speck N.A. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka M., Maruyama M., Satake M., Naito-Fujimoto M., Ito Y., Shigesada K. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology. 1993a;194:314–331. doi: 10.1006/viro.1993.1262. [DOI] [PubMed] [Google Scholar]
- Ogawa E., Maruyama M., Kagoshima H., Inuzuka M., Lu J., Satake M., Shigesada K., Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. U. S. A. 1993b;90:6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S.W., Grosveld G., Downing J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/S0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Ottersbach K. Endothelial-to-haematopoietic transition: an update on the process of making blood. Biochem. Soc. Trans. 2019;47:591–601. doi: 10.1042/BST20180320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel J.P., Gonen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg H.J., Harrison H., Rogerson C., Shore P. The RUNX transcriptional coregulator, CBFbeta, suppresses migration of ER(+) breast cancer cells by repressing ERalpha-mediated expression of the migratory factor TFF1. Mol. Cancer Res. 2019;17:1015–1023. doi: 10.1158/1541-7786.MCR-18-1039. [DOI] [PubMed] [Google Scholar]
- Pereira B., Chin S.F., Rueda O.M., Vollan H.K., Provenzano E., Bardwell H.A., Pugh M., Jones L., Russell R., Sammut S.J., et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planaguma J., Diaz-Fuertes M., Gil-Moreno A., Abal M., Monge M., Garcia A., Baro T., Thomson T.M., Xercavins J., Alameda F., et al. A differential gene expression profile reveals overexpression of RUNX1/AML1 in invasive endometrioid carcinoma. Cancer Res. 2004;64:8846–8853. doi: 10.1158/0008-5472.CAN-04-2066. [DOI] [PubMed] [Google Scholar]
- Planaguma J., Gonzalez M., Doll A., Monge M., Gil-Moreno A., Baro T., Garcia A., Xercavins J., Alameda F., Abal M., et al. The up-regulation profiles of p21WAF1/CIP1 and RUNX1/AML1 correlate with myometrial infiltration in endometrioid endometrial carcinoma. Hum. Pathol. 2006;37:1050–1057. doi: 10.1016/j.humpath.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Pozner A., Goldenberg D., Negreanu V., Le S.Y., Elroy-Stein O., Levanon D., Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 2000;20:2297–2307. doi: 10.1128/MCB.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozner A., Lotem J., Xiao C., Goldenberg D., Brenner O., Negreanu V., Levanon D., Groner Y. Developmentally regulated promoter-switch transcriptionally controls Runx1 function during embryonic hematopoiesis. BMC Dev. Biol. 2007;7:84. doi: 10.1186/1471-213X-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptasinska A., Assi S.A., Martinez-Soria N., Imperato M.R., Piper J., Cauchy P., Pickin A., James S.R., Hoogenkamp M., Williamson D., et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep. 2014;8:1974–1988. doi: 10.1016/j.celrep.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S., Ross K.N., Lander E.S., Golub T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Rennert J., Coffman J.A., Mushegian A.R., Robertson A.J. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol. Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggio A.I., Blyth K. The enigmatic role of RUNX1 in female-related cancers - current knowledge & future perspectives. FEBS J. 2017;284:2345–2362. doi: 10.1111/febs.14059. [DOI] [PubMed] [Google Scholar]
- Robinson H.M., Broadfield Z.J., Cheung K.L., Harewood L., Harris R.L., Jalali G.R., Martineau M., Moorman A.V., Taylor K.E., Richards S., et al. Amplification of AML1 in acute lymphoblastic leukemia is associated with a poor outcome. Leukemia. 2003;17:2249–2250. doi: 10.1038/sj.leu.2403140. [DOI] [PubMed] [Google Scholar]
- Rody A., Karn T., Liedtke C., Pusztai L., Ruckhaeberle E., Hanker L., Gaetje R., Solbach C., Ahr A., Metzler D., et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakura C., Hagiwara A., Miyagawa K., Nakashima S., Yoshikawa T., Kin S., Nakase Y., Ito K., Yamagishi H., Yazumi S., et al. Frequent downregulation of the runt domain transcription factors RUNX1, RUNX3 and their cofactor CBFB in gastric cancer. Int. J. Cancer. 2005;113:221–228. doi: 10.1002/ijc.20551. [DOI] [PubMed] [Google Scholar]
- Salarpour F., Goudarzipour K., Mohammadi M.H., Ahmadzadeh A., Faraahi S., Farsani M.A. Evaluation of CCAAT/enhancer binding protein (C/EBP) alpha (CEBPA) and runt-related transcription factor 1 (RUNX1) expression in patients with de novo acute myeloid leukemia. Ann. Hum. Genet. 2017;81:276–283. doi: 10.1111/ahg.12210. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yagi H., Bronson R.T., Tominaga K., Matsunashi T., Deguchi K., Tani Y., Kishimoto T., Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl. Acad. Sci. U. S. A. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheitz C.J., Lee T.S., McDermitt D.J., Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S., Dicker F., Kern W., Wendland N., Sundermann J., Alpermann T., Haferlach C., Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- Shang Y., Zhao X., Xu X., Xin H., Li X., Zhai Y., He D., Jia B., Chen W., Chang Z. CHIP functions an E3 ubiquitin ligase of Runx1. Biochem. Biophys. Res. Commun. 2009;386:242–246. doi: 10.1016/j.bbrc.2009.06.043. [DOI] [PubMed] [Google Scholar]
- Sood R., Kamikubo Y., Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J., Trakhtenbrot L., Najfeld V., Lipton J.M., Mathew S., Avet-Loiseau H., De Braekeleer M., Salem S., Baruchel A., Raimondi S.C., et al. Amplification of band q22 of chromosome 21, including AML1, in older children with acute lymphoblastic leukemia: an emerging molecular cytogenetic subgroup. Leukemia. 2003;17:1679–1682. doi: 10.1038/sj.leu.2403000. [DOI] [PubMed] [Google Scholar]
- Sroczynska P., Lancrin C., Kouskoff V., Lacaud G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood. 2009;114:5279–5289. doi: 10.1182/blood-2009-05-222307. [DOI] [PubMed] [Google Scholar]
- Sun C.C., Li S.J., Chen Z.L., Li G., Zhang Q., Li D.J. Expression and prognosis analyses of runt-related transcription factor family in human leukemia. Mol. Ther. Oncolytics. 2019;12:103–111. doi: 10.1016/j.omto.2018.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G., Baumann C., O'Rourke J., Giannoulatou E., Taylor S., Joshi A., Moignard V., Pina C., Bee T., Kokkaliaris K.D., et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nat. Commun. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M., Tezuka C., Muroi S., Nishimoto S., Katsumoto T., Nakajima A., Kitabayashi I., Taniuchi I. Phosphorylation of Runx1 at Ser249, Ser266, and Ser276 is dispensable for bone marrow hematopoiesis and thymocyte differentiation. Biochem. Biophys. Res. Commun. 2008;368:536–542. doi: 10.1016/j.bbrc.2008.01.124. [DOI] [PubMed] [Google Scholar]
- Tahirov T.H., Inoue-Bungo T., Morii H., Fujikawa A., Sasaki M., Kimura K., Shiina M., Sato K., Kumasaka T., Yamamoto M., et al. Structural analyses of DNA recognition by the AML1/Runx-1 runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/S0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- Takayama K., Suzuki T., Tsutsumi S., Fujimura T., Urano T., Takahashi S., Homma Y., Aburatani H., Inoue S. RUNX1, an androgen- and EZH2-regulated gene, has differential roles in AR-dependent and -independent prostate cancer. Oncotarget. 2015;6:2263–2276. doi: 10.18632/oncotarget.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kurokawa M., Ueki K., Tanaka K., Imai Y., Mitani K., Okazaki K., Sagata N., Yazaki Y., Shibata Y., et al. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 1996;16:3967–3979. doi: 10.1128/MCB.16.7.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J.L., Hou H.A., Chen C.Y., Liu C.Y., Chou W.C., Tseng M.H., Huang C.F., Lee F.Y., Liu M.C., Yao M., et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Ito Y. Runx1: no longer just for leukemia. EMBO J. 2012;31:4098–4099. doi: 10.1038/emboj.2012.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay L.S., Krishnan V., Sankar H., Chong Y.L., Chuang L.S.H., Tan T.Z., Kolinjivadi A.M., Kappei D., Ito Y. RUNX poly(ADP-Ribosyl)ation and BLM interaction facilitate the fanconi anemia pathway of DNA repair. Cell Rep. 2018;24:1747–1755. doi: 10.1016/j.celrep.2018.07.038. [DOI] [PubMed] [Google Scholar]
- Telfer J.C., Rothenberg E.V. Expression and function of a stem cell promoter for the murine CBFalpha2 gene: distinct roles and regulation in natural killer and T cell development. Dev. Biol. 2001;229:363–382. doi: 10.1006/dbio.2000.9991. [DOI] [PubMed] [Google Scholar]
- van Bragt M.P., Hu X., Xie Y., Li Z. RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife. 2014;3:e03881. doi: 10.7554/eLife.03881.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu L.P., Perna F., Wang L., Voza F., Figueroa M.E., Tempst P., Erdjument-Bromage H., Gao R., Chen S., Paietta E., et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 2013;5:1625–1638. doi: 10.1016/j.celrep.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Brugge J.S., Janes K.A. Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E803–E812. doi: 10.1073/pnas.1103423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Binder M., Marin-Padilla M., Sharpe A.H., Speck N.A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 1996a;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G., et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/S0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang Q., Crute B.E., Melnikova I.N., Keller S.R., Speck N.A. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 1993;13:3324–3339. doi: 10.1128/MCB.13.6.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee H.J., Voon D.C., Bae S.C., Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylation of RUNX1 and p300 by HIPK2: implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Kurokawa M., Imai Y., Izutsu K., Asai T., Ichikawa M., Yamamoto G., Nitta E., Yamagata T., Sasaki K., et al. AML1 is functionally regulated through p300-mediated acetylation on specific lysine residues. J. Biol. Chem. 2004;279:15630–15638. doi: 10.1074/jbc.M400355200. [DOI] [PubMed] [Google Scholar]
- Yan J., Liu Y., Lukasik S.M., Speck N.A., Bushweller J.H. CBFbeta allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nat. Struct. Mol. Biol. 2004;11:901–906. doi: 10.1038/nsmb819. [DOI] [PubMed] [Google Scholar]
- Yeh H.Y., Cheng S.W., Lin Y.C., Yeh C.Y., Lin S.F., Soo V.W. Identifying significant genetic regulatory networks in the prostate cancer from microarray data based on transcription factor analysis and conditional independency. BMC Med. Genomics. 2009;2:70. doi: 10.1186/1755-8794-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T., Hasegawa K., Ishitobi H., Osato M., Ema M., Ito Y., Yamamoto M., Takahashi S. Runx1 is involved in primitive erythropoiesis in the mouse. Blood. 2008;111:4075–4080. doi: 10.1182/blood-2007-05-091637. [DOI] [PubMed] [Google Scholar]
- Yonezawa T., Takahashi H., Shikata S., Liu X., Tamura M., Asada S., Fukushima T., Fukuyama T., Tanaka Y., Sawasaki T., et al. The ubiquitin ligase STUB1 regulates stability and activity of RUNX1 and RUNX1-RUNX1T1. J. Biol. Chem. 2017;292:12528–12541. doi: 10.1074/jbc.M117.785675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi M., Goyama S., Kawazu M., Nakagawa M., Ichikawa M., Imai Y., Kumano K., Asai T., Mulloy J.C., Kraft A.S., et al. Multiple phosphorylation sites are important for RUNX1 activity in early hematopoiesis and T-cell differentiation. Eur. J. Immunol. 2012;42:1044–1050. doi: 10.1002/eji.201040746. [DOI] [PubMed] [Google Scholar]
- Zeng Y., He J., Bai Z., Li Z., Gong Y., Liu C., Ni Y., Du J., Ma C., Bian L., et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 2019;29:881–894. doi: 10.1038/s41422-019-0228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Biggs J.R., Kraft A.S. Phorbol ester treatment of K562 cells regulates the transcriptional activity of AML1c through phosphorylation. J. Biol. Chem. 2004;279:53116–53125. doi: 10.1074/jbc.M405502200. [DOI] [PubMed] [Google Scholar]
- Zhao X., Jankovic V., Gural A., Huang G., Pardanani A., Menendez S., Zhang J., Dunne R., Xiao A., Erdjument-Bromage H., et al. Methylation of RUNX1 by PRMT1 abrogates SIN3A binding and potentiates its transcriptional activity. Genes Dev. 2008;22:640–653. doi: 10.1101/gad.1632608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Luo M., Cai W., Zhou S., Feng D., Xu C., Wang H. Runt-related transcription factor 1 (RUNX1) promotes TGF-beta-induced renal tubular epithelial-to-mesenchymal transition (EMT) and renal fibrosis through the PI3K subunit p110delta. EBioMedicine. 2018;31:217–225. doi: 10.1016/j.ebiom.2018.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]