Abstract
RUNX1 plays an important role in the regulation of normal hematopoiesis. RUNX1 mutations are frequently found and have been intensively studied in hematological malignancies. Germline mutations in RUNX1 cause familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML). Somatic mutations of RUNX1 are observed in various types of hematological malignancies, such as AML, acute lymphoblastic leukemia (ALL), myelodysplastic syndromes (MDS), myeloproliferative neoplasm (MPN), chronic myelomonocytic leukemia (CMML), and congenital bone marrow failure (CBMF). Here, we systematically review the clinical and molecular characteristics of RUNX1 mutations, the mechanisms of pathogenesis caused by RUNX1 mutations, and potential therapeutic strategies to target RUNX1-mutated cases of hematological malignancies.
Keywords: clinical incidence and prognosis, pathogenesis, RUNX1 mutations, targeted therapy
INTRODUCTION
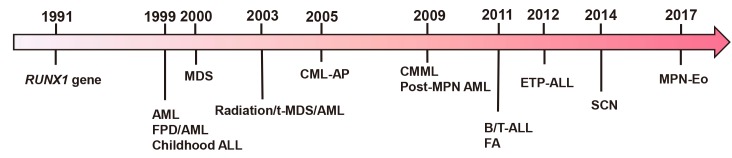
The RUNX1 transcription factor is a critical regulator of embryogenesis and definitive hematopoiesis in vertebrates. Since the somatic point mutation of RUNX1 was first identified two decades ago, RUNX1 has become known to be one of the most frequently mutated genes in a variety of hematological malignancies (Fig. 1) (Deltcheva and Nimmo, 2017; Hayashi et al., 2017; Osato et al., 1999). Despite the improvement of technology for the detection of mutations and a deeper understanding of the diseases, there are still unanswered questions about the functional consequences of RUNX1 mutations in hematological malignancies, such as (1) the frequency of different RUNX1 mutations in various subgroups of hematological malignancies and their impact on prognosis; (2) the mechanisms of how RUNX1 mutations contribute to pathogenesis; and (3) the potential mechanism-based therapeutic strategies. In this review article, we describe the clinical and molecular characteristics of RUNX1 mutations, the mechanisms of pathogenesis caused by its mutations, and potential therapeutic strategies for those RUNX1-mutated cases.
Fig. 1. The discovery procession of RUNX1 gene and its mutations in hematological malignancies.
GERMLINE MUTATION OF RUNX1 AND FPD/AML
Familial platelet disorder with predisposition to acute myeloid leukemia (FPD/AML) is an autosomal dominant disorder characterized by quantitative and qualitative platelet abnormalities and predisposition to AML (Online Mendelian Inheritance in Man [OMIM] No. #601399). To date, more than 70 families have been reported (Cavalcante de Andrade Silva et al., 2018; Latger-Cannard et al., 2016; Sood et al., 2017; Vormittag-Nocito et al., 2019). FPD/AML is caused by germline mutations of RUNX1, which is located at 21q22 and plays pivotal roles in the regulation of hematopoietic differentiation (Song et al., 1999). RUNX1 is essential for the development of hematopoietic stem cells (HSCs) in the embryonic stage. In adult hematopoiesis, however, it is dispensable for the maintenance of HSCs but required for megakaryocyte maturation and T lymphocyte-lineage differentiation (Ichikawa et al., 2004; Taniuchi et al., 2002). Loss-of-function or dominant-negative effect caused by mutated RUNX1 leads to the phenotype of FPD/AML (Cavalcante de Andrade Silva et al., 2018; Latger-Cannard et al., 2016; Vormittag-Nocito et al., 2019). Most of the mutations were clustered in the runt homology domain (RHD) and the c-terminal transactivation domain (TAD) with a few exceptions (Schlegelberger and Heller, 2017; Sood et al., 2017). FPD/AML was reported to transform to MDS/AML at a median onset age of 33 years old (Churpek et al., 2013). The median incidence rate of transformation is ranged from 35% to 44% in different studies (Godley, 2014; Owen et al., 2008a; 2008b). A few cases transformed to other types of leukemia, such as T-ALL (Nishimoto et al., 2010) or CMML (Shiba et al., 2012). Compared with loss-of-function mutations, dominant-negative mutations of RUNX1 are correlated to a higher risk of developing hematological malignancies (Latger-Cannard et al., 2016). However, these RUNX1 mutations by themselves are not sufficient for the development of leukemias. Additional mutations in RUNX1 (a second mutation), CDC25C, epigenetic modifiers, splicing factors, and tumor suppressors were reported to coordinately induce myeloid malignancies (Antony-Debre et al., 2016; Preudhomme et al., 2009; Yoshimi et al., 2014). Mutations in ASXL1, TET2, IDH1, CEBPD, RB1, MLL2, FLT3-ITD, WT1, and SRSF2 have also been detected by next-generation sequencing (Schlegelberger and Heller, 2017).
RUNX1 MUTATION-RELATED MDS AND MDS/MPN (CMML)
As one of the frequently mutated genes in MDS, somatic mutations of RUNX1 account for about 10% of the cases (Cazzola et al., 2013; Chen et al., 2007; Haferlach et al., 2014; Steensma et al., 2005; Tsai et al., 2015), while the frequency in childhood MDS is about 15% (Migas et al., 2011). The incidence of RUNX1 mutations in CMML is even higher at 32.1% to 37% (Kuo et al., 2009; Tsai et al., 2015). As in FPD/AML, most RUNX1 mutations are found in the RHD and the TAD (Kuo et al., 2009). Mutated RUNX1 is frequently accompanied by additional mutations of the genes ASXL1, SRSF2, TET2, SF3B1, and EZH2 in MDS (Stengel et al., 2019). Del(7)/del(7q) also coexists frequently with RUNX1 mutations in MDS patients (Chen et al., 2007; Xu et al., 2017). Notably, RUNX1 mutations are common in high-risk MDS (MDS-MLD/ MDS-EB) and are associated with poor clinical outcomes, especially higher risk and shorter latency for progression to secondary AML (Harada and Harada, 2015; Kuo et al., 2009; Steensma et al., 2005; Tsai et al., 2015). Shorter overall survival (OS) was also observed in MDS patients with RUNX1 mutations (Bejar et al., 2012; Chen et al., 2007).
RUNX1 MUTATION-RELATED AML
RUNX1 mutations are found in approximately 5.6-17.9% of cases in AML (Cancer Genome Atlas Research Network et al., 2013; Gaidzik et al., 2011; 2016; Grossmann et al., 2012; Tang et al., 2009), 3% in childhood AML patients (Migas et al., 2011), and about 27.7% in secondary AML transformed from MDS (Dicker et al., 2010). Besides being associated with older age and male gender (Gaidzik et al., 2016; Tang et al., 2009), the frequency of RUNX1 mutation was reported to be varied in different risk levels of patients and French-American-British (FAB) subtypes. For different risk levels of patient, the highest frequency of RUNX1 mutations was reported in intermediate-risk AML patients (7.2%-32.7%), followed by high-risk patients (9%), while RUNX1 mutations were absent in low-risk patients (Schnittger et al., 2011; Tang et al., 2009). The incidence of RUNX1 mutations was different in each FAB subtype; M0 (40%), M1 (17.5%), M2 (6.3%), M4 (15.1%), M5 (16%), and M6 (25%) (Tang et al., 2009). In AML patients with normal karyotype or with noncomplex chromosomal imbalances, patients of subtypes M0, M1, M2, and M4 showed even higher incidences; M0 (65.2%), M1 (30.2%), M2 (32.4%), and M4 (20%) Schnittger et al., 2011). However, RUNX1 mutations were not detected in M3 cases (Gaidzik et al., 2011). In particular M0 cases with RUNX1 mutations, 56.4-88.9% of them presented biallelic RUNX1 mutations (Osato, 2004; Preudhomme et al., 2000). The high incidence of biallelic mutations in this subtype suggests that the loss of RUNX1 activity affects hematopoietic cells at a very early undifferentiated stage.
At least one more additional mutation was observed in 40.8% to 95% of AML patients with RUNX1 mutations, such as Class I genes; FLT3-ITD/TKD and NRAS; Epigenetic factors; MLL-PTD, ASXL1, IDH1/IDH2, TET2, BCOR, and DNMT3A; Splicing factors; SRSF2 and SF3B1, and others including WT1 (Bullinger et al., 2017; Gaidzik et al., 2011; Haferlach et al., 2016; Schnittger et al., 2011; Stengel et al., 2018; Tang et al., 2009). Trisomy 8 and trisomy 13 showed a high incidence of RUNX1 mutations (Preudhomme et al., 2000; Schnittger et al., 2011). In contrast, RUNX1 mutations were negatively associated with NPM1 and CEBPA mutations (Gaidzik et al., 2011; Haferlach et al., 2016; Mendler et al., 2012; Schnittger et al., 2011; Tang et al., 2009).
In RUNX1-mutated AML, other aberrations were less frequent in MDS than de novo AML and secondary AML that progressed from MDS. Trisomy 13 was frequently found in de novo RUNX1-mutated AML, while trisomy 8 was exclusively harbored in RUNX1-mutated MDS and secondary AML. In RUNX1-mutated MDS, a higher frequency of ASXL1, TET2, and EZH2 mutations was observed, while DNMT3A and IDH2 mutations were more abundant in de novo RUNX1-mutated AML (Stengel et al., 2019).
AML with RUNX1 mutations predicts worse prognosis, resistance to chemotherapy, and inferior event free survival (EFS), relapse free survival (RFS), and OS. Mutated RUNX1 is an independent prognostic factor for EFS or OS (Gaidzik et al., 2011; Mendler et al., 2012; Schnittger et al., 2011; Tang et al., 2009). RUNX1 mutations associated with ASXL1 or SRSF2 mutations predicted particularly poorer prognosis (Bullinger et al., 2017).
RUNX1 MUTATION-RELATED ALL
RUNX1 mutations were detected in 15.5% to 18.3% of patients with T-ALL, 3.8% of patients with B-ALL (Grossmann et al., 2011a; 2013), and in 9.2% of childhood patients with T-ALL. The incidence was higher in patients with early T-cell precursor (ETP) ALL, reaching 15.6%. RUNX1 mutations were reported to be associated with ETP ALL, which is a high-risk subtype of ALL lacking several T cell surface markers and exhibiting aberrant expression of myeloid and stem cell markers (Zhang et al., 2012). Mutated RUNX1 in T-ALL was associated with older age and lower white blood cell count, but not platelet count, hemoglobin levels, gender or karyotype (Grossmann et al., 2011a). Mutated RUNX1 conferred a poor prognosis on early T-ALL patients with inferior OS (Grossmann et al., 2011a; 2013).
RADIATION-ASSOCIATED AND THERAPY-RELATED MDS/AML WITH RUNX1 MUTATIONS
Radiation-associated MDS/AML and therapy-related MDS/ AML (t-MDS/t-AML) are well-known complications after treatment with ionizing radiation, alkylating agents, and topoisomerase II inhibitors which can induce chromosome damages and cytogenetic abnormalities. The most common primary diseases are breast cancer and Hodgkin and non-Hodgkin lymphoma (Deltcheva and Nimmo, 2017; Ito et al., 2015; Pedersen-Bjergaard et al., 2006; Shih et al., 2013). The frequency of RUNX1 mutations in radiation-associated MDS/AML and t-MDS/t-AML varies from 15.7% to 39% (Christiansen et al., 2004; Harada et al., 2003; 2004; Singhal et al., 2019; Zharlyganova et al., 2008). The RUNX1 mutations in radiation-associated MDS/AML and t-MDS/t-AML includes missense, nonsense, and frameshift mutations, most of which are located in the RHD and the TAD (Christiansen et al., 2004). Besides RUNX1 mutations, additional mutations or cytogenetic abnormalities were found such as del(5)/del(5q), del(7)/del(7q), NRAS, TP53, and FLT3 (Niimi et al., 2006; Pedersen-Bjergaard et al., 2008). The prognosis of RUNX1-mutated cases are poorer than patients without mutations. Additionally, the survial of t-AML is shorter than t-MDS (3.5 vs 13.2 months) (Singhal et al., 2019).
RUNX1 MUTATIONS IN MPN DISEASE TRANSFORMATION
RUNX1 also plays important roles in MPN and its transformation to acute leukemias. Since 2009, RUNX1 mutations have been detected in 10.3% to 37.5% of post-MPN AML patients (Beer et al., 2010; Cerquozzi and Tefferi, 2015; Ding et al., 2009; Klampfl et al., 2011; Thoennissen et al., 2010). RUNX1 mutations also appear in several chronic myeloid leukemia (CML) studies (Branford et al., 2018). Since the first case of CML-AP (accelerated phase) with RUNX1 mutation was reported (Corm et al., 2005), RUNX1 mutations have been found in 12.9% to 33.3% of CML-AP/BC (blast crisis) patients in the followed-up study (Branford et al., 2018; Grossmann et al., 2011b; Roche-Lestienne et al., 2008; Schmidt et al., 2014; Zhao et al., 2012). In 2016, the WHO classification defines myeloid/lymphoid neoplasms associated with eosinophilia (MLN-Eo) with rearrangement of PDGFRA, PDGFRB, or FGFR1. RUNX1 mutations were positive in 5 out of 7 (71%) patients with FGFR1 rearrangement, and in a subsequent study, 6 out of 19 (32%) patients had RUNX1 mutations in FGFR1and PDGFRA-rearranged cases (Baer et al., 2018; Strati et al., 2018). In the MPN group, most of the mutations were detected in the RHD. Accompanied with RUNX1 mutations, the additional chromosome translocations (1q, 3q, 5q, 6p, 7p, 19q, and 22q) and mutations (ASXL1, NRAS, FLT3, TP53, TET2, CBL, etc.) were detected (Beer et al., 2010; Cerquozzi and Tefferi, 2015; Grossmann et al., 2011b; Klampfl et al., 2011). Regardless of the presence of the Ph chromosome or not, the prognosis is poor (Cerquozzi and Tefferi, 2015; Grossmann et al., 2011b).
RUNX1 MUTATIONS IN CBMF DISEASE PROGRESSION
Congenital bone marrow failure (CBMF) disorders are rare diseases characterized by peripheral blood cytopenia and hypoproliferation of one or more cell lineages in the BM (Gohring et al., 2007; Kutler et al., 2003). Individuals with Fanconi anemia (FA) have a high risk (30%-40%) of developing MDS and AML, yet the secondary somatic mutations leading to hematological malignancies remain to be elucidated (Quentin et al., 2011). RUNX1 mutations were detected in 20.7% to 31.25% of FA-associated MDS or MDS/AML (Chao et al., 2017; Quentin et al., 2011). The frequency of RUNX1 mutations in severe congenital neutropenia (SCN) was up to 64.5% (Skokowa et al., 2014). Mutated RUNX1 was also frequently associated with additional aberrations, such as -7/7q-, -5/5q-, and ASXL1, EZH2, KRAS, NRAS, SUZ12, CBL, FLT3-ITD, and TET2 mutations, in this group (Chao et al., 2017; Skokowa et al., 2014). Notably, in SCN-related MDS/ AML, the frequency of CSF3R mutations was as high as that of RUNX1 mutations (Skokowa et al., 2014). In the sections above, we briefly introduced and summarized the clinical and molecular characteristics of RUNX1 mutations in each of hematological malignancies (Table 1).
Table 1.
The frequency of RUNX1 mutations in various types of hematological malignancies
MECHANISMS
The RUNX1 transcription factor is a key regulator of normal hematopoiesis and its functional disruption by point mutations is one of the major factors for developing hematological malignancies (Deltcheva and Nimmo, 2017). There are two major subtypes of RUNX1 mutations in hematological malignancies: (1) the RHD, in which many mutations have been identified and are involved in residues at the DNA binding interface; (2) the TAD, in which most mutations result in production of the proteins lacking all or part of the TAD. Most of the RUNX1 mutations are mono-allelic, and different mutation types contribute to different biological properties of RUNX1 protein and presumably to disease phenotype as well (Mangan and Speck, 2011). We will describe the mechanisms of pathogenesis caused by RUNX1 mutations according to the biological function of RUNX1.
RUNX1 mutations on stem cells
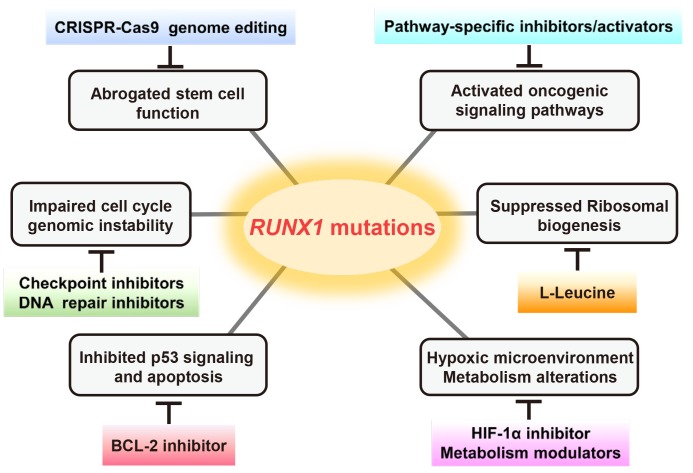
RUNX1 is required for the emergence of adult HSCs during embryonic development and for the maturation of different lineages from HSCs in adult BM (Hong et al., 2017). Loss of RUNX1 function is associated with a pre-leukemic state, probably owing to the expansion of HSCs and progenitor cells, as well as differentiation defects. To rescue RUNX1 mutations in HSCs, genome editing technologies such as CRISPR-Cas9 will hopefully accelerate the studies of the mutations, leading to a better understanding of the pathogenesis of leukemia and novel targeted treatments (Sood et al., 2017).
RUNX1 mutations on cell cycle and genomic instability
RUNX1 levels are increased at the G1-S phase and decreased during G2/M transition in hematopoietic cells. RUNX1 mutations may cause enhanced proliferation, attenuated mitotic checkpoint, and cell-cycle arrest. RUNX1 mutations can also cause genomic instability including increased DNA damage and impaired DNA repair. The potential therapeutic options for mutated RUNX1-associated abnormalities of cell cycle and genomic instability may come down to check point inhibitors and DNA repair inhibitors, which can bypass cells with DNA damage/impaired DNA repair to M phase (Goyama et al., 2015; Ito et al., 2015).
RUNX1 mutations on oncogenic signaling pathways
Mutations in RUNX1 are associated with alterations of various signaling pathways, such as WNT, BMP, TGF-β, RAS-ERK, Hippo-YAP1, and Notch, most of which have been described in cases of solid tumors. Notably, the possible involvement of RUNX1 mutations in WNT signaling has been shown in AML. WNT signaling controls cellular proliferation and differentiation and aberrant activation of WNT signaling has been reported in various tumors. RUNX1 mutations were closely associated with hypermethylation of the promoter of one of the WNT inhibitor gene, SFRP2, in AML. It is suggested that the WNT inhibitor hypermethylation might lead to aberrant activation of WNT signaling and interact with genetic alterations in the leukemogenesis (Hou et al., 2011).
RUNX1 mutations on p53 signaling and cell apoptosis
In response to the DNA damaging agent adriamycin, the RUNX1-p53 complex is recruited to the p53 target genes such as CDKN1A and BAX. RUNX1 increases the transcriptional activity of p53, probably by increasing p300-mediated acetylation of p53, and RUNX1 depletion attenuates p53-mediated apoptosis (Wu et al., 2013). Thus, abrogated function of mutated RUNX1 might lead to defects in p53-mediated apoptosis pathway/DNA repair/cell cycle regulation, resulting in tumorigenesis. Furthermore, it was shown that oncogenic Nras induces Runx1, which is required for induction of apoptosis and senescence, and Runx1 deficiency and oncogenic Nras cooperatively contribute to the clonal maintenance of leukemia-initiating cells (Ito et al., 2015; Motoda et al., 2007). Thus, loss-of-function mutations of RUNX1 may support the emergence of tumor-initiating cells in hematological malignancies partly by inhibiting p53 signaling and apoptosis.
RUNX1 mutation in ribosomal biogenesis
Loss-of-function mutations of RUNX1 were found to exhibit reduced ribosomal biogenesis in HSCs. RUNX1 directly binds to promoters of the genes encoding ribosomal RNA/proteins and regulates their transcription. Thus, RUNX1 mutations may cause low biosynthetic activity and confer stress resistance on HSCs, which provides a proliferative advantage to HSCs at the preleukemic stage (Cai et al., 2015; Deltcheva and Nimmo, 2017). In clinical trials, L-leucine is administrated to patients with Diamond–Blackfan anemia (DBA), which is caused by loss-of-function mutations in ribosomal protein genes. It has been shown that the treatment can improve anemia in the genetic DBA mouse models as well as DBA patients, possibly through mTOR activation, resulting in stimulation of protein translation (Ruggero and Shimamura, 2014). Thus, L-leucine might be a possible therapeutic option for RUNX1-mutated cases as well.
Hypoxic microenvironment
It has been reported that RUNX1 suppresses transactivation activity of hypoxia-inducible factor 1α (HIF-1α), while HIF-1α increases the activity of RUNX1 (Peng et al., 2008). HIF-1α is critical for cellular response to hypoxia and facilitates glycolysis but suppresses the TCA cycle. As most of the RUNX1 mutations cause loss of its function, RUNX1-mutated HSCs may have more activated HIF-1α pathway and glycolysis-biased metabolism. Metabolic rewiring to a hypoxia-like status is a hallmark of cancer as well as MDS and maintains stemness of tumor initiating cells (Hayashi et al., 2019). Thus, HIF-1α inhibitors or metabolic pathway modulators could be potential therapeutic strategies.
In conclusion, we summarized the mechanisms of pathogenesis caused by RUNX1 mutations and potential therapeutic strategies for RUNX1-mutated cases (Fig. 2).
Fig. 2. The key pathophysiological mechanisms related to RUNX1 mutations and potential therapeutic strategies.
CONCLUSIONS AND PERSPECTIVES
In this review, we briefly described the impact of RUNX1 mutations on clinical disease phenotypes and prognosis in hematological malignancies and the mechanisms of how RUNX1 mutations contribute to pathophysiology. RUNX1 mutations are frequently observed in various types of hematological malignancies and contribute to poor prognosis. RUNX1 is one of the most extensively studied molecules in hematopoiesis and leukemogenesis, and it functions in a variety of biological processes including cell differentiation, proliferation, cell cycle, DNA repair, apoptosis, ribosomal biogenesis, and metabolism. There are still many unknowns, such as how mutations affect this diverse function of RUNX1 and their clinical outcomes. Further elucidation is needed for a deeper understanding of RUNX1-associated hematological malignancies and for the development of better therapeutic strategies.
ACKNOWLEDGMENTS
This review work was supported partly by grants from Taub Foundation (to G.H.), National Institutes Health (NIH) (R01DK105014 to G.H.).
We apologize to all scientists in the field whose important work could not be cited in this report due to space limitation.
Footnotes
Disclosure
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Antony-Debre I., Duployez N., Bucci M., Geffroy S., Micol J.B., Renneville A., Boissel N., Dhedin N., Rea D., Nelken B., et al. Somatic mutations associated with leukemic progression of familial platelet disorder with predisposition to acute myeloid leukemia. Leukemia. 2016;30:999–1002. doi: 10.1038/leu.2015.236. [DOI] [PubMed] [Google Scholar]
- Baer C., Muehlbacher V., Kern W., Haferlach C., Haferlach T. Molecular genetic characterization of myeloid/lymphoid neoplasms associated with eosinophilia and rearrangement of PDGFRA, PDGFRB, FGFR1 or PCM1-JAK. Haematologica. 2018;103:e348–e350. doi: 10.3324/haematol.2017.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer P.A., Delhommeau F., LeCouedic J.P., Dawson M.A., Chen E., Bareford D., Kusec R., McMullin M.F., Harrison C.N., Vannucchi A.M., et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood. 2010;115:2891–2900. doi: 10.1182/blood-2009-08-236596. [DOI] [PubMed] [Google Scholar]
- Bejar R., Stevenson K.E., Caughey B.A., Abdel-Wahab O., Steensma D.P., Galili N., Raza A., Kantarjian H., Levine R.L., Neuberg D., et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford S., Wang P., Yeung D.T., Thomson D., Purins A., Wadham C., Shahrin N.H., Marum J.E., Nataren N., Parker W.T., et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood. 2018;132:948–961. doi: 10.1182/blood-2018-02-832253. [DOI] [PubMed] [Google Scholar]
- Bullinger L., Dohner K., Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J. Clin. Oncol. 2017;35:934–94. doi: 10.1200/JCO.2016.71.2208. [DOI] [PubMed] [Google Scholar]
- Cai X., Gao L., Teng L., Ge J., Oo Z.M., Kumar A.R., Gilliland D.G., Mason P.J., Tan K., Speck N.A. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell. 2015;17:165–17. doi: 10.1016/j.stem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., et al. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcante de Andrade Silva M., Krepischi A.C.V., Kulikowski L.D., Zanardo E.A., Nardinelli L., Leal A.M., Costa S.S., Muto N.H., Rocha V., Velloso E. Deletion of RUNX1 exons 1 and 2 associated with familial platelet disorder with propensity to acute myeloid leukemia. Cancer Genet. 2018;222-223:32–37. doi: 10.1016/j.cancergen.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Cazzola M., Della Porta M.G., Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerquozzi S., Tefferi A. Blast transformation and fibrotic progression in polycythemia vera and essential thrombocythemia: a literature review of incidence and risk factors. Blood Cancer J. 2015;5:e366. doi: 10.1038/bcj.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M.M., Thomay K., Goehring G., Wlodarski M., Pastor V., Schlegelberger B., Schindler D., Kratz C.P., Niemeyer C. Mutational spectrum of Fanconi anemia associated myeloid neoplasms. Klin. Padiatr. 2017;229:329–334. doi: 10.1055/s-0043-117046. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Lin L.I., Tang J.L., Ko B.S., Tsay W., Chou W.C., Yao M., Wu S.J., Tseng M.H., Tien H.F. RUNX1 gene mutation in primary myelodysplastic syndrome--the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br. J. Haematol. 2007;139:405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- Christiansen D.H., Andersen M.K., Pedersen-Bjergaard J. Mutations of AML1 are common in therapy-related myelodysplasia following therapy with alkylating agents and are significantly associated with deletion or loss of chromosome arm 7q and with subsequent leukemic transformation. Blood. 2004;104:1474–1481. doi: 10.1182/blood-2004-02-0754. [DOI] [PubMed] [Google Scholar]
- Churpek J.E., Lorenz R., Nedumgottil S., Onel K., Olopade O.I., Sorrell A., Owen C.J., Bertuch A.A., Godley L.A. Proposal for the clinical detection and management of patients and their family members with familial myelodysplastic syndrome/acute leukemia predisposition syndromes. Leuk. Lymphoma. 2013;54:28–35. doi: 10.3109/10428194.2012.701738. [DOI] [PubMed] [Google Scholar]
- Corm S., Biggio V., Roche-Lestienne C., Lai J.L., Yakoub-Agha I., Philippe N., Nicolini F.E., Facon T., Preudhomme C. Coexistence of AML1/RUNX1 and BCR-ABL point mutations in an imatinib-resistant form of CML. Leukemia. 2005;19:1991–1992. doi: 10.1038/sj.leu.2403931. [DOI] [PubMed] [Google Scholar]
- Deltcheva E., Nimmo R. RUNX transcription factors at the interface of stem cells and cancer. Biochem. J. 2017;474:1755–1768. doi: 10.1042/BCJ20160632. [DOI] [PubMed] [Google Scholar]
- Dicker F., Haferlach C., Sundermann J., Wendland N., Weiss T., Kern W., Haferlach T., Schnittger S. Mutation analysis for RUNX1, MLL-PTD, FLT3-ITD, NPM1 and NRAS in 269 patients with MDS or secondary AML. Leukemia. 2010;24:1528–1532. doi: 10.1038/leu.2010.124. [DOI] [PubMed] [Google Scholar]
- Ding Y., Harada Y., Imagawa J., Kimura A., Harada H. AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood. 2009;114:5201–5205. doi: 10.1182/blood-2009-06-223982. [DOI] [PubMed] [Google Scholar]
- Gaidzik V.I., Bullinger L., Schlenk R.F., Zimmermann A.S., Rock J., Paschka P., Corbacioglu A., Krauter J., Schlegelberger B., Ganser A., et al. RUNX1 mutations in acute myeloid leukemia: results from a comprehensive genetic and clinical analysis from the AML study group. J. Clin. Oncol. 2011;29:1364–1372. doi: 10.1200/JCO.2010.30.7926. [DOI] [PubMed] [Google Scholar]
- Gaidzik V.I., Teleanu V., Papaemmanuil E., Weber D., Paschka P., Hahn J., Wallrabenstein T., Kolbinger B., Kohne C.H., Horst H.A., et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30:2282. doi: 10.1038/leu.2016.207. [DOI] [PubMed] [Google Scholar]
- Godley L.A. Inherited predisposition to acute myeloid leukemia. Semin. Hematol. 2014;51:306–321. doi: 10.1053/j.seminhematol.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Gohring G., Karow A., Steinemann D., Wilkens L., Lichter P., Zeidler C., Niemeyer C., Welte K., Schlegelberger B. Chromosomal aberrations in congenital bone marrow failure disorders--an early indicator for leukemogenesis? Ann Hematol. 2007;86:733–739. doi: 10.1007/s00277-007-0337-z. [DOI] [PubMed] [Google Scholar]
- Goyama S., Huang G., Kurokawa M., Mulloy J.C. Posttranslational modifications of RUNX1 as potential anticancer targets. Oncogene. 2015;34:3483–3492. doi: 10.1038/onc.2014.305. [DOI] [PubMed] [Google Scholar]
- Grossmann V., Haferlach C., Weissmann S., Roller A., Schindela S., Poetzinger F., Stadler K., Bellos F., Kern W., Haferlach T., et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer. 2013;52:410–422. doi: 10.1002/gcc.22039. [DOI] [PubMed] [Google Scholar]
- Grossmann V., Kern W., Harbich S., Alpermann T., Jeromin S., Schnittger S., Haferlach C., Haferlach T., Kohlmann A. Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica. 2011a;96:1874–1877. doi: 10.3324/haematol.2011.043919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V., Kohlmann A., Zenger M., Schindela S., Eder C., Weissmann S., Schnittger S., Kern W., Muller M.C., Hochhaus A., et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia. 2011b;25:557–560. doi: 10.1038/leu.2010.298. [DOI] [PubMed] [Google Scholar]
- Grossmann V., Schnittger S., Kohlmann A., Eder C., Roller A., Dicker F., Schmid C., Wendtner C.M., Staib P., Serve H., et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120:2963–2972. doi: 10.1182/blood-2012-03-419622. [DOI] [PubMed] [Google Scholar]
- Haferlach T., Nagata Y., Grossmann V., Okuno Y., Bacher U., Nagae G., Schnittger S., Sanada M., Kon A., Alpermann T., et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T., Stengel A., Eckstein S., Perglerova K., Alpermann T., Kern W., Haferlach C., Meggendorfer M. The new provisional WHO entity 'RUNX1 mutated AML' shows specific genetics but no prognostic influence of dysplasia. Leukemia. 2016;30:2109–2112. doi: 10.1038/leu.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Harada Y. Recent advances in myelodysplastic syndromes: molecular pathogenesis and its implications for targeted therapies. Cancer Sci. 2015;106:329–336. doi: 10.1111/cas.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H., Harada Y., Niimi H., Kyo T., Kimura A., Inaba T. High incidence of somatic mutations in the AML1/RUNX1 gene in myelodysplastic syndrome and low blast percentage myeloid leukemia with myelodysplasia. Blood. 2004;103:2316–2324. doi: 10.1182/blood-2003-09-3074. [DOI] [PubMed] [Google Scholar]
- Harada H., Harada Y., Tanaka H., Kimura A., Inaba T. Implications of somatic mutations in the AML1 gene in radiation-associated and therapy-related myelodysplastic syndrome/acute myeloid leukemia. Blood. 2003;101:673–680. doi: 10.1182/blood-2002-04-1010. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Harada Y., Huang G., Harada H. Myeloid neoplasms with germ line RUNX1 mutation. Int. J. Hematol. 2017;106:183–188. doi: 10.1007/s12185-017-2258-5. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Yokota A., Harada H., Huang G. Hypoxia/pseudohypoxia-mediated activation of hypoxia-inducible factor-1alpha in cancer. Cancer Sci. 2019;110:1510–1517. doi: 10.1111/cas.13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D., Messier T.L., Tye C.E., Dobson J.R., Fritz A.J., Sikora K.R., Browne G., Stein J.L., Lian J.B., Stein G.S. Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget. 2017;8:17610–17627. doi: 10.18632/oncotarget.15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H.A., Kuo Y.Y., Liu C.Y., Lee M.C., Tang J.L., Chen C.Y., Chou W.C., Huang C.F., Lee F.Y., Liu M.C., et al. Distinct association between aberrant methylation of Wnt inhibitors and genetic alterations in acute myeloid leukaemia. Br. J. Cancer. 2011;105:1927–1933. doi: 10.1038/bjc.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- Ito Y., Bae S.C., Chuang L.S. The RUNX family: developmental regulators in cancer. Nat. Rev. Cancer. 2015;15:81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- Klampfl T., Harutyunyan A., Berg T., Gisslinger B., Schalling M., Bagienski K., Olcaydu D., Passamonti F., Rumi E., Pietra D., et al. Genome integrity of myeloproliferative neoplasms in chronic phase and during disease progression. Blood. 2011;118:167–176. doi: 10.1182/blood-2011-01-331678. [DOI] [PubMed] [Google Scholar]
- Kuo M.C., Liang D.C., Huang C.F., Shih Y.S., Wu J.H., Lin T.L., Shih L.Y. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia. 2009;23:1426–1431. doi: 10.1038/leu.2009.48. [DOI] [PubMed] [Google Scholar]
- Kutler D.I., Singh B., Satagopan J., Batish S.D., Berwick M., Giampietro P.F., Hanenberg H., Auerbach A.D. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- Latger-Cannard V., Philippe C., Bouquet A., Baccini V., Alessi M.C., Ankri A., Bauters A., Bayart S., Cornillet-Lefebvre P., Daliphard S., et al. Haematological spectrum and genotype-phenotype correlations in nine unrelated families with RUNX1 mutations from the French network on inherited platelet disorders. Orphanet J. Rare Dis. 2016;11:49. doi: 10.1186/s13023-016-0432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan J.K., Speck N.A. RUNX1 mutations in clonal myeloid disorders: from conventional cytogenetics to next generation sequencing, a story 40 years in the making. Crit. Rev. Oncog. 2011;16:77–91. doi: 10.1615/CritRevOncog.v16.i1-2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendler J.H., Maharry K., Radmacher M.D., Mrozek K., Becker H., Metzeler K.H., Schwind S., Whitman S.P., Khalife J., Kohlschmidt J., et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J. Clin. Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migas A., Savva N., Mishkova O., Aleinikova O.V. AML1/RUNX1 gene point mutations in childhood myeloid malignancies. Pediatr. Blood Cancer. 2011;57:583–587. doi: 10.1002/pbc.22980. [DOI] [PubMed] [Google Scholar]
- Motoda L., Osato M., Yamashita N., Jacob B., Chen L.Q., Yanagida M., Ida H., Wee H.J., Sun A.X., Taniuchi I., et al. Runx1 protects hematopoietic stem/progenitor cells from oncogenic insult. Stem Cells (Dayton, Ohio) 2007;25:2976–2986. doi: 10.1634/stemcells.2007-0061. [DOI] [PubMed] [Google Scholar]
- Niimi H., Harada H., Harada Y., Ding Y., Imagawa J., Inaba T., Kyo T., Kimura A. Hyperactivation of the RAS signaling pathway in myelodysplastic syndrome with AML1/RUNX1 point mutations. Leukemia. 2006;20:635–644. doi: 10.1038/sj.leu.2404136. [DOI] [PubMed] [Google Scholar]
- Nishimoto N., Imai Y., Ueda K., Nakagawa M., Shinohara A., Ichikawa M., Nannya Y., Kurokawa M. T cell acute lymphoblastic leukemia arising from familial platelet disorder. Int. J. Hematol. 2010;92:194–197. doi: 10.1007/s12185-010-0612-y. [DOI] [PubMed] [Google Scholar]
- Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23:4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- Osato M., Asou N., Abdalla E., Hoshino K., Yamasaki H., Okubo T., Suzushima H., Takatsuki K., Kanno T., Shigesada K., et al. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2alphaB gene associated with myeloblastic leukemias. Blood. 1999;93:1817–1824. doi: 10.1182/blood.V93.6.1817.406k36_1817_1824. [DOI] [PubMed] [Google Scholar]
- Owen C., Barnett M., Fitzgibbon J. Familial myelodysplasia and acute myeloid leukaemia--a review. Br. J. Haematol. 2008a;140:123–132. doi: 10.1111/j.1365-2141.2007.06909.x. [DOI] [PubMed] [Google Scholar]
- Owen C.J., Toze C.L., Koochin A., Forrest D.L., Smith C.A., Stevens J.M., Jackson S.C., Poon M.C., Sinclair G.D., Leber B., et al. Five new pedigrees with inherited RUNX1 mutations causing familial platelet disorder with propensity to myeloid malignancy. Blood. 2008b;112:4639–4645. doi: 10.1182/blood-2008-05-156745. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J., Andersen M.K., Andersen M.T., Christiansen D.H. Genetics of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2008;22:240–248. doi: 10.1038/sj.leu.2405078. [DOI] [PubMed] [Google Scholar]
- Pedersen-Bjergaard J., Christiansen D.H., Desta F., Andersen M.K. Alternative genetic pathways and cooperating genetic abnormalities in the pathogenesis of therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2006;20:1943–1949. doi: 10.1038/sj.leu.2404381. [DOI] [PubMed] [Google Scholar]
- Peng Z.G., Zhou M.Y., Huang Y., Qiu J.H., Wang L.S., Liao S.H., Dong S., Chen G.Q. Physical and functional interaction of Runt-related protein 1 with hypoxia-inducible factor-1alpha. Oncogene. 2008;27:839–847. doi: 10.1038/sj.onc.1210676. [DOI] [PubMed] [Google Scholar]
- Preudhomme C., Renneville A., Bourdon V., Philippe N., Roche-Lestienne C., Boissel N., Dhedin N., Andre J.M., Cornillet-Lefebvre P., Baruchel A., et al. High frequency of RUNX1 biallelic alteration in acute myeloid leukemia secondary to familial platelet disorder. Blood. 2009;113:5583–5587. doi: 10.1182/blood-2008-07-168260. [DOI] [PubMed] [Google Scholar]
- Preudhomme C., Warot-Loze D., Roumier C., Grardel-Duflos N., Garand R., Lai J.L., Dastugue N., Macintyre E., Denis C., Bauters F., et al. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 alpha B gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood. 2000;96:2862–2869. doi: 10.1182/blood.V96.8.2862. [DOI] [PubMed] [Google Scholar]
- Quentin S., Cuccuini W., Ceccaldi R., Nibourel O., Pondarre C., Pages M.P., Vasquez N., Dubois d'Enghien C., Larghero J., Peffault de Latour R., et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood. 2011;117:e161–e170. doi: 10.1182/blood-2010-09-308726. [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C., Deluche L., Corm S., Tigaud I., Joha S., Philippe N., Geffroy S., Lai J.L., Nicolini F.E., Preudhomme C. RUNX1 DNA-binding mutations and RUNX1-PRDM16 cryptic fusions in BCR-ABL+ leukemias are frequently associated with secondary trisomy 21 and may contribute to clonal evolution and imatinib resistance. Blood. 2008;111:3735–3741. doi: 10.1182/blood-2007-07-102533. [DOI] [PubMed] [Google Scholar]
- Ruggero D., Shimamura A. Marrow failure: a window into ribosome biology. Blood. 2014;124:2784–2792. doi: 10.1182/blood-2014-04-526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelberger B., Heller P.G. RUNX1 deficiency (familial platelet disorder with predisposition to myeloid leukemia, FPDMM) Semin. Hematol. 2017;54:75–80. doi: 10.1053/j.seminhematol.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Rinke J., Schafer V., Schnittger S., Kohlmann A., Obstfelder E., Kunert C., Ziermann J., Winkelmann N., Eigendorff E., et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia. 2014;28:2292–2299. doi: 10.1038/leu.2014.272. [DOI] [PubMed] [Google Scholar]
- Schnittger S., Dicker F., Kern W., Wendland N., Sundermann J., Alpermann T., Haferlach C., Haferlach T. RUNX1 mutations are frequent in de novo AML with noncomplex karyotype and confer an unfavorable prognosis. Blood. 2011;117:2348–2357. doi: 10.1182/blood-2009-11-255976. [DOI] [PubMed] [Google Scholar]
- Shiba N., Hasegawa D., Park M.J., Murata C., Sato-Otsubo A., Ogawa C., Manabe A., Arakawa H., Ogawa S., Hayashi Y. CBL mutation in chronic myelomonocytic leukemia secondary to familial platelet disorder with propensity to develop acute myeloid leukemia (FPD/AML) Blood. 2012;119:2612–2614. doi: 10.1182/blood-2011-02-333435. [DOI] [PubMed] [Google Scholar]
- Shih A.H., Chung S.S., Dolezal E.K., Zhang S.J., Abdel-Wahab O.I., Park C.Y., Nimer S.D., Levine R.L., Klimek V.M. Mutational analysis of therapy-related myelodysplastic syndromes and acute myelogenous leukemia. Haematologica. 2013;98:908–912. doi: 10.3324/haematol.2012.076729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal D., Wee L.Y.A., Kutyna M.M., Chhetri R., Geoghegan J., Schreiber A.W., Feng J., Wang P.P., Babic M., Parker W.T., et al. The mutational burden of therapy-related myeloid neoplasms is similar to primary myelodysplastic syndrome but has a distinctive distribution. Leukemia. 2019;33:2842–2853. doi: 10.1038/s41375-019-0479-8. [DOI] [PubMed] [Google Scholar]
- Skokowa J., Steinemann D., Katsman-Kuipers J.E., Zeidler C., Klimenkova O., Klimiankou M., Unalan M., Kandabarau S., Makaryan V., Beekman R., et al. Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood. 2014;123:2229–2237. doi: 10.1182/blood-2013-11-538025. [DOI] [PubMed] [Google Scholar]
- Song W.J., Sullivan M.G., Legare R.D., Hutchings S., Tan X., Kufrin D., Ratajczak J., Resende I.C., Haworth C., Hock R., et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Sood R., Kamikubo Y., Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129:2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma D.P., Gibbons R.J., Mesa R.A., Tefferi A., Higgs D.R. Somatic point mutations in RUNX1/CBFA2/AML1 are common in high-risk myelodysplastic syndrome, but not in myelofibrosis with myeloid metaplasia. Eur. J. Haematol. 2005;74:47–53. doi: 10.1111/j.1600-0609.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- Stengel A., Kern W., Meggendorfer M., Haferlach T., Haferlach C. RUNX1 mutations in MDS, s-AML, and de novo AML: differences in accompanying genetic alterations and outcome. Leuk. Lymphoma. 2019;60:1334–1336. doi: 10.1080/10428194.2018.1522439. [DOI] [PubMed] [Google Scholar]
- Stengel A., Kern W., Meggendorfer M., Nadarajah N., Perglerova K., Haferlach T., Haferlach C. Number of RUNX1 mutations, wild-type allele loss and additional mutations impact on prognosis in adult RUNX1-mutated AML. Leukemia. 2018;32:295–302. doi: 10.1038/leu.2017.239. [DOI] [PubMed] [Google Scholar]
- Strati P., Tang G., Duose D.Y., Mallampati S., Luthra R., Patel K.P., Hussaini M., Mirza A.S., Komrokji R.S., Oh S., et al. Myeloid/lymphoid neoplasms with FGFR1 rearrangement. Leuk. Lymphoma. 2018;59:1672–1676. doi: 10.1080/10428194.2017.1397663. [DOI] [PubMed] [Google Scholar]
- Tang J.L., Hou H.A., Chen C.Y., Liu C.Y., Chou W.C., Tseng M.H., Huang C.F., Lee F.Y., Liu M.C., Yao M., et al. AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood. 2009;114:5352–5361. doi: 10.1182/blood-2009-05-223784. [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/S0092-8674(02)01111-X. [DOI] [PubMed] [Google Scholar]
- Thoennissen N.H., Krug U.O., Lee D.H., Kawamata N., Iwanski G.B., Lasho T., Weiss T., Nowak D., Koren-Michowitz M., Kato M., et al. Prevalence and prognostic impact of allelic imbalances associated with leukemic transformation of Philadelphia chromosome-negative myeloproliferative neoplasms. Blood. 2010;115:2882–2890. doi: 10.1182/blood-2009-07-235119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.C., Shih L.Y., Liang S.T., Huang Y.J., Kuo M.C., Huang C.F., Shih Y.S., Lin T.H., Chiu M.C., Liang D.C. Biological activities of RUNX1 mutants predict secondary acute leukemia transformation from chronic myelomonocytic leukemia and myelodysplastic syndromes. Clin. Cancer Res. 2015;21:3541–3551. doi: 10.1158/1078-0432.CCR-14-2203. [DOI] [PubMed] [Google Scholar]
- Vormittag-Nocito E., Ni H., Schmidt M.L., Lindgren V. Thrombocytopenia and predisposition to acute myeloid leukemia due to mosaic ring 21 with loss of RUNX1: cytogenetic and molecular characterization. Mol. Syndromol. 2019;9:306–311. doi: 10.1159/000494645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Ozaki T., Yoshihara Y., Kubo N., Nakagawara A. Runt-related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J. Biol. Chem. 2013;288:1353–1364. doi: 10.1074/jbc.M112.402594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Wu L.Y., He Q., Wu D., Zhang Z., Song L.X., Zhao Y.S., Su J.Y., Zhou L.Y., Guo J., et al. Exploration of the role of gene mutations in myelodysplastic syndromes through a sequencing design involving a small number of target genes. Sci. Rep. 2017;7:43113. doi: 10.1038/srep43113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi A., Toya T., Kawazu M., Ueno T., Tsukamoto A., Iizuka H., Nakagawa M., Nannya Y., Arai S., Harada H., et al. Recurrent CDC25C mutations drive malignant transformation in FPD/AML. Nat. Commun. 2014;5:4770. doi: 10.1038/ncomms5770. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L.J., Wang Y.Y., Li G., Ma L.Y., Xiong S.M., Weng X.Q., Zhang W.N., Wu B., Chen Z., Chen S.J. Functional features of RUNX1 mutants in acute transformation of chronic myeloid leukemia and their contribution to inducing murine full-blown leukemia. Blood. 2012;119:2873–2882. doi: 10.1182/blood-2011-08-370981. [DOI] [PubMed] [Google Scholar]
- Zharlyganova D., Harada H., Harada Y., Shinkarev S., Zhumadilov Z., Zhunusova A., Tchaizhunusova N.J., Apsalikov K.N., Kemaikin V., Zhumadilov K., et al. High frequency of AML1/RUNX1 point mutations in radiation-associated myelodysplastic syndrome around Semipalatinsk nuclear test site. J. Radiat. Res. 2008;49:549–555. doi: 10.1016/j.ebiom.2018.04.023. [DOI] [PubMed] [Google Scholar]