Abstract
Numerous studies have demonstrated that microRNAs (miRNAs) play a key role in human carcinogenesis and metastasis. For example, miR-299-5p has previously been revealed to be dysregulated in several human cancers. However, the biological function of miR-299-5p in breast cancer remains unclear. The present study demonstrated that miR-299-5p was downregulated in breast cancer tissues and cell lines. The restoration of miR-299-5p expression suppressed cell migration and invasion, whereas inhibition of miR-299-5p promoted cell migration and invasion. In addition, in vivo studies demonstrated that miR-299-5p overexpression was able to inhibit tumour metastasis in nude mice. Mechanistically, through bioinformatics analysis and a dual-luciferase assay, it was confirmed that miR-299-5p directly targets serine/threonine kinase 39 (STK39). Silencing STK39 inhibited cell metastasis and suppressed epithelial-mesenchymal transition markers and matrix metalloproteinase expression, whereas restoration of STK39 expression was able to reverse miR-299-5p-inhibited cell migration and invasion. Collectively, the results of the present study demonstrated that miR-299-5p supresses breast cancer cell migration and invasion by targeting STK39. These findings may provide novel insights into miR-299-5p and its potential diagnostic and therapeutic benefits in breast cancer.
Keywords: miR-299-5p, serine/threonine kinase 39, breast cancer, migration, invasion, microRNA
Introduction
Breast cancer is among the most common types of cancer among women worldwide (1). The metastatic spread of breast cancer is generally associated with poor prognosis and increased disease-related mortality (2). The metastasis of breast cancer cells requires several essential steps, including epithelial-mesenchymal transition (EMT) (3) and extracellular matrix degradation (4). Studies over the past decade have demonstrated that aberrant microRNA (miRNA) expression is closely associated with a variety of biological processes in breast cancer, including cell proliferation (5,6), apoptosis (7,8), metastasis (9,10) and chemosensitivity (11,12). MicroRNAs (miRNAs) are single-stranded RNA molecules typically 21–25 nucleotides in length, that can negatively regulate target gene expression. miRNAs are evolutionarily conserved, and encoded by endogenous non-coding genes (13). It is well-known that miRNAs exert post-transcriptional genetic regulatory effects by binding to the 3′-untranslated region (UTR) of potential target mRNAs. Additionally, miRNAs may be utilized as diagnostic and prognostic biomarkers for breast cancer (14,15). For example, the levels of miR-214 in the peripheral blood may be used to detect mammary malignant tumours (16). In another study, miRNA expression profiling revealed that the expression levels of circulating miR-92a and miR-21 were correlated with tumour diameter and distant metastasis (17). The miR-299-5p gene belongs to a spanning DLK1-DIO3 region, which is one of the largest miRNA-containing clusters, located in chromosome 14q32 (18). Previously, miR-299 has been reported to be involved in several carcinogenic processes in thyroid cancer (19), glioblastoma (20) and prostate cancer (21). Additionally, it has been reported that miR-299-5p is downregulated in both the tumour and serum samples of breast cancer patients (22). However, the biological function of miR-299-5p in breast cancer remains unclear.
Using bioinformatics analysis (TargetScan and miRanda), it was predicted that miR-299-5p may bind to a potential site within the 3′-UTR of serine/threonine kinase 39 (STK39) in the present study, which provides evidence that miR-299-5p may regulate STK39 expression. STK39 belongs to the STE20 kinase family (23) and contains N-terminal proline and alanine repeats, a kinase catalytic domain, and a C-terminal region. STK39 is considered to activate p38α mitogen-activated protein kinase (MAPK), which participates in regulating various cellular biological activities, such as cell proliferation, differentiation, cytoskeletal rearrangement and ion transport (24–26). Previous studies reported that STK39 affects carcinogenesis in several types of cancer and is closely associated with tumour progression and metastasis, indicating that STK39 may act as an oncogene (27–29). However, the effects of miRNAs on the regulation of STK39 have not been extensively investigated.
In the present study, two human breast cancer cell lines, MDA-MB-231 and MCF-7, were used to evaluate the effect of miR-299-5p on cellular biological functions. Using bioinformatics algorithms, it was investigated whether STK39 is a direct target of miR-299-5p, and the results were further confirmed by dual-luciferase reporter assays and immunoblot analysis. The effects of STK39 knockdown by RNA interference (RNAi) on cancer cell metastasis were also investigated. The aim of the present study was to investigate the role of miR-299-5p and its potential target, STK39, in the regulation of breast cancer biological functions, and provide evidence that they may be used as novel targets for breast cancer diagnosis and therapy.
Materials and methods
Breast cancer sample and cell culture
A total of 30 paired formalin-fixed paraffin-embedded human breast cancer tissue samples and adjacent non-cancerous tissue samples were obtained from the Department of Pathology of The Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China) between May 2015 and December 2015. The patients were aged 35–79 years, with an average age of 59.0±12.7 years. No patients had received radiotherapy or chemotherapy prior to surgery. All the patients provided informed consent, and the study protocol was approved by the Ethics Committee of Xi'an Jiaotong University.
The human breast cancer cell lines, MCF-7 and MDA-MB-231, and the normal breast epithelial cell line MCF-10A, were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). MDA-MB-231 and MCF-7 cells were cultured in Dulbecco's modified Eagles medium (DMEM) supplemented with 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.). MCF-10A cells were maintained in DMEM/F12 medium (HyClone; GE Healthcare Life Sciences) with the addition of 10 µg/ml insulin, 20 ng/ml epidermal growth factor (Sigma-Aldrich; Merck KGaA) and 5% horse serum (Gibco; Thermo Fisher Scientific, Inc.). All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Cell transfection
miR-299-5p mimics, miR-299-5p inhibitor, small interfering RNAs (siRNAs) of STK39, STK39 overexpression plasmid, and their respective negative controls were chemically synthesized by Genepharm Technologies. Prior to transfection, breast cancer cells were incubated in a 6-well plate at a density of 2×105 cells/well. After a 24-h incubation, cells were transfected with miRNA mimics or plasmid using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. All oligonucleotides used in the present study are listed in Table SI.
Lentivirus infection
The packaged lentivirus for miR-299-5p overexpression was obtained from GeneChem and is referred to as LV-miR-299-5p hereafter. The lentiviral vector, LV-miR-ctrl, was used as a negative control. MDA-MB-231 cells were infected at a multiplicity of infection of 10 in the presence of 5 µg/ml polybrene. All steps were performed according to the manufacturer's instructions.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from each group of breast cancer cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. cDNA was synthesized from total RNA using the PrimerScript RT Reagent kit (Takara Bio, Inc.). The amount of 500 ng total RNA was used for reverse transcription with the following protocol: 42°C for 15 min and 85°C for 5 sec. Real-time PCR amplification reactions were conducted on a CFX96 Real Time PCR System (Bio-Rad Laboratories, Inc.) using SYBR Green Premix Ex Taq II (Takara Bio, Inc.). The relative expression of genes was calculated using the 2−ΔΔCq method and normalized to the expression of β-actin (30). For miR-299-5p quantitative detection, small nuclear RNA (U6) was used as an internal control. The primer sequences of the target genes used in the present study are listed in Table SI. The thermocycling conditions were as follows: 40 cycles with denaturation at 95°C for 30 sec, and annealing at 60°C for 30 sec.
Wound healing assay
Artificial wounds were created when cells were ~80% confluent. A sterile 200-µl pipette tip was used to scratch the cell monolayer. The width of the wound was measured, and images were captured using an inverted microscope at 0, 24, 48 and 72 h after wounding. The distance of cell migration was used to quantify the migration capacity.
Transwell assay
Following cell transfection, breast cancer cells were collected and suspended in serum-free medium. For the cell migration assay, a total of 3×104 cells were added into the upper chamber of Transwell plates, while the lower chamber was filled with 800 µl DMEM supplemented with 10% FBS. For the cell invasion assay, the upper surface of the membrane was precoated with a thin layer of Matrigel (BD Biosciences) and a total of 6×104 cells were added into the upper chamber of the Transwell plates. After incubation for 24 h, the non-invasive cells were gently removed from the upper chamber with a cotton swab. Invading cells that adhered to the bottom of the membrane were fixed at room temperature with 4% paraformaldehyde and stained with 0.1% crystal violet solution for 30 min. Images were randomly captured by an inverted microscope (IX73; Olympus Corp.). The number of invading cells was counted by using at least five fields for each membrane (magnification, ×200).
Bioinformatics analysis
The TargetScan (http://www.targetscan.org/vert_72) and miRanda (http://www.microrna.org/) online databases were employed to predict potential targets and their miR-299-5p binding sites.
Dual-luciferase reporter assay
The potential binding sites of miR-299-5p in STK39 mRNA 3′-UTR were constructed and cloned between the XhoI and NotI sites of the psiCHECK-2 dual-luciferase expression vector (Promega Corporation). psiCHECK2-STK39 wild-type (WT) and psiCHECK2-STK39 mutant (Mut) vectors were constructed. Subsequently, 293T cells were co-transfected with psiCHECK2-STK39-WT or psiCHECK2-STK39-Mut along with miR-299-5p mimics or miR-negative control. After 48 h of co-transfection, luciferase activity was detected using the Dual-Luciferase Reporter Assay System (Promega Corporation). Renilla luciferase activity was used for normalization.
Western blot assay
After 48 h of transfection, transfected breast cancer cells were lysed using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with a protease inhibitor cocktail (Roche Diagnostics). The protein concentration of each lysate was detected using a BCA assay kit (Beyotime Institute of Biotechnology). Equal amounts of cellular proteins (40 µg/lane) were separated by 10% SDS-PAGE and subsequently transferred to PVDF membranes (EMD Millipore). After blocking in 5% non-fat dry milk for 1 h, the membranes were incubated with diluted primary antibodies at 4°C overnight. After washing with Tris-buffered saline supplemented with 0.1% Tween-20, the membranes were incubated with horseradish peroxidase-conjugated (HRP) secondary antibodies (diluted in 1:20,000; cat no. 111-035-003 or 115-035-003; Jackson ImmunoResearch Laboratories, Inc.) for 1 h at room temperature. The washed membranes were incubated with ECL Western HRP Substrate (EMD Millipore) for chemiluminescence detection. The levels of β-actin were used to normalize the relative expression of proteins, and the protein band intensity was analysed using ImageJ software (version 1.48; National Institutes of Health). The primary antibodies used were as follows: STK39 (also known as SPAK) (diluted 1:2,000, product code ab128894), matrix metallopeptidase (MMP)-2 (diluted 1:2,000; ab92536), and MMP-9 (diluted 1:2,000; product code ab76003; all from Abcam), E-cadherin (diluted 1:1,000, product no. 3195) and N-cadherin (diluted 1:1,000; product no. 13116; both from Cell Signaling Technology Inc.), vimentin (diluted 1:1,000; product code ab92547; Abcam) and β-actin (diluted in 1:1,000; cat. no. sc47778; Santa Cruz Biotechnology, Inc.).
Xenograft assay
MDA-MB-231 cells stably overexpressing miR-299-5p were generated. Subsequently, MDA-MB-231 cells were suspended in phosphate-buffered saline at a density of 2×106 cells/ml. Female BALB/c nude mice (aged 4–5 weeks and weighing 18–20 g, purchased from Beijing Vital River Laboratory Animal Technology Co.) were injected with 0.1 ml of the cell suspension via the tail vein (n=10/group). All mice were euthanized by isoflurane after 7 weeks. The organs with metastatic foci were subjected to haematoxylin-eosin staining. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University.
Immunohistochemistry
Paraffin sections (4-µm) were deparaffinized with xylene and rehydrated through a graded ethanol series. Endogenous peroxidase was blocked with 3% hydrogen peroxide for 5 min at room temperature, and then antigen retrieval [in 110°C citrate buffer (pH 6.0) for 2 min] and blocking were performed. The sections were incubated with anti-STK39 antibody (diluted in 1:100) at 4°C overnight. Subsequently, the sections were incubated with HRP-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). Detection was performed using 3,3′-diaminobenzidine and haematoxylin. For each sample, the percentage of positive cells was counted to evaluate the expression of STK39.
Statistical analysis
Data are presented as the mean ± standard error of mean. All data were pooled from at least three independent experiments. Differences between two groups were analysed using the Student's t-test and differences among multiple groups were analysed using one-way ANOVA followed by Dunnett's post hoc test. The association between miR-299-5p or STK39 expression and clinicopathological characteristics of breast cancer patients was analysed using Fisher's exact probabilities test. All tests were two-sided, and P<0.05 was considered to indicate statistically significant differences. All statistical calculations were performed using SPSS 17.0 (SPSS Inc.), and all graphs were drawn with GraphPad Prism 5.0 (GraphPad Software, Inc.).
Results
miR-299-5p is downregulated in breast cancer clinical samples and cell lines
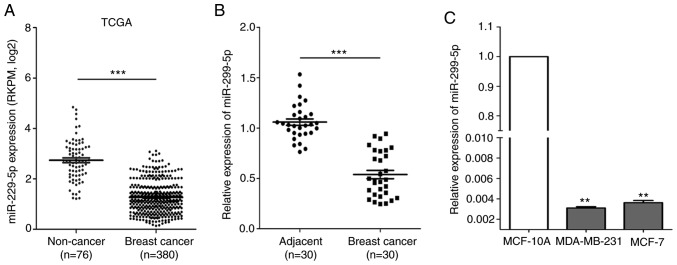
By searching The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), it was observed that miR-299-5p expression was significantly decreased in breast cancer tissues (n=380, P<0.001) compared with that in non-cancerous tissues (n=76) (Fig. 1A). miR-299-5p expression was evaluated in 30 pairs of human breast cancer tissue and adjacent non-cancerous tissue samples using RT-qPCR. The association between miR-299-5p expression and clinicopathological characteristics is presented in Table I. Decreased expression of miR-299-5p was revealed to be significantly correlated with lymph node metastasis (P=0.023). Compared with adjacent non-cancerous tissues, miR-299-5p was significantly downregulated in breast cancer tissues (Fig. 1B). The expression of miR-299-5p was also assessed in two different breast cancer cell lines and in normal human mammary epithelial cells using RT-qPCR. The results indicated that miR-299-5p was markedly downregulated in both breast cancer cell lines (Fig. 1C).
Figure 1.
miR-299-5p is significantly downregulated in breast cancer. (A) miR-299-5p expression in non-cancerous breast tissues and breast cancer tissues based on TCGA dataset. ***P<0.001. (B) miR-299-5p expression in 30 pairs of human breast cancer tissue and adjacent non-cancerous tissue samples. ***P<0.001. (C) Relative expression of miR-299-5p in a normal breast epithelial cell line and breast cancer cell lines. The expression of miR-299-5p was normalized to U6. Data are expressed as the mean ± standard error of mean. **P<0.01. TCGA, The Cancer Genome Atlas; RKPM, reads per kilobase per million mapped reads.
Table I.
Correlation between miR-299-5p expression and clinicopathological characteristics of breast cancer patients.
| miR-299-5p expression | ||||
|---|---|---|---|---|
| Characteristics | Patients (n=30) | Low (n=19) | High (n=11) | P-value |
| Age (years) | 0.707 | |||
| <60 | 16 | 11 | 5 | |
| ≥60 | 14 | 8 | 6 | |
| Tumor size (cm) | 0.238 | |||
| <2 | 11 | 5 | 6 | |
| ≥2 | 19 | 14 | 5 | |
| Lymph node metastasis | 0.023a | |||
| Negative | 13 | 5 | 8 | |
| Positive | 17 | 14 | 3 | |
| TNM stage | 0.708 | |||
| I+II | 17 | 10 | 7 | |
| III+IV | 13 | 9 | 4 | |
Fisher's exact probabilities test, P<0.05. TNM, T the size of primary tumor site; N the regional lymph node involvement; M the distant metastasis.
miR-299-5p inhibits breast cancer cell migration and invasion
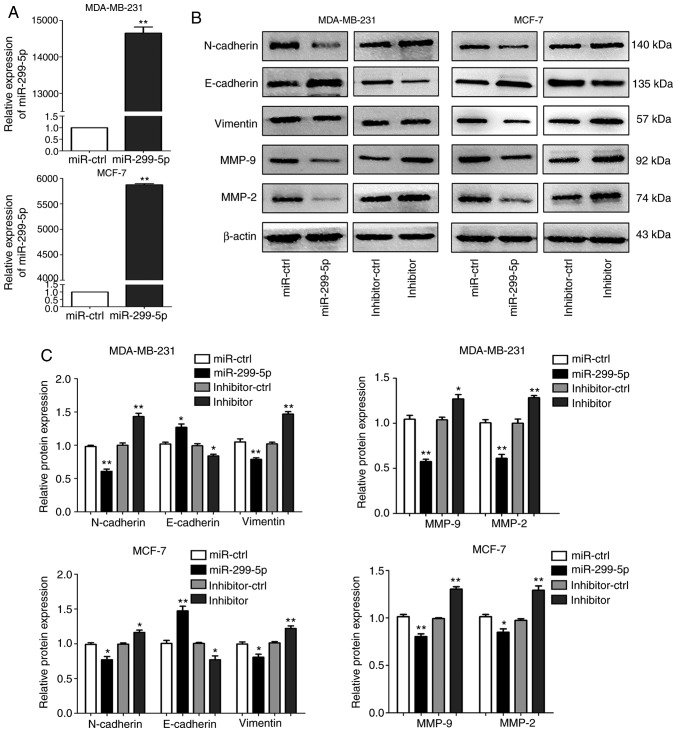
To explore the function of miR-299-5p in breast cancer, MDA-MB-231 and MCF-7 cell lines were transfected with miR-299-5p mimics or negative control (miR-ctrl). The expression levels of miR-299-5p in the two cell lines were significantly increased following transfection (Fig. 2A). However, the cell proliferation and apoptosis assays demonstrated that miR-299-5p exerted no effect on cell proliferation and apoptosis (Fig. S1). In addition, the in vivo experiment also demonstrated this point (Fig. S2).
Figure 2.
Overexpression of miR-299-5p affects the expression of EMT markers and MMPs. (A) The relative expression of miR-299-5p in MDA-MB-231 and MCF-7 cells following transfection with miR-299-5p mimics was assessed by reverse transcription-quantitative PCR. **P<0.01. (B) Western blotting detected the protein expression of EMT markers (N-cadherin, E-cadherin and vimentin) and MMP-2/9 in MDA-MB-231 and MCF-7 cells following transfection with miR-299-5p mimics or inhibitor. β-actin was used as an internal control. (C) Quantification of protein expression by grayscale analysis. *P<0.05, **P<0.01. EMT, epithelial-mesenchymal transition; MMP, matrix metallopeptidase.
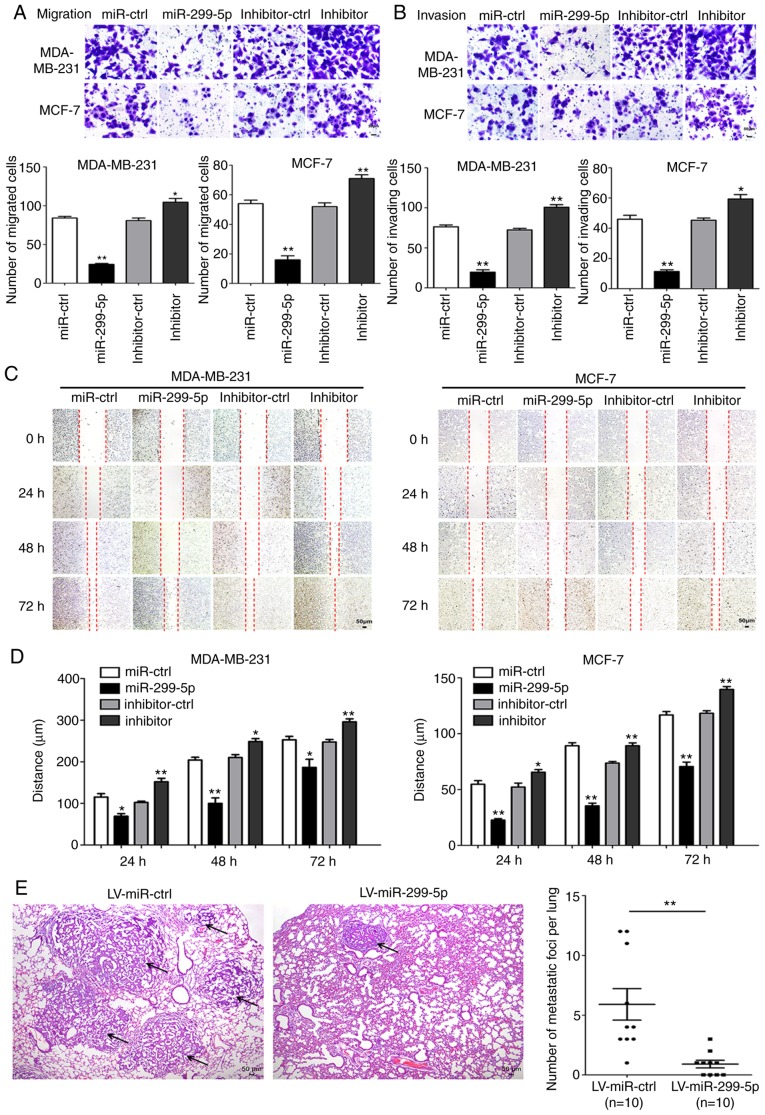
Next, the effects of miR-299-5p on cell migration and invasion were detected. Transwell and wound healing assays demonstrated that increased expression of miR-299-5p led to a marked decrease in the capacity for migration and invasion. By contrast, the suppression of miR-299-5p with a miRNA inhibitor promoted cancer cell migration and invasion (Fig. 3A and B). Wound healing assays yielded consistent results (Fig. 3C and D). Following transfection with miR-299-5p mimics and inhibitor, the expression of markers for EMT and extracellular matrix degradation was assessed, including E-cadherin, N-cadherin, vimentin, MMP-2 and MMP-9. Overexpression of miR-299-5p decreased the expression of N-cadherin, vimentin, MMP-2 and MMP-9 in MDA-MB-231 and MCF-7 cells. The expression of E-cadherin was markedly upregulated in the two cell lines (Fig. 2B and C). By contrast, miR-299-5p inhibitor exhibited the opposite results. These results indicated that miR-299-5p may inhibit breast cancer cell migration and invasion via regulation of EMT and extracellular matrix degradation.
Figure 3.
miR-299-5p inhibits the invasion and migration of breast cancer cells in vitro and in vivo. (A) Migration assays of MDA-MB-231 and MCF-7 cells following transfection with miR-299-5p mimics or inhibitor. The number of migrating cells was quantified using at least five random fields for each membrane (magnification, ×200). *P<0.05, **P<0.01. (B) Invasion assays of MDA-MB-231 and MCF-7 cells following transfection with miR-299-5p mimics or inhibitor. The number of invading cells was quantified using at least five random fields (magnification, ×200). *P<0.05, **P<0.01. (C) Wound healing assays of MDA-MB-231 and MCF-7 cells following transfection with miR-299-5p mimics or inhibitor. Images of the cells were captured every 24 h, and the width of the remaining gap was measured. (D) Quantification of the wound healing assay. The distance of cell migration was used to quantify the migratory capacity. *P<0.05, **P<0.01. (E) Representative images of H&E-stained sections of pulmonary metastases in nude mice (n=10) injected with miR-299-5p-overexpressing MDA-MB-231 cells compared with those injected with MDA-MB-231 negative control cells. **P<0.01. Data are expressed as the mean ± standard error of mean. LV, lentiviral; ctrl, control; H&E, haematoxylin and eosin.
To further validate the tumour suppressive effect of miR-299-5p in vivo, miR-299-5p-expressing MDA-MB-231 cells and control MDA-MB-231 cells were injected into the tail veins of nude mice. The mean number of pulmonary metastatic foci was significantly reduced in mice injected with miR-299-5p-expressing cells compared with the control group (Fig. 3E). The maximum diameter of metastatic foci in the LV-miR-ctrl group was 0.36 cm, and in the LV-miR-299-5p group it was 0.11 cm. These findings indicated that miR-299-5p may act as a tumour suppressor in breast cancer by inhibiting breast cancer metastasis in vitro and in vivo.
STK39 is a direct target of miR-299-5p in breast cancer
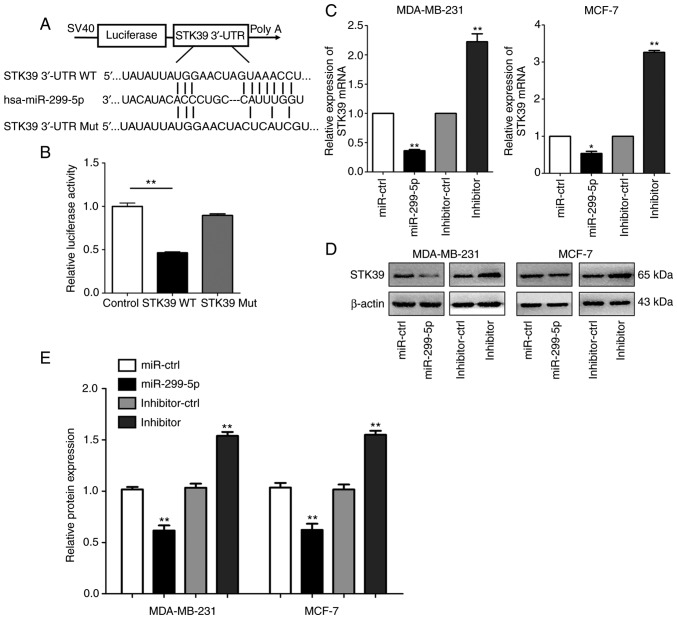
To investigate the underlying molecular mechanism through which miR-299-5p suppresses breast cancer cell metastasis, the TargetScan (http://www.targetscan.org/vert_72) and miRanda (http://www.microrna.org/) prediction databases were searched to identify miR-299-5p target genes. We were able to determine that the 3′-UTR of STK39 mRNA contains putative binding sites for miR-299-5p. To validate the association between miR-299-5p and STK39, STK39-WT and STK39-Mut 3′-UTR fragments were cloned into the psiCHECK-2 dual-luciferase reporter vector (Fig. 4A). miR-299-5p mimics and STK39-WT- or STK39-Mut-3′-UTR vectors were co-transfected into 293T cells. miR-299-5p significantly reduced the relative luciferase activity of the STK39-WT-3′-UTR vector in 293T cells, but did not affect the luciferase activity of the STK39-Mut-3′-UTR vector, indicating that miR-299-5p binds directly to 3′-UTR of STK39 (Fig. 4B). This result revealed that miR-299-5p exerts its effect by directly targeting the 3′-UTR of STK39. Moreover, the mRNA and protein levels of STK39 were revealed to be inversely correlated with miR-299-5p expression (Fig. 4C-E).
Figure 4.
STK39 is a direct target of miR-299-5p in breast cancer cells. (A) miR-299-5p is highly conserved across species and has binding sites within the 3′-UTR of human STK39. (B) Dual-luciferase reporter assays showing the suppression of the wild-type STK39 3′-UTR by miR-299-5p in 293T cells. **P<0.01. (C) The relative expression of STK39 mRNA was measured by reverse transcription-quantitative PCR following transfection with miR-299-5p mimics or inhibitor in MDA-MB-231 and MCF-7 cells. *P<0.05, **P<0.01. (D) The expression of STK39 was measured by western blotting following transfection with miR-299-5p mimics or inhibitor in MDA-MB-231 and MCF-7 cells. (E) Quantification of the protein expression by grayscale analysis. **P<0.01. Data are expressed as the mean ± standard error of mean. STK39, serine/threonine kinase 39. UTR, untranslated region; WT, wild-type; Mut, mutant.
STK39 is upregulated in breast cancer tissues and cell lines
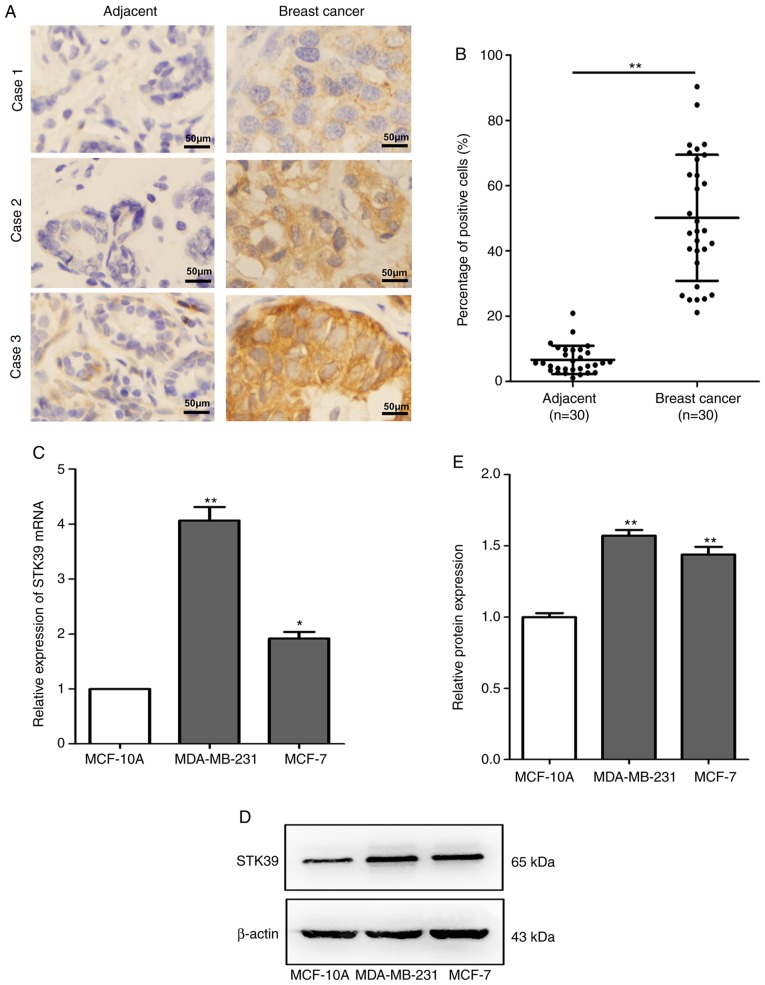
To determine whether STK39 is associated with miR-299-5p-mediated inhibition of breast cancer metastasis, STK39 expression was assessed in 30 pairs of human breast cancer tissue and adjacent non-cancerous tissue samples using immunohistochemistry. The association between STK39 expression and clinicopathological characteristics is presented in Table II. Increased expression of STK39 was revealed to be significantly correlated with lymph node metastasis (P=0.020). However, no association was observed between STK39 levels and age, tumour size, or TNM stage. Compared with non-cancerous tissues, STK39 was significantly upregulated (Fig. 5A and B) in breast cancer tissues. Moreover, the expression of STK39 at the mRNA and protein levels was also increased in breast cancer cell lines compared with that in normal breast epithelial cells (Fig. 5C-E).
Table II.
Correlation between STK39 expression and clinicopathological characteristics of breast cancer patients.
| STK39 expression | ||||
|---|---|---|---|---|
| Characteristics | Patients (n=30) | Low (n=9) | High (n=21) | P-value |
| Age (years) | 0.999 | |||
| <60 | 16 | 5 | 11 | |
| ≥60 | 14 | 4 | 10 | |
| Tumor size (cm) | 0.687 | |||
| <2 | 11 | 4 | 7 | |
| ≥2 | 19 | 5 | 14 | |
| Lymph node metastasis | 0.020a | |||
| Negative | 13 | 7 | 6 | |
| Positive | 17 | 2 | 15 | |
| TNM stage | 0.691 | |||
| I+II | 17 | 6 | 11 | |
| III+IV | 13 | 3 | 10 | |
Fisher's exact probabilities test, P<0.05. TNM, T the size of primary tumor site; N the regional lymph node involvement; M the distant metastasis; STK39, serine/threonine kinase 39.
Figure 5.
STK39 is upregulated in breast cancer. (A) STK39 expression levels were measured by immunohistochemistry in human breast cancer and adjacent non-cancerous tissue samples (magnification, ×400). (B) The percentage of positive cells was counted to evaluate the expression of STK39. **P<0.01. (C) The relative expression of STK39 mRNA in normal breast epithelial cell lines and breast cancer cell lines was measured by reverse transcription-quantitative PCR, and the expression of STK39 was normalized to β-actin. *P<0.05, **P<0.01. (D) The expression of STK39 in normal breast epithelial cells and breast cancer cell lines was measured by western blotting. β-actin was used as an internal control. (E) Quantification of protein expression by grayscale analysis. **P<0.01. Data are expressed as the mean ± standard error of mean. STK39, serine/threonine kinase 39.
Knockdown of STK39 inhibits breast cancer cell migration and invasion
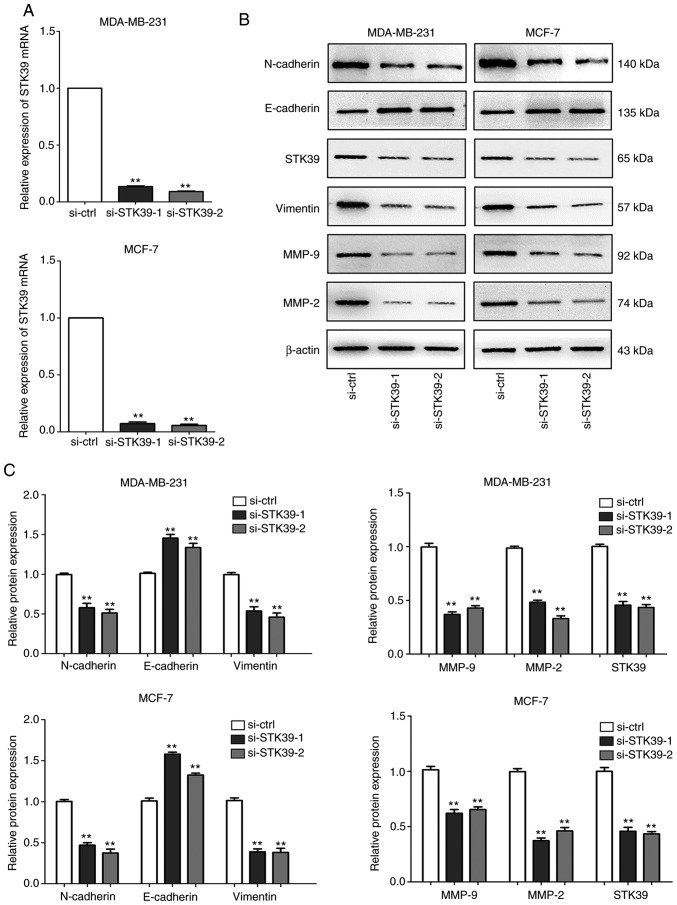
To further validate the role of STK39 in breast cancer, siRNAs were used to perform STK39 loss-of-function experiments. As revealed in Fig. 6A and B, following transfection, siRNA-mediated depletion of STK39 in MDA-MB-231 and MCF-7 cells reduced the levels of STK39 mRNA and protein.
Figure 6.
Knockdown of STK39 by siRNA transfection in breast cancer cells. (A) The relative expression of STK39 mRNA was measured by reverse transcription-quantitative PCR in MDA-MB-231 and MCF-7 cells after transfection with si-STK39, and the expression of STK39 was normalized to β-actin. **P<0.01. (B) Western blotting detected the expression of EMT markers (N-cadherin, E-cadherin and vimentin), MMP-2/9, and STK39 in MDA-MB-231 and MCF-7 cells after transfection with si-STK39 or si-ctrl. β-actin was used as an internal control. (C) Quantification of protein expression by grayscale analysis. **P<0.01. STK39, serine/threonine kinase 39; EMT, epithelial-mesenchymal transition; MMP, matrix metallopeptidase.
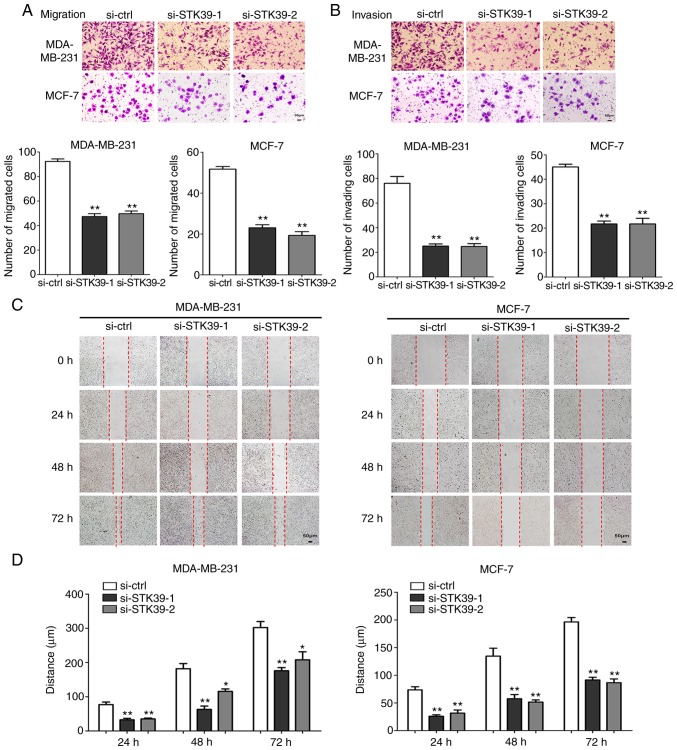
Cell migration and invasion abilities after STK39 knockdown were next examined. As revealed in Fig. 7A and B, MDA-MB-231 and MCF-7 cell migration and invasion were markedly inhibited in Transwell assays. In addition, the results of the wound healing assays were consistent (Fig. 7C and D). Furthermore, immunoblot assays revealed that the knockdown of STK39 led to increased expression of E-cadherin and decreased expression of vimentin, N-cadherin, MMP-2, and MMP-9 (Fig. 6B and C).
Figure 7.
STK39 affects migration and invasion in breast cancer cells. (A) Migration assays of MDA-MB-231 and MCF-7 cells following transfection with si-STK39 or si-ctrl. The number of migrated cells was quantified from at least five random fields for each membrane (magnification, ×200). **P<0.01. (B) Invasion assays of MDA-MB-231 and MCF-7 cells following transfection with si-STK39 or si-ctrl. The number of invading cells was counted from at least five random fields for each membrane (magnification, ×200). **P<0.01. (C) Wound healing assays of MDA-MB-231 and MCF-7 cells following transfection with si-STK39 or si-ctrl. Images of these cells were captured every 24 h, and the remaining unhealed gap was marked. (D) Quantification of the wound healing assay. *P<0.05, **P<0.01. Data are expressed as the mean ± standard error of mean. STK39, serine/threonine kinase 39; si-ctrl, small interfering RNA control; si-STK39, small interfering RNA of STK39; STK39, serine/threonine kinase 39.
STK39 reverses the effects of miR-299-5p on breast cancer cell lines
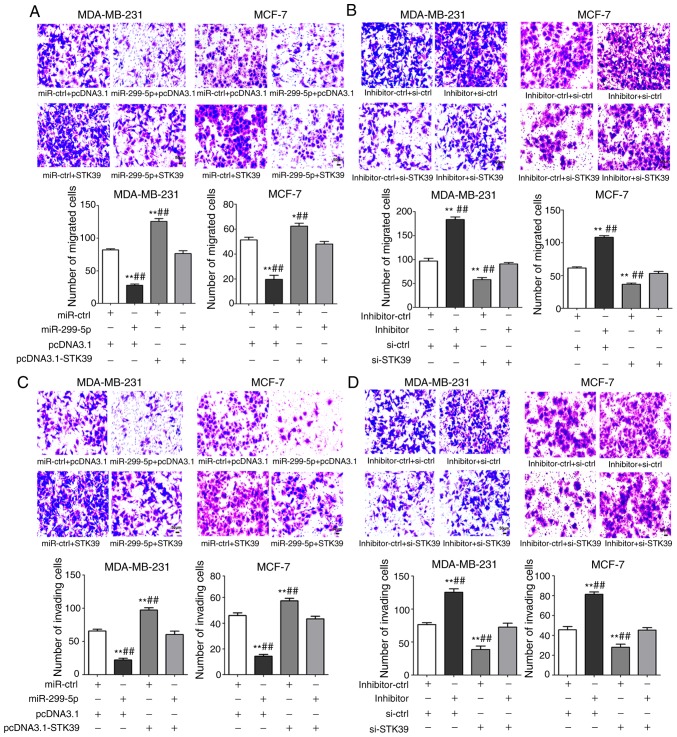
To confirm that STK39 is a functional target gene of miR-299-5p, rescue experiments in breast cancer cell lines were performed. MDA-MB-231 and MCF-7 cells were co-transfected with STK39-overexpression plasmid and miR-299-5p mimics. To ensure the efficiency of co-transfection, the levels of miR-299-5p and STK39 were evaluated after transfection. As revealed in Fig. 8A and B, STK39 overexpression reduced the tumour-suppressing effects of miR-299-5p on cell migration, and STK39 knockdown limited the effect of the miR-299-5p inhibitor on cell migration. Similar effects were also observed on cell invasion (Fig. 8C and D). Collectively, these results provide evidence that STK39 downregulation is indispensable for the tumour-suppressive role of miR-299-5p in breast cancer metastasis.
Figure 8.
STK39 reverses the effects of miR-299-5p on breast cancer cell migration and invasion. (A) STK39 overexpression reverses the decrease in cell migration induced by miR-299-5p upregulation (magnification, ×200). *P<0.05, **P<0.01 vs. miR-ctrl + pcDNA3.1. ##P<0.01 vs. miR-299-5p mimics + pcDNA3.1-STK39. (B) STK39 knockdown attenuates increased cell migration induced by the miR-299-5p inhibitor (magnification, ×200). **P<0.01 vs. inhibitor-ctrl + si-ctrl. ##P<0.01 vs. miR-299-5p inhibitor + si-STK39. (C) STK39 overexpression reverses the decrease in cell invasion induced by miR-299-5p upregulation (magnification, ×200). **P<0.01 vs. miR-ctrl + pcDNA3.1. ##P<0.01 vs. miR-299-5p mimics + pcDNA3.1-STK39. (D) STK39 knockdown attenuates the increase in cell invasion induced by miR-299-5p inhibition (magnification, ×200). **P<0.01 vs. inhibitor-ctrl + si-ctrl. ##P<0.01 vs. miR-299-5p inhibitor + si-STK39. Data are expressed as the mean ± standard error of mean. miR-ctrl, miRNA mimics control; si-ctrl, small interfering RNA control; si-STK39, small interfering RNA of STK39; STK39, serine/threonine kinase 39.
Discussion
The carcinogenesis and metastasis of breast cancer is a multifactorial process that includes expression changes in various oncogenes and tumour suppressors. Accordingly, numerous cancer-related genes and their biological functions have been identified, and the regulatory effects of thousands of unique non-coding RNAs are currently being investigated (31). Specifically, miRNAs are known to be widely involved in cancer development and progression, and several studies have suggested that miRNAs are dysregulated in various cancers (32), and regulate diverse biological functions in tumour cells.
The majority of studies on miR-299-5p have identified it as a tumour suppressor that is downregulated in different type of cancers (19–21). van Schooneveld et al observed that miR-299-5p was downregulated in both breast cancer tissue and serum samples compared with healthy individuals. They compared the expression level of miR-299-5p and other miRNAs in serum from patients with metastatic breast cancer receiving treatment, patients with untreated metastatic breast cancer, and healthy individuals, and found that the lowest expression value of miR-299-5p was observed in patients with metastatic breast cancer, whereas the expression level returned to normal with treatment. Moreover, the expression level of miR-299-5p exhibited a negative association with patient age at diagnosis (22). In the present study, miR-299-5p was revealed to be significantly downregulated in breast cancer tissues and cell lines, which was consistent with the results from clinical samples. These data indicated that miR-299-5p inhibited breast cancer cell migration and invasion, whereas it was not involved in cell proliferation and apoptosis. The restoration of miR-299-5p expression inhibited cell migration and invasion in breast cancer cells, while miR-299-5p inhibition promoted cell migration and invasion.
Previous studies have reported that the targets of miR-299-5p, include RAD21 (33) and osteopontin (34), which increase the expression of mesenchymal markers in EMT (35). Using bioinformatics analysis, STK39 was predicted to be a direct target of miR-299-5p. STK39 (also referred to as SPAK) is a member of the SPS1 subfamily of STE20 kinases (23). In previous studies, STK39 was demonstrated to activate the p38 MAPK pathway, which indicates that STK39 is involved in cellular stress responses (26). In addition, STK39 has been revealed to play a critical role in the regulation of ionic and osmotic cellular homeostasis (36,37). Recent studies have demonstrated that STK39 has carcinogenic functions in several cancer types (27–29). Single nucleotide polymorphisms of STK39 are correlated with prognosis in early-stage non-small cell lung cancer (38). Stanton et al identified STK39 as an early-stage antigen in breast cancer, and its overexpression in women with breast cancer predicted worse prognosis (39). In another study, the gene expression of STK39 was upregulated in approximately half of patients with ductal carcinoma in situ and invasive ductal carcinomas compared with normal breast tissue. Furthermore, the autoantibody of STK39 provided a feasible tool for human breast cancer diagnostics (40). The results of the present bioinformatics analysis and dual-luciferase assays demonstrated that miR-299-5p directly targeted STK39. To the best of our knowledge, this is the first study to demonstrate that miR-299-5p targets STK39 in breast cancer. It was observed that the upregulation of STK39 promoted breast cancer cell migration and invasion and demonstrated that STK39 may be involved in the EMT process and MMP expression during breast cancer metastasis.
Breast cancer is a heterogeneous disease characterized by diverse tumour biology, disparate response to therapeutics, and wide variations in the expression of hormone receptors and epidermal growth factor receptor (41). The established breast cancer cell lines belong to different subtypes of human breast cancer, which may lead to differences in biological behaviours. The triple-negative breast cancer cell line, MDA-MB-231, exhibits a high propensity to metastasize. MCF-7 is a hormone receptor positive, luminal epithelium-like, non-aggressive tumour cell line. In the present study, both cell lines produced similar results when subjected to miR-299-5p and STK39 functional experiments. Thus, the effect of miR-299-5p and STK39 on breast cancer is consistent in different biological contexts.
In conclusion, the present study provided evidence that miR-299-5p acts as an anti-tumour miRNA in breast cancer. miR-299-5p inhibited the migration and invasion capabilities of breast cancer cells by directly targeting a novel target gene, STK39. STK39 may play a role in the EMT process and in the expression of MMPs, both of which facilitate cell metastasis in breast cancer. As a novel biomarker, miR-299-5p not only evaluates the efficacy of the treatment in metastatic breast cancer, but also takes part in the metastasis of breast cancer, which facilitates its potential application in diagnosis and therapy. The findings of the present study demonstrated that the association between miR-299-5p and STK39 regulates breast cancer metastasis and highlights that it may lead to the development of new diagnostic and therapeutic approaches to breast cancer.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JZ conceived and designed the study. CL and AW performed the experiments. CL, YC and YL analyzed the data. CL wrote the manuscript. YC, HZ and JZ reviewed and edited the manuscript. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The experiment was approved by the Ethics Committee of Xi'an Jiaotong University and all patients provided informed consent. All experimental procedures involving the use of animals were approved by the Institutional Animal Care and Use Committee of Xi'an Jiaotong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso F, Costa A, Senkus E, Aapro M, André F, Barrios CH, Bergh J, Bhattacharyya G, Biganzoli L, Cardoso MJ, et al. 3rd ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 3) Ann Oncol. 2017;28:3111. doi: 10.1093/annonc/mdw544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaffer CL, San Juan BP, Lim E, Weinberg RA. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35:645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 4.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie F, Hosany S, Zhong S, Jiang Y, Zhang F, Lin L, Wang X, Gao S, Hu X. MicroRNA-193a inhibits breast cancer proliferation and metastasis by downregulating WT1. PLoS One. 2017;12:e0185565. doi: 10.1371/journal.pone.0185565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y, Xu W, Zhao W, Wang W, Zhang D, Wu P. miR-381 inhibited breast cancer cells proliferation, epithelial-to-mesenchymal transition and metastasis by targeting CXCR4. Biomed Pharmacother. 2017;86:426–433. doi: 10.1016/j.biopha.2016.12.051. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, He T, Yang L, Yang G, Chen Y, Zhang X. The role of miR-100 in regulating apoptosis of breast cancer cells. Sci Rep. 2015;5:11650. doi: 10.1038/srep11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breunig C, Pahl J, Küblbeck M, Miller M, Antonelli D, Erdem N, Wirth C, Will R, Bott A, Cerwenka A, Wiemann S. MicroRNA-519a-3p mediates apoptosis resistance in breast cancer cells and their escape from recognition by natural killer cells. Cell Death Dis. 2017;8:e2973. doi: 10.1038/cddis.2017.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G, Zhang W, Li B, Stringer-Reasor E, Chu C, Sun L, Bae S, Chen D, Wei S, Jiao K, et al. MicroRNA-200c and microRNA-141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 2017;19:73. doi: 10.1186/s13058-017-0858-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, et al. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res. 2017;23:1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L, Li YX, Gao P. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239:459–472. doi: 10.1002/path.4743. [DOI] [PubMed] [Google Scholar]
- 12.Xue J, Chi Y, Chen Y, Huang S, Ye X, Niu J, Wang W, Pfeffer LM, Shao ZM, Wu ZH, Wu J. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. 2016;35:448–458. doi: 10.1038/onc.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nassar FJ, Nasr R, Talhouk R. MicroRNAs as biomarkers for early breast cancer diagnosis, prognosis and therapy prediction. Pharmacol Ther. 2017;172:34–49. doi: 10.1016/j.pharmthera.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Schwarzenbach H, Milde-Langosch K, Steinbach B, Muller V, Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res Treat. 2012;134:933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
- 17.Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, Hu C. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–229. doi: 10.1007/s00432-012-1315-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benetatos L, Hatzimichael E, Londin E, Vartholomatos G, Loher P, Rigoutsos I, Briasoulis E. The microRNAs within the DLK1-DIO3 genomic region: Involvement in disease pathogenesis. Cell Mol Life Sci. 2013;70:795–814. doi: 10.1007/s00018-012-1080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, He L, Sun W, Qin Y, Dong W, Zhang T, Zhang P, Zhang H. miRNA-299-5p regulates estrogen receptor alpha and inhibits migration and invasion of papillary thyroid cancer cell. Cancer Manag Res. 2018;10:6181–6193. doi: 10.2147/CMAR.S182625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng Y, He X, Chen H, Duan H, Shao B, Yang F, Li H, Yang P, Zeng Y, Zheng J, et al. Inhibition of microRNA-299-5p sensitizes glioblastoma cells to temozolomide via the MAPK/ERK signaling pathway. Biosci Rep. 2018;38 doi: 10.1042/BSR20181051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Formosa A, Markert EK, Lena AM, Italiano D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino G, Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 22.van Schooneveld E, Wouters MC, Van der Auwera I, Peeters DJ, Wildiers H, Van Dam PA, Vergote I, Vermeulen PB, Dirix LY, Van Laere SJ. Expression profiling of cancerous and normal breast tissues identifies microRNAs that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012;14:R34. doi: 10.1186/bcr3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallolu Kankanamalage S, Karra AS, Cobb MH. WNK pathways in cancer signaling networks. Cell Commun Signal. 2018;16:72. doi: 10.1186/s12964-018-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagnon KB, Delpire E. Molecular physiology of SPAK and OSR1: Two Ste20-related protein kinases regulating ion transport. Physiol Rev. 2012;92:1577–1617. doi: 10.1152/physrev.00009.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delpire E, Gagnon KB. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem J. 2008;409:321–331. doi: 10.1042/BJ20071324. [DOI] [PubMed] [Google Scholar]
- 26.Alessi DR, Zhang J, Khanna A, Hochdorfer T, Shang Y, Kahle KT. The WNK-SPAK/OSR1 pathway: Master regulator of cation-chloride cotransporters. Sci Signal. 2014;7:re3. doi: 10.1126/scisignal.2005365. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Zhu W, Xiong L, Yu X, Chen X, Lin Q. Role of high expression levels of STK39 in the growth, migration and invasion of non-small cell type lung cancer cells. Oncotarget. 2016;7:61366–61377. doi: 10.18632/oncotarget.11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang T, Zhou Y, Cao Y, Tao J, Zhou ZH, Hang DH. STK39, overexpressed in osteosarcoma, regulates osteosarcoma cell invasion and proliferation. Oncol Lett. 2017;14:4599–4604. doi: 10.3892/ol.2017.6728. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Zhao Q, Zhu Y, Liu L, Wang H, Jiang S, Hu X, Guo J. STK39 blockage by RNA interference inhibits the proliferation and induces the apoptosis of renal cell carcinoma. Onco Targets Ther. 2018;11:1511–1519. doi: 10.2147/OTT.S153806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38:485–491. doi: 10.1093/carcin/bgx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 33.Yan M, Xu H, Waddell N, Shield-Artin K, Haviv I, kConFab authors. McKay MJ, Fox SB. Enhanced RAD21 cohesin expression confers poor prognosis in BRCA2 and BRCAX, but not BRCA1 familial breast cancers. Breast Cancer Res. 2012;14:R69. doi: 10.1186/bcr3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shevde LA, Metge BJ, Mitra A, Xi Y, Ju J, King JA, Samant RS. Spheroid-forming subpopulation of breast cancer cells demonstrates vasculogenic mimicry via hsa-miR-299-5p regulated de novo expression of osteopontin. J Cell Mol Med. 2010;14:1693–1706. doi: 10.1111/j.1582-4934.2009.00821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong Q, Zhu X, Dai C, Zhang X, Gao X, Wei J, Sheng Y, Zheng Y, Yu J, et al. Osteopontin promotes epithelial-mesenchymal transition of hepatocellular carcinoma through regulating vimentin. Oncotarget. 2016;7:12997–13012. doi: 10.18632/oncotarget.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Karimy JK, Delpire E, Kahle KT. Pharmacological targeting of SPAK kinase in disorders of impaired epithelial transport. Expert Opin Ther Targets. 2017;21:795–804. doi: 10.1080/14728222.2017.1351949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X, Naguro I, Ichijo H, Watanabe K. Mitogen-activated protein kinases as key players in osmotic stress signaling. Biochim Biophys Acta. 2016;1860:2037–2052. doi: 10.1016/j.bbagen.2016.05.032. [DOI] [PubMed] [Google Scholar]
- 38.Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, Haugen A, Wu MC, Wang Z, Su L, et al. Genome-wide analysis of survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2009;27:2660–2667. doi: 10.1200/JCO.2008.18.7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stanton SE, Gad E, Corulli LR, Lu H, Disis ML. Tumor-associated antigens identified early in mouse mammary tumor development can be effective vaccine targets. Vaccine. 2019;37:3552–3561. doi: 10.1016/j.vaccine.2019.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao J, Ladd J, Gad E, Rastetter L, Johnson MM, Marzbani E, Childs JS, Lu H, Dang Y, Broussard E, et al. Mining the pre-diagnostic antibody repertoire of TgMMTV-neu mice to identify autoantibodies useful for the early detection of human breast cancer. J Transl Med. 2014;12:121. doi: 10.1186/1479-5876-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivenbark AG, O'Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine. Am J Pathol. 2013;183:1113–1124. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.