Abstract
Changes in mitochondrial morphology by dysregulated mitochondrial fission-fusion proteins have been increasingly recognized as a hallmark of cancer. MiD49 (mitochondrial dynamics protein of 49 kDa) is a newly identified mitochondrial fission protein involved in the dynamic regulation of mitochondrial morphology. However, the expression pattern and biological functions of MiD49 in human cancers remain largely unexplored, especially in pancreatic cancer (PC). In the present study, the expression and clinical significance of MiD49 was firstly determined by RT-qPCR and western blot analyses in PC cell lines and tumor tissues. In addition, the biologic functions of MiD49 in PC cell growth and metastasis were investigated using gain- and loss-of-function assays both in vitro and in vivo. Moreover, the underlying mechanisms by which MiD49 regulates PC cell growth and metastasis were further explored. Our results showed that MiD49 was markedly downregulated in both PC cell lines and human PC specimens. Forced expression of MiD49 suppressed PC cell growth and metastasis both in vitro and in vivo, while knockdown of MiD49 exhibited the opposite effect. Mechanistic exploration demonstrated that the tumor-suppressive effect of MiD49 was mediated by decreased mitochondrial fission and subsequent reduced ROS production in PC cells. Our findings suggest a critical tumor-suppressive role played by MiD49 in pancreatic cancer.
Keywords: MiD49, tumor cell growth, metastasis, reactive oxygen species, pancreatic cancer
Introduction
Pancreatic cancer (PC) is one of the deadliest human cancer types, ranking as the seventh leading cause of cancer-related deaths worldwide (1,2). The 5-year survival rate of PC patients remains less than 8%, despite the fact that surgical and adjuvant treatment approaches have improved greatly during recent years (3,4). Identification of the key mechanisms involved in the development and progression of PC is still urgently needed.
Mitochondria are highly dynamic organelles that constantly change their morphology by fission and fusion to adapt to the cellular environment (5,6). Several recent studies have revealed close links between unbalanced mitochondrial dynamics and cancers (7–9). In addition, deregulated expression of mitochondrial fission and fusion proteins such as dynamin-related protein 1 (DRP1), mitofusin 1 (MFN1) and mitofusin 2 (MFN2) have also been observed in human cancer of the liver (10,11), lung (12), thyroid (13), and breast (14). MiD49 (mitochondrial dynamics protein of 49 kDa) is a newly identified mitochondrial anchored protein, which is involved in the regulation of mitochondrial fission (15–17). In the present study, we conducted the first study to elucidate the expression pattern and biological functions of MiD49 in PC.
Materials and methods
PC cell lines and tissue samples
Six human PC cell lines (BxPC-3, SW1990, PANC-1, PaCa-2, AsPC-1 and HPAC) and one normal pancreatic cell line hTERT-HPNE were obtained from The American Type Culture Collection (ATCC), and cultured in Dulbecco's modified Eagle's medium (DMEM) (Hyclone Laboratories, Inc.) supplemented with 10% fetal bovine serum (FBS) (Hyclone Laboratories, Inc.). All cells were cultured in a humidified atmosphere at 37°C with 5% CO2.
In addition, tumor and adjacent normal pancreatic tissues were obtained from 20 PC patients (13 males and 7 females) between March 2016 and July 2017 from the Department of Hepato-Biliary Surgery, The First Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China). The median age was 73 years (ranging from 63 to 85). Written informed consent was obtained from the patients for their tissues to be used for biomedical research. The specimens were stored at −80°C prior to quantitative real-time PCR (qPCR) and western blot analysis. All experimental protocols were approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University and carried out in accordance with the Declaration of Helsinki.
Oncomine data analysis
The Oncomine database (https://www.oncomine.org) (18), which compiles a large amount of previously published microarray data, was employed to compare the mRNA expression levels of MiD49 in tumor and normal tissue of pancreatic cancer.
Knockdown or forced expression of target genes in PC cells
PC cells were transfected with the MiD49 expression vector or siRNAs by using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) following the manufacturer's instructions. In addition, synthetic miR-424 precursor (pre-miR-424; Ambion; Thermo Fisher Scientific, Inc.) was used to study the function of miR-424. The MiD49 RNA interference sequences were 5′-ACACCTAAGTTCAGCACTATAGCAC-3′ (siMiD49#1) and 5′-GCCATGCCTTGAAGATGTGAATAAA-3′ (siMiD49#2). A scramble control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) was also used. A pSilencer™ 3.1-H1 puro vector (Ambion; Thermo Fisher Scientific, Inc.) was used for generation of shRNA expression vectors targeting MiD49. For MiD49 overexpression, the coding sequence of MiD49 was amplified from cDNA derived from BxPC-3 cells and subsequently cloned into a pcDNA™ 3.1 (+) vector (Invitrogen; Thermo Fisher Scientific, Inc., V790-20).
Quantitative real-time PCR (qPCR) analysis
Total RNA from PC tissues or cell lines was extracted by using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Then, a PrimeScript RT Reagent Kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used for the reverse transcription of mRNA into cDNA. Finally, qPCR was carried out using a SYBR Premix Ex Taq™ (Takara, Japan) on a Corbett 6200. The primer sequences used in the study are listed in Table I.
Table I.
Primers used in qPCR analysis.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| MiD49 | CAGAAACGGGGGAAGCGG | CACCAGGAGACGCACATGG |
| miR-424 | GGCTAGTCAGCAGCAATTCATGT | GTGCAGGGTCCGAGGT-3 |
| E-cadherin | ATTTTTCCCTCGACACCCGAT | TCCCAGGCGTAGACCAAGA |
| N-cadherin | TCAGGCGTCTGTAGAGGCTT | ATGCACATCCTTCGATAAGACTG |
| Vimentin | CCTGAACCTGAGGGAAACTAA | GCAGAAAGGCACTTGAAAGC |
| ZO-1 | CACGCAGTTACGAGCAAG | TGAAGGTATCAGCGGAGG |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
MiD49, mitochondrial dynamics protein of 49 kDa.
Western blot analysis
RIPA buffer was used for the extraction of whole protein from both PC cell lines and tissue samples. A total of 20 µg extracted proteins were loaded per lane and separated by 10% SDS-polyacrylamide gels and transferred onto the polyvinylidene fluoride (PVDF) membrane. After blocked with 5% BSA (SABC, Shanghai, China) for 1 h at room temperature, the membranes were immunostained with the primary antibody overnight at 4°C and the secondary antibody for 1 h at room temperature. Equal amount of protein sample loading was monitored using an anti-β-actin antibody. Blots were detected by an enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.) and images were captured with a Molecular Imager VersaDoc™ MP 5000 system (Bio-Rad Laboratories). The primary antibodies used and their dilutions are listed in Table II.
Table II.
Primary antibodies used in the western blot and IHC analyses.
| Antibody | Company (cat. no.) | Working concentration dilutions |
|---|---|---|
| MiD49 | Proteintech (16413-1-AP) | WB: 1/8 IHC: 1/150 |
| Caspase-9 | Proteintech (66169-1-Ig) | WB: 1/800 |
| Caspase-3 | Proteintech (25546-1-AP) | WB: 1/800 |
| Cytochrome c | Proteintech (10993-1-AP) | WB: 1/1,000 |
| COX IV | Abgent (#AP9153a) | WB: 1/1,000 |
| E-cadherin | Proteintech (20874-1-AP) | WB: 1/1,000 |
| N-cadherin | Cell Signaling (13116) | WB: 1/1,000 |
| Vimentin | Proteintech (10366-1-AP) | WB: 1/1,000 |
| ZO-1 | Proteintech (21773-1-AP) | WB: 1/1,000 |
| Ki-67 | MAB (MAB-0129) | IHC:1/200 |
| β-actin | Beijing TDY (TDY051F) | WB: 1/3,000 |
IHC, immunohistochemical; WB, western blotting; MiD49, mitochondrial dynamics protein of 49 kDa.
Immunohistochemical analysis
An IHC detection kit (Invitrogen; Thermo Fisher Scientific, Inc.) was used for immunohistochemical (IHC) analysis. Sections (4 µm) from paraffin-embedded tissues were deparaffinized and rehydrated and then treated with boiling sodium citrate buffer (pH 6.0) under pressure to unmask epitopes. Samples were blocked with 5% goat serum and then incubated with primary antibodies, including MiD49 and Ki-67 overnight at 4°C. The color was developed using DAB and counterstained with hematoxylin. Images were captured using a a light microscope (Olympus Corp.) at ×400 magnification. Semi-quantitative IHC staining scores were evaluated on a scale of 1 to 12 according to both the intensity and percentage of positive staining. Briefly, the staining intensities were graded on a scale of 0–3 as follows: 0 for no staining, 1 for weak staining, 2 for moderate staining, and 3 for strong staining. The percentages of the stained area were graded on a scale of 0–4 as follows: 0 is 0%, 1 (0–25%), 2 (25–50%), 3 (51–75%), and 4 (75–100%). The intensity grade multiplied by the percentage grade was calculated to equal the final IHC staining score.
MTS cell proliferation assay
Cell proliferation ability was determined by the MTS assay (Promega, G3581) according to the manufacturer's protocol. Briefly, 2,000 PC cells were seeded onto 96-well plates and cell viability was determined by the absorbance at 490 nm.
Colony formation assay
Two hundred PC cells were seeded into 6-well plates and cultured for 14 days. After that, the colonies were fixed with 4% paraformaldehyde and stained with crystal violet for 20 min at 28°C. Images were captured using a microscope with a light microscope (Olympus Corp.) at ×400 magnification. Finally, the number of colonies in each well was counted.
EdU (5-ethynyl-2′-deoxyuridine) incorporation assay
For EdU staining, 50 mM EdU in culture media was added to PC cells and incubated for 2 h at 37°C. Cells were then fixed with 4% paraformaldehyde for 20 min and treated with 0.5% Triton X-100 for 30 min. Afterwards, cells were incubated with DAPI at 28°C for 20 min, followed by observation under an Olympus confocal microscope (Olympus Corp.).
Flow cytometric analysis for cell apoptosis and cell cycle distribution
An Annexin V and PI apoptosis kit (F-6026, US Everbright Inc.) was used for cell apoptosis analysis according to the manufacturer's instructions. Briefly, 5 µl Annexin 5-FITC and 2 µl propidium iodide (PI) solutions were added to PC cells and incubated on ice for 15 min. Data were analyzed with a flow cytometer (Beckman Coulter).
A cell cycle kit from Everbright Inc. (C6031) was used for analysis of cell cycle distribution according to the manufacturer's instructions. Briefly, PC cells were fixed overnight in 80% cold ethanol at −20°C, followed by staining with 0.5 ml PI for 20 min at 4°C. Finally, data were analyzed with a flow cytometer (Beckman Coulter).
Scratch wound healing and Matrigel invasion assays
Cell migration ability was assessed by the scratch wound healing assay. Briefly, PC cells (2×106) were seeded into a 6-well plate. A single vertical scratch was made in the middle of the wells with a plastic pipette tip when cells had grown to 90% confluence. Finally, the PC cells that had migrated into the wounded area were photographed under a light microscope (Olympus Corp.) at ×400 magnification.
For the Transwell Matrigel invasion assay, 1×105 PC cells were seeded into the top of a Transwell chamber containing a Matrigel-coated membrane and cultured for 48 h. Penetrated PC cells were stained with crystal violet for 20 min at 28°C and counted under a light microscope (Olympus Corp.) at ×400 magnification.
In vivo tumor growth and metastasis assays
A total of 24 mice (12 for tumor growth and 12 for tumor metastasis) with body weight ranging from 17 to 21 g were used in our study. The nude mice were purchased from the Kunming Institute of Zoology, Chinese Academy of Sciences (Kunming, Yunnan, China) and group-housed (3 mice per cage) in plastic shoebox cages with autoclaved bedding and filtered air with access to sterilized food and water with a 12-h light/dark cycle at 28°C. Animal health and behavior were monitored daily. For in vivo tumor growth assay, 1×107 PC cells were subcutaneously injected into the right flanks of 6-week-old male BALB/c nude mice (one tumor per mice, six mice per group). Tumor volume (V) was calculated according to the formula: V=0.52 × L × W2 (L, length; W, width) every week for 5 weeks. At the end of the experiment, the mice were euthanized with CO2 rodent euthanasia chamber with a flow rate of CO2 not more than 30% of the chamber volume/min, according to the University of Minnesota guidelines. The death was verified by the absence of respiratory movement and heart beat. For in vivo metastasis assay, 1×106 PC cells were injected intravenously into the mice. The mice were euthanized on day 28 after injection and their lungs were removed. The metastatic nodes in the lung were counted. Images were captured and photographed under a light microscope (Olympus Corp.) at ×400 magnification. All animal experiments were performed in accordance with the relevant guidelines and regulations of the Animal Ethics Committee of the First Affiliated Hospital of Xian Jiaotong University.
Mitochondrial morphology analysis
MitoTracker Green staining was used for mitochondrial morphology analysis. Briefly, PC cells with different MiD49 levels were seeded into a confocal dish and cultured overnight. Then, cells were fixed in 4% paraformaldehyde for 20 min and incubated with MitoTracker green probe (Molecular Probes, M7514) for 1.5 h. An Olympus confocal microscope (Olympus) was used for the photography of immunofluorescence images. Mitochondrial length was measured with an ImageJ software (National Institutes of Health).
Detection of reactive oxygen species (ROS) levels
Cellular ROS levels were detected by the fluorescent probe DCFH-DA (Beyotime Institute of Biotechnology) following to the manufacturer's instructions. Briefly, a final concentration of 10 mM of DCFH-DA in serum-free medium was added to PC cells in each group. Cell suspensions in each group were incubated with DCFH-DA at 37°C for 30 min. Finally, the fluorescence was assessed by flow cytometry.
Statistical analysis
Experiments were performed independently at least three times and the results are expressed as mean ± SEM. Paired or unpaired student's t-test was used for comparisons between two groups. One-way analysis of variance (ANOVA) followed by Dunnett's t-test was used for comparisons between multiple groups. Correlations between measured variables were tested by Spearman's rank correlation analysis. The SPSS 17.0 software program (SPSS, Inc.) was used for statistical analysis and a P-value <0.05 was considered statistically significant.
Results
MiD49 is downregulated in pancreatic cancer cell lines and tissues
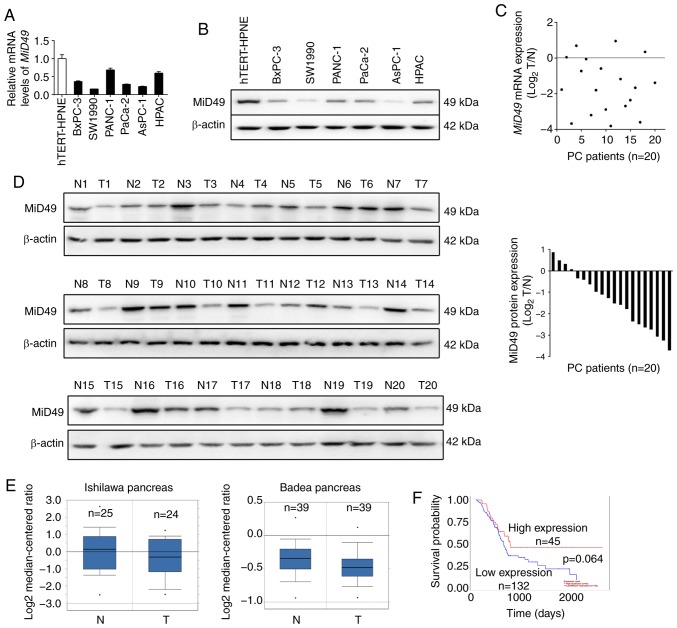
We firstly determined the expression of MiD49 in six pancreatic cancer (PC) cell lines and one immortalized normal pancreatic cell line hTERT-HPNE by quantitative real-time PCR (qPCR) and western blot analyses. As shown in Fig. 1A and B, MiD49 expression was significantly lower in all of the six PC cell lines when compared with that noted in the nonmalignant hTERT-HPNE cells, especially in the SW1990 and AsPC-1 cell lines with high metastatic potential. To determine whether MiD49 is also downregulated in tumor tissues of human pancreatic cancer, MiD49 expression was determined in 20-paired pancreatic tumor and adjacent non-tumor tissues by qPCR and western blot analyses. In concordance with the results from the PC cell lines, MiD49 expression was significantly decreased in the pancreatic tumor tissues than that found in the adjacent non-tumor tissues (Fig. 1C and D). In addition, two datasets (19,20) from the Oncomine platform consistently indicated that the level of MiD49 was significantly lower in tumor tissues of PC than in their normal counterparts (Fig. 1E). Moreover, prognostic significance analysis using the online web portal UALCAN (21) (http://ualcan.path.uab.edu) indicated that PC patients with a low expression level of MiD49 had clearly poorer overall survival than those with high MiD49 (Fig. 1F), although the difference was not significant (P=0.064). These findings indicate that MiD49 is significantly downregulated in PC, indicating a potential tumor-suppressive role in PC.
Figure 1.
MiD49 is downregulated in pancreatic cancer (PC) cell lines and tissues. (A and B) The expression levels of MiD49 were determined by qPCR and western blot analyses in six PC cell lines (BxPC-3, SW1990, PANC-1, PaCa-2, AsPC-1 and HPAC) and nonmalignant pancreatic hTERT-HPNE cells. (C and D) The expression levels of MiD49 were determined by qPCR and western blot analyses in 20-paired PC tumor tissues (T1-T20) and adjacent non-tumor (N1-N20) tissues. The data were log2-transformed. (E) MiD49 expression analysis based on datasets from Oncomine (N, adjacent non-tumor tissues; T, tumor tissues). (F) Data mining for prognostic significance of MiD49 in PC patients using the online web portal UALCAN. MiD49, mitochondrial dynamics protein of 49 kDa.
MiD49 suppresses PC growth by inducing G1-S phase cell cycle arrest and cell apoptosis
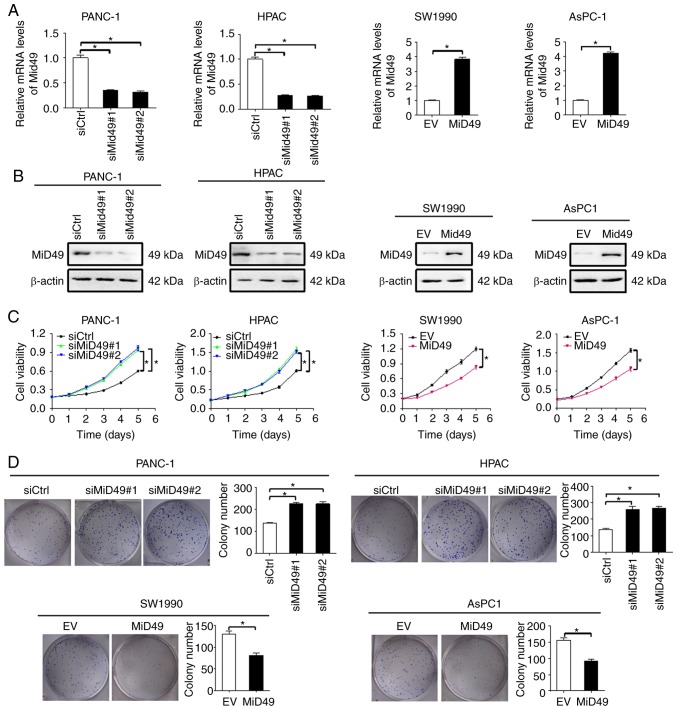
To investigate the functional roles of MiD49 in PC cells, gain- and loss-of-function studies were applied in human PC cell lines. Efficient knockdown of MiD49 in PANC-1 and HPAC cells and overexpression of MiD49 in SW1990 and AsPC-1 cells was confirmed and is shown in Fig. 2A and B. MTS cell viability and colony formation assays indicated that downregulation of MiD49 significantly promoted PC cell growth in the PANC-1 and HPAC cells, whereas forced expression of MiD49 suppressed PC cell growth in the SW1990 and AsPC-1 cell lines (Fig. 2C and D).
Figure 2.
MiD49 suppresses the growth of pancreatic cancer (PC) cells. (A and B) Knockdown (PANC-1 and HPAC) or overexpression of MiD49 (SW1990 and AsPC-1) in these PC cells was confirmed by qPCR and western blot analyses (siMiD49, siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector). (C and D) MTS cell viability and colony formation assays in PC cells with different levels of MiD49 as indicated. *P<0.05. MiD49, mitochondrial dynamics protein of 49 kDa.
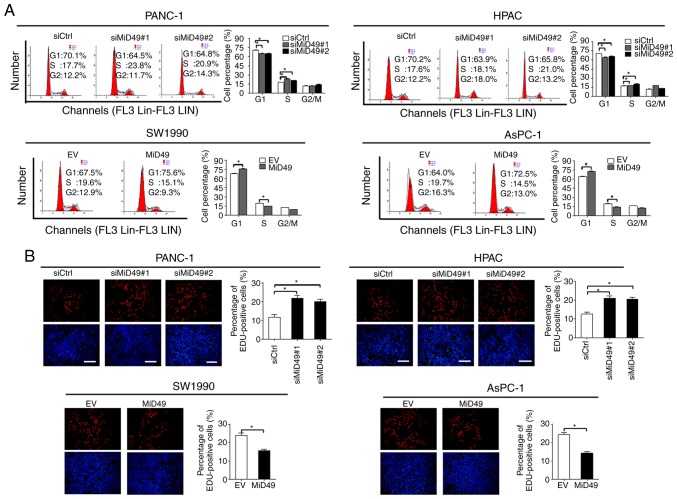
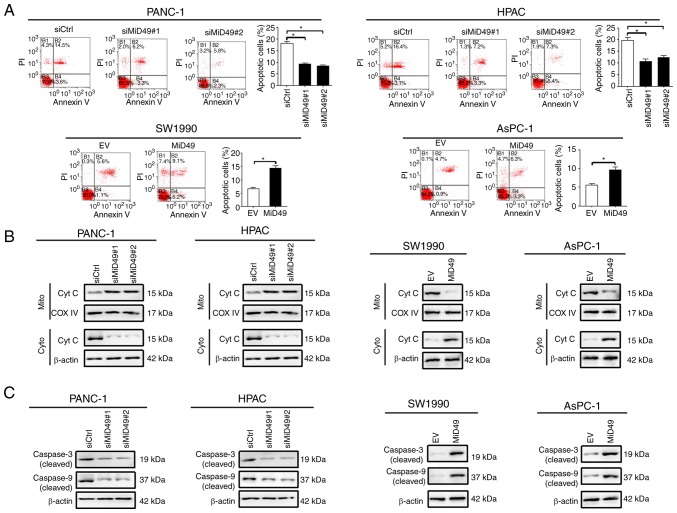
Increased cell growth could be caused by decreased apoptosis or accelerated cell cycle progression, or both. To determine the underlying mechanism by which MiD49 suppresses PC cell growth, the effects of MiD49 on apoptosis and cell cycle distribution were analyzed. Knockdown of MiD49 significantly decreased the number of cells in the G1 phase but increased the number of cells in the S phase in PANC-1 and HPAC cells. Conversely, forced expression of MiD49 arrested the cell cycle at the G1-S transition in SW1990 and AsPC-1 cells (Fig. 3A). Consistently, significantly more proliferating cells were detected by the EdU (5-ethynyl-2′- deoxyuridine) incorporation assay in PANC-1 and HPAC cells when MiD49 was knocked down, whereas fewer proliferating cells were observed in the SW1990 and AsPC-1 cells when MiD49 was overexpressed (Fig. 3B). Cell apoptosis analysis by flow cytometry revealed that knockdown of MiD49 significantly suppressed PC cell apoptosis, whereas forced expression of MiD49 exhibited an opposite effect (Fig. 4A). Consistently, knockdown of MiD49 suppressed the release of cytochrome c and the cleavage of caspase 3 and caspase 9, whereas forced expression of MiD49 had the opposite effect (Fig. 4B and C). Collectively, these results indicated that MiD49 suppresses the growth of PC cells by inducing G1-S cell cycle arrest and cell apoptosis.
Figure 3.
MiD49 induces G1-S phase cell cycle arrest in pancreatic cancer (PC) cells. (A) Cell cycle distribution was determined by flow cytometry in PC cells with different levels of MiD49 as indicated. (B) The percentage of proliferating cells was determined by EdU incorporation assay in PC cells with different levels of MiD49 as indicated. *P<0.05. MiD49, mitochondrial dynamics protein of 49 kDa; siMiD49, siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector.
Figure 4.
MiD49 induces pancreatic cancer (PC) cell apoptosis. (A) Cell apoptosis was determined by Annexin V/PI staining using flow cytometry in PC cells with different levels of MiD49 as indicated. (B) Levels of cytochrome c (Cyt c) in the cytoplasm (Cyto) and mitochondria (Mito) were determined by western blot analysis in PC cells with different levels of MiD49 as indicated. (C) Levels of cleaved caspase 3 and cleaved caspase 9 were determined by western blot analysis in PC cells with different levels of MiD49 as indicated. *P<0.05. MiD49, mitochondrial dynamics protein of 49 kDa; siMiD49, siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector.
MiD49 suppresses the migration and invasion of PC cells through inhibition of epithelial-mesenchymal transition (EMT)
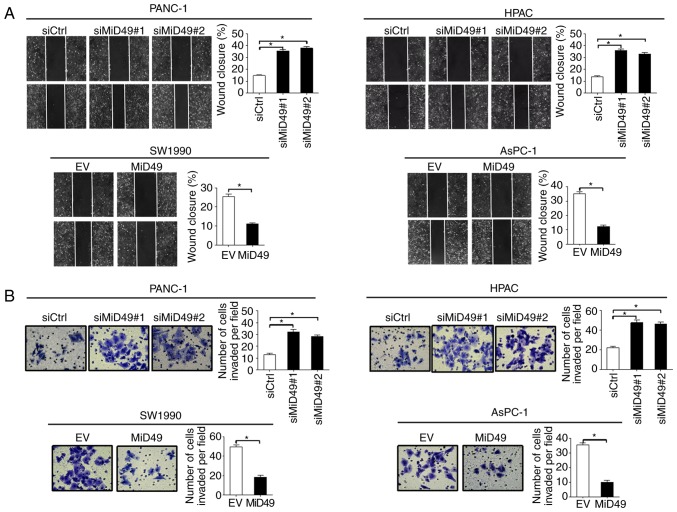
To further evaluate the role of MiD49 in the metastasis of PC cells, scratch wound healing and Transwell invasion assays were performed. Downregulation of MiD49 significantly increased the wound closure of PANC-1 and HPAC cells, whereas overexpression of MiD49 significantly decreased the migration abilities of the SW1990 and AsPC-1 cells (Fig. 5A). Similarly, the invasion potential was also significantly increased when MiD49 was knocked down, while the invasion potential was significantly decreased upon MiD49 was overexpressed (Fig. 5B).
Figure 5.
MiD49 suppresses the migration and invasion of pancreatic cancer (PC) cells. (A) Cell migration ability was determined by scratch wound-healing assay in PC cells with different levels of MiD49 as indicated. (B) Cell invasion ability was determined by Transwell matrigel invasion assay in PC cells with different levels of MiD49 as indicated. MiD49, mitochondrial dynamics protein of 49 kDa; siMiD49, siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector. *P<0.05.
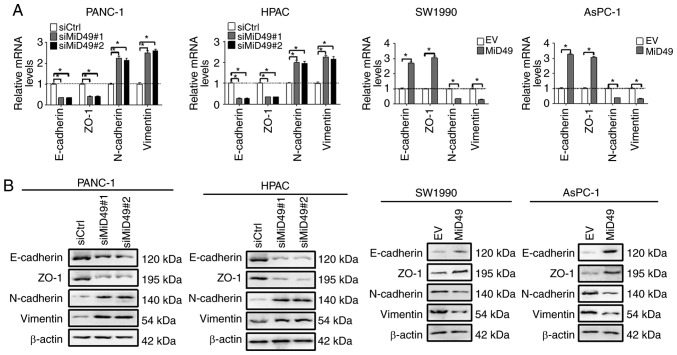
Epithelial-mesenchymal transition (EMT) is one of the major mechanisms underlying tumor metastasis (22). To determine the underlying mechanism by which MiD49 suppresses PC cell metastasis, the expression levels of key EMT markers were examined in PC cells with different MiD49 level. As shown in Fig. 6A and B, knockdown of MiD49 significantly increased the expression levels of mesenchymal markers, N-cadherin and vimentin, and significantly reduced the expression levels of epithelial markers E-cadherin and ZO-1, indicating an induction of EMT. In contrast, forced expression of MiD49 inhibited the EMT of PC cells.
Figure 6.
MiD49 suppresses the migration and invasion of pancreatic cancer (PC) cells through inhibition of epithelial-mesenchymal transition. (A) The expressions of EMT markers E-cadherin, ZO-1, N-cadherin, and vimentin were determined by qPCR analysis in PC cells with different levels of MiD49 as indicated. (B) The expression of EMT markers E-cadherin, ZO-1, N-cadherin, and vimentin were determined by western blot analysis in PC cells with different levels of MiD49 as indicated. MiD49, mitochondrial dynamics protein of 49 kDa; siMiD49, siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector. *P<0.05.
MiD49 suppresses the growth and metastasis of PC cells in vivo
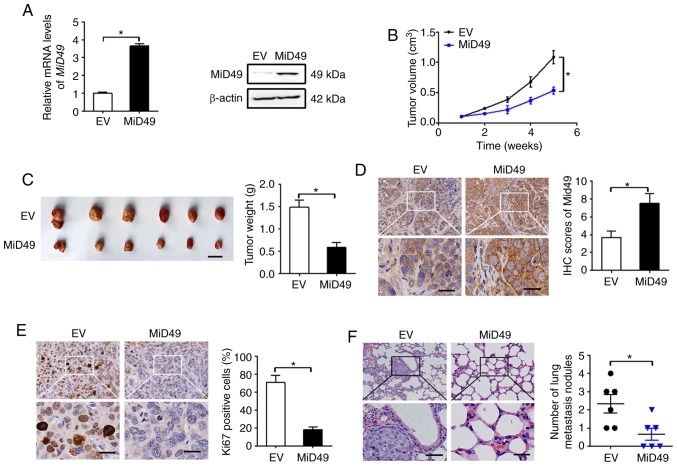
To study the role of MiD49 in PC growth in vivo, SW1990 cells overexpressing MiD49 (Fig. 7A) were injected into the right flanks of nude mice. As shown in Fig. 7B, tumors developed from SW1990 cells with stable MiD49 overexpression grew much slower than those developed from control cells. Consistently, markedly reduced tumor volume (length, width and volume of the largest tumor was 2.82 cm, 0.95 cm and 1.32 cm3, respectively) and wet weight were also observed in the MiD49 overexpression group (Fig. 7C). Immunohistochemical (IHC) analysis showed significantly higher expression of MiD49 in tumors developed from MiD49-overexpressing SW1990 cells than in the controls (Fig. 7D), implying that the tumor growth inhibitory effects were exerted by forced expression of MiD49. Similar to the in vitro experiments, significantly fewer proliferating cells were observed in the MiD49-overexpressing xenografts, as indicated by Ki-67 staining assay (Fig. 7E). Moreover, forced expression of MiD49 in the SW1990 cells significantly suppressed lung metastasis compared to the control cells.
Figure 7.
MiD49 suppresses the growth and metastasis of pancreatic cancer (PC) cells in vivo. (A) MiD49 expression was determined by qPCR and western blot analyses in SW1990 cells (MiD49, expression vector encoding MiD49; EV, empty vector). (B) Tumor growth curves of the two groups of MiD49 and EV are shown. (C) Harvested tumors and their weights were compared between the two groups. Scale bar, 1 cm. (D) The expression level of MiD49 was determined by immunohistochemical (IHC) staining in tumor tissues from the two groups of MiD49 and EV. Scale bars, 10 µm. (E) Ki-67 staining by IHC analysis in tumor tissues from the two groups of MiD49 and EV. Scale bars, 10 µm. (F) Incidence of pulmonary metastasis in the two groups of MiD49 and EV. MiD49, mitochondrial dynamics protein of 49 kDa. *P<0.05.
Downregulation of MiD49 is mainly mediated by elevated miR-424 level in PC cells
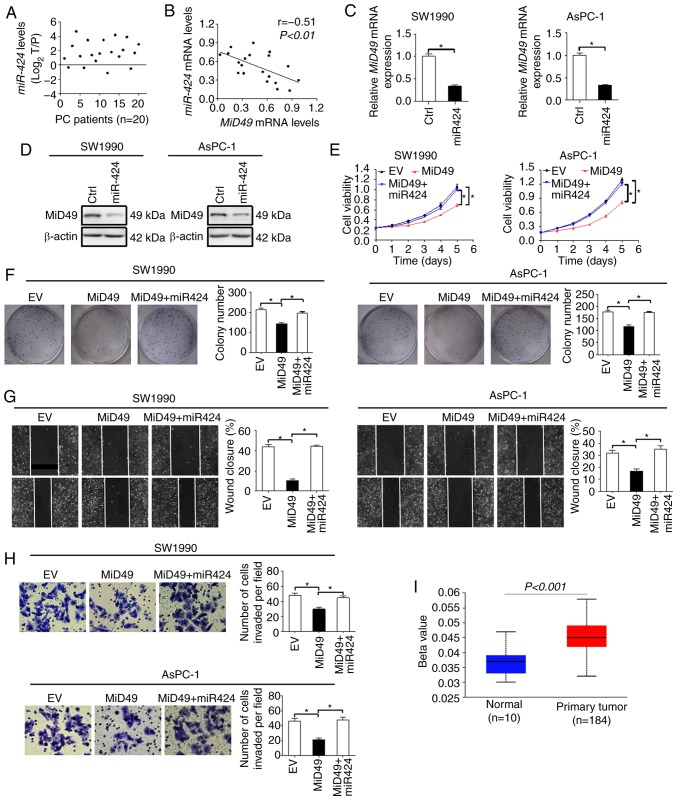
MicroRNAs are important regulators of gene expression at the post-transcriptional level. We thus explored the possibility that downregulation of MiD49 in PC cells was mediated by miRNA. Using the publicly available database miRDIP (microRNA Data Integration Portal) (23), we identified miR-424, an aberrantly overexpressed miRNA in PC (24), that could potentially target MiD49 (data not shown). We firstly evaluated the level of miR-424 in 20-paired tumor tissues and adjacent non-tumor tissues of PC. As shown in Fig. 8A, the level of miR-424 was significant higher in tumor tissues of PC compared with that observed in the adjacent non-tumor tissues. In addition, a significant inverse correlation (r=−0.51, P<0.01) between the expression of miR-424 and MiD49 was observed in the PC tumor tissues (Fig. 8B). To further test whether miR-424 is involved in the downregulation of MiD49 in PC cells, synthetic precursor of miR-424 was transfected into SW1990 and AsPC-1 cells. As shown in Fig. 8C and D, introduction of the synthetic precursor of miR-424 significantly decreased the expression of MiD49 at both the mRNA and protein levels in SW1990 and AsPC-1 cells. Moreover, miR-424 significantly reversed the tumor growth and metastasis-suppressive effects of MiD49 in SW1990 and AsPC-1 cells (Fig. 8E-H). Considering that genetic changes could also contribute to the downregulation of MiD49 in PC cells, we further applied promoter methylation profile analysis using the online web portal UALCAN (21). The promoter methylation of MiD49 is significantly higher in primary PC tumor tissues than that in normal pancreatic tissues (Fig. 8I), suggesting that promoter hypermethylation may also contribute to the downregulation of the expression of MiD49 in PC cells.
Figure 8.
Downregulation of MiD49 is mediated by an elevated miR-424 level in pancreatic cancer (PC) cells. (A) The level of miR-424 was determined by qPCR analysis in 20-paired tumor and adjacent non-tumor tissues of PC. (B) Correlation between the mRNA expression levels of MiD49 and miR-424 in tumor tissues from 20 PC patients. (C and D) The expression of MiD49 was determined by qPCR and western blot analyses in SW1990 and AsPC-1 cells transfected with synthetic precursor of miR-424. (E and F) MTS cell viability and colony formation assays in SW1990 and AsPC-1 cells transfected with synthetic precursor of miR-424. (G and H) Scratch wound-healing and Transwell invasion assays in SW1990 and AsPC-1 cells transfected with synthetic precursor of miR-424. (I) The promoter methylation profile analysis of MiD49 using the online web portal UALCAN. MiD49, mitochondrial dynamics protein of 49 kDa. *P<0.05.
MiD49 suppresses PC growth and metastasis through decreased ROS production
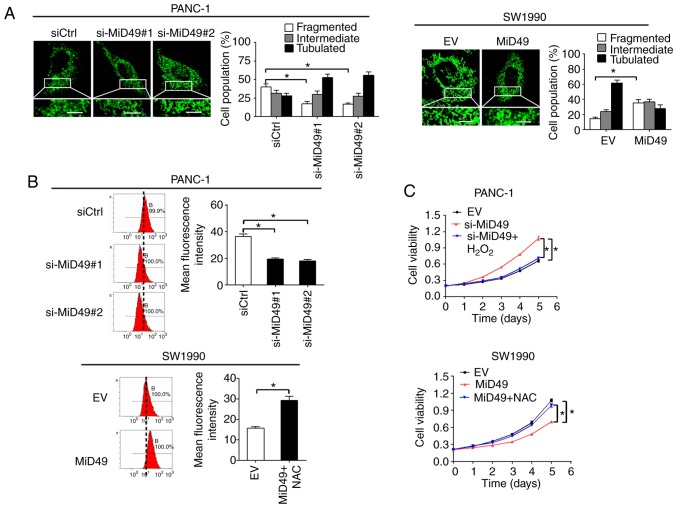
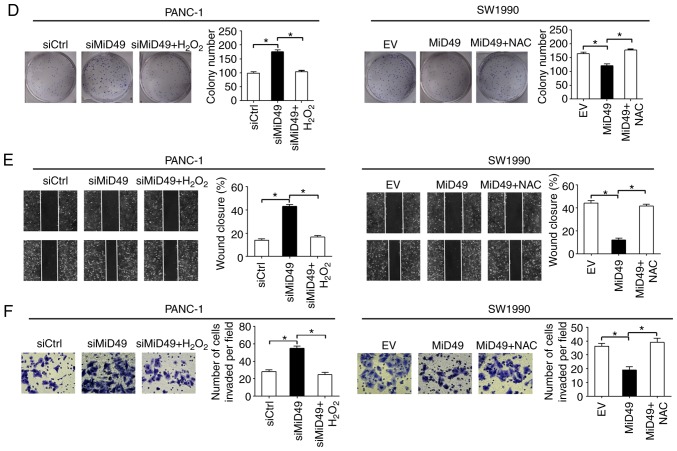
To explore the mechanistic basis of the tumor-suppressive effects of MiD49 in PC, the changes in mitochondria morphological were firstly investigated in PC cells when MiD49 was overexpressed or knocked down. MitoTracker Green staining results indicated that knockdown of MiD49 resulted in an extensive elongation of mitochondria in the PANC-1 cells, while overexpression of MiD49 induced a marked fragmentation of mitochondria in the SW1990 cells (Fig. 9A). Mitochondria are a major source of reactive oxygen species (ROS) production, which contributes to tumor development and progression through activation of multiple oncogenic signaling pathways (25,26). To explore the potential role of ROS in MiD49-mediated suppressed tumor growth and metastasis in PC, the production of ROS was evaluated in PC cells with different MiD49 expression levels. Knockdown of MiD49 significantly decreased the production of ROS, whereas overexpression of MiD49 significantly increased ROS levels in the PC cells (Fig. 9B). Moreover, treatment with H2O2 significantly inhibited the growth and metastasis of PANC-1 cells induced by MiD49 knockdown, whereas N-acetyl-L-cysteine (NAC) (a ROS scavenger) treatment reversed the growth and metastasis of SW1990 cells suppressed by MiD49 overexpression (Fig. 9C-F). These results collectively indicate that MiD49 suppresses the growth and metastasis of PC cells through decreased ROS production.
Figure 9.
MiD49 suppresses pancreatic cancer (PC) growth and metastasis through decreased ROS production. (A) Left panel: Representative images of mitochondria in PC cells with different MiD49 levels as indicated. Scale bars, 1 µm. Right panel: proportions of PC cells with elongated, intermediate, and fragmented mitochondria were quantified. (B) The level of intracellular ROS was determined by flow cytometry in PC cells with different MiD49 levels as indicated. (C and D) MTS cell viability and colony formation assays in PANC-1 cells treated with 90 µM H2O2 or SW1990 cells treated with 30 mM N-acetyl-L-cysteine (NAC) for 12 h as indicated. (E and F) Scratch wound-healing and Transwell invasion assays in PANC-1 cells treated with 90 µM H2O2 or SW1990 cells treated with 30 mM NAC for 12 h as indicated. MiD49, mitochondrial dynamics protein of 49 kDa; ROS, reactive oxygen species; siRNA against MiD49; siCtrl, control siRNA; MiD49, expression vector encoding MiD49; EV, empty vector. *P<0.05.
Discussion
In the present study, we demonstrated that mitochondrial dynamics protein of 49 kDa (MiD49) is commonly downregulated in pancreatic cancer (PC) cell lines and tissues mainly due to elevated miR-424 expression. Consistently, abnormal expression of other mitochondrial dynamic proteins, including dynamin-related protein 1 (DRP1), mitofusin 1 (MFN1) and mitofusin 2 (MFN2), have also been observed in several types of human cancers (11,12,14). For example, Zhan et al reported that the mitochondrial fission protein DRP1 was significantly upregulated and the mitochondrial fusion protein MFN1 was downregulated in liver cancer, both of which contributed to poor prognosis in liver cancer patients (10). These observations support the notion that dysfunction of mitochondrial dynamics plays a crucial role in tumor progression. However, considering that tumors are heterogeneous tissues composed of a mixture of cancer and non-tumor cells, we cannot rule out the possibility that mid49 is off in tumor cells but on in infiltrating non-tumor cells, which still needs further confirmation by immunohistochemical staining of mid49 in clinical PC tissues.
Given the common downregulation of MiD49 in PC cells, we hypothesized that MiD49 functions as a tumor suppressor in pancreatic carcinogenesis. However, the biological function of MiD49 in tumor progression remains largely unexplored. Our present study showed that MiD49 overexpression suppressed PC growth and metastasis both in vitro and in vivo, whereas knockdown of MiD49 exhibited the opposite effects. In agreement with our results, previous studies in several other cancer types have also demonstrated that dysregulation of mitochondrial dynamics proteins are involved in the promotion of tumor growth and metastasis (10,14,27). Additionally, we found that MiD49 suppressed PC growth and metastasis through induction of G1-S phase cell cycle arrest and inhibition of epithelial-mesenchymal transition.
MicroRNAs are crucial post-transcriptional regulators of gene expression. We thus explored the possibility that downregulation of MiD49 in PC cells was mediated by miRNA. Using the publicly available database miRDIP, we proposed that miR-424, an aberrantly overexpressed miRNA in PC (24), could potentially target MiD49. However, the target of miR-424 and its biological functions in PC cells are still unclear. Our present study demonstrated that MiD49 is a novel target of miR-424 in PC cells and downregulation of MiD49 is mainly due to the overexpression of miR-424, suggesting that miR-424 plays a crucial oncogenic role in PC. In addition to miR-424, other mechanisms such as promoter hypermethylation may also contribute to the downregulation of MiD49 in PC cells, which still needs further investigation.
MiD49 functions as a receptor on the outer membrane of mitochondria that recruits Drp1 to facilitate mitochondrial fission (17). Expectedly, our results indicated a marked fragmentation of mitochondria when MiD49 was overexpressed in PC cells. Mounting evidence indicates that mitochondria are a main source of reactive oxygen species (ROS), which contributes to tumor development and progression through activation of multiple oncogenic signaling pathways (25). We revealed that overexpression of MiD49 significantly increased ROS production in PC cells. Additionally, we found that increased ROS production is involved in the suppression of PC growth and metastasis by MiD49. However, a previous study in liver cancer has reported a promoting effect of mitochondrial fragmentation-induced ROS production on tumor growth and metastasis (11). This contradictory finding may be explained by the fact that moderate levels of ROS function as signals to promote tumor growth and metastasis, while excessive ROS induces tumor cell death.
In summary, our data indicate for the first time that MiD49 is frequently downregulated in PC cells, which contributes to the growth and metastasis of PC, suggesting a crucial tumor-suppressive role of MiD49 in pancreatic cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Natural Science Basic Research Plan in Shaanxi Province of China (program no. 2017JM8071).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
LB and EL designed the experiments. LB and JL and LL collected, analyzed the data and prepared the figures. LB and JL wrote the manuscript. All authors reviewed the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Written informed consent was obtained from the patients for their tissues to be used for biomedical research. All experimental protocols were approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University and carried out in accordance with the Declaration of Helsinki. All procedures involving animals study were approved by the Animal Care Committee of Xi'an Jiao Tong University in accordance with institutional requirements and Chinese government guidelines for animal experiments.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari D, Tingstedt B, Andersson B, Holmquist F, Sturesson C, Williamsson C, Sasor A, Borg D, Bauden M, Andersson R. Pancreatic cancer: Yesterday, today and tomorrow. Future Oncol. 2016;12:1929–1946. doi: 10.2217/fon-2016-0010. [DOI] [PubMed] [Google Scholar]
- 3.Gupta R, Amanam I, Chung V. Current and future therapies for advanced pancreatic cancer. J Surg Oncol. 2017;116:25–34. doi: 10.1002/jso.24623. [DOI] [PubMed] [Google Scholar]
- 4.Loc WS, Smith JP, Matters G, Kester M, Adair JH. Novel strategies for managing pancreatic cancer. World J Gastroenterol. 2014;20:14717–14725. doi: 10.3748/wjg.v20.i40.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, Yoon Y. Mitochondrial fission and fusion. Biochem Soc Trans. 2016;44:1725–1735. doi: 10.1042/BST20160129. [DOI] [PubMed] [Google Scholar]
- 6.van der Bliek AM, Shen Q, Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol. 2013;5:a011072. doi: 10.1101/cshperspect.a011072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trotta AP, Chipuk JE. Mitochondrial dynamics as regulators of cancer biology. Cell Mol Life Sci. 2017;74:1999–2017. doi: 10.1007/s00018-016-2451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maycotte P, Marín-Hernández A, Goyri-Aguirre M, Anaya- Ruiz M, Reyes-Leyva J, Cortés-Hernández P. Mitochondrial dynamics and cancer. Tumour Biol. 2017;39:1010428317698391. doi: 10.1177/1010428317698391. [DOI] [PubMed] [Google Scholar]
- 9.Prieto J, Torres J. Mitochondrial dynamics: In cell reprogramming as it is in cancer. Stem Cells Int. 2017;2017:8073721. doi: 10.1155/2017/8073721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhan L, Cao H, Wang G, Lyu Y, Sun X, An J, Wu Z, Huang Q, Liu B, Xing J. Drp1-mediated mitochondrial fission promotes cell proliferation through crosstalk of p53 and NF-κB pathways in hepatocellular carcinoma. Oncotarget. 2016;7:65001–65011. doi: 10.18632/oncotarget.11339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo X, Zhang J, Ji L, Ren T, An J, et al. Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NF-KB and TP53 pathways. Autophagy. 2016;12:999–1014. doi: 10.1080/15548627.2016.1166318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira-da-Silva A, Valacca C, Rios E, Pópulo H, Soares P, Sobrinho-Simões M, Scorrano L, Máximo V, Campello S. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PLoS One. 2015;10:e0122308. doi: 10.1371/journal.pone.0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkins K, Dasgupta A, Chen KH, Mewburn J, Archer SL. The role of Drp1 adaptor proteins MiD49 and MiD51 in mitochondrial fission: Implications for human disease. Clin Sci (Lond) 2016;130:1861–1874. doi: 10.1042/CS20160030. [DOI] [PubMed] [Google Scholar]
- 16.Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loson OC, Meng S, Ngo H, Liu R, Kaiser JT, Chan DC. Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1. Protein Sci. 2015;24:386–394. doi: 10.1002/pro.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 20.Ishikawa M, Yoshida K, Yamashita Y, Ota J, Takada S, Kisanuki H, Koinuma K, Choi YL, Kaneda R, Iwao T, et al. Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Sci. 2005;96:387–393. doi: 10.1111/j.1349-7006.2005.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L, Li W. Epithelial-mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I. mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res. 2018;46:D360–D370. doi: 10.1093/nar/gkx1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, Ohtsuka T, Ueda J, Nagai E, Hashizume M, Tanaka M. MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol. 2011;18:2381–2387. doi: 10.1245/s10434-011-1602-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles' heel? Nat Rev Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rigoulet M, Yoboue ED, Devin A. Mitochondrial ROS generation and its regulation: Mechanisms involved in H(2)O(2) signaling. Antioxid Redox Signal. 2011;14:459–468. doi: 10.1089/ars.2010.3363. [DOI] [PubMed] [Google Scholar]
- 27.Sun X, Cao H, Zhan L, Yin C, Wang G, Liang P, Li J, Wang Z, Liu B, Huang Q, Xing J. Mitochondrial fission promotes cell migration by Ca2+/CaMKII/ERK/FAK pathway in hepatocellular carcinoma. Liver Int. 2018;38:1263–1272. doi: 10.1111/liv.13660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.