Abstract
Ewing sarcoma (ES) is a primary bone marrow tumor that very rarely develops in extra-osseous tissues, such as lung. The hallmark of ES tumors is a translocation between chromosomes 11 and 22, resulting in a fusion protein, commonly referred to as EWS-FLI1. The epigenetic profile (histone acetylation and methylation enrichment of the promoter region) that may regulate the expression of the aberrant transcription factor EWS-FLI1, remains poorly studied and understood. Knowledge of epigenetic patterns associated with covalent histone modification and expression of enzymes associated with this process, can contribute to the understanding of the molecular basis of the disease, as well as to the identification of possible molecular targets involved in expression of the EWS-FLI1 gene, so that therapeutic strategies may be improved in the future. In the present study, the transcriptional activation and repression of the EWS-FLI1 fusion gene in ES was accompanied by selective deposition of histone markers on its promoter. The EWS-FLI1 fusion gene was evaluated in two patients with ES using conventional cytogenetic, fluorescence in situ hybridization and nested PCR assays, which revealed that the aberrant expression of the EWS-FLI1 gene is accompanied by enrichment of H3K4Me3, H3K9ac and H3K27ac at the promoter region.
Keywords: Ewing's sarcoma, epigenetics, histone, FISH, translocation
Introduction
Ewing sarcoma (ES) is a rare disease characterized by tumors originating in the bone marrow of long flat bones of the body. ES commonly affects the pelvis (19%), long bones (47%) and ribs (12%) (1). However, this disease can develop in any other tissue or organ (extra-osseous ES) (2,3).
Generally, ES occurs during the second decade of life and rarely arises before 5 years of age or after 30 years of age (4). In Colombia, the incidence rate of ES is approximately 0.1 cases per 100,000 inhabitants, with similar rates in Chile, Ecuador and Costa Rica (5), while 225 cases have been reported in individuals less than 20 years of age in North America (6).
ES tumors are histologically characterized by a greyish, bright, translucent mass, with necrotic and hemorrhaging areas with several cavities and marked vascularization (7). The presence of small round cells with narrow cytoplasm and oval hyperchromatic and granulated nuclei constitutes another key feature of ES tissue (8). Immunohistochemical analyses usually reveal positivity for CD99 membrane glycoprotein, and in at least 90% of cases the tumor is caused by the chromosomal translocation t(11;22), although it is possible that other translocations are also involved in its pathology (10% of cases) (8). The t(11;22) translocation is responsible for the generation of the EWS-FLI1 fusion gene that expresses the EWS-FLI1 transcription factor. This fusion gene is often used to diagnose the pathological condition, whereby ES molecular profiling is performed, quantifying EWS-FLI1 and FLI1-EWS expression by quantitative polymerase chain reaction (qPCR), and detecting chromosomal rearrangements by fluorescence in situ hybridization (FISH) (9,10).
EWS-FLI1 functions as an aberrant transcription factor and is believed to be an initiator of tumorigenic events (11). The study by Matsumoto et al (12) reported that the EWS-FLI1 transcription factor affects the expression of regulatory genes of the G1 stage of the cell cycle, including cyclin G1, cyclin D1, p21 and p27 (13). Other genes that have been identified as transcriptional targets of EWS-FLI1 are homeobox protein NKX2-2, nuclear receptor subfamily 0 group B member 1 (NR0B1), and histone-lysine N-methyltransferase EZH2 (14). The function of EWS-FLI1 is not only restricted to transcriptional regulation, but also includes modulating the expression of microRNAs (miRNAs) and long non-coding RNAs.
The epigenetic events described within the pathogenic context of ES include the participation of the EWS-FLI1 protein in aberrant chromatin remodeling processes (15). EWS-FLI1 has specific binding domains that recognize microsatellites with GGAA sequences embedded in specific genes; this binding leads to the recruitment of the p300 protein (acetyltransferase) to the nucleosome, resulting in loosening the packaging of DNA, facilitating the transcriptional mechanisms to reach the gene (15). It has been reported that the aforementioned epigenetic mode of action is involved in the activation of the NR0B1 genes (15).
Cases of ES have been reported in which the EWS-FLI1 protein fulfills a role of transcriptional repressor of miRNA-22, whose expression participates in the inhibition of cell proliferation programs. In addition, lysine-specific demethylase 3A has been reported to cause this silencing in tumorigenic ES scenarios (16).
The contribution of altered DNA promoter methylation to ES development is beginning to come into focus (14), whereas the epigenetic profile (histone acetylation and methylation enrichment of the promoter) that may be regulating the expression of aberrant transcription factor EWS-FLI1, remains poorly studied and understood. The in-depth study of the molecular mechanisms that regulate the expression of aberrant transcription factors such as EWS-FLI1 is fundamental and necessary (17), because this knowledge will allow us to understand and develop novel therapeutic approaches to this pathology, such as epi-drugs.
Materials and methods
General
The present study was carried out under the 1993 guidelines of the Colombian Ministry of Health and Social Protection (resolution no. 008430), and adhered to the ethics principles for medical investigation with human beings, according to the Ethical Committee of the School of Medicine, Javeriana Pontifical University (Bogota, Colombia), and the Helsinki Declaration of 1975. The code assigned by the ethics committee for the execution of this study is FM-CIE-8148-14 (2014) Pontificia Universidad Javeriana / Hospital Universitario San Ignacio.
The study was conducted at the San Ignacio University Hospital (Bogota, Colombia), and included two patients in whom malignant pulmonary lesions were suspected (Table I). These patients had a lung biopsy and histopathological examinations between October and December 2016. Informed consent was obtained to access each of the patient samples.
Table I.
Clinical characteristics of all the patients with cancer.
| Case | 1 | 2 | |
|---|---|---|---|
| Age | 49 | 19 | |
| Sex | F | M | |
| Pathology | Cancer | Yes | Yes |
| Pathological diagnosis | Ewing sarcoma extraoseus | Poorly differentiated adenocarcinoma | |
| Origin | Primary | Metastatic | |
| Origin | Lung | No | No |
| Other | Femur Ewing sarcoma | Bone | |
| TNM classification | T | 1 | 1 |
| N | 1 | 0 | |
| M | 1b | 1a | |
| G | 2 | 3 | |
| Stage | IV | IV | |
| Comorbidity | Absent | Absent | |
| One year survival | No | Yes | |
| Smoker | No | Yes | |
| Treatment | No | Yes |
Samples and cell culture
Samples were obtained during open lung biopsy and submerged in base C culture medium containing antibiotics and 5% fetal bovine serum (cat. no. A3840201; Gibco; Thermo Fisher Scientific, Inc.) in a sterile 24-well culture plate (18). Using a stereoscope, solid tumors were fractionated mechanically with scissors or a scalpel. The cell suspensions were subdivided into 2–3 wells of the 24-well culture platform, and incubated at 37°C and in 5% CO2 conditions. Cell culture medium renewal was carried out every 48 h, until a confluent monolayer was obtained from the tumor fragments. Cells were enzymatically dissociated using trypsin/EDTA (cat. no. 2520056; Gibco; Thermo Fisher Scientific, Inc.), and then the passages were transferred to Petri dishes of 35, 60 and 100 mm (18). Once 100% confluency was reached, the cells were used for immunohistochemistry, gene expression, cytogenetic and epigenetic analysis.
Cell lines
ES A673 cell line [CRL-1598; American Type Culture Collection (ATCC)] was cultured in Dulbecco's modified Eagle's medium (DMEM; cat. no. 12491; Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum and antibiotics (cat. no. 15140122; Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated under conditions of 37°C and 5% CO2 (ATCC: The Global Bioresource Center). Hs 1.Tes (CRL-7002; ATCC) is a non-tumor (NT) human testicular cell line that does not carry the EWS-FLI1 fusion gene, and was used as a negative control. This cell line was cultured in DMEM and supplemented with 10% fetal bovine serum, according to the instructions from the ATCC.
Immunohistochemistry
Samples from two patients were available for immunohistochemical analysis of CD99, Friend leukemia integration 1 transcription factor (FLI1), CD57 (HNK-1), Vimentin, enolase, Chromogranin, Synaptophysin, S-100, CD45, CD117, TdT, Desmin, Myogenin, Cytokeratin (AE/AE3). List of antibodies used in immunhistochemistry assays are shown in Table SI.
The paraffin-embedded sections were rehydrated and incubated for 55 min at 20°C in methanol 10% H2O2 to block endogenous peroxidase (EnVisionTM FLEX + Dako kit). Sections were pretreated to facilitate antigen retrieval and increase membrane permeability to antibodies with the same kit and then incubated with the primary antibody (19). Positive reaction was visualized by 3.3′-diaminobenzidine (DAB) peroxidation according to the protocol. The sections were counterstained with Harris's hematoxylin, dehydrated, coverslipped, and observed under an Olympus optical microscope BX53. Positive and negative controls were performed and validated for each antibody.
Analysis of the immunohistochemical markers was carried out at the San Ignacio University Hospital as part of the histopathological diagnosis protocol necessary to process the samples of the patients.
Cytogenetic analysis and FISH
Cells were seeded on Knittel Microscope slides and cultured until 80% confluency was reached. Colchicine (1 µg/ml) was added and incubated for 2 h. Then, the cells were harvested by treatment with 0.05 M KCl (cat. no. 7477-40-7; Sigma-Aldrich; Merck KGaA) followed by fixation with 3:1 methanol (cat. no. 67-56-1; Sigma-Aldrich; Merck KGaA) and glacial acetic acid (cat. no. 100063; Merck KGaA). Finally, the slides were evaluated using the G-banding protocol (20). A total of 30 metaphases were evaluated for each patient and each cell line. Molecular cytogenetic analysis using FISH was performed to evaluate the presence of the translocation between EWSR1 (ES region 1) and any of the ETS transcription factor family genes, in particular FLI1, located in chromosomes 22 and 11, respectively. The Cytocell Aquarius® kit was used, containing the EWSR1 Breakapart Probe (Cytocell, Ltd.), with a 392-kb red probe and a 631-kb green probe to be placed on either side of the EWSR1 gene. The Carl Zeiss fluorescence microscope (Axio-Scope A1) with image capture software was used for the evaluation. The presence of fluorescent signals from the different probes in both metaphase (25 metaphase) and nucleus (200 nucleus) chromosomes were analyzed, and the findings were described according to the International Cytogenetic Nomenclature System (21).
Molecular analysis: RNA isolation, PCR and nested PCR
Total RNA was extracted using TRIzol reagent (cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol, and cDNA was generated by reverse transcription of 2 µg RNA using the ProtoScript® II First Strand cDNA Synthesis kit (cat. no. E6560S; New England BioLabs, Inc.). PCR reactions were performed with Taq DNA Polymerase products (cat. no. M0273S; New England BioLabs, Inc.) on the T100™ Thermal Cycler (Bio-Rad Laboratories, Inc.).
Conventional PCR was performed to detect the presence of genes EWS, FLI1 and reciprocal fusion gene FLI1-EWS. The primers used to amplify these genes were: EWS forward, 5′-CAGCCTCCCACTAGTTACCC-3′ and reverse, 5′-GTTCTCTCCTGGTCCGGAAA-3′; FLI1 forward, 5′-AATACAACCTCCACACCGA-3′ and reverse, 5′-CTTACTGATCGTTTGTGCCCC-3′; and FLI1-EWS forward, 5′-GTGCTGTTGTCACACCTCAG-3′ and reverse, 5′-GTTCTCTCCTGGTCCGGAAA-3′ (22). Amplifications were performed according to conditions established for each set of primers: EWS with an initial denaturation step at 94°C for 30 sec, and 42 cycles of denaturation at 59°C for 30 sec and annealing at 72°C for 60 sec for 15 sec; FLI1 with an initial denaturation step at 94°C for 30 sec, and 42 cycles of denaturation at 58°C for 30 sec and annealing at 72°C for 60 sec; FLI1-EWS with an initial denaturation step at 94°C for 30 sec, and 42 cycles of denaturation at 62.4°C for 30 sec and annealing at 72°C for 60 sec. Nested PCR was performed to detect the EWS-FLI1 fusion gene, using 5 µl cDNA from each sample and two sets of primers. Nested PCR is a modification of PCR that was designed to improve sensitivity and specificity. The first round of amplification was performed using EWS 22.8 forward, 5′-CCCACTAGTTACCCACCCCAAA-3′, and FLI1 reverse, 5′-AGGGTTGGCTAGGCGACTGCT-3′, and the second round of amplification was performed using 5 µl of the first PCR product and primers EWS 22.3 forward, 5′-TCCTACAGCCAAGCTCCAAGTC-3′, and FLI1 reverse, 5′-GTCGGGCCCAGGATCTGATAC-3′ (23). The thermocycling conditions used for the first and second round of amplification included an initial denaturation step at 94°C for 30 sec, and 42 cycles of denaturation at 64.5°C for 30 sec and annealing at 72°C for 60 sec. GAPDH was used as an internal control.
Chromatin immunoprecipitation (ChIP) assay
ChIP asssays were performed in cross-linked cromagin samples. The cell were incubated for 10 min with 1% formaldehyde with gentle agitation at room temperature, and washed three times with 10 ml PBS. The formaldehyde cross-linked cells were incubated for 1 h at room temperature with gentle agitation, washed three times with cold PBS, resuspended in 1 ml cell lysis buffer (5 mM Hepes, pH 8.0, 85 mM KCl, Triton X-100, and proteinase inhibitors), and homogenized with a Dounce homogenizer (approximately 25 times using a tight pestle). The cell extract was collected by centrifugation at 3,000 × g for 5 min, resuspended in 0.5 ml sonication buffer (50 mM Hepes, pH 7.9, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate acid, 0.1% SDS, and a mixture of proteinase inhibitors) and incubated for 10 min on ice. Next, the samples were sonicated at high power for four pulses of 10 min each and centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was collected and stored at −80°C and chromatin size was confirmed by electrophoretic analysis in 1% agarose gel.
Samples were precleared by incubating with 2–4 µg of normal IgG and 50 µl of protein A/G-agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C with agitation. Chromatin was centrifuged at 4000 × g for 5 min, and the supernatant was collected and immunoprecipitated with specific antibodies (2 µg/ml) for 12–16 h at 4°C. The immune complexes were recovered with the addition of 50 µl of protein A or G-agarose beads followed by incubation for 1 h at 4°C with gentle agitation. Immunoprecipitated complexes were washed once with sonication buffer, twice with LiCl buffer (100 mM Tris-EDTA buffer, pH 8.0 500 mM LiCl, 0,1% Nonidet P-40, and 0,1% deoxycholic acid), and once with Tris-EDTA buffer, pH 8.0 (2 mM EDTA and 50 mM Tris-HCl, pH 8.0), each time for 5 min at 4°C. The protein-DNA complexes were eluted by incubation with 100 µl of elution buffer (50 mM NaHCO3 and 1% SDS) for 15 min at 65°C to reverse the crosslinking. Proteins were digested with 100 µg/ml proteinase K for 2 h at 50°C, and the DNA was recovered by phenol/Chloroform extraction and ethanol precipitation using glycogen (20 µg/ml). The qPCR primers used to evaluate the EWS-FLI1 promoter region were: set1EWS: Forward, 5′-CCGTAAACCTCCTCCTGCAT-3; and reverse, 5′-AAGCCCTTCACCCTTGCTAA-3, directed towards the sequence of the promoter (24). The primer sequences used in this study were taken from the study carried out by Jacques et al (25).
In order to quantify the ChIP experiments, qPCR (quantitative polymerase chain reaction) analysis was performed using FastStart Essential DNA SYBR-Green Master; cat. no. 06402712001; Roche Diagnostics. The results were analyzed using the percentage Input Method according to Haring et al (26). Results are expressed as % input ± SEM using normal IgG as a specificity control.
The antibodies used in the ChIP assays were: H3K27me3 (cat. no. 07-449), H3K9ac (cat. no. 06942) (both Merck KGaA), H3K4me3 (cat. no. ab8580), H3K9Me3 (cat. no. ab8898) and H3K27ac (cat. no. ab4729) (all from Abcam).
Statistical analysis
In order to perform statistical analysis, NT control cells (testis Hs 1.Tes cell line) were used for the comparison with the ES samples. For ChIP assays, we used a one-way analysis of variance followed by the Dunnett's post hoc test to compare significant changes with respect to control. A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics of patients
Table I lists the study subjects with their clinical characteristics. Patient 1 was a 49-year-old female patient, with no history of pathological conditions, who underwent a consultation following two months of respiratory issues, including cough, dyspnea and functional class deterioration associated with left hemithorax pain. During the physical examination, decreased respiratory sounds were identified in the left lung. A requested X-ray revealed enlargement of the left pulmonary hilum, and contrast CT demonstrated a decrease in the filling of the left pulmonary artery, and the presence of a lobulated mass around the left pulmonary artery and left bronchus. The patient underwent surgery, but complete tumor resection could not be achieved; instead only biopsy samples were collected.
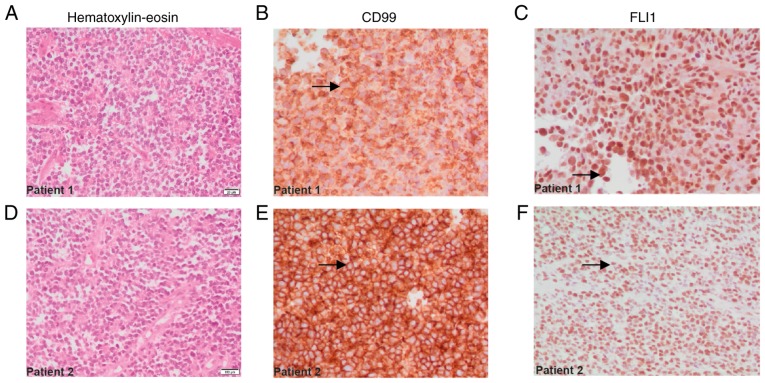
Patient 2 was an 18-year-old female with advanced-stage ES in the left femur. Following tumor resection, extension studies revealed a pulmonary nodule in the left upper lobe. A wedge biopsy was performed, detecting visceral pleura and lymphovascular tissue compromise due to metastatic ES, positive for immunohistochemical markers FLI1 and CD99 (Fig. 1E-F), and the cell proliferation index (Ki-67) was >90%.
Figure 1.
Immunohistochemical analysis of lung tissue biopsy sections from patients with Ewing sarcoma. The samples of patient 1 are seen in A, B and C, and those of patient 2 are seen in D, E and F. (A and D) Small round cells, characteristic of tumors of the Ewing family, with hyperchromatic nuclei and small cytoplasmic space, as observed by hematoxylin and eosin staining. (B and E) Membranous staining for the CD99 protein (arrows). (C and F) Positive staining for the FLI1 transcription factor located in the nucleus (arrows). Magnification, ×40.
ES immunohistochemical and cytogenetic analysis
For patients 1 and 2, immunohistochemical analysis results revealed positive membranous staining for CD99 (Fig. 1B and E, arrows), and nuclear positivity for FLI1 (Fig. 1C and F, arrows), consistent with the characteristics of ES family tumors. Other markers, including S-100, CD45, desmin, enolase and myogenin, were also investigated (Table II).
Table II.
Immunohistochemical markers of patients with Ewing sarcoma. The table lists 12 immunohistochemical markers assayed in samples from two patients.
| Case number | 1 | 2 |
|---|---|---|
| CD99 | + | + |
| FLI-1 | + | + |
| CD57(HNK-1) | + | + |
| Vimentin | + | NA |
| Enolase | NA | − |
| Chromo-granin | − | NA |
| Synapto-physin | − | NA |
| S-100 | − | + |
| CD45 | − | − |
| CD117 | − | NA |
| TdT | − | NA |
| Desmin | − | − |
| Miogenin | NA | − |
| Cytokeratin (AE/AE3) | − | NA |
The histological description of each case was also carried out, based on the images captured of the tissues treated with hematoxylin and eosin dyes. This staining made it possible to demonstrate the presence of small, round, blue cells with hyperchromatic nuclei and decreased cytoplasm, all characteristics of ES (Fig. 1A and D).
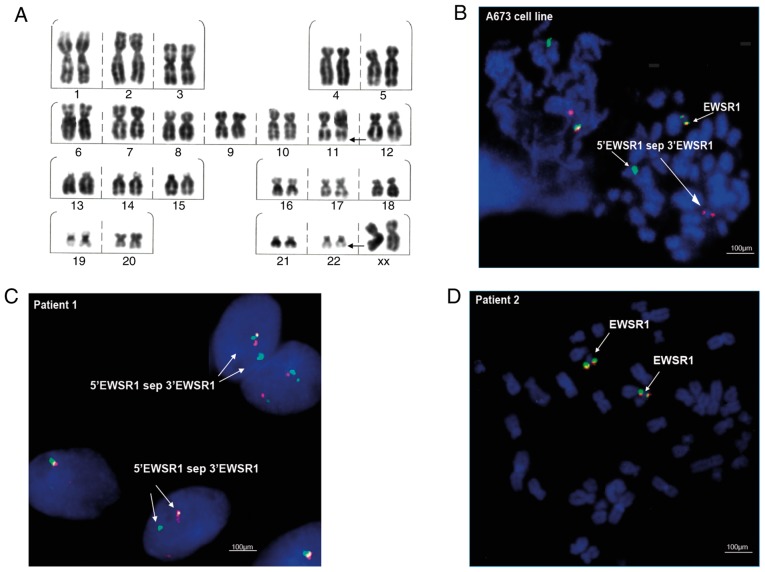
Cytogenetic investigation included G-banding analysis to detect chromosomal abnormalities, and FISH for the analysis of EWSR1 gene translocation on chromosome 22. In patient 1, it was possible to observe clonal chromosomal alterations such as del(22)(q13) (in 14 metaphases), del(20)(q13.2) (in 5 metaphases) and del(16)(q22) (in 4 metaphases). In addition, translocation t(11;22)(q22;q12), characteristic of ES, was observed at a low frequency (2%; Fig. 2A), as well as translocation t(1;16)(q21;p13), also reported for this type of tumor (Table III). By contrast, the presence of the translocation t(1;16)(q21;p13) was not detected in the samples of patient 2.
Figure 2.
Cytogenetic analysis. (A) Karyotype in patient 1: 46,XX,t(11;22)(q22;q12). FISH analysis with Cytocell Aquarius EWSR1 Breakapart Probes in (B) the A673 cell line, positive for EWSR1 translocation; (C) patient 1, positive for EWSR1 translocation, and (D) patient 2, with EWSR1 in metaphase without translocation: ish 22q12(EWSR1×2). Red probes, 3′EWSR1; green probes, 5′EWSR1. EWSR1, Ewing sarcoma region 1.
Table III.
Cytogenetic findings obtained from the G-banding technique.
| Samples | Chromosomal alterations | Frequency [%] | FISH EWSR1 translocation |
|---|---|---|---|
| A673 cell line | Multiple alterations | 90 | Positive |
| Hs-Tes cell line | None | 0 | Negative |
| Patient 1 | t(11;22)(q22;q12) | 2 | Positive (2%) |
| t(1;16)(q21;p13) | 1 | ||
| del(22)(q13) | 14 | ||
| del(20)(q13.2) | 5 | ||
| del(16)(q22) | 4 | ||
| Patient 2 | del(17)(p10) | 2 | Negative |
| del(7)(p10) | 1 |
The FISH assays in the ES A673 cell line, revealed the EWSR1 translocation ish 22q12(EWSR1×2)(5′EWSR1 sep 3′EWSR1×1) (Fig. 2B), represented by the separation of the probe and the visualization of separate green and red signals (Fig. 2B; indicated with arrows and the label 5′EWSR1 sep 3′EWSR1). In the clinical samples, the FISH cytogenetic analysis showed the presence of the EWSR1 translocation nuc ish(EWSR1×2)(5′EWSR1 sep 3′EWSR1×1) in patient 1, indicated as separated signals in 10% of the interphase nuclei analyzed (Fig. 2C). By contrast, the translocation was not detected in the samples of patient 2 (Fig. 2D).
EWS-FLI1 fusion gene expression profile
The normal functionality of the EWSR1 and FLI1 genes can be affected following reciprocal chromosomal translocation events that lead to the synthesis of the fusion genes responsible for the pathological condition of ES. The chimeric proteins EWS-FLI1 and FLI1-EWS commonly characterizes the molecular profile of the disease (22).
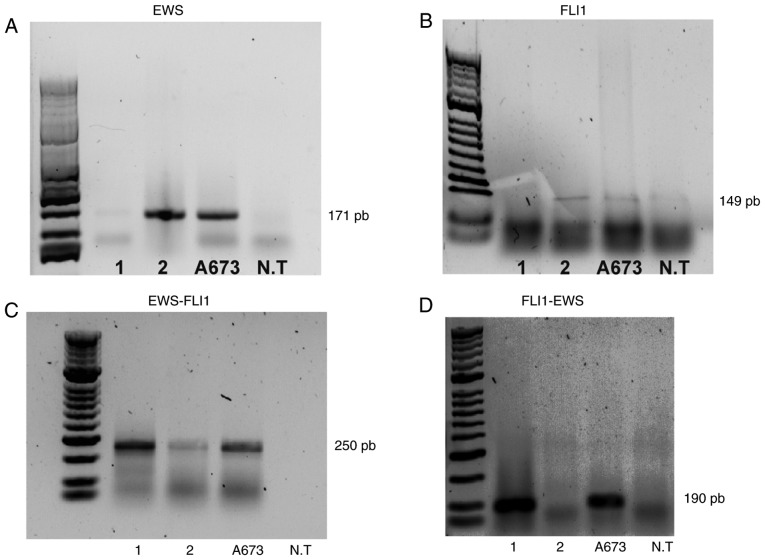
Conventional and nested PCR tests were performed on lung samples from patients 1 and 2, using ES A673 cells as a positive control and an NT cell line as a negative control for all experiments (Fig. 3). Expression of the EWS and FLI1 genes was observed in patient 2 and the A673 cells (Fig. 3A and 3B). On the other hand, when performing two rounds of amplification using two sets of EWS-FLI1 primers, the detection of the EWS-FLI1 fusion gene was possible in the A673 cells and the two patients (Fig. 3C). Additionally, the reciprocal fusion gene FLI1-EWS was detected in the cell line A673 and patient 1 (Fig. 3D).
Figure 3.
PCR analysis. The conventional and nested PCR products from A673 cells, NT cells and patients 1 and 2 demonstrate the presence of the EWS-FLI1 fusion gene in the patient samples and in the Ewing sarcoma A673 cell line. GAPDH serves as a loading control. NT=non-tumor. (A) EWS; (B) FLI1; (C) EWS-FLI1; (D) FLI1-EWS.
Notably, the mRNA levels of the fusion gene were lower in patient 2 than those detected in patient 1 and in the A673 cell line (Fig. 3C), explaining why patient 2 exhibited higher expression of the non-fusion EWS and FL11 genes compared with patient 1 (Fig. 3A and B).
Epigenetic modifications in the EWS-FLI1 fusion gene
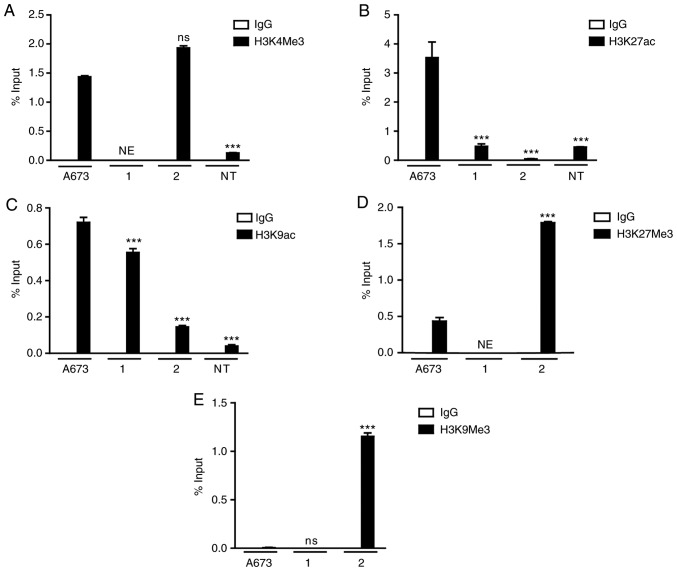
To elucidate the contribution of epigenetic mechanisms on the transcriptional control of the EWS-FLI1 fusion gene and, therefore, on the cellular processes controlled by EWS-FLI1 expression in ES, ChIP assays were performed, analyzing histone H3 covalent modifications in the EWS promoter, which controls the transcription of the EWS-FLI1 fusion gene (Fig. 4). Tumor cells obtained from patient 1, and the A673 cell line exhibited enrichment at H3K4Me3 and H3K9ac, and decreased levels of H3K9Me3 (Fig. 4A, C and E). Surprisingly, H3K27ac was present only in the A673 cell line (Fig. 4B), whereas decreased levels of H3K27Me3 were detected in the A673 cells as well as in patient 1 (Fig. 4D). On the other hand, the samples from patient 2 exhibited enrichment of repressive markers H3K27Me3 and H3K9Me3 in the EWS promoter (Fig. 4D and E).
Figure 4.
Epigenetic covalent modifications in EWS-FLI1. Histone post-translational modifications at the EWS promoter region in primary cancer cells, A673 cells and NT Hs 1.Tes cells. Primary cancer cells from two patients (1 and 2) and cells lines A673 and Hs 1.Tes were used to perform chromatin immunoprecipitation assays. Antibodies against (A) H3K4Me3, (B) H3K27ac, (C) H3K9ac, (D) H3K27Me3 and (E) H3K9Me3 were used. Results are expressed as % input ± SEM using normal IgG as a specificity control. Statistical analyses were performed compared with the NT cells. ***P<0.001. ns, non-significant differences; NE, not evaluated; NT, non-tumor.
In the NT cells, the ChIP results revealed the presence of low levels of the activating markers H3K4Me3, H3K27ac and H3K9ac (Fig. 4A-C), consistent with the absence of the detection of EWS-FLI1 fusion gene expression.
Discussion
Detection and analysis of the EWS-FLI1 fusion gene, together with immunohistochemical markers CD99 and FLI1, constitutes an important part of the molecular characterization that must be considered when making a correct diagnosis of ES, given the symptomatic and histopathological similarities that this condition may have with other soft tissue sarcomas (8,11). Therefore, immunohistochemical, cytogenetic and molecular tests confirmed the suspicion of two ES cases examined within the present study.
Sarcomas, by definition, are rare tumors with primary origin in soft and bony tissues of the body (1,2). In addition, they display high genetic and clonal heterogeneity, which, in some cases, may make diagnosis challenging. This means that the same clinical picture can be the product of different genotypes. Sarcomas in general are caused by chromosomal rearrangements, including translocations, deletions, duplications and inversions, and in certain cases by genetic mutations (8). Therefore, it is not surprising that, in a single ES sample, a genetic mosaic can be responsible for the condition, since there are at least 16 structural possibilities of gene fusions that may cause this neoplasm, formed by different combinations between the EWS gene and other genes belonging to the ETS transcription factors family (1). In this study, lung tissue samples analyzed from patient 1 were obtained from a biopsy performed by open surgery as a consequence of the suspicion of a possible lung adenocarcinoma. On the other hand, samples from patient 2, also pulmonary tissue, were included in the study after having undergone histopathological analyses that suggested a case of ES with primary origin in the left femur and lung metastasis. Both samples were subjected to FISH analysis and initial results demonstrated the presence of the characteristic ES translocation t(11;22)(q22;q12) in patient 1 (in only 2 of 200 analyzed nuclei). However, this chromosomal rearrangement was not detected in the samples of patient 2. This represents a possible reason for the negative results obtained during the FISH analyses of the samples of patient 2, since this case could be made up of clonal and genetic mosaic patterns different from the known characteristic translocation, or the samples provided may have contained a large proportion of healthy cells, as the FISH analysis detected several signals referring to cells apparently free of chromosomal rearrangements. However, the possibility that the cell samples contained expression profiles for this gene that were too low to be detected by the probe, cannot be excluded.
On the other hand, molecular analyses confirmed the initial suspicion of ES in both samples (patient 1 and 2), which showed expression of the EWS-FLI1 fusion gene, the molecular hallmark of ES. A higher expression of EWS-FLI1 gene was evident in patient 1, compared with the expression observed in patient 2, providing a possible explanation to the tenuously expressed band in the FISH results of the latter. In addition, it was also clear that the EWS and FLI1 genes were present in this sample (patient 2), leading to the conclusion that the proportion of cells with the fusion gene in this case could be significantly smaller than that without the translocation. On the other hand, these two genes (EWS and FLI1) were not detectable in the samples of patient 1, reflecting the prevalence of cells carrying the characteristic chromosomal translocation of the disease. In addition, expression of the reciprocal fusion gene FLI1-EWS, whose presence is associated with diagnosis of ES cases with more aggressive behavior, was quantified. This protein collaborates with EWS-FLI1 to deregulate the normal expression of genes associated with differentiation, proliferation and cell survival processes (22). Expression of the FLI1-EWS mature transcript expression was demonstrated in patient 1; however, the translocated chromosome encoding FLI1-EWS is likely lost in a small subset of cells, contributing to the genetic heterogeneity of the sarcoma (22). As a complement to the cytogenetic and molecular assays that contributed to the formation of the differential diagnosis of the disease, immunohistochemical analysis was also carried out. Immunohistochemical labeling for CD99 and FLI1 was positive in both cases, in agreement with the main hypothesis that both samples were framed in a cellular context of ES. The clinical-molecular analysis ended up ruling out the fact that both samples are clear cases of ES, particularly patient 1, whose tumor corresponded to ES with primary origin in lung tissue, a case rarely reported in the literature.
ChIP assays were performed using a set of primers (set1EWS) directed to the EWS1 gene promoter. The primers were designed as part of the study by Jacques et al (25), a product of the identification of sections in EWS-FLI1 gene highly enriched by transcription factors following a ChIPseq analysis. This analysis involved the detection of covalent modifications of histone H3, including methylation (H3K4Me3) and acetylation events (H3K27ac and H3K9ac), characteristic markers of gene activation, as well as trimethylation of lysine 27 (H3K27Me3) and trimethylation of lysine 9 (H3K9Me3), two transcriptional repressor markers.
A significant enrichment of H3K4Me3 was found in the ES A673 cell line and patient 2, compared with that detected in the NT cells, indicating its possible contribution to gene activation in pathological contexts. This modification could not be evaluated in patient 1, as not enough chromatin was available to perform the analysis. On the other hand, H3K27ac proved to be an abundant modification in the region amplified from the A673 cell line, and was also found in patient 1, providing evidence for its possible role in EWS-FLI1 fusion gene activation. H3K9ac was enriched in A673 cells and in the sample of patient 1, compared with that observed in the NT cells. A previous study indicated a probable contribution in the activation of the EWS-FLI1 gene by the covalent histone modifications evaluated in this study (25).
Furthermore, repressive covalent histone modifications H3K27Me3 and H3K9Me3 were also analyzed. Interestingly, low levels of H3K27Me3 were detected in the A673 cell line and in patient 1. However, cells from patient 2 exhibited significant enrichment for this modification, possibly explaining why the expression levels of EWS-FLI1 in this case were lower than in patient 1, noting that clonal heterogeneity of the sample also contributes to this condition. Finally, H3K9Me3, which was completely absent in the A673 cells and patient 1, was highly detected in patient 2 compared with the other samples, reinforcing the arguments regarding the reason EWS-FLI1 was expressed less in this sample.
In general, it is possible to predict the role that histone modifications H3K27ac, H3K9ac and H3K4Me3 may have in the transcriptional activation of EWS-FLI1 fusion gene, since it was possible to detect significant enrichment levels when the promoter was transcriptionally active. However, the enrichment levels of all activating modifications were lower in patient 2 compared with the other ES groups. This result was in agreement with the EWS-FLI1 mRNA expression levels, which were low compared with those in patient 1, as well as with the high genetic and cytogenetic heterogeneity exhibited in the cells analyzed. This also corresponded with the enrichment of two repressive histone modifications (H3K27Me3 and H3K9Me3), which were present to a greater proportion in patient 2 compared with those in the A673 cells and patient 1, where the enrichment levels were low.
Previous findings have established that epigenetic regulation of the EWS-FLI1 promoter is mediated by the presence of reader-type proteins belonging to the bromodomain and extra-terminal domain (BET) family (25). This type of peptide possesses a functional domain, known as a bromodomain, responsible for the identification of highly acetylated sections of the genome. After detection of the activating modification, a set of molecular events is initiated, concluding in the recruitment of transcription factors that promote expression of the fusion gene. Studies using ChIP assays have demonstrated the enrichment of enzymes belonging to the BET family on the same sequences of the EWS-FLI1 gene promoter analyzed during the development of the present study (25). In this context, the covalent histone modifications present on the EWS promoter reported here, especially acetylation, may regulate the transcription of the EWS-FLI1 gene through recruitment of BET family proteins, which are responsible for triggering the first activation signaling cascades of the EWS-FLI1 gene, as described by Jacques et al (25).
The hallmark of ES tumors is a translocation between chromosomes 11 and 22, resulting in a fusion protein, commonly referred to as EWS-FLI1. The expression of the aberrant EWS-FLI1 transcription factor is responsible for the activation of signaling pathways involved in cancer (11). Herein, it is reported that the transcriptional activation and repression of the EWS-FLI1 fusion gene is accompanied by selective deposition and elimination of histone markers during ES disease. These epigenetic profiles are mediated by the enrichment of H3K4Me3, H3K9ac and H3K27ac when the promoter is active. By contrast, when the promoter is repressed, there is H3K27Me3 and H3K9Me3 enrichment.
The present study is a product of transdisciplinary work by individuals belonging to areas of clinical research and basic science, two bodies of knowledge that complement each other and contribute to the realization of more accurate diagnoses.
The present study demonstrates the possible potential of clinical-molecular analysis in the understanding of ES, in order to develop more efficient treatments that direct their therapeutic strategies to the molecular level.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from Hospital Universitario San Ignacio (2017–2018) and Pontificia Universidad Javeriana (PUJ 7711 and PUJ 8530).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CM, LR and AR designed and performed the experiments and analyzed the data. DT, MJF and AC performed the clinical analysis. JR performed the pathological analysis. OM supervised Cytogenetic Analysis. BH and AR designed and supervised the ChIP experiments. A.R. supervised the research and analyzed the data. C.M., A.R., B.H., and O.M. wrote the main parts of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was conducted under the 1993 guidelines established by the Colombian Ministry of Health and Social Protection (resolution no. 008430), and following the ethical principles for medical investigation with human beings, according to the Ethical Committee of the School of Medicine, Javeriana Pontifical University, Bogotá, Colombia (FM-CIE.8148-14).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Desai KI, Nadkarni TD, Goel A, Muzumdar DP, Naresh KN, Nair CN. Primary Ewing's sarcoma of the cranium. Neurosurgery. 2000;46:62–68. doi: 10.1097/00006123-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Deokar K, Nana GK, Shivhari G. Primary ewings sarcoma of the lung. J Clin Diagn Res. 2015;9:XD01–XD03. doi: 10.7860/JCDR/2015/10946.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Alava E. Patología molecular de los srcomas. Oncología (Barcelona) 2005;28:22–38. doi: 10.4321/S0378-48352005000900003. (In Spain) [DOI] [Google Scholar]
- 4.Kumar N, Gupta B. Global incidence of primary malignant bone tumors. Curr Orthopaedic Practice. 2016;27:5. doi: 10.1097/BCO.0000000000000405. [DOI] [Google Scholar]
- 5.Soto C, Gómez L, Criollo F, Romo R, Messa Ó, Arbeláez P. Sarcoma de Ewing de la falange proximal del meñique. Reporte de caso. Revista Colombiana de Cancerología. 2014;18:137–142. doi: 10.1016/j.rccan.2014.04.005. (In Spain) [DOI] [Google Scholar]
- 6.Dynamed, corp-author. Ewing sarcoma in children. https://www.dynamed.com/condition/ewing-sarcoma-in-children Available at:
- 7.Desai SS, Jambhekar NA. Pathology of Ewing's sarcoma/PNET: Current opinion and emerging concepts. Indian J Orthop. 2010;44:363–368. doi: 10.4103/0019-5413.69304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko Y, Kobayashi H, Handa M, Satake N, Maseki N. EWS-ERG fusion transcript produced by chromosomal insertion in a Ewing sarcoma. Genes Chromosomes Cancer. 1997;18:228–231. doi: 10.1002/(SICI)1098-2264(199703)18:3<228::AID-GCC9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Kim SK, Park YK. Ewing sarcoma: A chronicle of molecular pathogenesis. Hum Pathol. 2016;55:91–100. doi: 10.1016/j.humpath.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Balamuth NJ, Womer RB. Ewing's sarcoma. Lancet Oncol. 2010;11:184–192. doi: 10.1016/S1470-2045(09)70286-4. [DOI] [PubMed] [Google Scholar]
- 11.Franzetti GA, Laud-Duval K, Bellanger D, Stern MH, Sastre-Garau X, Delattre O. MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor. Oncogene. 2012;32:3915–3921. doi: 10.1038/onc.2012.403. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto Y, Tanaka K, Nakatani F, Matsunobu T, Matsuda S, Iwamoto Y. Downregulation and forced expression of EWS-Fli1 fusion gene results in changes in the expression of G(1)regulatory genes. Br J Cancer. 2001;84:768–775. doi: 10.1054/bjoc.2000.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Randall L, Calvert G, Spraker H, Lessnick S, Sarcoma de Ewing. Diagnóstico, tratamiento y pronóstico. http://sarcomahelp.org/translate/es-sarcoma-ewing.html. [Jan 18;2018 ];Liddy Shriver Sarcoma Initiative. Available at:
- 14.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: Promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavalheiro dos Santos D, da Cruz Evangelista L, Kerche-Silva L. Genetic alterations and diagnosis in Ewing sarcoma: A review. http://www.oatext.com/genetic-alterations-and-diagnosis-in-ewing-sarcoma-a-review.php. [Apr 5;2018 ];Oatext.com (2017) Available at:
- 16.Parrish JK, Sechler M, Winn RA, Jedlicka P. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing sarcoma. Oncogene. 2013;34:257–262. doi: 10.1038/onc.2013.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocchi A, Manara MC, Sciandra M, Zambelli D, Nardi F, Nicoletti G, Garofalo C, Meschini S, Astolfi A, Colombo MP, et al. CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis. J Clin Invest. 2010;120:668–680. doi: 10.1172/JCI36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herreño AM, Fernández MJ, Rey L, Mejia JA, Cañas A, Moreno OM, Henríquez B, Montecino MA, Rojas AP. Primary lung cancer cell culture from transthoracic needle biopsy samples. Cogent Med. 2018;5:1. doi: 10.1080/2331205X.2018.1503071. [DOI] [Google Scholar]
- 19.Vural C, Uluoğlu O, Akyürek N, Oğuz A, Karadeniz C. The evaluation of CD99 immunoreactivity and EWS/FLI1 translocation by fluorescence in situ hybridization in central PNETs and Ewing's sarcoma family of tumors. Pathol Oncol Res. 2011;17:619–625. doi: 10.1007/s12253-010-9358-3. [DOI] [PubMed] [Google Scholar]
- 20.Moorhead PS, Nowell PC, Mellman WJ, Battips DM, Hungerford DA. Chromosome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res. 1960;20:613–616. doi: 10.1016/0014-4827(60)90138-5. [DOI] [PubMed] [Google Scholar]
- 21.McGowan-Jordan J, Simons A, Schmid M, editors. An International System for Human Cytogenomic Nomenclature (2016) Karger; Basel: 2016. ISCN 2016. [DOI] [Google Scholar]
- 22.Elzi DJ, Song M, Houghton PJ, Chen Y, Shiio Y. The role of FLI-1-EWS, a fusion gene reciprocal to EWS-FLI-1, in Ewing sarcoma. Genes Cancer. 2015;6:452–461. doi: 10.18632/genesandcancer.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Przybyl J, Kozak K, Kosela H, Falkowski S, Switaj T, Lugowska I, Szumera-Cieckiewicz A, Ptaszynski K, Grygalewicz B, Chechlinska M, et al. Gene expression profiling of peripheral blood cells: New insights into Ewing sarcoma biology and clinical applications. Med Oncol. 2014;31:109. doi: 10.1007/s12032-014-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abcam (2011). A-beginners-guide-to-ChIP. http://docs.abcam.com/pdf/chromatin/A-beginners-guide-to-ChIP.pdf. [Jan 22;2018 ]; Available at.
- 25.Jacques C, Lamoureux F, Baud'huin M, Rodriguez Calleja L, Quillard T, Amiaud J, Tirode F, Rédini F, Bradner JE, Heymann D, Ory B. Targeting the epigenetic readers in Ewing Sarcoma inhibits the oncogenic transcription factor EWS/Fli1. Oncotarget. 2016;7:24125–24140. doi: 10.18632/oncotarget.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.