Abstract
Actinobacteria is a goldmine for the discovery of abundant secondary metabolites with diverse biological activities. This study explores antimicrobial biosynthetic potential and diversity of actinobacteria from Pobitora Wildlife Sanctuary and Kaziranga National Park of Assam, India, lying in the Indo-Burma mega-biodiversity hotspot. A total of 107 actinobacteria were isolated, of which 77 exhibited significant antagonistic activity. 24 isolates tested positive for at least one of the polyketide synthase type I, polyketide synthase type II or non-ribosomal peptide synthase genes within their genome. Their secondary metabolite pathway products were predicted to be involved in the production of ansamycin, benzoisochromanequinone, streptogramin using DoBISCUIT database. Molecular identification indicated that these actinobacteria predominantly belonged to genus Streptomyces, followed by Nocardia and Kribbella. 4 strains, viz. Streptomyces sp. PB-79 (GenBank accession no. KU901725; 1313 bp), Streptomyces sp. Kz-28 (GenBank accession no. KY000534; 1378 bp), Streptomyces sp. Kz-32 (GenBank accession no. KY000536; 1377 bp) and Streptomyces sp. Kz-67 (GenBank accession no. KY000540; 1383 bp) showed ~89.5% similarity to the nearest type strain in EzTaxon database and may be considered novel. Streptomyces sp. Kz-24 (GenBank accession no. KY000533; 1367 bp) showed only 96.2% sequence similarity to S. malaysiensis and exhibited minimum inhibitory concentration of 0.024 µg/mL against methicilin resistant Staphylococcus aureus ATCC 43300 and Candida albicans MTCC 227. This study establishes that actinobacteria isolated from the poorly explored Indo-Burma mega-biodiversity hotspot may be an extremely rich reservoir for production of biologically active compounds for human welfare.
Subject terms: Drug discovery, Ecology, Microbiology
Introduction
Since ancient times, mankind has been exploring nature for bioactive organisms to treat different ailments. More than 100000 natural products have been identified in the last 150 years, which include highly assorted chemical classes such as polyketides, alkaloids, non-ribosomal peptides, isoprenoids or phenylpropanoids1,2. Microbial biodiversity can provide us with the richest and the most versatile reservoir of potentially active natural products. A large portion of genomes from beneficial microbes are dedicated to the production of these valuable natural products. A single microbe is capable of making 30–50 natural product compounds3. There are approximately 1 million natural products out of which about 25% are biologically active showing either positive activity or toxicity. Among the prospective sources of natural products, bacteria are prolific sources and a vast majority of these products are produced by a phylum of Gram-positive bacteria known as the “Actinobacteria”4–6.
Actinobacteria are diverse group of bacteria with high content of guanosine-cytosine (65–75%) ranging from 2.5–9.7 Mb genome size7. Among actinobacteria, about 75% of antibiotics such as ivermectin, streptomycin, nystatin and tetracycline are produced by the microbial world’s richest antibiotic-producing family known as “Streptomycetes” 8,9. Drug resistance has been found in many bacterial pathogens due to the misuse or overuse of antibiotics over a prolonged period. The number of fungal infections is also increasing. The economic burden associated with these infections is immense and pharmaceutical industries have been reluctant to invest in antibiotic research and development10,11. For the development of novel pharmaceuticals, new biologically active compounds have to be found. This necessity has led to the search for new bioactive compounds among poorly explored habitats which can efficiently target these life-threatening pathogens12. Hence, actinobacteria may be a potential solution to these problems. In order to find new microbial products derived from newly identified activities of actinobacteria, the search has to be shifted from routinely explored ecological niches to unexplored ones13,14.
Northeast India, a part of the Indo-Burma mega-biodiversity hotspot, is well known for its rich biodiversity. The region has diverse climatic, edaphic and altitudinal variations resulting in wide ecological habitats. Northeast India is the connection between the Indian, Indo-Malayan and Indo-Chinese bio-geographic regions. It is home to a wide spectrum of India’s flora and fauna15. Unlike the floral and faunal diversity, the microbial diversity is also relatively unexplored in this part of the world. The local environment may influence the evolution of novel secondary metabolic pathways in organisms found in the biodiversity hotspots16. Forests are considered to be the most bio-diversified terrestrial ecosystems on earth. They are the largest possible resource available to obtain novel microorganisms and their valuable natural products17,18. Quite a few researchers have reported actinobacteria from the forest ecosystems of Northeast India for the search of natural products endowed with antimicrobial activity19–26. However, the literature review reveals that Pobitora Wildlife Sanctuary, located in Assam, India is yet an unexplored forest ecosystem and we had previously reported regarding the bioactivity prospective of its microflora24. Thus, this forest ecosystem can be considered as an unexplored source of actinobacteria producing bioactive metabolites. Regarding the study of actinobacteria from Kaziranga National Park of Assam, this forest has been previously explored by few researchers and as such, holds a lot of promise for isolation of actinobacteria having pharmaceutical potential19,20.
In the light of the above studies, the present investigation was undertaken with an aim to isolate actinobacteria from Pobitora Wildlife Sanctuary (26 °12′ to 26 °16′N and 91 °58′ to 92 °05′E) and Kaziranga National Park (26 °30′ to 26 °45′N; 93 °08′ to 93 °36′E) of Assam, India and screening them against an array of microbial pathogens responsible for human pathogenesis. Emphasis was given for screening of the isolates for the presence of antibiotic biosynthetic genes and the different chemical classes of antimicrobial compounds they produce were also predicted using DoBISCUIT. Analysis of genetic diversity of the actinobacteria isolates was carried out by 16S rDNA-ARDRA. An attempt was made to study a promising actinobacteria exhibiting potent antimicrobial potential against a wide range of microorganisms.
Results
Isolation of actinobacteria
A total of 107 presumptive actinobacteria of different phenotypes were isolated from different environmental sites of Pobitora Wildlife Sanctuary (n = 54) and Kaziranga National Park (n = 53) of Assam, India. The details of the nature of soil samples, its pH and the number of actinobacteria isolated are given in Table 1. They were associated with scanty to profuse sporulating capacity and showed the presence of distinctive colonial morphology, mycelia colour and pigment production (See Supplementary Table S1). The white colour series was found to be the most dominant one (n = 48; 44.8%). The light microscopy results showed the spiral chain morphology of the aerial mycelia (See Supplementary Fig. S1).
Table 1.
Description of soil samples and number of actinobacteria isolated from each sample.
| Sample No. | Type of soil sample | Soil pH | No. of actinobacteria |
|---|---|---|---|
| Pobitora Wildlife Sanctuary, Assam, India | |||
| Sample 1 | Grass rhizosphere soil | 5.7 | 13 |
| Sample 2 | Leaf litter soil | 4.6 | 8 |
| Sample 3 | Tree rhizosphere soil | 4.9 | 17 |
| Sample 4 | Sediment soil | 4.5 | 16 |
| Kaziranga National Park, Assam, India | |||
| Sample A | Leaf litter soil | 5.3 | 10 |
| Sample B | Tree rhizosphere soil | 4.5 | 9 |
| Sample C | Sediment soil | 5.6 | 10 |
| Sample D | Tree rhizosphere soil | 6.0 | 8 |
| Sample E | Leaf litter soil | 5.6 | 16 |
| Total presumptive actinobacteria | 107 | ||
Antimicrobial assay of actinobacteria
During preliminary screening, all the 107 actinobacteria were assessed against four test microorganisms for their potential to produce antimicrobials. 77 actinobacteria (71.9%) exhibited positive activity out of which 39 isolates are from Pobitora Wildlife Sanctuary and 38 isolates are from Kaziranga National Park. A total of 51 isolates (66%) showed antimicrobial activity against Staphylococcus aureus MTCC 96 with maximum zone of inhibition (70 ± 1.3) mm by Kz-32. 49 isolates (64%) exhibited antimicrobial activity against methicilin resistant Staphylococcus aureus (MRSA) ATCC 43300 with maximum zone of inhibition of (56 ± 1) mm by Kz-24. Against Escherichia coli MTCC 40, 59 isolates (77%) showed antimicrobial activity with highest zone of inhibition of (56 ± 0.8) mm diameter by PB-65. 60 isolates (78%) exhibited antimicrobial activity against Candida albicans MTCC 227 where highest inhibition zone was observed by Kz-24 with (52 ± 1.8) mm. Furthermore, 29 isolates (37.6%) showed antimicrobial activity against all the four test microorganisms. Results of antimicrobial activity screening of actinobacteria by spot inoculation method are shown in Table 2.
Table 2.
In vitro antimicrobial activity of actinobacteria isolated from forest ecosystems of Assam, India by spot inoculation method.
| Sl. No. | Isolate code | *Zone of inhibition (mm) | |||
|---|---|---|---|---|---|
| Bacterial test microorganisms | Fungal test microorganism | ||||
| S. aureus MTCC 96 | MRSA ATCC 43300 | E. coli MTCC 40 | C. albicans MTCC 227 | ||
| 1. | PB-9 | 11a ± 0.5 | ND | 12ab ± 1.2 | ND |
| 2. | PB-10 | ND | ND | ND | 13abcdef ± 1 |
| 3. | PB-15 | 26l ± 1 | 14efgh ± 2 | 18hi ± 1.5 | 20n°p ± 1 |
| 4. | PB-17 | 31mn ± 1 | ND | ND | 11a ± 1 |
| 5. | PB-19 | 14 cdefg ± 1.5 | 11ab ± 1.6 | ND | 13abcdef ± 2 |
| 6. | PB-20 | ND | ND | 12ab ± 0.7 | ND |
| 7. | PB-21 | 32n ± 0.7 | ND | 55x ± 1.5 | 20nop ± 1 |
| 8. | PB-22 | ND | ND | ND | 15efghij ± 1.5 |
| 9. | PB-25 | 20ij ± 1.2 | 11abc ± 1.7 | ND | 15cdefghi ± 1.5 |
| 10. | PB-26 | 13abc ± 1 | ND | 15ef ± 1.7 | ND |
| 11. | PB-27 | ND | ND | 30tu ± 2 | ND |
| 12. | PB-28 | 19hi ± 2 | 20° ± 0.7 | 35v ± 0.5 | 18lmn ± 2.7 |
| 13. | PB-31 | ND | ND | 46w ± 3 | ND |
| 14. | PB-32 | ND | ND | 20ij ± 0.7 | 12abc ± 1 |
| 15. | PB-33 | 18h ± 0.7 | 17jkl ± 1.3 | 20ij ± 1.2 | 12abcd ± 1.2 |
| 16. | PB-39 | 14cde±1 | 12abcd ± 2 | 28qrs ± 0.5 | 14bcdefgh ± 1.7 |
| 17. | PB-43 | 26l ± 1.5 | 18lmn ± 1 | 27pqrs ± 2 | 12ab ± 0.7 |
| 18. | PB-46 | ND | ND | ND | 36x ± 1 |
| 19. | PB-47 | 14bcd ± 1.5 | ND | ND | 16ghijkl ± 1.5 |
| 20. | PB-48 | 15defg ± 1 | 19no ± 1.2 | 30tu ± 1.5 | 40z ± 1.7 |
| 21. | PB-50 | 38p ± 0.7 | ND | 15ef ± 1 | ND |
| 22. | PB-51 | 14cdef ± 1.2 | 15ghij ± 2 | 28rs ± 1 | 17hijklm ± 1.4 |
| 23. | PB-52 | 36o ± 0.8 | 30r ± 0.8 | 31 u ± 1.2 | 27u ± 0.4 |
| 24. | PB-54 | 15cdefg ± 1.2 | 12abcd ± 1.6 | 36 v ± 1 | 14bcdefg ± 2 |
| 25. | PB-55 | ND | ND | 13abc ± 1.3 | 15fghijk ± 1 |
| 26. | PB-56 | 15cdefg ± 2.5 | ND | 14cdef ± 0.5 | 18lmn ± 1 |
| 27. | PB-64 | 26l ± 1.2 | 25p ± 0.5 | 14bcde ± 0.5 | 17ijklm±1.5 |
| 28. | PB-65 | 41q ± 1 | 28q ± 0.7 | 56x±0.8 | 15fghijk ± 1.2 |
| 29. | PB-66 | ND | ND | ND | 37xy ± 1 |
| 30. | PB-68 | 60s ± 0.5 | 13def ± 1.7 | 20ij ± 1 | 18lmn ± 2.7 |
| 31. | PB-70 | 31n ± 1.5 | 30r ± 1.5 | 55x ± 0.7 | 14bcdef ± 2 |
| 32. | PB-75 | 19hi ± 1.7 | ND | 15def ± 1.5 | 19mno ± 1.7 |
| 33. | PB-76 | 16fg ± 1 | 20o ± 1.2 | 13abcde ± 1.7 | 13abcde ± 1.3 |
| 34. | PB-79 | 12ab ± 1 | ND | 25nop ± 2 | ND |
| 35. | PB-81 | 16efg ± 0.7 | 31r ± 1.5 | 19hij ± 1.2 | ND |
| 36. | PB-82 | ND | ND | 16fg ± 1 | ND |
| 37. | PB-83 | 14cdef ± 1.2 | 10a ± 2 | 27opqr ± 1.5 | 17jklm ± 1 |
| 38. | PB-85 | 21jk ± 0.7 | 13bcde ± 1.3 | 46w ± 1.2 | 12abc ± 1 |
| 39. | PB-86 | 19hi ± 1 | 30r ± 1.2 | 30tu ± 2 | 22pqr ± 0.8 |
| 40. | Kz-2 | ND | 13cde ± 1 | 14cdef ± 1.5 | 15fghijk ± 0.5 |
| 41. | Kz-10 | ND | 17klm ± 1.2 | 12a ± 1.3 | 18lmn ± 0.7 |
| 42. | Kz-11 | ND | 15ghij | 16fg ± 0.7 | 17klm ± 1 |
| 43. | Kz-12 | 25l ± 1.5 | 13defg ± 1.5 | 35v ± 0.7 | ND |
| 44. | Kz-13 | 15defg ± 1 | 14efgh ± 1 | 21jk ± 0.7 | 18lmn ± 2 |
| 45. | Kz-14 | ND | 16ijkl ± 1.2 | 26nopq ± 0.7 | 23rs ± 1 |
| 46. | Kz-18 | ND | 16ijkl ± 1.5 | 31tu ± 0.7 | 20nopq ± 1.2 |
| 47. | Kz-21 | ND | 19mno ± 0.5 | 26nopq ± 0.7 | 21opqr ± 1.5 |
| 48. | Kz-23 | 16g ± 1.2 | ND | 20ij | 23qrs ± 0.8 |
| 49. | Kz-24 | 42q ± 2 | 56t ± 1 | 26nopq ± 0.5 | 52Ψ ± 1.8 |
| 50. | Kz-27 | ND | ND | 20ij ± 1 | 25tu ± 1.2 |
| 51. | Kz-28 | 29m ± 0.5 | 25p ± 1 | 24lmn ± 2 | 31v ± 0.7 |
| 52. | Kz-29 | 31mn ± 0.5 | 25p ± 1.5 | 27opqr ± 2 | ND |
| 53. | Kz-31 | ND | ND | ND | 17jklm ± 0.7 |
| 54. | Kz-32 | 70t ± 1.3 | 12abcd ± 0.7 | ND | 30v ± 1.5 |
| 55. | Kz-36 | 51r ± 1 | ND | 29st ± 0.7 | 34w ± 1 |
| 56. | Kz-38 | ND | 15ghij ± 1 | ND | 15defghij ± 0.5 |
| 57. | Kz-41 | 31n ± 1.2 | 16hijk ± 1 | 20ij ± 0.2 | 21opqr ± 1 |
| 58. | Kz-42 | ND | ND | ND | 12ab ± 1.7 |
| 59. | Kz-44 | 20ij ± 1 | 14efghi ± 1.5 | 20ij ± 0.7 | 14cdefghi ± 1.5 |
| 60. | Kz-47 | 20ij ± 2 | 10a | 20ij ± 1 | 40yz ± 1.2 |
| 61. | Kz-49 | 15defg ± 1 | 15ghij ± 1.2 | ND | ND |
| 62. | Kz-52 | 13abc ± 2 | 25p ± 0.5 | ND | 15efghij ± 1.5 |
| 63. | Kz-55 | 31mn ± 0.7 | 16hijk ± 0.8 | 20hij ± 1 | 17ghijkl ± 1.5 |
| 64. | Kz-56 | 21jk ± 1 | 15hijk ± 1.5 | 25mno ± 0.5 | ND |
| 65. | Kz-58 | ND | 16hijk ± 1 | 23lm ± 1.7 | ND |
| 66. | Kz-61 | ND | ND | 18hij ± 0.5 | ND |
| 67. | Kz-62 | 31n ± 1.5 | 37s ± 0.7 | ND | ND |
| 68. | Kz-66 | 19hi ± 1 | 12bcd | 19hij ± 2 | 20nop ± 1.7 |
| 69. | Kz-67 | 22k ± 0.5 | 14efgh ± 1.7 | 26opqr ± 1.2 | 19mno ± 1 |
| 70. | Kz-72 | ND | ND | ND | 21opqr ± 1 |
| 71. | Kz-73 | ND | ND | ND | 22pqrs ± 2 |
| 72. | Kz-74 | 25l ± 1.2 | 15fghij ± 1.7 | 23kl ± 0.8 | 26u ± 1.5 |
| 73. | Kz-75 | ND | ND | ND | 24st ± 1.2 |
| 74. | Kz-76 | 26l ± 0.8 | 12bcd ± 1 | 18gh ± 1 | ND |
| 75. | Kz-78 | 25l | 13cde ± 0.7 | 18gh ± 1.5 | 30v ± 2 |
| 76. | Kz-79 | ND | 15ghij ± 1.2 | 13abcd ± 1 | 17jklm ± 1.6 |
| 77. | Kz-80 | 21ij ± 1.8 | 38s ± 1.2 | 19hij ± 1 | 20nop ± 2 |
The average size of colony of the actinobacteria isolates in GLM agar were (7 ± 2) mm in diameter after 5–10 days of incubation at 28 °C.
*Zone of inhibition by spot inoculation method on GLM agar medium.
Zone of inhibition values are given as mean ± SD (n = 3). Values having different superscripts (a–z, Ψ) differ significantly (P < 0.05).
ND: not detectable.
Based on the preliminary antimicrobial activity, 19 antagonistic actinobacteria were tested against ten test microorganisms for secondary antimicrobial screening in liquid media. The result of antimicrobial activity of ethyl acetate extracted product of actinobacteria is presented in Supplementary Table S2. From the result, it was observed that all the 19 isolates had the ability to inhibit S. aureus MTCC 96, Micrococcus luteus MTCC 1538, E. coli MTCC 40, Pseudomonas aeruginosa MTCC 741 and C. albicans MTCC 227. However, 12 isolates, i.e. PB-15, PB-28, PB-43, PB-48, PB-52, PB-64, PB-65, PB-68, PB-76, Kz-13, Kz-55 and Kz-74 had the ability to inhibit all the test microorganisms. 10% DMSO which served as negative control did not show any antimicrobial activity. Antimicrobial activity of the isolates by spot inoculation method and disc diffusion method against test microorganisms is shown in Supplementary Fig. S2.
Extracellular enzymes production
Out of the 77 antagonistic actinobacteria, 63 (82%) produced amylase, 56 isolates (73%) produced cellulase, 53 isolates (69%) produced protease, 59 isolates (77%) produced lipase and 58 isolates (75%) produced esterase (See Supplementary Table S3). Interestingly, 24 isolates (31%) produced all the five enzymes tested. The detailed data of enzymes production by the isolates is represented by Venn diagram in Supplementary Fig. S3.
Detection and analysis of PKS-I, PKS-II and NRPS genes for prediction of chemical classes
All the 77 antagonistic actinobacteria were evaluated for their biosynthetic potential in terms of natural product drug discovery. 24 isolates indicated the presence of at least one of the PKS-I, PKS-II or NRPS genes. PKS-I genes were detected in 6 isolates, PKS-II in 20 isolates and NRPS genes were detected in 2 isolates. The partial gene sequences of PKS-I, PKS-II and NRPS were deposited in GenBank under the following accession numbers KY073865-KY073869, KY235144-KY235162, KU721842, KU721843, KY271082 and KY274457 (Table 3).
Table 3.
Amino acid sequence similarities of the PKS-I, PKS-II and NRPS genes of the actinobacteria and predicted chemical classes for functional genes.
| Isolate | GenBank accession no. | Top blast match (GenBank accession no) | Similarity (%) | *Predicted pathway product (by DoBISCUIT) | Product classification | Activity reported |
|---|---|---|---|---|---|---|
| PKS-I | ||||||
| PB-10 | KY073865 | Modular polyketide synthase of S. neyagawaensis ATCC 27449 (AAZ94386) | 55 | Concanamycin A | Macrocyclic lactone | Antifungal, Antiprotozoal, Antitumor, Antiviral57 |
| PB-32 | KY073866 | Type I modular polyketide synthase of S. spiroverticillatus (ABW96540) | 55 | Tautomycin | Tetronic Acid Derivative | Antibacterial, Antifungal, Antitumor58 |
| PB-47 | KY073867 | ChlA1 polyketide synthase of S.antibioticus DSM 40725 (AAZ77693) | 58 | Chlorothricin | Tetronic acid derivative | Antibacterial119 |
| PB-52 | KU721843 | NanA8 polyketide synthase of S. nanchangensis NS3226 (AAP42874) | 56 | Nanchangmycin | Polyether | Antibacterial, Insecticidal120, Ionophore121 |
| PB-64 | KY073868 | Modular polyketide synthase of S. avermitilis ATCC 31267 (BAB69192) | 58 | Oligomycin | Macrocyclic lactone | Antifungal, Antitumor27 |
| Kz-24 | KY073869 | RifA polyketide synthase of Amycolatopsis mediterranei S699 (AAC01710) | 68 | Rifamycin | Ansamycin | Antibacterial122 |
| PKS-II | ||||||
| PB-9 | KY235144 | β-ketoacyl synthase of S. echinatus Tu303 (ABL09959) | 71 | Aranciamycin | Anthracycline | Antibacterial, Collagenase inhibitor123 |
| PB-10 | KY271082 | Ketoacyl synthase of S. violaceoruber Tu22 (CAA09653) | 81 | Granaticin | Benzoisochromanequinone | Antibacterial124 |
| PB-15 | KY235145 | Putative ketoacyl synthase of S. fradiae Tu2717 (CAA60569) | 74 | Urdamycin | Angucycline | Antibacterial, Antitumor59 |
| PB-22 | KY235146 | β-ketoacyl synthase of S. arenae DSM 40737 (AAD20267) | 72 | Naphthocyclinone | Naphthoquinone, Isochromanequinone | Antibacterial125 |
| PB-33 | KY235147 | β-ketoacyl synthase of S. coelicolor A3(2) (CAA45043) | 73 | Actinorhodin | Benzoisochromanequinone | Antibacterial126 |
| PB-47 | KY235148 | Ketoacyl synthase of S. argillaceus ATCC 12956 (CAA61989) | 78 | Mithramycin | Aureolic acid | Antibacterial, Antitumor127 |
| PB-48 | KY235149 | Jadomycin polyketide ketosynthase of S. venezuelae ATCC 10712 (AAB36562) | 72 | Jadomycin B | Angucycline | Antibacterial128 |
| PB-64 | KY235150 | Jadomycin polyketide ketosynthase of S. venezuelae ATCC 10712 (AAB36562) | 94 | Jadomycin B | Angucycline | Antibacterial128 |
| PB-65 | KY235151 | Putative ketoacyl synthase of Streptomyces sp. SCC-2136 (CAH10117) | 89 | Sch 47554 | Angucycline | Antifungal8 |
| PB-66 | KY235152 | Putative ketoacyl synthase of S. fradiae Tu2717 (CAA60569) | 95 | Urdamycin | Angucycline, Benzanthraquinone | Antibacterial, Antitumor59 |
| PB-68 | KY274457 | β-ketoacyl synthase of Streptomyces sp. AM-7161 (BAC79045) | 89 | Medermycin | Benzoisochromanequinone | Antibacterial, Antitumor129 |
| PB-70 | KY235153 | AlnL ketoacyl synthase of Streptomyces sp. CM020 (ACI88861) | 74 | Alnumycin | Naphthoquinone, Benzoisochromanequinone related | Antitumor, Topoisomerase inhibitory130 |
| PB-75 | KY235154 | β-ketoacyl synthase of S. nogalater ATCC 27451 (CAA12017) | 79 | Nogalamycin | Anthracycline | Antibacterial, Antitumor131 |
| PB-81 | KY235155 | β-ketoacyl-ACP synthase homolog of S. cyanogenus S136 (AAD13536) | 83 | Landomycin | Angucycline | Antitumor132 |
| Kz-12 | KY235157 | ChaA β-ketoacyl synthase of S. chartreusis HKI-249 (CAH10161) | 68 | Chartreusin | Aromatic polyketide glycoside | Antibacterial, Antitumor60 |
| Kz-13 | KY235158 | β-ketoacyl synthase of S. arenae DSM 40737 (AAD20267) | 75 | Naphthocyclinone | Naphthoquinone, Isochromanequinone | Antibacterial133 |
| Kz-28 | KY235159 | β-ketoacyl synthase I of Streptomyces sp. R1128 (AAG30189) | 70 | R1128 | Anthraquinone | Estrogen receptor antagonist134 |
| Kz-55 | KY235160 | Jadomycin polyketide ketosynthase of S. venezuelae ATCC 10712 (AAB36562) | 84 | Jadomycin B | Angucycline | Antibacterial135 |
| Kz-66 | KY235161 | 3-ketoacyl-ACP synthase of S. griseus ATCC 49344 (AAQ08916) | 70 | Fredericamycin | — | Antibacterial, Antifungal, Antitumor136 |
| Kz-74 | KY235162 | BenA β-ketoacyl synthase of Streptomyces sp. A2991200 (CAM58798) | 71 | Benastatin | Pentangular polyketide | Antibacterial, Apoptosis inducer, glutathione-S-transferase inhibitor137 |
| NRPS | ||||||
| PB-52 | KU721842 | NRPS for virginiamycin S of S. virginiae MAFF 10-06014 (BAF50720) | 40 | Virginiamycin | Streptogramin | Antibacterial138 |
| PB-64 | KY235156 | NRPS peptide synthetase of S. albus JA3453 (ABS90470) | 53 | Oxazolomycin | Polyene-type alkaloid | Antibacterial, Antitumor, Antivirus, Ionophore139 |
These genes were translated to amino acid sequences and the secondary metabolite pathway products were identified using DoBISCUIT database. The genes of all the isolates showed similarities to the phylum actinobacteria at the amino acid level. PKS-I sequences shared 56–68% similarity with their closest matches at the amino acid level in BLASTP search. The amino acid sequences of PB-10, PB-32, PB-47, PB-52, PB-64 and Kz-24 matched with the KS domain and are closely related to the gene product involved in concanamycin A, tautomycin, chlorothricin, nanchangmycin, oligomycin and rifamycin production respectively. PKS-II sequences of 20 actinobacteria shared 68–95% similarity with their closest matches at the amino acid level. Their closest match pathway products were predicted as actinorhodin, naphthocyclinone, jadomycin B, landomycin. These compounds belonged to diverse polyketide groups such as glycosides, anthracycline, naphthoquinone, angucycline, benzoisochromanequinone. Highest sequence identity (95%) at amino acid level was observed in PB-66 which was most closely involved in the synthesis of urdamycin. Adenylation domain of NRPS genes was screened to be positive in PB-52 and PB-64. At the amino acid level, NRPS fragments derived from PB-52 and PB-64 showed 40–53% similarity to their closest relatives. The closest match pathway products of PB-52 and PB-64 isolates were analyzed as virginiamycin and oxazolomycin belonging to streptogramin and polyene-type alkaloid group of compounds respectively (Table 3). Interestingly, PB-64 harbors all the three antibiotic biosynthetic genes, i.e. PKS-I, PKS-II and NRPS. This isolate had the ability to inhibit all the test microorganisms during in vitro antimicrobial screening.
Amplified Ribosomal DNA Restriction Analysis (ARDRA)
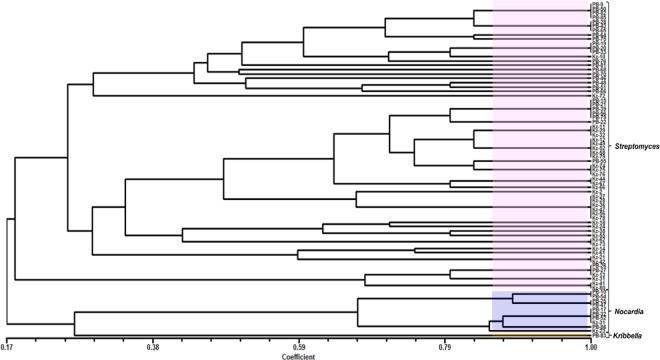
The restriction digestion profile of the 77 antagonistic actinobacteria were analysed by ARDRA fingerprinting analysis using HinfI restriction endonuclease. Digestion with Hinf1 showed different restriction banding patterns and the dendrogram was constructed. Fragments which were smaller than 100 bp in size could not be reproducibly visualized and so not considered. Critical analysis of the dendrogram obtained from Unweighted Pair Group Method with Arithmetic Mean (UPGMA) revealed three major clusters consisting of Streptomyces sp. Nocardia sp. and Kribbella sp. Overall analysis of the soil samples (Sample 1–4 and A–E) from the two forest ecosystems, indicated Streptomyces as the dominant genus in these habitats (Fig. 1).
Figure 1.
UPGMA dendrogram showing the clustering of 77 antagonistic actinobacteria isolates based on the 16S rDNA generated from ARDRA by Jaccard’s coefficient using NTSYS-pc 2.02e. The scale on the x-axis refers to the similarity coefficient.
16S rDNA sequence analysis
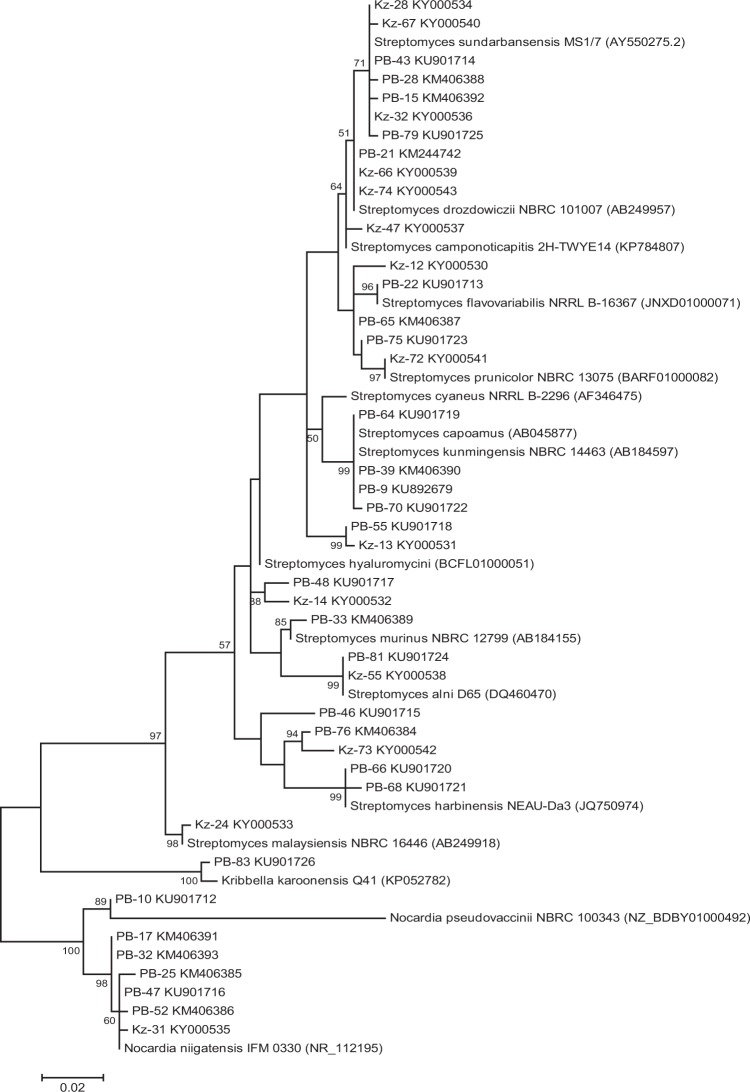
The 16S rDNA of 41 representative actinobacteria (600–1388 bp) exhibited low to high level of sequence similarity (89–100%) to the 16S rDNA of their closest homolog match using EzTaxon database. 4 strains, namely, Streptomyces sp. PB-79 (GenBank accession no. KU901725; 1313 bp), Streptomyces sp. Kz-28 (GenBank accession no. KY000534; 1378 bp), Streptomyces sp. Kz-32 (GenBank accession no. KY000536; 1377 bp) and Streptomyces sp. Kz-67 (GenBank accession no. KY000540; 1383 bp) showed very low levels of sequence similarities (~89.5%) to their nearest type strain. Taxonomic identification of the representative isolates revealed that Streptomyces was the predominant genus (n = 33), followed by Nocardia (n = 7) and Kribbella (n = 1). The phylogenetic tree was constructed using MEGA (Molecular Evolutionary Genetics Analysis) version 6 based on maximum likelihood method represented in Fig. 2. The partial 16S rDNA sequences of the actinobacteria have been deposited in GenBank database under the following accession numbers: KM406384-KM406393, KU901712-KU901726, KY000530-KY000543, KM244742 and KU892679.
Figure 2.
Phylogenetic tree of actinobacteria isolated from forest ecosystems and the closest type strains based on the 16S rDNA sequences by maximum likelihood method using Kimura-2 parameter model. Numbers at branches indicate bootstrap values in 1,000 replicates. Bar, 0.02 substitutions per nucleotide position.
Kz-24 was selected based on its promising antimicrobial potential against test microorganisms and the partial 16S rDNA sequence (1367 bp) was deposited to NCBI GenBank with accession number KY000533. The strain indicated maximum 16S rDNA similarity (96.2%) with S. malaysiensis NBRC 16446 (AB249918). The phylogenetic tree also showed the closest similarity to S. malaysiensis based on maximum likelihood method (Fig. 2). The phenotypic and genomic data showed that Kz-24 belonged to genus Streptomyces and was thus, referred to as Streptomyces sp. strain Kz-24.
Minimum inhibitory concentration (MIC) of EA-Kz-24
Broth dilution method was used to determine MIC of EA-Kz-24 ranging from 100-0.024 μg/mL against all test microorganisms (Table 4). EA-Kz-24 exhibited lowest MIC against MRSA ATCC 43300 and C. albicans MTCC 227 (0.024 μg/mL) whereas highest was recorded against S. marcescens MTCC 97 (50 μg/mL). According to Clinical and Laboratory Standards Institute (CLSI) recommendations for MIC, S. marcescens MTCC 97 were found to be resistant to EA-Kz-24 (MIC: 50 μg/mL), as ≤8 μg/mL was taken as susceptible, ≤16 μg/mL as intermediate and ≥32 μg/mL as resistant.
Table 4.
MIC (µg/mL) of EA-Kz-24 by broth dilution method.
| Test microorganisms | MIC of EA-Kz-24 (µg/mL) | MIC of Rif (µg/mL) | MIC of Strep (µg/mL) | MIC of Amp B (µg/mL) |
|---|---|---|---|---|
| Gram-positive bacteria | ||||
| S. aureus MTCC 96 | >0.098 | >1.95 | >6.25 | NA |
| S. aureus MTCC 3160 | >0.195 | >1.95 | >12.5 | NA |
| S. epidermidis MTCC 435 | >0.78 | >3.125 | >6.25 | NA |
| B. subtilis MTCC 441 | >0.048 | >1.95 | >6.25 | NA |
| B. cereus MTCC 1272 | >0.78 | >6.25 | >12.5 | NA |
| B. megaterium MTCC 8075 | >0.048 | >3.12 | >3.12 | NA |
| M. luteus MTCC 1538 | >0.39 | >0.97 | >6.25 | NA |
| MRSA ATCC 43300 | >0.024 | >25 | >50 | NA |
| Gram-negative bacteria | ||||
| E. coli MTCC 40 | >0.39 | >6.25 | >3.12 | NA |
| E. coli MTCC 739 | >1.56 | >50 | — | NA |
| S. marcescens MTCC 97 | >50 | >12.5 | >3.12 | NA |
| K. pneumoniae MTCC 3384 | >6.25 | >25 | — | NA |
| K. pneumonia ATCC 13883 | >25 | >50 | — | NA |
| P. aeruginosa MTCC 741 | >3.125 | >25 | >25 | NA |
| P. aeruginosa MTCC 424 | >12.5 | — | >12.5 | NA |
| P. aeruginosa MTCC 2582 | >3.125 | >50 | >25 | NA |
| P. vulgaris MTCC 426 | >25 | >25 | >6.25 | NA |
| Yeast | ||||
| C. albicans MTCC 227 | >0.024 | NA | NA | >0.97 |
| C. tropicalis MTCC 2208 | >0.78 | NA | NA | >0.48 |
| C. albicans ATCC 10231 | >0.098 | NA | NA | >1.95 |
Rif: Rifampicin (antibacterial agent); Strep: Streptomycin (antibacterial agent); Amp B: Amphotericin B (antifungal agent); NA: not applicable; -: No activity.
SEM analysis
SEM was performed for the assessment of antibacterial and anti-candidal activity of EA-Kz-24 against MRSA ATCC 43300 and C. albicans MTCC 227. SEM indicated significant morphological changes in the cells of the test microorganisms including cell deformity and shrinkage leading to prominent loss and integrity of cell shape after treatment with 1 × MIC EA-Kz-24. 10% DMSO-treated control cells appeared smooth with intact cell surface (Fig. 3).
Figure 3.
Scanning electron micrograph showing the effect of 1×MIC EA-Kz-24 against MRSA ATCC 43300 (A) without treatment, (B) treatment with EA-Kz-24; and against C. albicans MTCC227 (C) without treatment, (D) treatment with EA-Kz-24.
GC-MS analysis
Chemical composition of EA-Kz-24 was characterized with GC-MS. Based on the retention time, molecular weight and molecular formula, thirteen chemical compounds were identified by comparing their mass spectra with the NIST library as shown in Table 5. The peak area of the compound is directly proportional to its antimicrobial metabolite quantity (See Supplementary Fig. S4).
Table 5.
Chemical compounds detected in EA-Kz-24 by GC-MS analysis.
| Sl no | Compound name | RT (min) | MW (g/mol) | Formula | Abundance% | Nature of compound | Activity |
|---|---|---|---|---|---|---|---|
| 1 | (Z)-3-Tridecene | 18.27 | 182.34 | C13H26 | 1.23 | Alkene | No activity reported |
| 2 | 3,5-bis(1,1-dimethylethyl)-phenol | 19.93 | 206.32 | C14H22O | 6.72 | Phenolic compound | Antimicrobial24 |
| 3 | Benzoic acid,4-ethoxy-ethyl ester | 20.13 | 194.22 | C11H14O3 | 1.66 | Benzoic acid ester | Antimicrobial91 |
| 4 | (Z)-3-Tetradecene | 20.94 | 196.37 | C14H28 | 2.61 | Alkene | Antimicrobial88 |
| 5 | Dodecyl acrylate | 22.13 | 240.38 | C15H28O2 | 12.39 | Ester | Antibacterial90 |
| 6 | Propanoic acid, decyl ester | 22.21 | 214.34 | C13H26O2 | 3.06 | Ester | No activity reported |
| 7 | N-acetyl-3-methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione | 22.56 | 210.22 | C10H14N2 O3 | 2.01 | Piperazine | No activity reported |
| 8 | Hexahydro-pyrrolo[1,2-a]pyrazine-1,4-dione | 23.12, 29.46 | 154.16 | C7H10N2O2 | 22.91 | Pyrrolizidine | Antimicrobial140 |
| 9 | (E)-9-Octadecene | 23.32 | 252.47 | C18H36 | 1.71 | Alkene | Antimicrobial87,89 |
| 10 | Hexahydro-3-(2-methylpropyl)-pyrrolo[1,2-a]pyrazine-1,4-dione | 23.77, 24.07, 24.83, 25.01, 25.11, 25.18 | 210.27 | C11H18N2O2 | 37.27 | Pyrrolizidine | Antimicrobial140 |
| 11 | Propanoic acid,3-mercapto-dodecyl ester | 25.74 | 274.46 | C15H30O2S | 1.01 | Ester | No activity reported |
| 12 | 3-(phenylmethyl)-2,5-piperazinedione | 27.58 | 204.22 | C11H12N2O2 | 1.41 | Piperazinedione | No activity reported |
| 13 | Hexahydro-3-(phenylmethyl)-pyrrolo[1,2-a]pyrazine-1,4-dione | 29.07 | 244.28 | C14H16N2O2 | 6.00 | Pyrrolizidine | Antimicrobial85,140 |
RT is retention time; MW is molecular weight of compounds.
Discussion
Actinobacteria have been recognized as the most proliferant producers of natural bioactive compounds like antimicrobials, wide range of enzymes and valuable secondary metabolites with incredible diversity of biological activities27–29. Our primary goal was to study the functionality of culturable actinobacteria from the Indo-Burma mega-biodiversity hotspot with key emphasis towards understanding their biosynthetic potential.
In total, 107 presumptive actinobacteria were isolated from nine varied soil samples collected from Pobitora Wildlife Sanctuary and Kaziranga National Park of Assam, India. Forest is considered as a wealthy biological diversity with millions of animals, plants and microorganisms30,31. Soil samples are known to be rich in organic matter and microorganisms where most of the biological activities occur. A total of 34 actinobacteria were isolated from leaf litter soil and tree rhizosphere soil each, 26 from sediment soil and only 13 isolates from grass rhizosphere soil. The pH range of the soil samples was found to be in the range 4.5–6.0 implying to acidic soil pH of the forest ecosystems. Forest soils are previously reported to be of low pH32. Actinomycetes isolation agar, Streptomyces agar and GLM agar were used for actinobacteria isolation which was reported previously12,19. Maximum numbers of actinobacteria (n = 52; 49%) were recovered from Actinomycetes isolation agar as this media contains glycerol and asparagine which most actinobacteria use as a source of carbon and nitrogen respectively. Additionally, it also contains sodium propionate which makes it most suitable for the isolation of actinobacteria because it acts as an antifungal agent19. The use of amending the isolation media with amphotericin B and rifampicin has been confirmed to be a good strategy for promotion of growth of slow growing actinobacteria in the absence of fast growing contaminants19,33,34.
Morphological differentiation of all the 107 actinobacteria was done based on their colony morphology, aerial and substrate mycelium colour and diffusible pigment production. The chromogenicity of aerial mycelium is a significant character for grouping of actinobacteria35. Furthermore, 38 actinobacteria produced diffusible pigment which is considered as a characteristic feature for identification and classification of Streptomyces36.
Actinobacteria have been screened from diverse habitats for the search of novel bioactive compounds from last few years37–39. Yet there are very few reports regarding the exploration of actinobacteria from forest ecosystems for production of bioactive compounds. In the preliminary screening by spot inoculation method, 77 actinobacteria (72%) were found to be potential antagonists against at least one of the test microorganisms. A total of 51 isolates exhibited antagonistic action against S. aureus MTCC 96, 49 isolates against MRSA ATCC 43300, 59 isolates against E. coli MTCC 40 and 60 isolates against C. albicans MTCC 227. As reported by Berdy4, while many antibiotic compounds display antimicrobial activity against Gram-positive bacteria, only about 1.5% of those are effective against Gram-negative bacteria. There are limited effective antifungal agents for treating life-threatening fungal infections, which is a major challenge to the pharmaceutical industry40. In the present study, the result of significant antimicrobial activity against Gram-negative bacteria and fungus strongly suggests that forest ecosystems of Assam are a good source of antagonistic actinobacteria exhibiting promising antimicrobial activity.
Out of 77 antagonistic actinobacteria, 63, 56, 53, 59 and 58 number of isolates produced amylase, cellulase, protease, lipase and esterase enzymes respectively. Interestingly, 24 isolates produced all the five enzymes. Ramesh and Mathivanan41 and Meena et al.42 isolated actinobacteria from marine sources with multi-enzyme activity. As per the findings of Ramesh and Mathivanan41, they isolated actinobacteria from marine sources and found that majority of the isolates produced lipase which plays an important role in the degradation of polymers in oceans for adaptation in the extreme environment. However, in this study, 56 actinobacteria isolated from the forest ecosystems produced cellulase. The population of cellulase producing actinobacteria is reasonably high in forest floors and soil for the purpose of soil cycling and decomposition of tough plant materials and woody stems. These actinobacteria are largely responsible for the breakdown of large biopolymers like cellulose, hemicellulose, lignin and chitin32,43,44.
Biosynthetic gene clusters PKS-I, PKS-II and NRPS play a fundamental role in the biosynthesis of microbial natural products45. Out of the 77 actinobacteria exhibiting antimicrobial activity, 24 isolates tested positive for at least one of these biosynthetic genes. This result indicated that 31% of the isolates with antimicrobial potential from the forest ecosystems possessed one of the biosynthetic genes. A total of 6 isolates (8%) were found positive for the presence of PKS-I genes while 20 isolates (26%) were positive for PKS-II genes and NRPS genes were detected in only 2 isolates (3%). These findings were in concurrence with the reports of Lee et al.46, who isolated actinobacteria from mangrove forest soil in Malaysia and reported that PKS-II genes were found to be most frequent among the actinobacteria endowed with antimicrobial activity in forest ecosystems, followed by PKS-I and NRPS genes. Lack of amplification of biosynthetic gene sequences in some of the isolates may be due to absence of these genes in their genome47,48. The absence of PKS and NRPS genes does not detriment the antagonistic activity of the isolates, signifying that additional biosynthetic mechanisms or types of bioactive agents may be involved in the production of antimicrobial activity49. Genus Streptomyces and Nocardia are previously reported as recognized producers of polyketides and nonribosomal peptides24,38. Earlier reports confirm that Streptomyces strains possess multiple copies of PKS and NRPS genes the functions of which are still not explored fully50–52. In the antimicrobial screening, Streptomyces sp. PB-64 exhibited bioactivity against all the test microorganisms and interestingly it indicated the presence of all the three biosynthetic genes i.e. PKS-I, PKS-II and NRPS. Challis53 reported that microbial genome sequences harvest many orphan or cryptic biosynthetic gene clusters which have the capability to direct the synthesis of novel, structurally complex natural products. The secondary metabolite pathway products of these biosynthetic gene clusters were predicted using DoBISCUIT database54. In recent years, prediction of chemical classes have been applied effectively for the discovery of type I and type II polyketides and nonribosomal peptides38,55,56. The top BLAST match of the PKS-I gene sequences of the six actinobacteria isolates PB-10, PB-32, PB-47, PB-52, PB-64 and Kz-24 yielded different pathway products such as concanamycin A, tautomycin, chlorothricin, nanchangmycin, oligomycin and rifamycin respectively classified under macrocyclic lactone, tetronic acid derivative, ansamycin and polyether group of compounds. These compounds are reported to be antimicrobials in nature effective against a wide range of bacterial and fungal pathogens along with their antitumor properties27,57,58. Top BLAST match of the PKS-II gene sequences of the actinobacteria isolates PB-9, PB-10, PB-15, PB-22, PB-33, PB-47, PB-48, PB-64, PB-65, PB-66, PB-68, PB-70, PB-75, PB-81, Kz-12, Kz-13, Kz-28, Kz-55, Kz-66 and Kz-74 yielded significant aromatic polyketides such as naphthocyclinone, jadomycin B, urdamycin, nogalamycin, actinorhodin. classified under the angucycline, anthracycline, naphthoquinone group of compounds. The compounds are reported to possess excellent antimicrobial activity against human disease causing pathogens and antitumor property8,59,60. Top BLAST match pathway products of the NRPS gene sequences of PB-52 and PB-64 yielded virginiamycin and oxazolomycin classified under streptogramin and polyene-type alkaloid group of compounds. Virginiamycin is a PKS/NRPS hybrid showing closest amino acid sequence similarity to PB-52 isolate which indicated the presence of both PKS-I and NRPS during gene-targeted PCR amplification. Additionally, oxazolomycin is also a PKS/NRPS hybrid showing closest amino acid sequence similarity to PB-64 isolate and interestingly, this isolate was positive for the presence of all the three biosynthetic genes. These compounds are reported to be antibacterial in nature61,62.
16S rDNA-ARDRA is an excellent molecular genome typing method for classification of actinobacteria at the genus level63. Comparative diversity analysis of the 77 antagonistic actinobacteria through ARDRA using Hinf1 revealed significant difference among the isolates indicating its true diversity study. The genetic variation among the isolates might be due to mutation or recombination64,65. Natural populations of soil bacterium may exhibit genetic diversity due to variable habitat conditions and soil properties66. Also, the genetic composition and diversity of actinobacteria is influenced by variety in frequency and intensity of competition among locations of isolation67. ARDRA has proved its use in differentiating bacterial species within same genus and bacterial strains within same species68,69. Genetic diversity of the forests soil consisted of Streptomyces, Nocardia and Kribbella sp. which was determined by sequencing of 16S rDNA. Based on ARDRA fingerprinting analysis, strong antimicrobial activity and presence of antibiotic biosynthetic genes (PKS-I, PKS-II and NRPS), 41 isolates were selected as representatives and partially identified by 16S rDNA sequencing. BLAST search results implied that these 41 isolates belonged to the genus Streptomyces (33 isolates), Nocardia (7 isolates) and Kribbella (1 isolate). From the sequencing result, it was clear that Streptomyces was found to be the dominant genus in the soil of protected forest ecosystems which was also reported previously12,19,70,71.
Kz-24 was selected based on its promising antimicrobial potential against the test microorganisms. MIC of EA-Kz-24 support the popular notion that antimicrobial metabolites extracted from Streptomyces sp. at very low concentrations can be one of the finest sources of potent antimicrobials for treatment of infectious diseases especially those caused by clinically resistant pathogens, such as P. aeruginosa, MRSA, C. albicans72. Our finding is similar with the result of Kumar et al.73 where the crude ethyl acetate extract of S. lavendulae SCA5 showed potent antimicrobial action with an MIC of 125 μg/mL against bacteria and MIC against fungi was 31.25 μg/mL. Kz-24 showed 96.2% 16S rDNA sequence similarity with S. malaysiensis NBRC 16446 (AB249918). The aerial mycelium of Kz-24 was brown in colour in GLM agar containing yeast extract/malt extract while S. malaysiensis NBRC 16446 was repoted to be dark grey in a medium with same composition74. S. malaysiensis was previously reported to exhibit strong antifungal activity and low antibacterial activity against B. subtilis NCIB 361074–76. This is the first report on its antibacterial activity against E. coli MTCC 40.
SEM experiments further confirmed that the strong antimicrobial activity of EA-Kz-24 led to significant morphological changes in the selected test microorganisms leading to cell shrinkage and cytosolic loss. These results are in symmetry with the findings of Sharma et al.24, Supaphon et al.77, Nurkanto and Julistiono78.
Actinobacteria are significant producers of bioactive secondary metabolites with different biological activity. There are many reports available for the use of GC-MS to analyze microbial metabolites chemically79–81. In this study, GC-MS analysis was performed on EA-Kz-24 and thirteen chemical compounds with different retention time and abundance were detected. The compounds identified were esters, alkenes, a phenolic compound, diketopiperazine and pyrrolidinopiperazine. Phenolic compounds are known to be powerful antimicrobials and antioxidants because they can reduce free radicals by hydrogen-donating ability82. Studies led by Balachandran et al.83 and Kumar et al.84 exhibited maximum antimicrobial action with highest phenolic compounds in GC-MS fractions. The antimicrobial activity of 3,5-bis(1,1-dimethylethyl)-phenol by Nocardia sp. PB-52 was reported for the first time by our group24. The pyrrolizidine compounds present in EA-Kz-24 included hexahydro-pyrrolo[1,2-a]pyrazine-1,4-dione (22.91%), hexahydro-3-(2-methylpropyl)-pyrrolo[1,2-a]pyrazine-1,4-dione (37.27%) and hexahydro-3-(phenylmethyl)- pyrrolo[1,2-a]pyrazine-1,4-dione (6%). Previous findings conclude that these compounds possess promising antimicrobial activity85,86. These compounds could be the key contributors for potent antimicrobial action of EA-Kz-24. A subset of researchers reported the antagonistic potential of alkenes such as (Z)-3-tetradecene and (E)-9-octadecene against an array of test pathogens87–89 but (Z)-3-tridecene has not been reported for any antimicrobial activity till date. Manilal et al.90 reported antibacterial action of dodecyl acrylate against multidrug-resistant clinical pathogens by Falkenbergia hillebrandii, a red algae. Hussain and Kumaresan91 reported the antifungal nature of benzoic acid,4-ethoxy-ethyl ester. The compounds detected in GC-MS are well known for their ability to inhibit the growth and proliferation of test microorganisms and together may be responsible for the broad-spectrum antimicrobial activity of EA-Kz-24. However, till date, there have been no report on the antimicrobial effect of propanoic acid decyl ester, propanoic acid,3-mercapto-dodecyl ester, 3-(phenylmethyl)-2,5-piperazinedione and N-acetyl-3-methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione which are the key components of EA-Kz-24. Propanoic acid decyl ester and propanoic acid,3-mercapto-dodecyl ester are esters in nature, 3-(phenylmethyl)-2,5-piperazinedione is a piperazinedione compound and N-acetyl-3-methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione lies in the piperazine group of compounds. These compounds together constitute 7.49% of the total constituents present in EA-Kz-24. Previous report by Manilal et al.90 reported strong antimicrobial activity of ester compounds. Niku-Paavola et al.92 reported that a piperazinedione compound, 3-(2-methylpropyl)-2,5-piperazinedione, from Lactobacillus plantarum strongly inhibited the growth of Pantoea agglomerans and Fusarium avenaceum. Musthafa et al.93 reported the effect of 2,5-piperazinedione in reducing the production of quorum sensing dependent factors in Pseudomonas aeruginosa PAO1 both in vivo and in vitro. According to the reports by Jain et al.94, 1-(4-chlorophenyl)-1-propyl piperazine and 1-(4-methylphenyl)-1-propyl piperazine exhibited excellent inhibitory activity against S. aureus and P. aeruginosa respectively. Jadhav et al.95 reported strong antibacterial and antifungal activity by different piperazine group of compounds. Thus, it can be concluded that propanoic acid decyl ester, propanoic acid,3-mercapto-dodecyl ester, 3-(phenylmethyl)-2,5-piperazinedione and N-acetyl-3-methyl-1,4-diazabicyclo[4.3.0]nonan-2,5-dione present in EA-Kz-24 might have a significant role to play in its inhibitory effect against a wide range of test microorganisms which is reported for the first time. The combinatorial effect of bioactive compounds found in GC-MS analysis was demonstrated previously79–81. We, therefore, suggest that these compounds might be the key contributing factor for the antimicrobial action of EA-Kz-24. The forest ecosystems of biodiversity hotspots represent diverse and largely underexplored ecosystem for the isolation of actinobacteria producing effective antimicrobial compounds. It can be inferred from our findings that actinobacteria can be the fundamental sources for the discovery of valuable antibiotic compounds of high industrial and commercial values for human welfare.
Materials and Methods
Sample collection and measurement of soil pH
Soil samples were collected from Pobitora Wildlife Sanctuary and Kaziranga National Park of Assam, India (Fig. 4). Soil samples each weighing ∼50 g were collected randomly in sterile zip-lock plastic bags within 50 m2 area from a depth of 5 to 20 cm after removing the top soil. Four samples from each site were bulked and homogenized to prepare composite samples. pH of the soil samples was also measured41.
Figure 4.
Sampling sites for isolation of actinobacteria are denoted. Soil samples were collected from different environmental sites of Pobitora Wildlife Sanctuary and Kaziranga National Park of Assam, India. (Map has been adapted from https://d-maps.com/index.php?lang = en).
Selective isolation and preliminary identification of actinobacteria
The composite soil samples were suspended in physiological saline (NaCl 9 g/L), thoroughly homogenized by stirring and serial dilutions up to 10−4 were plated on three isolation media: Actinomycetes isolation agar, Streptomyces agar and GLM agar (Yeast extract, 3 g; malt extract, 3 g; peptone Type I, 5 g; starch, 10 g; agar, 20 g; distilled water, 1000 mL) supplemented with amphotericin B (75 µg/mL) and rifampicin (2.5 µg/mL)19,24. The inoculated plates were incubated at 28 °C and examined regularly for the appearance of actinobacteria colonies until 4 weeks.
The pure isolates were grouped according to their colony morphology, colour of aerial and substrate mycelium, colour of diffusible pigments and spore chain morphology96,97. The spore chain morphological features were observed by light microscopy.
Screening for antimicrobial potential
The following test microorganisms were used for the experiment. Staphylococcus aureus MTCC 96, S. aureus MTCC 3160, S. epidermidis MTCC 435, MRSA ATCC 43300, Micrococcus luteus MTCC 1538, Bacillus cereus MTCC 1272, B. subtilis MTCC 441, B. megaterium MTCC 8075 (Gram-positive bacteria); Escherichia coli MTCC 40, E. coli MTCC 739, Klebsiella pneumoniae MTCC 3384, K. pneumoniae ATCC 13883, Serretia marcescens MTCC 97, Proteus vulgaris MTCC 426, Pseudomonas aeruginosa MTCC 741, P. aeruginosa MTCC 2582, P. aeruginosa MTCC 424 (Gram-negative bacteria); Candida albicans MTCC 227, C. albicans ATCC 10231 and C. tropicalis MTCC 2208 (yeasts). MTCC cultures were procured from Microbial Type Culture Collection (CSIR-IMTECH), India and ATCC cultures from HiMedia, India.
The preliminary antimicrobial screening was done by spot inoculation method98 using four test microorganisms. Gram-positive bacterium: Staphylococcus aureus MTCC 96; Gram-positive resistant bacterium: Methicilin resistant Staphylococcus aureus (MRSA) ATCC 43300; Gram-negative bacterium: Escherichia coli MTCC 40; yeast: Candida albicans MTCC 227. The diameter of inhibition zones was determined after 24 hrs99. The experiment was repeated for three times.
The actinobacteria found to be promising in the preliminary antimicrobial screening were subjected to secondary screening by disc diffusion method (Bauer et al., 1966). The following test microorganisms were used for this study: S. aureus MTCC 96, S. epidermidis MTCC 435, Bacillus subtilis MTCC 441, Micrococcus luteus MTCC 1538, MRSA ATCC 43300 (Gram-positive bacteria); E. coli MTCC 40, Serretia marcescens MTCC 97, Klebsiella pneumoniae MTCC 3384, Pseudomonas aeruginosa MTCC 741 (Gram-negative bacteria); and C. albicans MTCC 227 (yeast). Crude antimicrobial extracts were recovered from the culture filtrate by solvent extraction using ethyl acetate (1:1, v/v). The diameters of zone of inhibition were determined after 24 hours. All samples were assayed in triplicate.
Screening for the production of extracellular enzymes
All the 77 antagonistic actinobacteria were screened qualitatively for the production of 5 important extracellular enzymes, i.e. amylase, cellulase, protease, lipase and esterase. Each actinobacteria was spot inoculated on agar plates amended with the respective substrates such as starch, carboxyl methyl cellulose, casein and Tween 80 and Tween 20 and was incubated for up to 10 days at 28 °C100–103.
PCR amplification of biosynthetic genes PKS-I, PKS-II and NRPS and analysis of biosynthetic genes for prediction of chemical classes
Genomic DNA isolation of actinobacteria and PCR amplification of PKS-I and NRPS was performed as described previously24,104. Degenerate primers KSαF (5′-TSG CST GCT TCG AYG CSA TCA-3′) and KSαR (5′-TGG AAN CCG CCG AAB CCG CT-3′) targeting ketosynthase gene of the minimal PKS cluster were used for amplification of PKS-II105. PCR reactions were performed in a final volume of 50 μl in Proflex PCR System (Applied Biosystems, USA). The reaction mixture comprised of template DNA (50 ng), each dNTP (0.2 mM), 1X Taq DNA polymerase buffer, MgCl2 (1.5 mM), each primer (0.2 µM) and 1 U Taq DNA polymerase. The profile used for amplification of PKS-II genes were programmed as: initial denaturation at 94 °C for 5 mins; followed by 35 cycles at 95 °C for I min, 65 °C for 1 min, 72 °C for 2 mins and a final extension at 72 °C for 10 mins. The size of the amplicons was 613 bp (KSαF/ KSαR). The amplified products were determined by 1.8% (w/v) agarose gel electrophoresis and partially sequenced by automated DNA sequencer at Scigenome Labs Pvt. Ltd. (India).
The nucleotide sequences of PKS-I, PKS-II and NRPS were translated into amino acid sequences using the web tool ORF FINDER (http://www.ncbi.nlm.nih.gov/ projects/gorf/). The deduced amino acid sequences were used as queries to search related gene products in the NCBI and DoBISCUIT (Database of BIoSynthesis clusters CUrated and InTegrated, http://www. bio.nite.go.jp/pks/)54 using the BLASTP algorithm106 with default parameters. The secondary metabolites pathway products of these biosynthetic genes were identified using DoBISCUIT.
ARDRA
16S rDNA PCR amplification was carried out as previously described24,107. ARDRA was performed for the identification of number of polymorphic groups and then select the representative actinobacteria isolates among these groups108. 50 ng of purified 16S rDNA PCR products were digested with 1.5 U of Hinf1109,110 following the manufacturer’s protocol (New England Biolabs, UK). The mixture was incubated at 37 °C for 4 hours. Fully digested restriction fragments together with 100 bp and 1 kb markers were resolved by 2% (w/v) agarose gel electrophoresis at 100 volts for 90 min containing 10 μg/mL ethidium bromide. Different ARDRA banding patterns were observed and the isolates were grouped accordingly. ARDRA banding pattern in binary data was graded visually where “1” indicated the presence of band and “0” indicated for absence of band. This binary data was useful for construction of the dendrogram. Similarities among these actinobacteria were calculated by Jaccard’s coefficient in the SimQual program. Similarity index matrix was used to cluster the actinobacteria with SAHN tool based on UPGMA method and the TreePlot program of NTSYS-pc 2.02e software package111.
Identification of actinobacteria by 16S rDNA sequence analysis
Based on the antimicrobial potential, presence of biosynthetic genes and ARDRA, actinobacteria were identified based on the 16S rDNA sequencing using the facility at Scigenom Labs Pvt. Ltd. (India) and Molbiogen (India). Identification of nearest phylogenetic neighbours of sequenced 16S rDNA was carried out using EzTaxon database (http://www. eztaxon. org/)112. The 16S rDNA gene sequences used in the phylogenetic analysis was retrieved from NCBI GeneBank. The aligned sequences were used to reconstruct the phylogeny using maximum likelihood method algorithm by MEGA version 6113. Bootstrap analysis carried out with 1000 replications determined the support of each clade114.
Determination of MIC of Kz-24
MIC of Kz-24 was performed according to CLSI115 and Andrews116 using broth dilution method with slight modifications. To 5 mL of Mueller Hinton broth (for bacterial test organisms) and Sabouraud dextrose broth (for yeasts), 1×105cfu/mL inoculum of test microorganisms (log phase culture) was added in different test tubes and incubated at 37 °C for bacteria (24 hours) and 25 °C for yeasts (48 hours). 10% DMSO was used to dissolve EA-Kz-24 (1 mg/mL) and the extract was prepared for MIC screening with two fold serial dilution (100-0.024 μg/mL). MIC was determined in presence of EA-Kz-24 after 24–48 hours. 10 μL test microorganisms were spread on Mueller Hinton agar /sabouraud dextrose agar plates and observed after 24–48 hours for MIC determination. Control with no antimicrobial agent was turbid (negative control) and the control with standard antibiotic such as rifampicin, streptomycin and amphotericin B was clear (positive control).
Study of antimicrobial effect of EA-Kz-24 by SEM analysis
EA-Kz-24 was studied by SEM for its potential effects on MRSA ATCC 43300 and C. albicans MTCC 227 according to Sharma et al.24. Test microorganisms were treated with 1×MIC EA-Kz-24.
GC-MS analysis of EA-Kz-24
GC-MS was used for the detection of chemical compounds present in EA-Kz-24 as previously described24. The peaks were identified by comparing the mass spectra with the National Institute of Standards and Technology (NIST, USA) library.
Data analysis
All experiments were conducted in triplicate and repeated three times. The data were presented as the mean of three replicates ± standard deviation of mean. Duncan’s multiple range test was done using SPSS (SPSS 18.0, SPSS Inc., Chicago, IL, USA) in the antimicrobial activity data to compare that the sample means differ at a significant level P < 0.05 from each other117. Venn diagram is used for the representation of enzymes production by actinobacteria by using multiple dataset analysis features of VENNTURE software118.
Supplementary information
Acknowledgements
P.S. and D.T. are thankful to the Director, Institute of Advanced Study in Science and Technology (IASST), Assam, India, for providing facilities for this work. P.S. is thankful to Department of Science and Technology (DST), Government of India for the fellowship. This work was partly supported by the Department of Biotechnology (DBT), Govt. of India project (Grant No. BT/PR25029/NER/95/967/2017).
Author contributions
D.T. guided the experimental design and supervised the research work. P.S. conducted the experiments, analyzed the data. P.S. and D.T. prepared the manuscript.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-60968-6.
References
- 1.Micallef ML, D’Agostino PM, Al-Sinawi B, Neilan BA, Moffitt MC. Exploring cyanobacterial genomes for natural product biosynthesis pathways. Mar Genomics. 2015;21:1–12. doi: 10.1016/j.margen.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Carbonell P, et al. Bioinformatics for the synthetic biology of natural products: integrating across the Design-Build-Test cycle. Nat. Prod. Rep. 2016;33:925–932. doi: 10.1039/C6NP00018E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katz L, Baltz RH. Natural product discovery: Past, present and future. J. Ind. Microbiol. Biotechnol. 2016;43:155–176. doi: 10.1007/s10295-015-1723-5. [DOI] [PubMed] [Google Scholar]
- 4.Berdy J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012;65:385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 5.Demain AL. Importance of microbial natural products and the need to revitalize their discovery. J. Ind. Microbiol. Biotechnol. 2014;41:185–201. doi: 10.1007/s10295-013-1325-z. [DOI] [PubMed] [Google Scholar]
- 6.Guo X, et al. Red soils harbor diverse culturable actinomycetes that are promising sources of novel secondary metabolites. Appl. Environ. Microbiol. 2015;81:3086–3103. doi: 10.1128/AEM.03859-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miao V, Davies J. Actinobacteria: The good, the bad, and the ugly. Anton. Leeuw. 2010;98:143–150. doi: 10.1007/s10482-010-9440-6. [DOI] [PubMed] [Google Scholar]
- 8.Basnet DB, et al. Angucyclines Sch 47554 and Sch 47555 from Streptomyces sp. SCC- 2136: cloning, sequencing and characterization. Mol. Cells. 2006;22:154–62. [PubMed] [Google Scholar]
- 9.Hopwood, D. A. Streptomyces in nature and medicine in The antibiotic makers (New York, USA: Oxford University Press, 2007).
- 10.Baltz RH. Marcel Faber Roundtable: Is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 2006;33:507–513. doi: 10.1007/s10295-005-0077-9. [DOI] [PubMed] [Google Scholar]
- 11.Simpkin VL, Renwick MJ, Kelly R, Mossialos E. Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J Antibiot. 2017;70:1087–1096. doi: 10.1038/ja.2017.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari K, Upadhyay DJ, Mosker E, Sussmuth R, Gupta RK. Culturable bioactive actinomycetes from the Great Indian Thar Desert. Ann. Microbiol. 2015;65:1901–1914. doi: 10.1007/s13213-014-1028-3. [DOI] [Google Scholar]
- 13.Debnath R, et al. Psychrotolerant antifungal Streptomyces isolated from Tawang, India and the shift in chitinase gene family. Extremophiles. 2013;17:1045–1059. doi: 10.1007/s00792-013-0587-8. [DOI] [PubMed] [Google Scholar]
- 14.Manikkam R, Venugopal G, Subramaniam B, Ramasamy B, Kumar V. Bioactive potential of actinomycetes from less explored ecosystems against Mycobacterium tuberculosis and other non-mycobacterial pathogens. Int. Sch. Res. Not. 2014;812974:1–9. doi: 10.1155/2014/812974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers N, Russel MA, Cristina MG, Gustavo ABF, Jennifer. K. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 16.Glover, J. B. Applications of fungal ecology in the search of new bioactive natural products. In The Mycota IV: Environmental, Microbial Relationships (eds. Wicklow, D. T.) 249–268 (Berlin Heidelberg: Springer-Verlag, 1995).
- 17.Strobel G, Daisy B, Castillo U, Harper J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004;67:257–268. doi: 10.1021/np030397v. [DOI] [PubMed] [Google Scholar]
- 18.Kim H, Lee J-Y, Lee K-K. Thermal characteristics and bacterial diversity of forest soil in the Haean Basin of Korea. Sci. World J. 2014;2014:247401. doi: 10.1155/2014/247401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thakur D, Yadav A, Gogoi BK, Bora TC. Isolation and screening of Streptomyces in soil of protected forest areas from the states of Assam and Tripura, India, for antimicrobial metabolites. J. Med Mycol. 2007;17:242–249. doi: 10.1016/j.mycmed.2007.08.001. [DOI] [Google Scholar]
- 20.Talukdar M, et al. Bioprospecting Micromonospora from Kaziranga National Park of India and their anti- infective potential. World. J. Microbiol. Biotechnol. 2012;28:2703–2712. doi: 10.1007/s11274-012-1080-8. [DOI] [PubMed] [Google Scholar]
- 21.Sharma P, Das R, Kalita MC, Thakur D. Investigation of extracellular antifungal proteinaceous compound produced by Streptomyces sp. 5K10. Afr. J. Microbiol. Res. 2014;8:986–993. doi: 10.5897/AJMR2013.6264. [DOI] [Google Scholar]
- 22.Gohain A, et al. Antimicrobial biosynthetic potential and genetic diversity of endophytic actinomycetes associated with medicinal plants. FEMS Microbiol. Lett. 2015;362:fnv158. doi: 10.1093/femsle/fnv158. [DOI] [PubMed] [Google Scholar]
- 23.Passari AK, Mishra VK, Saikia R, Gupta VK, Singh BP. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 2015;6:273. doi: 10.3389/fmicb.2015.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma P, Kalita MC, Thakur D. Broad spectrum antimicrobial activity of forest-derived soil actinomycete, Nocardia sp. PB-52. Front. Microbiol. 2016;7:347. doi: 10.3389/fmicb.2016.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Passari AK, et al. Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci. Rep. 2017;7:11809. doi: 10.1038/s41598-017-12235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das R, Romi W, Das R, Sharma HK, Thakur D. Antimicrobial potentiality of actinobacteria isolated from two microbiologically unexplored forest ecosystems of Northeast India. BMC Microbiol. 2018;18:71. doi: 10.1186/s12866-018-1215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omura S, et al. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Nat. Acad. Sci. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berdy J. Bioactive microbial metabolites: a personal view. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 29.Medema, et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lami N, Osada H. Screening of biologically active secondary metabolites from Zairian (Africa) microorganisms. RIKEN. Review. 1993;3:15–16. [Google Scholar]
- 31.Onifade AK. Preliminary studies on the bioactivity of secondary metabolites from Aureobasidium pullulans and Emericela regulosa. J. Anim. Vet. Adv. 2006;5:930–934. [Google Scholar]
- 32.Golinska P, Dahm H. Occurrence of actinomycetes in forest soil. Dendrobiol. 2011;66:3–13. [Google Scholar]
- 33.Seong CN, Choi JH, Baik K-S. An improved selective isolation of rare actinomycetes from forest soil. J. Microbiol. 2001;39:17–23. [Google Scholar]
- 34.Sripreechasak P, et al. Streptomyces siamensis sp. nov., and Streptomyces similanensis sp. nov., isolated from Thai soils. J. Antibiot. 2013;6:633–640. doi: 10.1038/ja.2013.60. [DOI] [PubMed] [Google Scholar]
- 35.Pridham, T. G. & Tresner, H. D. Family VII. Streptomycetaceae. In Bergey’s Manual of Determinative Bacteriology (eds. Buchanan, R. E. & Gibbons, W. E.) 747–748 (Baltimore: Williams & Wilkins, 1974).
- 36.Arai T, Mikami Y. Chromogenicity of Streptomyces. Appl. Microbiol. 1972;23:402–406. doi: 10.1128/AEM.23.2.402-406.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jadoon WA, Nakai R, Naganuma T. Biogeographical note on Antarctic microflorae: Endemism and cosmopolitanism. Geosci. Frontiers. 2012;4:633–646. doi: 10.1016/j.gsf.2012.11.002. [DOI] [Google Scholar]
- 38.Li J, Dong JD, Yang J, Luo XM, Zhang S. Detection of polyketide synthase and nonribosomal peptide synthetase biosynthetic genes from antimicrobial coral associated actinomycetes. Anton. Leeuw. Int. J. 2014;106:623–635. doi: 10.1007/s10482-014-0233-1. [DOI] [PubMed] [Google Scholar]
- 39.Riquelme C, et al. Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Front. Microbiol. 2015;6:1342. doi: 10.3389/fmicb.2015.01342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vicente MF, Basilio A, Cabello A, Pelaez F. Microbial natural products as a source of antifungals. Clin. Microbiol. Infect. 2003;9:15–32. doi: 10.1046/j.1469-0691.2003.00489.x. [DOI] [PubMed] [Google Scholar]
- 41.Ramesh S, Mathivanan N. Screening of marine actinomycetes isolated from the Bay of Bengal, India for antimicrobial activity and industrial enzymes. World J. Microbiol. Biotechnol. 2009;25:2103–2111. doi: 10.1007/s11274-009-0113-4. [DOI] [Google Scholar]
- 42.Meena B, Rajan LA, Vinithkumar NV, Kirubagaran R. Novel marine actinobacteria from emerald Andaman & Nicobar Islands: A prospective source for industrial and pharmaceutical by-products. BMC Microbiol. 2013;13:145. doi: 10.1186/1471-2180-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitra A, Santra SC, Mukherjee J. Distribution of actinomycetes, their antagonistic behaviour and the physic-chemical characteristics of the world’s largest tidal mangrove forest. Appl. Microbiol. Biotechnol. 2008;80:685–695. doi: 10.1007/s00253-008-1626-8. [DOI] [PubMed] [Google Scholar]
- 44.Chanthasena P, Nantapong N. Biodiversity of antimicrobial-producing actinomycetes strains isolated from dry dipterocarp forest soil in northeast Thailand. Braz. Arch. Biol. Technol. 2016;59:e16150674. doi: 10.1590/1678-4324-2016150674. [DOI] [Google Scholar]
- 45.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 46.Lee L-H, et al. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014;2014:1–14. doi: 10.1155/2014/698178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hornung A, Bertazzo M, Dziarnowski A, Schneider K, Welzel K. A genomic screening approach to the structure-guided identification of drug candidates from natural sources. Chem. Bio. Chem. 2007;8:757–766. doi: 10.1002/cbic.200600375. [DOI] [PubMed] [Google Scholar]
- 48.Qin S, et al. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microb. 2009;75:6176–6186. doi: 10.1128/AEM.01034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu L, et al. Diversity of culturable thermophilic actinobacteria in hot springs in Tengchong, China and studies of their biosynthetic gene profiles. Microb. Ecol. 2016;72:150–162. doi: 10.1007/s00248-016-0756-2. [DOI] [PubMed] [Google Scholar]
- 50.Bentley SD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 51.Ikeda H, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 52.Doroghazi JR, Metcalf WW. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics. 2013;14:611–623. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Challis GL. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiol. 2008;154:1555–1569. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- 54.Ichikawa N, et al. DoBISCUIT: a database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2013;41:D408–D414. doi: 10.1093/nar/gks1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gontang EA, Gaudencio SP, Fenical W, Jensen PR. Sequence based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 2010;76:2487–2499. doi: 10.1128/AEM.02852-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodges TW, Slattery M, Olson JB. Unique actinomycetes from marine caves and coral reef sediments provide novel PKS and NRPS biosynthetic gene clusters. Mar. Biotechnol. 2012;14:270–280. doi: 10.1007/s10126-011-9410-7. [DOI] [PubMed] [Google Scholar]
- 57.Haydock SF, et al. Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiol. 2005;151:3161–3169. doi: 10.1099/mic.0.28194-0. [DOI] [PubMed] [Google Scholar]
- 58.Ju J, et al. Functional characterization of ttmM unveils new tautomycin analogs and insight into tautomycin biosynthesis and activity. Org. Lett. 2009;11:1639–1642. doi: 10.1021/ol900293j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Decker H, Haag S. Cloning and characterization of a polyketide synthase gene from Streptomyces fradiae Tu2717, which carries the genes for biosynthesis of the angucycline antibiotic urdamycin A and a gene probably involved in its oxygenation. J. Bacteriol. 1995;177:6126–6136. doi: 10.1128/JB.177.21.6126-6136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Z, Jakobi K, Welzel K, Hertweck C. Biosynthesis of the antitumor agent chartreusin involves the oxidative rearrangement of an anthracyclic polyketide. Chem. Biol. 2005;12:579–588. doi: 10.1016/j.chembiol.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 61.Namwat W, Kamioka Y, Kinoshita H, Yamada Y, Nihira T. Characterization of virginiamycin S biosynthetic genes from Streptomyces virginiae. Gene. 2002;286:283–90. doi: 10.1016/S0378-1119(02)00424-9. [DOI] [PubMed] [Google Scholar]
- 62.Zhao C, et al. Oxazolomycin biosynthesis in Streptomyces albus JA3453 featuring an “acyltransferase-less” type I polyketide synthase that incorporates two distinct extender units. J. Biol. Chem. 2010;285:20097–20108. doi: 10.1074/jbc.M109.090092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cook AE, Meyers PR. Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S rRNA gene restriction fragment patterns. Int. J. Syst. Evol. Microbiol. 2003;53:1907–1915. doi: 10.1099/ijs.0.02680-0. [DOI] [PubMed] [Google Scholar]
- 64.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 65.Oliver A, Canton R, Campo P, Baquero F, Blazquez J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–1254. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- 66.McArthur JV, Kovacic DA, Smith MH. Genetic diversity in natural populations of a soil bacterium across a landscape gradient. Proc. Natl. Acad. Sci. 1988;85:9621–9624. doi: 10.1073/pnas.85.24.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davelos AL, Xiao K, Samac DA, Martin AP, Kinkel LL. Spatial variation in Streptomyces genetic composition and diversity in a Prairie soil. Microb. Ecol. 2004;448:601–612. doi: 10.1007/s00248-004-0031-9. [DOI] [PubMed] [Google Scholar]
- 68.Gurtler V, Wilson VA, Mayall BC. Classification of medically important Clostridia using restriction endonuclease site differences of PCR amplified 16S rDNA. J. Gen. Microbiol. 1991;137:2673–2679. doi: 10.1099/00221287-137-11-2673. [DOI] [PubMed] [Google Scholar]
- 69.Kohler G, Ludwig W, Schleifer KH. Differentiation of lactococci by rRNA gene restriction analysis. FEMS Microbiol. Lett. 1991;84:307–312. doi: 10.1111/j.1574-6968.1991.tb04615.x. [DOI] [PubMed] [Google Scholar]
- 70.Hop DV, et al. Taxonomic and ecological studies of actinomycetes from Vietnam: isolation and genus- level diversity. J. Antibiot. 2011;64:599–606. doi: 10.1038/ja.2011.40. [DOI] [PubMed] [Google Scholar]
- 71.Ding D, et al. Culturable actinomycetes from desert ecosystem in northeast of Qinghai-Tibet Plateau. Ann Microbiol. 2012;63:259–266. doi: 10.1007/s13213-012-0469-9. [DOI] [Google Scholar]
- 72.Oskay M, Tamer AU, Azeri C. Antibacterial activity of some actinomycetes isolated from farming soils of Turkey. Afr. J. Biotechnol. 2004;3:441–446. doi: 10.5897/AJB2004.000-2087. [DOI] [Google Scholar]
- 73.Kumar PS, et al. In vitro antimicrobial, antioxidant and cytotoxic properties of Streptomyces lavendulae strain SCA5. BMC Microbiol. 2014;14:291–302. doi: 10.1186/s12866-014-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Tai A, Kim B, Kim SB, Manfio GP, Goodfellow M. Streptomyces malaysiensis sp. nov., a new streptomycete species with rugose, ornamented spores. Int. J. Syst. Evol. Microbiol. 1999;49:1395–1402. doi: 10.1099/00207713-49-4-1395. [DOI] [PubMed] [Google Scholar]
- 75.Li W, et al. Malayamycin, a new streptomycete antifungal compound, specifically inhibits sporulation of Stagonospora nodorum (Berk) Castell and Germano, the cause of wheat glume blotch disease. Pest Manag. Sci. 2008;64:1294–1302. doi: 10.1002/ps.1632. [DOI] [PubMed] [Google Scholar]
- 76.Cheng J, et al. Azalomycin F complex is an antifungal substance produced by Streptomyces malaysiensis MJM1968 isolated from agricultural soil. J. Korean Soc. Appl. Biol. Chem. 2010;53:545–552. doi: 10.3839/jksabc.2010.084. [DOI] [Google Scholar]
- 77.Supaphon P, Phongpaichit S, Rukachaisirikul V, Sakayaroj J. Antimicrobial potential of endophytic fungi derived from three seagrass species: Cymodoceaserrulata, Halophilaovalis and Thalassiahemprichii. PLoS ONE. 2013;8:72520. doi: 10.1371/journal.pone.0072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nurkanto A, Julistiono H. Screening and study of antifungal activity of leaf litter actinomycetes isolated from Ternate Island, Indonesia. Asian Pac. J. Trop. Med. 2014;7:S238–S243. doi: 10.1016/S1995-7645(14)60239-X. [DOI] [PubMed] [Google Scholar]
- 79.Ser H, et al. Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 2015;6:854. doi: 10.3389/fmicb.2015.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ser H, et al. Evaluation of antioxidative and cytotoxic activities of Streptomyces pluripotens MUSC 137 isolated from mangrove soil in Malaysia. Front. Microbiol. 2015;6:1398. doi: 10.3389/fmicb.2015.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Teng_hern, et al. Investigation of antioxidative and anticancer potentials of Streptomyces sp. MUM256 isolated from Malaysia mangrove soil. Front. Microbiol. 2015;6:1316. doi: 10.3389/fmicb.2015.01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yogeswari S, Ramalakshmi S, Neelavathy R, Muthumary J. Identification and comparative studies of different volatile fractions from Monochaetia kansensis by GCMS. Global J. Pharmacol. 2012;6:65–71. [Google Scholar]
- 83.Balachandran C, Duraipandiyan V, Balakrishna K, Ignacimuthu S. Petroleum and polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalene metabolism in Streptomyces sp. (ERI-CPDA-1) isolated from oil contaminated soil. Bioresour. Technol. 2012;112:83–90. doi: 10.1016/j.biortech.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 84.Kumar PS, Duraipandiyan V, Ignacimuthu S. Isolation, screening and partial purification of antimicrobial antibiotics from soil Streptomyces sp. SCA 7. Kaohsiung J. Med. Sci. 2014;30:435–446. doi: 10.1016/j.kjms.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 85.Dashti Y, Grkovic T, Abdelmohsen UR, Hentschel U, Quinn RJ. Production of induced secondary metabolites by a co-culture of sponge-associated actinomycetes, Actinokineospora sp. EG49 and Nocardiopsis sp. RV163. Mar. Drugs. 2014;12:3046–3059. doi: 10.3390/md12053046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Devi NN, Wahab F. Antimicrobial properties of endophytic fungi isolated from medicinal plant Camellia sinesis. Int. J. Pharma. Bio. Sci. 2012;3:420–427. [Google Scholar]
- 87.Okwu DE, Ighodaro BU. GC-MS evaluation of bioactive compounds and antibacterial activity of the oil fraction from the leaves of Alstonia boonei De Wild. Der. Pharma. Chemica. 2010;2:261–272. [Google Scholar]
- 88.Natarajan V, Dhas ASAG. Phytochemical composition and in vitro antimicrobial, antioxidant activities of ethanolic extract of Leptadenia reticulata [W&A] leaves. Middle East J. Sci. Res. 2014;21:1698–1705. [Google Scholar]
- 89.Cao G, Xu Z, Wu X, Li Q, Chen X. Capture and identification of the volatile components in crude and processed herbal medicines through on-line purge and trap technique coupled with GC × GC-TOF MS. Nat. Prod. Res. 2014;28:1607–12. doi: 10.1080/14786419.2014.929129. [DOI] [PubMed] [Google Scholar]
- 90.Manilal A, Sujith S, Selvin J, Shakir C, Kiran GS. Antibacterial activity of Falkenbergia hillebrandii (Born) from the Indian coast against human pathogens. Int. J. Expt. Botany. 2009;78:161–166. [Google Scholar]
- 91.Hussain AZ, Kumaresan S. GC-MS studies and phytochemical screening of Sesbania grandiflora L. J. Chem. Pharm. Res. 2014;6:43–47. [Google Scholar]
- 92.Niku‐Paavola M‐L, Laitila A, Mattila‐Sandholm T, Haikara A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J. Appl. Microbiol. 1999;86:29–35. doi: 10.1046/j.1365-2672.1999.00632.x. [DOI] [PubMed] [Google Scholar]
- 93.Musthafa KS, Balamurugan K, Pandian SK, Ravi AV. 2,5-piperazinedione inhibits quorum sensing-dependent factor production in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 2012;52:679–686. doi: 10.1002/jobm.201100292. [DOI] [PubMed] [Google Scholar]
- 94.Jain VK, Jain B, Sharma UK, Saha D. Synthesis, characterization and antimicrobial screening of some 4-substituted-1-(4-substituted phenyl) piperazine derivatives. Int. J. Curr. Pharma. Res. 2011;3:1–5. [Google Scholar]
- 95.Jadhav RP, Raundal HN, Patil AA, Bobade VD. Synthesis and biological evaluation of a series of1,4-disubstituted 1,2,3-triazole derivatives as possible antimicrobial agents. J. Saudi Chem. Soc. 2017;21:152–159. doi: 10.1016/j.jscs.2015.03.003. [DOI] [Google Scholar]
- 96.Shirling EB, Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16:313–340. doi: 10.1099/00207713-16-3-313. [DOI] [Google Scholar]
- 97.Zhao H, Parry RL, Ellis DI, Griffith GW, Goodacre R. The rapid differentiation of Streptomyces isolates using fourier transform infrared spectroscopy. Vib. Spectrosc. 2006;40:213–218. doi: 10.1016/j.vibspec.2005.09.006. [DOI] [Google Scholar]
- 98.Shomurat T, Yoshida J, Amano S, Kojina M, Niida T. Studies on Actinomycetales producing antibiotics only in agar culture. I. Screening taxonomy and morphology-productivity relationship of Streptomyces halstedii strain SF-1993. J. Antibiot. 1979;32:427–435. doi: 10.7164/antibiotics.32.427. [DOI] [PubMed] [Google Scholar]
- 99.Grammer, A. Antibiotic sensitivity and assay test in Microbiological Methods. (eds. Collins, C. H. & Lyne, P. N.) 235 (Butterworths, London, 1976).
- 100.Chu W-H. Optimization of extracellular alkaline protease production from species of Bacillus. J. Ind. Microbiol. Biotechnol. 2007;34:241–245. doi: 10.1007/s10295-006-0192-2. [DOI] [PubMed] [Google Scholar]
- 101.Kar S, Datta TK, Ray RC. Optimization of Thermostable α- Amylase production by Streptomyces erumpens MTCC 7317 in solid-state fermentation using cassava fibrous residue. Braz. Arch. Biol. Technol. 2010;53:301–309. doi: 10.1590/S1516-89132010000200008. [DOI] [Google Scholar]
- 102.Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using Gram’s iodine. Curr. Microbiol. 2008;57:503–507. doi: 10.1007/s00284-008-9276-8. [DOI] [PubMed] [Google Scholar]
- 103.Kumar D, et al. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch. Appl. Sci. Res. 2012;4:1763–1770. [Google Scholar]
- 104.Ayuso-Sacido A, Genilloud O. New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 2005;49:10–24. doi: 10.1007/s00248-004-0249-6. [DOI] [PubMed] [Google Scholar]
- 105.Metsa-Ketela M, et al. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 1999;180:1–6. doi: 10.1016/S0378-1097(99)00453-X. [DOI] [PubMed] [Google Scholar]
- 106.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 107.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991;173:697–703. doi: 10.1128/JB.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heyndrickx M, Vauterin L, Vandamme P, Kersters K, De Vos P. Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J. Microbiol. Methods. 1996;26:247–259. doi: 10.1016/0167-7012(96)00916-5. [DOI] [Google Scholar]
- 109.Jose PA, Jebakumar SRD. Phylogenetic appraisal of antagonistic, slow growing actinomycetes isolated from hypersaline inland solar salterns at Sambhar salt Lake, India. Front. Microbiol. 2015;4:190. doi: 10.3389/fmicb.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nimaichand S, Devi AM, Tamreihao K, Ningthoujam DS, Li W-J. Actinobacterial diversity in limestone deposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front. Microbiol. 2015;6:413. doi: 10.3389/fmicb.2015.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rohlf, F. J. NTSYS-pc. Numerical taxonomy and multivariate analysis system, version 2.02. (ed. New York, USA: Exeter Software, Setauket, 1998).
- 112.Chun J, et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2007;57:2259–2261. doi: 10.1099/ijs.0.64915-0. [DOI] [PubMed] [Google Scholar]
- 113.Tamura K, Dudley J, Nei M, Kumar S. MEGA 4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 114.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 115.Clinical and Laboratory Standards Institute (CLSI) in Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement (M100-S22; Wayne, PA: CLSI, 2012).
- 116.Andrews M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001;48:5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 117.Gomez, K. A., & Gomez, A. A. A statistical procedure for agricultural research (ed. New York, USA: John Willy and Sons, 1984).
- 118.Martin B, et al. VENNTUREA novel Venn diagram investigational tool for multiple pharmacological dataset analysis. PLoS ONE. 2012;7:1–17. doi: 10.1371/journal.pone.0036911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jia XY, et al. Genetic characterization of the chlorothricin gene cluster as a model for spirotetronate antibiotic biosynthesis. Chem. Biol. 2006;13:575–85. doi: 10.1016/j.chembiol.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 120.Liu T, Lin X, Zhou X, Deng Z, Cane DE. Mechanism of thioesterase-catalyzed chain release in the biosynthesis of the polyether antibiotic nanchangmycin. Chem. Biol. 2008;15:449–458. doi: 10.1016/j.chembiol.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun Y, et al. A complete gene cluster from Streptomyces nanchangensis NS3226 encoding biosynthesis of the polyether ionophore nanchangmycin. Chem. Biol. 2003;10:431–441. doi: 10.1016/S1074-5521(03)00092-9. [DOI] [PubMed] [Google Scholar]
- 122.August PR, et al. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem. Biol. 1998;5:69–79. doi: 10.1016/S1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 123.Sianidis G, et al. Cloning, purification and characterization of a functional anthracycline glycosyltransferase. J. Biotechnol. 2006;125:425–33. doi: 10.1016/j.jbiotec.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 124.Sherman DH, et al. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber Tü22. EMBO J. 1989;8:2717–25. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brünker P, McKinney K, Sterner O, Minas W, Bailey JE. Isolation and characterization of the naphthocyclinone gene cluster from Streptomyces arenae DSM 40737 and heterologous expression of the polyketide synthase genes. Gene. 1999;227:125–135. doi: 10.1016/S0378-1119(98)00618-0. [DOI] [PubMed] [Google Scholar]
- 126.Fernández-Moreno, M. A., Martínez, E., Boto, L., Hopwood, D. A. & Malpartida, F. Nucleotide sequence and deduced functions of a set of cotranscribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J. Biol. Chem. 267, 19278–19290 (1992). [PubMed]
- 127.Lombó, F., Blanco, G., Fernández, E., Méndez, C. & Salas, J. A. Characterization of Streptomyces argillaceus genes encoding a polyketide synthase involved in the biosynthesis of the antitumor mithramycin. Gene. 172, 87–91 (1996). [DOI] [PubMed]
- 128.Kharel Madan K., Pahari Pallab, Shepherd Micah D., Tibrewal Nidhi, Nybo S. Eric, Shaaban Khaled A., Rohr Jürgen. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012;29(2):264–325. doi: 10.1039/C1NP00068C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ichinose, K., Ozawa, M., Itou, K., Kunieda, K. & Ebizuka, Y. Cloning, sequencing and heterologous expression of the medermycin biosynthetic gene cluster of Streptomyces sp. AM-7161: towards comparative analysis of the benzoisochromanequinone gene clusters. Microbiol. 149, 1633–1645 (2003). [DOI] [PubMed]
- 130.Oja Terhi, Palmu Kaisa, Lehmussola Hanna, Leppäranta Outi, Hännikäinen Kati, Niemi Jarmo, Mäntsälä Pekka, Metsä-Ketelä Mikko. Characterization of the Alnumycin Gene Cluster Reveals Unusual Gene Products for Pyran Ring Formation and Dioxan Biosynthesis. Chemistry & Biology. 2008;15(10):1046–1057. doi: 10.1016/j.chembiol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 131.Torkkell, S., Ylihonko, K., Hakala, J., Skurnik, M. & Mäntsälä, P. Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol. Gen. Genet. 256, 203–209 (1997). [DOI] [PubMed]
- 132.Luzhetskyy Andriy, Liu Tao, Fedoryshyn Marta, Ostash Bohdan, Fedorenko Viktor, Rohr Jürgen, Bechthold Andreas. Function of lanGT3, a Glycosyltransferase Gene Involved in Landomycin A Biosynthesis. ChemBioChem. 2004;5(11):1567–1570. doi: 10.1002/cbic.200400123. [DOI] [PubMed] [Google Scholar]
- 133.Brünke, P., Sterner, O., Bailey, J. E. & Minas, W. Heterologous expression of the naphthocyclinone hydroxylase gene from Streptomyces arenae for production of novel hybrid polyketides. Anton. Leeuw. 79, 235–245 (2001). [DOI] [PubMed]
- 134.Marti, T., Hu, Z., Pohl, N. L., Shah, A. N. & Khosla, C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. Insights into an unusual priming mechanism. J. Biol. Chem. 275,33443–33448 (2000). [DOI] [PubMed]
- 135.Han, L., Yang, K., Ramalingam, E., Mosher, R. H. & Vining, L. C. Cloning and characterization of polyketide synthase genes for jadomycin B biosynthesis in Streptomyces venezuelae ISP5230. Microbiol. 140,3379–3389 (1994). [DOI] [PubMed]
- 136.Das A, Szu PH, Fitzgerald JT, Khosla C. Mechanism and engineering of polyketide chain initiation in fredericamycin biosynthesis. J. Am. Chem. Soc. 2010;132:8831–8833. doi: 10.1021/ja102517q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lackner G, et al. Biosynthesis of pentangular polyphenols: deductions from the benastatin and griseorhodin pathways. J. Am. Chem. Soc. 2007;129:9306–9312. doi: 10.1021/ja0718624. [DOI] [PubMed] [Google Scholar]
- 138.Lee, C. K., Kamitani, Y., Nihira, T. & Yamada, Y. Identification and in vivo functional analysis of a virginiamycin S resistance gene (varS) from Streptomyces virginiae. J. Bacteriol. 181,3293–3297 (1999). [DOI] [PMC free article] [PubMed]
- 139.Zhao C., Ju J., Christenson S. D., Smith W. C., Song D., Zhou X., Shen B., Deng Z. Utilization of the Methoxymalonyl-Acyl Carrier Protein Biosynthesis Locus for Cloning the Oxazolomycin Biosynthetic Gene Cluster from Streptomyces albus JA3453. Journal of Bacteriology. 2006;188(11):4142–4147. doi: 10.1128/JB.00173-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Narasaiah, B. C. et al. Isolation and structural confirmation of bioactive compounds produced by the strain Streptomyces albus CN-4. IOSR. J. Pharm. Biol. Sci. 9, 49–54 (2014).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).