Abstract
The molecular mechanism of oral submucous fibrosis (OSF) is yet to be fully elucidated. The identification of reliable signature genes to screen patients with a high risk of OSF and to provide oral cancer surveillance is therefore required. The present study produced a filtering criterion based on network characteristics and principal component analysis, and identified the genes that were involved in OSF prognosis. Two gene expression datasets were analyzed using meta-analysis, the results of which revealed 1,176 biologically significant genes. A co-expression network was subsequently constructed and weighted gene modules were detected. The pathway and functional enrichment analyses of the present study allowed for the identification of modules 1 and 2, and their respective genes, SPARC (osteonectin), cwcv and kazal like domain proteoglycan 1 (SPOCK1) and kruppel like factor 6 (KLF6), which were involved in the occurrence of OSF. The results revealed that both genes had a prominent role in epithelial to mesenchymal transition during OSF progression. The genes identified in the present study require further exploration and validation within clinical settings to determine their roles in OSF.
Keywords: meta-analysis, singular value decomposition, weighted gene co-expression network analysis, module, protein-protein interaction, eigengene, epithelial to mesenchymal transition
Introduction
Oral submucous fibrosis (OSF) is a subtle chronic disease involving the oral cavity and sometimes the pharynx. It predominantly arises in India and South East Asia exhibiting an incremental prevalence over the past four decades from 0.03 to 6.42% (1). It has the potential to transform into head and neck squamous cell carcinoma (HNSCC) with a rate of 2-8% (2). Thus, early diagnosis of this potentially malignant disorder would be very effective for prognosis as well as treatment. The pathophysiological consequences of the disease begin with a juxta-epithelial inflammatory response leading to the formation of vesicles and fibro-elastically altered lamina propria, and finally resulting in epithelial atrophy causing oral mucosa stiffness, trismus and eating inability. This is evident from the juxta-epithelial inflammatory reaction followed by immediate fibrotic contraction (3). The emerging stiffness in oral mucosa and other deep tissues gradually confines mouth opening and tongue protrusion, causing eating, swallowing and speaking predicaments. However, epithelial atrophy is a more pronounced feature of the leading stage of the disease. Molecular pathogenesis of OSF include epithelial-mesenchymal transition (EMT), extracellular matrix (ECM) remodeling and fibroblast changes, escalated expression of growth factors and inflammatory cytokines (3). Areca nut alkaloid arecoline is defined as the causative factor of OSF (4). Traditionally, OSF medications included therapies that are empirical and symptomatic in nature. Mostly, combined drug treatment is performed but based on the clinical staging, physical therapy and/or surgery may be added to drug therapy (5).
Several attempts have been made to unravel the molecular mechanisms behind the disease. But, to uncover the complete mechanism, a greater understanding of differential gene expression between normal and diseased tissues is required. Khan et al reported the role of two biomarkers, BMP7, and TGF-β in the pathogenesis of OSF along with their validation (6). Yang et al (7), found upregulation of type I plasminogen activator inhibitor in OSF specimens which they validated by RT-PCR and western blot validation. Moreover, gene expression profiling experiments are restricted by individual analysis approaches using a small number of samples and hence are not very reliable (8). Hence, meta-analysis may be the ultimate solution to all these problems. It is a robust analytical and statistical tool that improvises the statistical significance of result by merging the results of several studies across the same platform. Meta-analysis of OSF datasets covers limitations of individual expression profiling since its statistical power detects consistent changes across the multiple datasets. Another, the collection of genes working in a coordinated fashion are responsible for the progression of any disorder rather than a single gene. Identification of the highly co-expressed genes and elucidation of their biological significance is the most alluring topic in the field of network biology. Network biology is an area where we represent any complex system in terms of graph (network). The network consists of nodes and edges (the connection between nodes). In this case, the nodes are the genes and their connection depend on the correlation between them. Nowadays, several attempts have been made to elucidate the biological problem using this concept of network theory. For example, the Gene Co-expression Networks (GCN) assist to identify dense communities or functional gene modules (9). The modules and their key genes may be involved in vital pathways and thereby act as a suitable biomarker for early diagnosis (10). Mahapatra et al (11), adopted a dense sub graph-based methodology to find the putative genes from microarray data. Their proposed setup discovers highly co-expressed gene modules and further amalgamates it with protein-protein interaction (PPI) to find strongly connected modules. Lin et al (12), also identified FN1 and CCNA2 as important genes via network-based module analysis in oral squamous cell carcinoma (OSCC) from microarray datasets. Shah and Braun (13), introduced a tool named as GeneSurrounder that discovers genes by combining gene expression data and pathway network information. Although Pant et al (14), reported role of important genes in the manifestation of OSF by treating the hGF cells with areca nut (5H), TGF-β (T), and areca nut with TGF-β (5H+T) followed by pathway analysis and qPCR respectively. Our study emphasized on screening of unique signature genes associated with OSF progression via rigorous statistical analysis and network-based module approach followed by functional enrichment analysis. In this study, we assumed that the pathogenesis of OSF is occurred by the perturbation of intercellular and intracellular connections of molecules. Overall the molecular mechanism is very complex in nature. To solve this problem, we adopted the well-established network-based approach in the field of biological science, weighted gene co-expression network analysis (WGCNA) to identify groups of highly co-expressed (modules) genes associated to OSF. The differentially expressed genes (DEGs) were obtained from the meta-analysis of gene expression data. A total of 4 significant modules were discovered which was followed by the pathway and functional enrichment analysis for each module. The first module was enriched in immune response and Phagosome pathway (hsa04145), while the second module was enriched in muscle structural development and muscle contraction (R-SHA-397014) pathway. These results showed high relevancy with the progression events of OSF.
Further, we analyzed that the highly co-expressed module leads to the identification of a potential biomarker for OSF that will help to understand the mechanism of OSF. This analysis comprised intersection from four gene subsets namely Set-A, B, C and D of each module. Set-A was based on the PPI centrality measures of corresponding module genes. Set-B was based on significantly enriched Gene Ontology (GO) terms, Set-C was based on highly correlated eigengene (biologically significant gene within the module) and Set-D was based on highly significant functional enrichment of whole DEGs. There were 47 unique signature genes filtered from all four modules using this newly developed filtering criterion.
Finally, we suggested that KLF6 and SPOCK1 were found to be associated with significantly enriched GO terms and pathways such as immune response and muscle contraction, making them the most promising signature genes. Belonging to the largest two modules, both the genes had a considerably high combined effect size (ES) score and helped to drive EMT: A major event activated during OSF progression. Moreover, no clinical data was available for the patients' samples related to the role of SPOCK1 and KLF6 in OSF on GEO. Thereby, no further expression analysis was possible with patients' samples. Although several research articles have carried out survival analysis for different biomarkers in other diseases, but no literature is available on such kind of analysis for OSF.
Materials and methods
Extraction of eligible OSF gene expression datasets
The gene expression data of OSF was retrieved from Gene Expression Omnibus (GEO) (15), an open genomic repository of National Center for Biotechnology Information (NCBI). The suitable keywords and their mixtures were used as follows: ‘Oral submucous fibrosis microarray’ for searching OSF datasets in GEO.
Inclusion criteria
Gene expression datasets related to OSF and normal tissue samples were taken for the study. The expression datasets obtained from standard microarray platforms were used for the study (16).
Exclusion criteria
Datasets with cell line study and without the inclusion of normal samples were excluded.
Datasets (Series matrix files) with accession numbers GSE20170 and GSE64216 were retrieved based on platforms GPL6480 Agilent and GPL10558 Illumina, respectively. The dataset with GSE64216 had 8 samples (2 normal and 6 OSF cases) and GSE20170 had 10 OSF tissues against 8 pooled normal tissues.
Individual study analysis
HGNC (HUGO Gene Nomenclature Committee) gene symbol(s) (17) were allotted to each probe in the series matrix expression file based on the respective platform type. This was performed using several key tools and databases such as Synergizer (18), gprofiler (19), GEO2R, AbIDconvert (20), bioDBnet:db2db (21), DAVID (22), omniBiomarker (23), NCBI Entrez Browser (24) and Ensembl 92 Genome Browser (25). Duplicates for a given gene symbol were removed by averaging the expression values from various probes corresponding to the same gene (16). Computation of P-values in Illumina dataset was done utilizing the two-sample t-test function mattest (MATLAB R2018a) between normal and OSF expression level for each gene. For Agilent dataset, one-sample t-test function ttest (MATLAB R2018a) was used for P-value computation across all the samples because this dataset had pooled expression levels of OSF vs. normal tissues respectively.
Meta-analysis of OSF datasets
Common genes between both datasets were found with intersect function (MATLAB R2018a) followed by the combined P-value computation based on Fisher's χ2-based method (26). Expression data corresponding to common genes was used for computing z-score, z-difference, z-ratio, combined ES score and fold change (FC). To screen significant genes, a two-step screening process was used. In step-I, a gene is differentially expressed if FC >1.5 and P-value <0.05(27). In step-II, z-ratio of ±1.96 is regarded as a significant threshold to screen final DEGs as they are in the non-rejection region at significance level α=0.05(28). For filtering upregulated and downregulated genes from DEGs, z-score >0.5 and FDR <0.05 and z-score <-0.5 and FDR <0.05 was used as the preferred threshold, respectively (29).
Weighted gene co-expression network construction and module detection
Correlation among expression level of genes along with all the diseased sample tissues was used to know their functionality. The network construction is conceptually straight forward and thus it is easy to know the nodes and their connections. In our case, nodes of the network are genes and two nodes are connected to each other if they are co-expressed across all the tissue samples. Based on the correlation coefficient or other hard threshold values, it is difficult to infer whether two genes are correlated or not. Hence, to accomplish this, there has to be a soft threshold value driving the absolute value of the correlation coefficient and following the network scale-free property (30). In this context, WGCNA developed by Steve Horvath is of relevance. This is a complete package suite in R being utilized for constructing a weighted network and discovering modules of highly correlated genes (31). Designing a weighted co-expression gene network depends strongly on the value of the soft threshold power, β to which co-expression similarity is raised to compute adjacency. Function pickSoftThreshold (R 3.5.0) assists in selecting a proper β value. For choosing the best value of β, R2 is the criterion to affirm the fitness of co-expression network with scale-free property (generally, R2>0.80). We considered the best value of β when the relation between R2 and β reached at saturation (32). To minimize the impact of noise and errors, we transform the adjacency into topological overlap Matrix (TOM). Computation of the corresponding dissimilarity is performed utilizing hierarchical clustering, thereby producing dendrogram of genes. Identification of individual branches (cutting the branches off the dendrogram) is the main ideology behind module identification. dynamicTreeCut (R 3.5.0) function distinguishes modules with very similar expression profiles and as their genes are co-expressed, it is reasonable to merge such modules. TOM plot enables to visualize a weighted gene network where every row and column correspond to a single gene representing adjacencies.
Pathway and functional enrichment analysis
To gain insights into the biological role of module genes, GO term enrichment and pathway analysis was done. Functional enrichment provides a common descriptive framework, functional annotation, and classification to analyze the gene datasets (33). The GO term enrichment analysis was performed using DAVID 6.8 (Database for Annotation, Visualization, and Integrated Discovery) web-based annotation tool (34,35) and ToppFun web-based application (36). We classified the module genes into different functional categories of biological process (BP) and molecular function (MF) with a significance threshold of P-value <0.05 based on the Bonferroni correction method. Top 20% of enriched GO terms (BP and MF) were filtered to select significantly enriched GO terms. The pathway enrichment analysis for each module was performed using WebGestalt 2017 (WEB-based Gene Set Analysis Toolkit) web tool (37) and ToppFun. Both of them integrated information from different sources like PANTHER 11 (Protein ANalysis THrough Evolutionary Relationships) (38), KEGG (Kyoto Encyclopedia of Genes and Genomes) (39), BioCarta (40) and Reactome (41), respectively. Top 20% of enriched pathways were filtered to select significantly enriched pathways at a significance threshold of P-value <0.01 based on the Bonferroni correction method.
Signature gene identification
For the identification of signature genes, three gene sub-sets (pre-filtering sets) from each module, namely Set-A, Set-B, Set-C and a post-filtering set (Set-D) were defined as follows. Then, we filtered only the genes which were common across all the four sets. This was performed by Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Engineering of pre-filtering SET-A (based on centrality measures)
PPI network for genes of the corresponding module was constructed from the STRING v10.5 database (42). The distribution of these genes was checked in the top 15% ranked genes by each centrality measure (Degree, Stress, Eccentricity, Radiality, Closeness and Betweenness). These centrality rankings were computed using software Cytoscape 3.6.1(43). Unique pooled genes consist of unique genes from all the listed centralities combined at 15% cutoff and were conserved for Set-A.
Engineering of pre-filtering SET-B (based on significant GO enrichment analysis)
There are many GO terms with false positives and thus a criterion is required to lessen the entire quantity of GO terms to increment prediction efficiency. The significantly enriched GO terms were attained with P-value <0.001 from DAVID (level 5) database. Out of these, the terms that were associated with fewer than 3 and greater than 50 genes (3 < = G< = 50, G = quantity of genes) were removed to eliminate non-specific ones (44). Genes associated with the remaining significant GO terms were conserved for Set-B.
Engineering of pre-filtering SET-C (based on eigengene correlation)
Singular value decomposition (SVD) assists in factorization of a matrix and let the vertices to be symbolized as vectors. It induces linear transformation of expression data from genes x arrays space into compact ‘eigengenes’ x ‘eigenarrays’ space (45). Here, the data are diagonalized, such that every eigengene is expressed solely in the corresponding eigenarray, with relative ‘eigenexpression’ values describing their comparative significance. Using SVD function (MATLAB), the module expression data is decomposed into three resultant matrices namely U, D and V. The first column of V is known as the module eigengene or first principal component (46) and is regarded as the most representative or influenced gene of the whole module. Positive correlation of this vector with the expression values across module genes was carried out. Highly correlated genes (correlation coefficient >=0.95) were conserved for Set-C.
Engineering of SET-D (post-filtering set)
The present study can be divided into two parts, where we studied the up and downregulated genes in the first part (methodology described in section ‘Materials and methods’) and in the second part, we studied module identification of the DEGs. Set-D was constructed from first part of the study by applying Set-B criterion on it, whereas, Set-A, B, C have been constructed from second part of the study.
The reason for formulation of Set D can be elaborated in the following manner: suppose M and N are two sets of genes where M is a subset of N. Another assumption is that J and K set of genes were screened after applying criterion of Set-B on M and N respectively. Then, J may not necessarily be the subset of K. Hence, Set-D was important for finding the functionally significant genes in the set of up and downregulated DEGs which were identified in first part of the study.
Results
Individual analysis
In the present study, we gathered a total of two gene expression datasets in accordance with the exclusion and inclusion criteria specified. Then, we performed mapping of probe IDs to HGNC gene symbol(s) for both datasets followed by removal of duplicate gene symbols. From this, we got a total of 27800 and 30864 HGNC gene symbols in Illumina and Agilent expression datasets respectively. Computation of P-values from expression values of both datasets was performed using the appropriate statistical t-test. These P-values were used for further DEGs screening.
Meta-analysis of OSF datasets
A total of 17,792 common genes across both the datasets were derived and combined P-value of these genes was computed using fisher's χ2-based method. Finally, 1,176 DEGs were screened after applying the two-step screening process. In step-I, 1,219 genes were obtained out of which 43 genes were removed based on step-II screening threshold to yield the final DEGs. Filtering of 238 upregulated and 902 downregulated genes using appropriate z-score threshold was performed.
Co-expression network construction and module detection
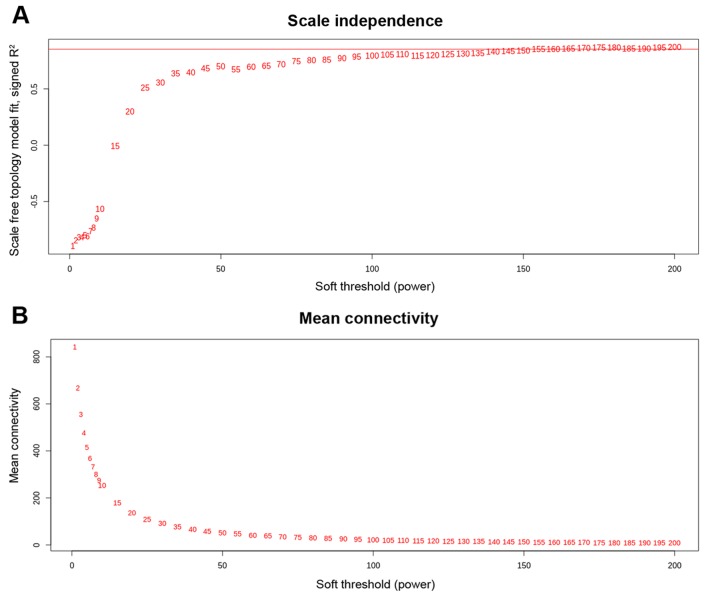
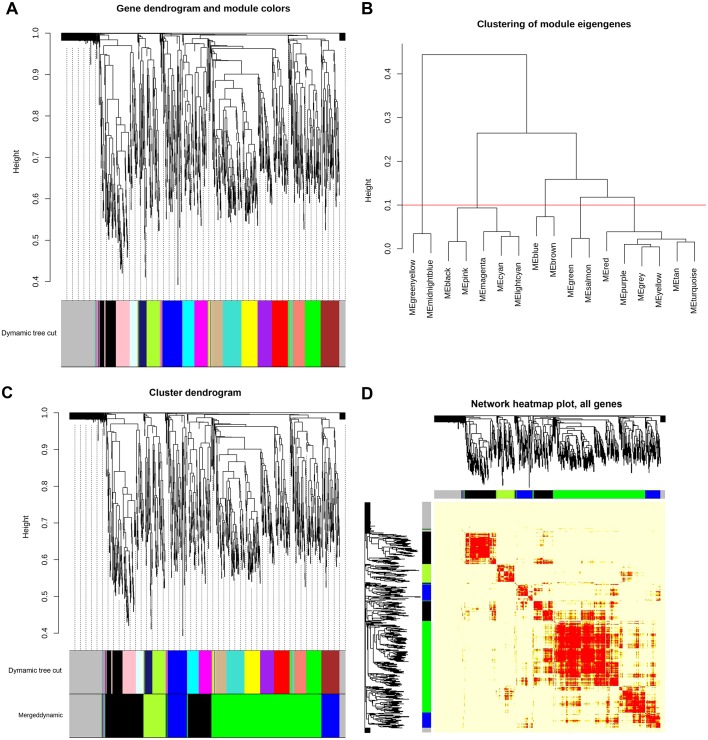
After using WGCNA as specified in the materials and methods section for calculating the soft threshold (β), we found an interesting scenario as shown in Fig. 1. We can see in the left and right panel of the Fig. 1 that the curve trades off at β=155 with R2=0. 85300, hence we utilized the power (s,β=155) to construct adjacency matrix of the weighted network. Then we computed TOM which is a biologically meaningful measure of node similarity, and the genes were clustered hierarchically utilizing 1-TOM as the distance measure. The modules were identified by picking a height cut-off of 0.90 (as guided by the TOM plot) for the resulting dendrogram (Fig. 2A and B). Highly analogous modules were characterized by clustering and merging together (Fig. 2C). Finally, four significant modules were identified and classified according to the specific color-coding scheme, each of which had co-ordinately expressed genes (Table I). TOM plot of the weighted network is shown in Fig. 2D for all 1,176 genes and it depicts that modules are highly robust with respect to the network construction method utilized. Out of 1,176 genes, module-1 had the highest number of genes, i.e., 478, module-2 had almost half of module-1, similarly, module-3 and module-4 had 160 and 91 genes, respectively (a summary can be seen in Table I). Remaining 176 genes were not able to significantly correlate with any of these modules. The list of genes with belongingness to different modules is shown in Table SI.
Figure 1.
Network topology analysis of several soft-threshold powers in weighted gene co-expression network analysis, where (A) depicts R2 as a function of β and (B) depicts mean connectivity as a function of β.
Figure 2.
WGCNA co-expression network and module detection analysis. (A) represents clustering dendrogram of genes with dissimilarity based on topological overlap. A total of 16 distinct modules are presented with different assigned colors in the horizontal bar below the dendrogram, with grey representing unassigned genes in any module. (B) Hierarchical clustering of 16 module eigengenes. The distance (1-TOM) is denoted by the y-axis and different MEs are denoted by the x-axis (labled as color with ME prefixed in each color module). The horizontal red line (1-TOM=0.10) depicts the benchmark for defining meta-modules. The green, blue, black and green-yellow modules illustrate meta-modules, while grey colored eigengenes were not assigned to any meta-module. (C) Original and merged co-expression modules with assigned original module colors and merged module colors. Genes were clustered based on a dissimilarity measure (1-TOM). The branches correspond to modules of highly correlated or interconnected groups of genes. Colors in the horizontal bar depict the modules before and after merging. Cluster dendrograms of genes with dissimilarity based on topological overlap is presented above the modules. A total of 16 original modules were merged to obtain 4 highly significant meta-modules denoted by the colors green, black, blue and green-yellow. Grey colored modules represent unassigned genes. (D) Heatmap plot of the weighted gene co-expression network. The plot indicates the TOM among all genes analyzed. Genes in columns and their corresponding rows are hierarchically clustered by cluster dendrograms, which are presented along the top and left side of the plot. Color-coded module membership is presented with colored bars (green, black, blue, green-yellow and grey) below and to the right of dendrograms. Green, black, blue and green-yellow colors signify the 4 significant meta-modules and grey represents the insignificant module. Progressively light and darker red colors in the matrix signify lower and higher overlap among genes. High co-expression interconnectedness are indicated by progressively more saturated yellow to red colors. TOM, topological overlap matrix; ME, module eigengene.
Table I.
Characteristics of identified modules using WGCNA.
| Module no. | Nodes (Genes) | Color coding by WGCNA |
|---|---|---|
| 01 | 478 | GREEN |
| 02 | 271 | BLACK |
| 03 | 160 | BLUE |
| 04 | 91 | GREENYELLOW |
| 05 | INSIGNIFICANT | GREY |
WGCNA, weighted gene co-expression network analysis.
Pathway and functional enrichment analysis
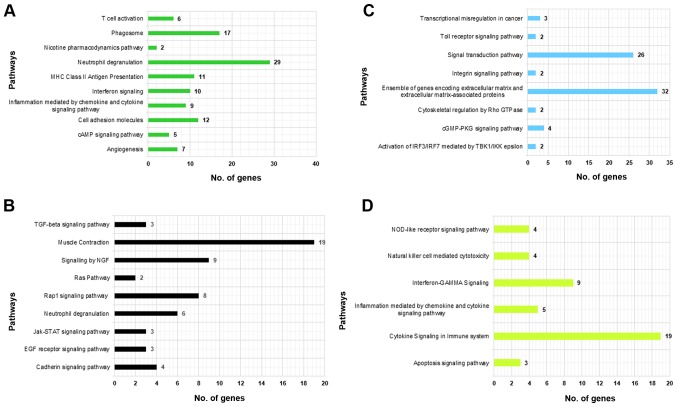
Pathway and GO term enrichment analysis was performed to characterize the module function. The GO term analysis (BP & MF) of module-1 genes can be seen in Fig. 3A, which was depicting a large range of biological functions and highly enriched terms including Immune Response (GO:0006955; adjacent P-value=3.788E-10) and MHC class II receptor activity (GO:0032395; adjacent P-value=4.00E-03). The most significantly enriched pathways of module-1 were Phagosome (hsa04145) and MHC class II antigen presentation (R-HSA-2132295; Fig. 4A). Highly significantly enriched terms of module-2 genes were muscle structure development (GO:0061061; adjacent P-value=3.483E-12) and structural constituent of muscle (GO:0008307; adjacent P-value=9.212E-08), can be seen in Fig. 3B. Highly significant enriched pathways of module-2 genes were muscle contraction (R-HSA-397014) and neutrophil degranulation (R-HSA-6798695) with a maximum number of genes being involved in the former pathway (Fig. 4B). Module-3 comprised of highly significant terms like defense response (GO:0006952; adjacent P-value=1.248E-05) and dolecular transducer activity (GO:0060089; adjacent P-value=4.674E-04). Highly significant enriched pathways of module-3 were ensemble of genes encoding ECM and extracellular matrix-associated proteins (M5889) and activation of IRF3/IRF7 mediated by TBK1/IKK epsilon (R-HSA-936964) (Figs. 3C and 4C). Genes of module-4 were involved in immune response (GO:0006955; adjacent P-value=9.770E-18) and interleukin-2 receptor activity (GO:0004911; adjacent P-value=4.300E-02). Highly significant pathways of module-4 were cytokine signaling in the immune system (R-HSA-1280215) and interferon-GAMMA signaling (R-HSA-877300; Figs. 3D and 4D).
Figure 3.
Representation of the top 20% significantly enriched GO terms. Top 20% of significantly enriched gene ontology terms (molecular function and biological process) using DAVID and ToppFun software, presented as pie-charts for (A) Module-1, (B) Module-2, (C) Module-3 and (D) Module-4.
Figure 4.
Representation of the top 20% significantly enriched pathways. Top 20% significantly enriched pathways obtained using WebGestalt and ToppFun platforms, presented as clustered bar charts for (A) Module-1, (B) Module-2, (C) Module-3 and (D) Module-4.
Signature gene identification and analysis
Pre-filtering Set-A
Significance of a node is governed by each centrality in a different manner for the network under consideration. In the present study PPI network for genes of the corresponding module was constructed from the STRING database and top 15% genes were ranked by each centrality. Unique pooled genes from all centralities were reserved for Set-A. The top 15% unique genes filtered from module-1, module-2, module-3, and module-4 were 63, 22, 15 and 5, respectively.
Pre-filtering Set-B
DAVID (level 5) was used for the identification of significantly enriched (P-value <0.001) GO terms for each module. If a term is associated with ≤3 and ≥50 genes, then it was removed and the genes associated with rest of the terms were reserved for Set-B. Using this criterion, we found 240, 159, 122, and 64 genes in module-1, module-2, module-3, and module-4, respectively.
Pre-filtering Set-C
SVD also known as principal component analysis (PCA) in statistics is a highly robust technique for matrix factorization and dimensionality reduction. The matrix decomposition using SVD leads to U, D and V matrices and the columns of U and V are left and right-singular vectors, respectively. Here, the first column of V is referred to as the representative eigengene of the module and its positive correlation with expression values leads to highly correlated genes for Set-C. The number of highly correlated genes were 310, 158, 91 and 80 in module-1, module-2, module-3, and module-4, respectively.
Post-filtering Set-D
The total number of significant and specific GO terms associated genes filtered from up and downregulated DEGs were 647.
Intersection of pre-filtering sets and post-filtering set
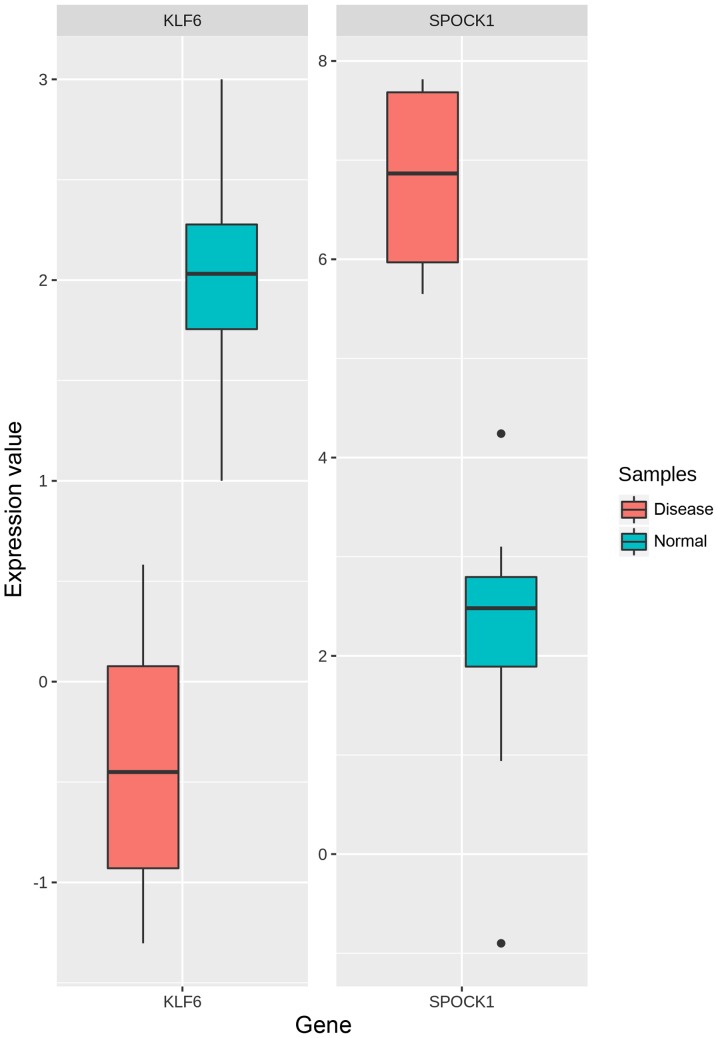
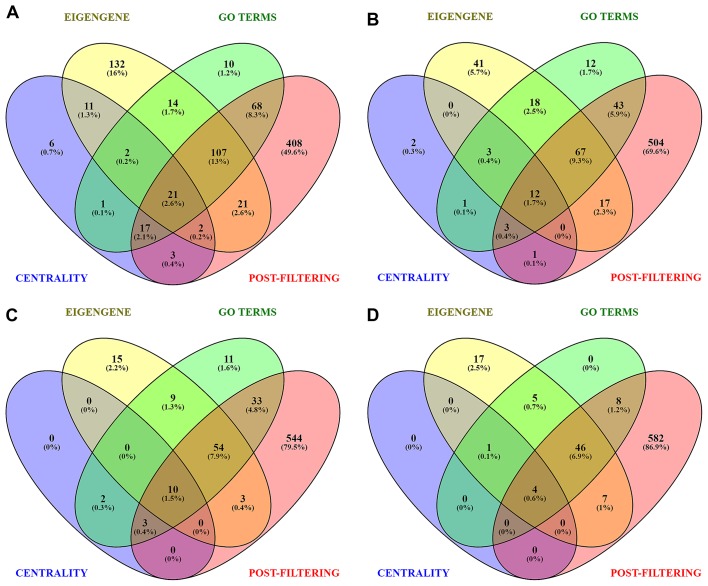
The summary of filtered genes from each set and their intersection can be seen in Fig. 5A-D for module-1, module-2, module-3, and module-4, respectively. Genes common between the pre-filtering sets (A, B, C) and post-filtering set (D) were regarded as signature genes. We found a total of 47 genes where 21, 12, 10 and 4 genes were from module-1, module-2, module-3, and module-4, respectively (Table SII). Out of these, we listed the top 10 upregulated and 10 downregulated genes each based on the combined ES score which can be seen in Table II. As SPOCK1 (upregulated) and KLF6 (downregulated) were involved in significantly enriched pathways and GO terms, hence they were regarded as the most promising signature genes. Also, they belonged to the largest two modules (module-1 and module-2). The box-and-whisker plot for comparison of KLF6 and SPOCK1 gene expressions in OSF and normal cases respectively is illustrated in Fig. 6.
Table II.
Top 10 upregulated and downregulated signature genes.
| Upregulated genes | Combined ES score | Chromosome (Band) | Downregulated genes | Combined ES score | Chromosome (Band) |
|---|---|---|---|---|---|
| CD14 | 1.9460 | 5 (5q31.3) | KLF6 | -0.7532 | 10 (10p15.2) |
| GYG1 | 1.4059 | 3 (3q24) | PODXL | -0.1044 | 7 (7q32.3) |
| ALOX5AP | 0.8693 | 13 (13q12.3) | GNG7 | -0.0608 | 19 (19p13.3) |
| PLAT | 0.3894 | 8 (8p11.21) | TUSC3 | -0.0448 | 8 (8p22) |
| TPP1 | 0.2160 | 11 (11p15.4) | BAG4 | -0.0217 | 8 (8p11.23) |
| OASL | 0.0979 | 12 (12q24.31) | GCNT2 | -0.0167 | 6 (6p24.3-p24.2) |
| SPOCK1 | 0.0901 | 5 (5q31.2) | SCP2 | -0.0043 | 1 (1p32.3) |
| IVD | 0.0382 | 15 (15q15.1) | PODXL2 | 0.0052 | 3 (3q21.3) |
| TINAGL1 | 0.0064 | 1 (1p35.2) | NRXN1 | 0.0064 | 2 (2p16.3) |
| ACAA2 | -0.2064 | 18 (18q21.1) | TNFRSF8 | 0.0070 | 1 (1p36.22) |
ES, effect size.
Figure 6.
Box-and-whisker plot comparing the relative gene expression of KLF6 and SPOCK1 in OSF and normal samples. The centre line of each box indicates the median result. SPOCK1, SPARC (osteonectin), cwcv and kazal like domain proteoglycan 1; KLF6, kruppel like factor 6. OSF, oral submucous fibrosis.
Discussion
The present study presents an integrated meta-analysis of OSF gene expression datasets with the objective of finding reliable signature genes. We observed a total of 1,176 DEGs using meta-analysis of 2 OSF datasets and by implying a specific threshold for filtering. Further, a module-based technique was adopted to delineate the DEGs into modules. The gene expression analysis can be done better with the aid of co-expression networks as it lets for analysis of expression data from a system's perspective (47). Weighted gene co-expression network expresses the relation amongst modules as it preserves the nature of co-expression information. Module detection techniques are a boon in the analysis of massive gene expression studies. With the advent of non-overlapping clustering approach, now there is an ease of rapidly presenting a global portrait of the dataset, exposing the main expression and functional relevance amongst the various samples within the dataset (48). And, WGCNA is especially right for such an analysis, outperforming other clustering techniques on the majority of datasets. It is an agglomerative hierarchical clustering technique that enables assembling of functionally related genes to implicitly detect modules. The most vital factor on which WGCNA depends is the Pearson's correlation coefficient that quantifies the extent of linear dependency between the two expression profiles irrespective of absolute expression level differences. The whole functional role of a module remains the same unlike individual gene expression which fluctuates, thereby modules are considered as stable units in systems biology. Modularity of the biological system enables to gain an insight into its components independently and to establish a mutual connection between modules and genes. It also assists in interpreting the progression of any disorder in a more elaborate manner (49). Module-based analysis from WGCNA resulted in the identification of 4 modules (color-coded by green, black, blue and greenyellow to distinguish them according to their nodes). Pathway and functional enrichment analysis showed that genes of module-1 and 2 were involved in the processes and pathways that correlate with OSF. Hence, both these modules were regarded as important ones. Module-1 was found to be linked with immune response and MHC activities, making it a suitable candidate for OSF pathogenesis. The major role of immune response is the identification of the foreign antigen. This process will eventually lead to hyperactivity of B cells and abnormal lymphocyte function. The upsurge in the levels of IgA, IgM, and IgG is also noticed in OSF patients (50). OSF has an association with autoimmunity which may be caused by areca nut elements. Genes located in the HLA region are significant determinants of genetic susceptibility in OSF (51). Module-2 is mostly associated with muscle structure development and muscle-related processes, which in turn have a sync with OSF. Masseter muscle is the toughest and largest one of the face. Because of continuous areca nut chewing for prolonged intervals of time, the masseter muscle becomes a victim of unnecessary stress. This results in escalated thicknesses of the muscle causing a major transformation in masseter muscle fiber size and composition (52). Restricted mouth opening in OSF cases relies solely not only on subepithelial fibrosis but also on the degree of muscle destruction. There is a progressive decline observed in the muscle fibers from the epithelial surface as the advancement of OSF occurs (53). Also, features suggestive of muscle atrophy and necrosis have been demonstrated in moderate and advanced OSF cases (54). The light microscopic study of OSF has revealed varying degrees of alterations involving the muscle fibers as the disease progresses (55).
The main crux of this whole work remains to be the identification of potent signature genes and studying their relevance in OSF progression. For this, we constructed 3 gene sub-sets (pre-filtering sets) for each module and a post-filtering set. Biological network comprises some of the crucial genes as they are involved in regulating the crucial processes. These genes are often found amongst the disease genes. As the importance of a protein is not judged in a complex PPI network, therefore the centrality measures assist in ranking the network components according to their importance. We calculated six different centrality measures to assess the importance of a protein in the PPI network for each module. Prominently, we evaluated the genes which are ranked (top 15%) higher in the PPI network and has major importance to the disorder. Biological importance of genes can be estimated via GO terms to which they were related as it gives information for protein functions, processes, and localization. To report genes linked with OSF, significantly enriched GO terms were characterized, and further pruning of these identified terms resulted in a set of more enriched terms for each module. To summarize the expression profile of a module, the module eigengene needs to be accessed. It is the first right-singular vector of the standardized module expression data and most influential gene of whole module. Categorizing the data according to correlations of genes (and arrays) with eigengenes (and eigenarrays) provides a summarized snapshot of the gene expression dynamics. SVD or PCA is a robust and fast matrix factorization algorithm and offers a beneficial mathematical framework for processing and modeling genome-wide expression data, where both the operations and mathematical variables may be allotted biological meaning (45). It can be united with unsupervised clustering and dimensionality reduction to reveal community structures of weighted or unweighted, bipartite or unipartite graphs in a computationally effective manner (56). Intersection amongst all the four sets revealed 21, 12, 10 and 4 genes from each module respectively. The results lay a foundation that a blend of GO, network centralities and eigengene play an important role in the detection of signature genes. SPOCK1 and KLF6 were found to have a prominent role in the EMT process-one of the major events of OSF and were regarded as reliable biomarkers. Location of KLF6 and SPOCK1 genes are on human chromosomes 10 and 5 respectively.
KLF6 (Kruppel-like factor 6) is a pervasive transcription factor with several targets and activities and is highly expressed in skin epithelial cells, lung, thymus, cornea, testis, lymphocytes and cardiac myocytes. It belongs to a family of conserved zinc-finger proteins that regulates a number of cellular events such as cell proliferation, differentiation, apoptosis and neoplastic transformation (57). It primarily works by enhancing p21 expression by a pathway independent of p53, and hence acts as a tumor-suppressor gene (58). It is downregulated in several human cancers and its reduced expression has been correlated with cellular proliferation and poor prognosis in oral cancer (59). One of the studies has demonstrated its role in metastasis and EMT (60). Downregulation of KLF6 in our study suggests its role in the EMT process of OSF. Expression of KLF6 decreases in chronic inflammation which is a common feature in OSF. It binds to the promoter region of MMP-9 (matrix metalloproteinase-9) and inhibits its expression in endothelial cells (61). Expression and activity levels of MMP-9 was significantly upregulated in primary keratinocytes derived from healthy gingival epithelium treated with arecoline (62). MMP-9 also shows positive stromal expression in OSF cases (63). Thus, targeting KLF6 would be highly effective in improving the prognosis of OSF.
On the other hand, SPOCK1, an extracellularly secreted proteoglycan originally called ‘TESTICAN’, as it was first isolated from human testis (64). Altered regulation of SPOCK1 has been observed in multiple tissues and organs, including lymphocytes, brain cartilage, vascular endothelium and neuromuscular junctions (65-67). It has been reported to be overexpressed in distinct cancer types. In addition, SPOCK-1 induces EMT to facilitate cancer cell migration. EMT plays a role in OSF and its malignant transformation. During this transition, epithelial cells are degraded, and they acquire mesenchymal properties, thereby causing an increase in the expression of mesenchymal markers such as N-cadherin, snail, and vimentin. Moreover, the glycan portion of SPOCK1 has an interplay with matrix architecture, growth factors and chemokines (68). These properties are utilized by cancer cells to invade tissues and achieve survival advantage over the organism. Based on our study, we infer that SPOCK1 can be a crucial gene for early detection of OSF.
We have identified two putative signature genes, SPOCK1, and KLF6 which are aberrantly expressed in the case of OSF. Dysregulation of these signature genes might probably lead to the development of oral cancer. Thus, more detailed analysis of mechanisms and pathways associated with these genes would provide us a deep insight into the process of oncogenesis and would prove to be an effective guide in designing novel therapeutic strategies against oral cancer. In this context, the present study provides a road map leading to a nearly ideal situation where putative results may be transformed into a robust tool for routine prognosis.
Supplementary Material
Figure 5.
Venn plots for Set-A, B, C and D. (A) Identification of signature genes using top ranked unique genes (Centrality set), significant GO terms (GO Terms set), eigengene based on principal component analysis correlation (Eigengene set) and Post-filtering set. The genes shared by all four sets revealed potent signature genes from (A) Module-1(21), (B) Module-2(12), (C) Module-3(10) and (D) Module-4(4). Blue represents centrality Set-A, green represents significant GO term Set-B, yellow represents eigengene Set-C and red represents post-filtering Set-D. GO, gene ontology.
Acknowledgements
The authors would like to thank Jamia Millia Islamia for providing infrastructure, journal access and internet facility. Shweta Arora would like to thank Indian Council of Medical Research for providing her Senior Research Fellowship.
Funding
The present study was supported by the Science and Engineering Research Board (grant no. EEQ/2016/000509) and partially supported by the Department of Biotechnology (grant no. DBT/PR12828/AAQ/1/622/2015), Government of India.
Availability of data and materials
Data associated with this study is available at Gene Expression Omnibus (GEO) under the accession numbers GSE20170 and GSE64216.
Authors' contributions
PS and RD performed statistical analysis. AR and SAr participated in the literature search. SAl, SP and MAS conceived the present study and prepared and revised the manuscript. SAr, SAl, AR, RD and PS designed the present study, critically revised the discussion and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bari S, Metgud R, Vyas Z, Tak A. An update on studies on etiological factors, disease progression, and malignant transformation in oral submucous fibrosis. J Cancer Res Ther. 2017;13:399–405. doi: 10.4103/0973-1482.179524. [DOI] [PubMed] [Google Scholar]
- 2.Ray JG, Ranganathan K, Chattopadhyay A. Malignant transformation of oral submucous fibrosis: Overview of histopathological aspects. Oral Surg Oral Med Oral Pathol Oral Radiol. 2016;122:200–209. doi: 10.1016/j.oooo.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Ekanayaka RP, Tilakaratne WM. doi: 10.1016/j.oooo.2015.12.018. Oral submucous fibrosis: Review on mechanisms of pathogenesis and malignant transformation. J Carcinog Mutagen, 2013. Available from: https://www.omicsonline.org/oral-submucous-fibrosis-a-clinico-histopathological-comparative-study-in-population-of-southern-rajasthan-2157-2518-S5-002.php?aid=19130. [DOI] [PubMed] [Google Scholar]
- 4.Prabhu RV, Prabhu V, Chatra L, Shenai P, Suvarna N, Dandekeri S. Areca nut and its role in oral submucous fibrosis. J Clin Exp Dent. 2014;6:e569–e575. doi: 10.4317/jced.51318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wollina U, Verma SB, Ali FM, Patil K. Oral submucous fibrosis: An update. Clin Cosmet Investig Dermatol. 2015;8:193–204. doi: 10.2147/CCID.S80576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan I, Kumar N, Pant I, Narra S, Kondaiah P. Activation of TGF-β pathway by areca nut constituents: A possible cause of oral submucous fibrosis. PLoS One. 2012;7(e51806) doi: 10.1371/journal.pone.0051806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang SF, Wang YH, Su NY, Yu HC, Wei CY, Yu CH, Chang YC. Changes in prevalence of precancerous oral submucous fibrosis from 1996 to 2013 in Taiwan: A nationwide population-based retrospective study. J Formos Med Assoc. 2018;117:147–152. doi: 10.1016/j.jfma.2017.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(e184) doi: 10.1371/journal.pmed.0050184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruan J, Dean AK, Zhang W. A general co-expression network-based approach to gene expression analysis: Comparison and applications. BMC Syst Biol. 2010;4(8) doi: 10.1186/1752-0509-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LX, Li Y, Chen GZ. Network-based co-expression analysis for exploring the potential diagnostic biomarkers of metastatic melanoma. PLoS One. 2018;13(e0190447) doi: 10.1371/journal.pone.0190447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahapatra S, Mandal B, Swarnkar T. Biological networks integration based on dense module identification for gene prioritization from microarray data. Gene Rep. 2018;12:276–288. doi: 10.1016/j.genrep.2018.07.008. [DOI] [Google Scholar]
- 12.Lin T, Zhang B, He H. Identification of genes correlated with oral squamous cell carcinoma. J Cancer Res Ther. 2018;14 (Suppl):S675–S679. doi: 10.4103/0973-1482.202893. [DOI] [PubMed] [Google Scholar]
- 13.Shah SD, Braun R. GeneSurrounder: Network-based identification of disease genes in expression data. BMC Bioinformatics. 2019;20(229) doi: 10.1186/s12859-019-2829-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pant I, Kumar N, Khan I, Rao SG, Kondaiah P. Role of areca nut induced TGF-β and epithelial-mesenchymal interaction in the pathogenesis of oral submucous fibrosis. PLoS One. 2015;10(e0129252) doi: 10.1371/journal.pone.0129252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thangaraj SV, Shyamsundar V, Krishnamurthy A, Ramani P, Ganesan K, Muthuswami M, Ramshankar V. Molecular portrait of oral tongue squamous cell carcinoma shown by integrative meta-analysis of expression profiles with validations. PLoS One. 2016;11(e0156582) doi: 10.1371/journal.pone.0156582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray KA, Seal RL, Tweedie S, Wright MW, Bruford EA. A review of the new HGNC gene family resource. Hum Genomics. 2016;10(6) doi: 10.1186/s40246-016-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berriz GF, Roth FP. The Synergizer service for translating gene, protein and other biological identifiers. Bioinforma Oxf Engl. 2008;24:2272–2273. doi: 10.1093/bioinformatics/btn424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, Vilo J. g:Profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohammad F, Flight RM, Harrison BJ, Petruska JC, Rouchka EC. AbsIDconvert: An absolute approach for converting genetic identifiers at different granularities. BMC Bioinformatics. 2012;13(229) doi: 10.1186/1471-2105-13-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mudunuri U, Che A, Yi M, Stephens RM. bioDBnet: The biological database network. Bioinformatics. 2009;25:555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Stephens R, Baseler MW, Lane HC, Lempicki RA. DAVID gene ID conversion tool. Bioinformation. 2008;2:428–430. doi: 10.6026/97320630002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan JH, Young AN, Wang MD. omniBiomarker: A web-based application for knowledge-driven biomarker identification. IEEE Trans Biomed Eng. 2013;60:3364–3367. doi: 10.1109/TBME.2012.2212438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez gene: Gene-centered information at NCBI. Nucleic Acids Res. 2005;33 (Database Issue):D54–D58. doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aken BL, Achuthan P, Akanni W, Amode MR, Bernsdorff F, Bhai J, Billis K, Carvalho-Silva D, Cummins C, Clapham P, et al. Ensembl 2017. Nucleic Acids Res. 2017;45:D635–D642. doi: 10.1093/nar/gkw1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Assen MALM, van Aert RC, Wicherts JM. Meta-analysis using effect size distributions of only statistically significant studies. Psychol Methods. 2015;20:293–309. doi: 10.1037/met0000025. [DOI] [PubMed] [Google Scholar]
- 27.Dalman MR, Deeter A, Nimishakavi G, Duan ZH. Fold change and p-value cutoffs significantly alter microarray interpretations. BMC Bioinformatics. 2012;13 (Suppl 2)(S11) doi: 10.1186/1471-2105-13-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho KH, Chen PH, Hsi E, Shih CM, Chang WC, Cheng CH, Lin CW, Chen KC. Identification of IGF-1-enhanced cytokine expressions targeted by miR-181d in glioblastomas via an integrative miRNA/mRNA regulatory network analysis. Sci Rep. 2017;7(732) doi: 10.1038/s41598-017-00826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabási AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 31.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9(559) doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(Article17) doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Dong W, Qu X, Zhao H, Wang S, Hao Y, Li Q, Zhu J, Ye M, Xiao W. Molecular dysexpression in gastric cancer revealed by integrated analysis of transcriptome data. Oncol Lett. 2017;13:3177–3185. doi: 10.3892/ol.2017.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(R183) doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 36.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37 (Web Server Issue):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45 (D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishimura D. BioCarta. Biotech Softw Internet Rep. 2001;2:117–120. doi: 10.1089/152791601750294344. [DOI] [Google Scholar]
- 41.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43 (Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar R, Samal SK, Routray S, Dash R, Dixit A. Identification of oral cancer related candidate genes by integrating protein-protein interactions, gene ontology, pathway analysis and immunohistochemistry. Sci Rep. 2017;7(2472) doi: 10.1038/s41598-017-02522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alter O, Brown PO, Botstein D. Singular value decomposition for genome-wide expression data processing and modeling. Proc Natl Acad Sci USA. 2000;97:10101–10106. doi: 10.1073/pnas.97.18.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1(54) doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pontén F, Gry M, Fagerberg L, Lundberg E, Asplund A, Berglund L, Oksvold P, Björling E, Hober S, Kampf C, et al. A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol. 2009;5(337) doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D'haeseleer P. How does gene expression clustering work? Nat Biotechnol. 2005;23:1499–1501. doi: 10.1038/nbt1205-1499. [DOI] [PubMed] [Google Scholar]
- 49.Alsina L, Israelsson E, Altman MC, Dang KK, Ghandil P, Israel L, von Bernuth H, Baldwin N, Qin H, Jin Z, et al. A narrow repertoire of transcriptional modules responsive to pyogenic bacteria is impaired in patients carrying loss-of-function mutations in MYD88 or IRAK4. Nat Immunol. 2014;15:1134–1142. doi: 10.1038/ni.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah N, Kumar R, Shah MK. Immunological studies in oral submucous fibrosis. Indian J Dent Res. 1994;5:81–87. [PubMed] [Google Scholar]
- 51.Canniff JP, Batchelor JR, Dodi IA, Harvey W. HLA-typing in oral submucous fibrosis. Tissue Antigens. 1985;26:138–142. doi: 10.1111/j.1399-0039.1985.tb00946.x. [DOI] [PubMed] [Google Scholar]
- 52.Kiliaridis S, Mahboubi PH, Raadsheer MC, Katsaros C. Ultrasonographic thickness of the masseter muscle in growing individuals with unilateral crossbite. Angle Orthod. 2007;77:607–611. doi: 10.2319/101105-360. [DOI] [PubMed] [Google Scholar]
- 53.Chawla H, Urs Ab, Augustine J, Kumar P. Characterization of muscle alteration in oral submucous fibrosis-seeking new evidence. Med Oral Patol Oral Cirugia Bucal. 2015;20:e670–e677. doi: 10.4317/medoral.20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sumathi M, Balaji N, Malathi N. A prospective transmission electron microscopic study of muscle status in oral submucous fibrosis along with retrospective analysis of 80 cases of oral submucous fibrosis. J Oral Maxillofac Pathol. 2012;16:318–324. doi: 10.4103/0973-029X.102474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rooban T, Saraswathi TR, Al Zainab FHI, Devi U, Eligabeth J, Ranganathan K. A light microscopic study of fibrosis involving muscle in oral submucous fibrosis. Indian J Dent Res. 2005;16:131–134. doi: 10.4103/0970-9290.29909. [DOI] [PubMed] [Google Scholar]
- 56.Sarkar S, Dong A. Community detection in graphs using singular value decomposition. Phys Rev E Stat Nonlin Soft Matter Phys. 2011;83(046114) doi: 10.1103/PhysRevE.83.046114. [DOI] [PubMed] [Google Scholar]
- 57.Hsu LS, Huang RH, Lai HW, Hsu HT, Sung WW, Hsieh MJ, Wu CY, Lin YM, Chen MK, Lo YS, Chen CJ. KLF6 inhibited oral cancer migration and invasion via downregulation of mesenchymal markers and inhibition of MMP-9 activities. Int J Med Sci. 2017;14:530–535. doi: 10.7150/ijms.19024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Botella LM, Sanz-Rodriguez F, Komi Y, Fernandez-L A, Varela E, Garrido-Martin EM, Narla G, Friedman SL, Kojima S. TGF-beta regulates the expression of transcription factor KLF6 and its splice variants and promotes co-operative transactivation of common target genes through a Smad3-Sp1-KLF6 interaction. Biochem J. 2009;419:485–495. doi: 10.1042/BJ20081434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Liu M, Su Y, Zhou X, Liu Y, Zhang X. The Janus-faced roles of Krüppel-like factor 4 in oral squamous cell carcinoma cells. Oncotarget. 2015;6:44480–44494. doi: 10.18632/oncotarget.6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L, Li W. Epithelial-mesenchymal transition in human cancer: Comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther. 2015;150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Das A, Fernandez-Zapico ME, Cao S, Yao J, Fiorucci S, Hebbel RP, Urrutia R, Shah VH. Disruption of an SP2/KLF6 repression complex by SHP is required for farnesoid X receptor-induced endothelial cell migration. J Biol Chem. 2006;281:39105–39113. doi: 10.1074/jbc.M607720200. [DOI] [PubMed] [Google Scholar]
- 62.Uehara O, Takimoto K, Morikawa T, Harada F, Takai R, Adhikari BR, Itatsu R, Nakamura T, Yoshida K, Matsuoka H, et al. Upregulated expression of MMP-9 in gingival epithelial cells induced by prolonged stimulation with arecoline. Oncol Lett. 2017;14:1186–1192. doi: 10.3892/ol.2017.6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajendran R, Rajeesh MP, Shaikh S, Shanthi , Pillai MR. Expression of matrix metalloproteinases (MMP-1, MMP-2 and MMP-9) and their inhibitors (TIMP-1 and TIMP-2) in oral submucous fibrosis. Indian J Dent Res. 2006;17:161–166. doi: 10.4103/0970-9290.29870. [DOI] [PubMed] [Google Scholar]
- 64.Miao L, Wang Y, Xia H, Yao C, Cai H, Song Y. SPOCK1 is a novel transforming growth factor-β target gene that regulates lung cancer cell epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2013;440:792–797. doi: 10.1016/j.bbrc.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 65.Hartmann U, Hülsmann H, Seul J, Röll S, Midani H, Breloy I, Hechler D, Müller R, Paulsson M. Testican-3: A brain-specific proteoglycan member of the BM-40/SPARC/osteonectin family. J Neurochem. 2013;125:399–409. doi: 10.1111/jnc.12212. [DOI] [PubMed] [Google Scholar]
- 66.Hausser HJ, Decking R, Brenner RE. Testican-1, an inhibitor of pro-MMP-2 activation, is expressed in cartilage. Osteoarthritis Cartilage. 2004;12:870–877. doi: 10.1016/j.joca.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 67.Cifuentes-Diaz C, Alliel PM, Charbonnier F, De La Porte S, Molgó J, Goudou D, Rieger F, Périn JP. Regulated expression of the proteoglycan SPOCK in the neuromuscular system. Mech Dev. 2000;94:277–282. doi: 10.1016/s0925-4773(00)00285-9. [DOI] [PubMed] [Google Scholar]
- 68.Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao Y, Wang XA, Zhang F, Xiang SS, Li HF, et al. SPOCK1 as a potential cancer prognostic marker promotes the proliferation and metastasis of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol Cancer. 2015;14(12) doi: 10.1186/s12943-014-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study is available at Gene Expression Omnibus (GEO) under the accession numbers GSE20170 and GSE64216.