Abstract
E2F transcription factors are associated with the development of cancer. However, the E2F family genes have not yet been studied in a comprehensive manner. Using The Cancer Genome Atlas, the present study analyzed the functions of the E2F family genes across different types of tumor. It was revealed that compared with normal tissues, the E2F family genes are highly expressed in several types of tumor tissue. Furthermore, E2F transcription factors were significantly enriched in tumor samples across different types of tumor. The high expression levels of E2F family genes were associated with an unfavorable prognosis in liver hepatocellular carcinoma (LIHC) and lung adenocarcinoma (LUAD). Furthermore, patients with pathological T1 stage and iCluster2 molecular subtype of LIHC expressed particularly low levels of E2F family genes. The present study demonstrated that hypo-DNA methylation, DNA amplification and TP53 mutation contributed to the high expression levels of E2F family genes in cancer cells. Finally, the present study revealed that, compared with other types of tumor, the E2F family genes were specifically downregulated in patients with LIHC. The expression levels and prognostic effects of the E2F family genes were validated using the Gene Expression Omnibus database. The results of the present study revealed the biological functions of E2F family genes in the development of cancer and provided potential biomarkers for further therapeutic studies, particularly for patients with LIHC and LUAD.
Keywords: The Cancer Genome Atlas, E2F transcription factors, liver hepatocellular carcinoma, lung adenocarcinoma, pan-cancer
Introduction
E2F transcription factors include eight genes, E2F1-8 (1), which play critical roles in the regulation of cell proliferation and cell cycle progression through the mediated transcriptional activation of specific downstream target genes (2–4). E2F family genes also regulate autophagy (5), mitochondrial functions (6) and the DNA damage response (7). Increased expression levels of the E2F family genes are observed in a number of different types of cancer. For example, E2F1 and E2F3 are increased in liver cancer, and the overexpression of E2F1 or E2F3 could induce spontaneous liver cancer development in mice (8,9). Furthermore, knockout of E2F8 could protect mice against the development of liver cancer (10). E2F8 is also a therapeutic target for lung cancer. Compared with normal lung cells, lung cancer cells exhibit upregulated E2F8 levels (11). Loss of E2F2 expression could decrease the level of susceptible breast tumor progression by alleviating the Myc-mediated proliferative effects (12). The global E2F signature is associated with clinical outcomes of patients with pancreatic adenocarcinoma (13). Restoring the balance between E2F1 and E2F7 is a therapeutic strategy in head and neck squamous cell carcinomas (14). These findings indicate the universal functions of the E2F family genes in the development of cancer.
However, previous studies on the E2F family genes have focused on a single E2F gene in a limited number of cancer types (8–14). Therefore, there is an urgent need to comprehensively analyze E2F family genes across tumor types. The Cancer Genome Atlas (TCGA) project contains gene expression data and DNA methylation profiles of normal and malignant tissues across several types of cancer (15–18). In addition, genetic aberrations, including gene mutations and DNA amplification, are documented in TCGA database (19–23). The majority of data in the present study was derived from TCGA, allowing the functions of E2F family genes to be investigated in a comprehensive manner.
The present study compared the expression levels of the E2F family genes between normal and cancer tissues in each tumor type. The present study also analyzed the prognostic effects of the E2F family genes. The expression and methylation level of the E2F family genes in different pathological stages and molecular subtypes of different types of tumor were studied. Correlation efficiency and multivariate cox regression analysis were used to determine the associations between E2F family genes. Furthermore, the present study identified key genomic mutations, DNA amplification or epigenetic DNA methylation that contributed to the increased expression levels of the E2F family genes. Overall, the analysis of TCGA database in the present study allowed an improved understanding of the functions of the E2F family genes. The results also indicated the E2F family genes that may serve as potential biomarkers for further therapeutic studies.
Materials and methods
Data acquisition
TCGA data were downloaded from TCGA hub (tcga.xenahubs.net). Pathological and molecular subtype information of patients with liver hepatocellular carcinoma (LIHC) (24) and lung adenocarcinoma (LUAD) (25) was obtained from previously published studies. Oncoprints of the E2F family genes in different types of tumor were downloaded from cbioportal (version 3.2.0; www.cbioportal.org/index.do).
The gene expression series matrix of normal and cancerous liver tissues was downloaded from the Gene Expression Omnibus (GEO) website (www.ncbi.nlm.nih.gov/geo), and included the GEO datasets GSE45436 (26) and GSE55092 (27). The gene expression series matrix of normal and cancerous lung tissues were downloaded from the GSE18442 (28), GSE19188 (29), GSE19804 (30) and GSE27262 (31) datasets. The gene expression and survival data of patients with lung cancer were downloaded from the GSE30219 dataset (32).
TCGA data analysis
Gene expression profiles across cancer types were analyzed using RNA-sequencing data (TCGA HiSeqV2 data). TP53 mutations were analyzed using DNA-sequencing data (TCGA mutation broad data). The DNA methylation profiles were analyzed using HumanMethylation450 microarray data (Methylation_Preprocess.Level3).
iCluster classification of patients with LIHC
iCluster is a novel classification system of patients with LIHC, proposed by TCGA network (24). The DNA copy number, DNA methylation, mRNA expression, miRNA expression and protein expression of 363 patients with LIHC were integrated to divide the patients into three iClusters using a joint multivariate regression (24). The iCluster information of each patient with LIHC was obtained from a previous study (24). The expression levels of the E2F family genes were further determined in each iCluster subtype of patients with LIHC.
GEO data analysis
The expression profiles obtained from the GEO datasets were processed separately. The expression values were averaged if multiple probes corresponded to the same gene symbol using the ‘plyr’ package (version 1.8.5; cran.r-project.org/web/packages/plyr/index.html) in R software (version 3.5.0; www.r-project.org). The paired Student's t-test was used to determine differences in gene expression between normal and cancer samples in R.
Kyoto Encyclopedia of Gens and Genomes signaling pathway enrichment analysis
The signaling pathway enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (version 6.8; david.ncifcrf.gov).
Gene set enrichment analysis (GSEA)
The GSEA of the E2F transcription factors was performed using GSEA software (version 2.0; www.broad.mit.edu/gsea/index.html). Genes were ranked by the signal-to-noise ratio, and statistical significance was determined by 1,000 gene set permutations.
Survival analysis
The Kaplan-Meier Plotter (kmplot.com/analysis/index.php?p=service) was used to identify the association between the expression levels of the E2F family genes and overall survival in patients with LIHC and LUAD derived from TCGA database and GEO datasets. The associations between the expression levels of the E2F family genes and overall survival in patients with lung cancer from the GSE30219 dataset were determined using the ‘survival’ package (version 3.1; cran.r-project.org/web/packages/survival/index.html) in R. Patients were divided into high and low expression groups using the mean expression level of E2F family genes. P-values were determined by the log-rank test.
Heatmaps
Heatmaps were created using the ‘pheatmap’ package (version 1.0.12; cran.r-project.org/web/packages/pheatmap/index.html) in R. The clustering scale was determined by the ‘average’ method. The clustering distance was determined by the ‘correlation’ method.
Statistical analysis
Box plots and contingency graphs were generated using GraphPad Prism software (version 5.0; GraphPad Software, Inc.). Statistical analysis was performed using the paired Student's t-test or χ2 test using R software. Multivariate cox regression analyses of the E2F family genes were performed using the ‘survival’ package (version 3.1; cran.r-project.org/web/packages/survival/index.html) in R. Correlation plots of the E2F family genes were created using the ‘corrplot’ package (version 0.84; cran.r-project.org/web/packages/corrplot/index.html) in R. The Spearman's correlation test was used to demonstrate the correlation between E2F family genes. P<0.05 was considered to indicate a statistically significant difference.
Results
E2F family genes are upregulated in several types of tumor
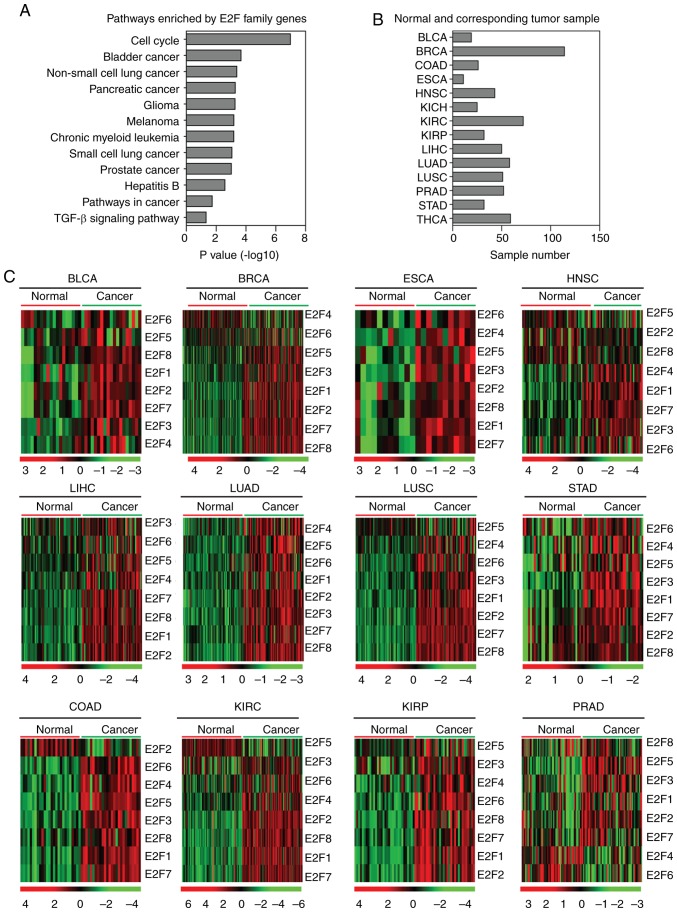
The E2F transcription factor family contains eight members, E2F1-8 (3). The present study identified the enriched signaling pathways of the eight E2F family genes, and the results revealed that the cell cycle signaling pathway was the most highly enriched (Fig. 1A). Notably, the majority of the enriched pathways were tumor-specific-associated pathways, for example, bladder cancer, non-small cell lung cancer and pancreatic cancer pathways (Fig. 1A). E2F family genes were also associated with the transforming growth factor β (TGFβ) signaling pathway (Fig. 1A). All the enriched signaling pathways highlighted the universal functions of the E2F family genes in cancer development in several types of tumor. However, the E2F family genes have not yet been studied in a comprehensive pan-cancer manner.
Figure 1.
Expression of E2F family genes is increased in several tumor types. (A) Functional pathway enrichment analysis of the E2F family genes using the Database for Annotation, Visualization and Integrated Discovery. The most significantly enriched pathways were shown. (B) Box plots demonstrated the number of normal and corresponding tumor samples used in the present study. (C) Heatmaps demonstrated the expression levels (log2 count) of E2F family genes in normal and tumor samples in BLCA, BRCA, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC, PRAD and STAD. Upregulated (red), downregulated (green) and unchanged (black) genes were delineated. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma.
The present study used TCGA database to integrate the analysis of the E2F family genes across different types of cancer. The expression levels of the E2F family genes in normal tissues and corresponding tumor tissues were investigated. As illustrated in the heatmaps (Fig. 1B), the majority of the E2F family genes were upregulated in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), LIHC, LUAD, lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD) tumor tissues compared with normal tissues (Fig. 1C). In particular, compared with the normal tissues, all the E2F family genes were highly expressed in LIHC, LUAD and STAD tumor tissues (Fig. 1C). However, it was revealed that the majority of the E2F family genes were not altered in prostate adenocarcinoma (PRAD; Fig. 1C). For the other types of tumor, the E2F family genes were all highly expressed in the corresponding tumor samples, excluding E2F4 in BRCA, E2F2 in COAD, and E2F5 in HNSC, KIRC, KIRP and LUSC.
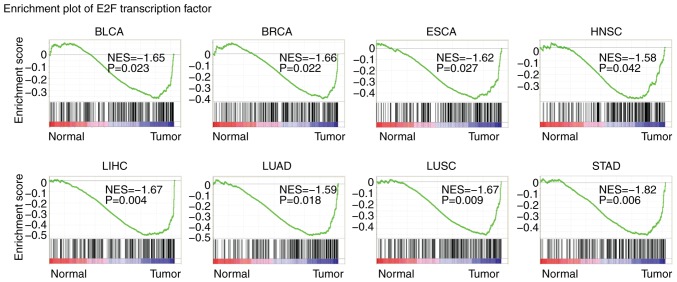
E2F transcription factors are highly enriched in several types of tumors
The present study identified the transcription factors enriched in tumors using GSEA (33). Among the transcription factors, E2F was significantly enriched in BLCA, BRCA, ESCA, HNSC, LIHC, LUAD, LUSC and STAD tumors, representing the most frequently enriched transcription factor (Fig. 2). These results indicated the universal importance of E2F transcription factors in the development of cancer.
Figure 2.
E2F transcription factors are highly enriched in several types of tumor. Enrichment plots demonstrated the E2F transcriptions factors in BLCA, BRCA, ESCA, HNSC, LIHC, LUAD, LUSC and STAD. Enrichment of NES and P-values are presented. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; STAD, stomach adenocarcinoma; NES, normalized enrichment score.
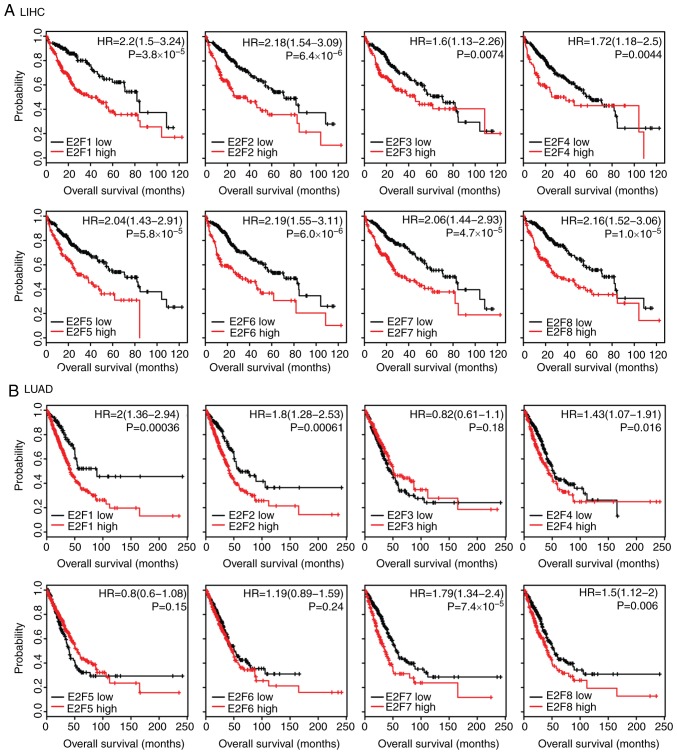
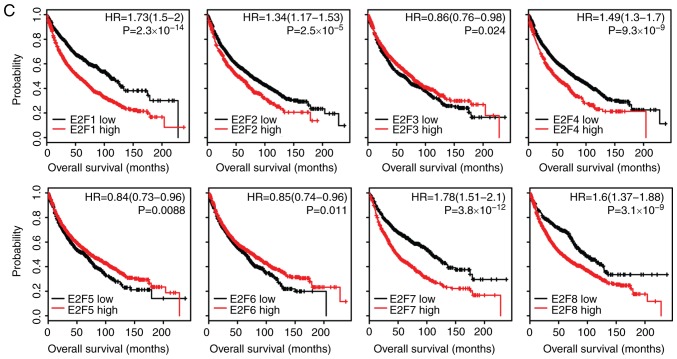
Expression levels of E2F family genes are associated with the overall survival in patients with LIHC and LUAD
The present study assessed whether the E2F family genes had prognostic effects in tumor progression. Using the Kaplan-Meier Plotter (34), the present study identified the association between the expression levels of the E2F family genes and overall survival in several types of cancer. The results revealed that, high expression levels of the E2F family genes were associated with an unfavorable prognosis, particularly in LIHC and LUAD (Fig. 3). Patients with LIHC with higher expression levels of E2F1-8 had worse prognosis than patients with low expression (Fig. 3A). Furthermore, the Kaplan-Meier survival analysis revealed that E2F1, E2F2, E2F4, E2F7 and E2F8 were all associated with overall survival in patients with LUAD (Fig. 3B). However, the E2F family genes had no or little prognostic effect in other types of tumor, including LUSC (Fig. S1A) or STAD (Fig. S1B). These results suggested that although the E2F family genes were highly expressed across tumor types, the E2F family genes were more important in the development of LIHC and LUAD compared with other tumor types.
Figure 3.
Expression levels of E2F family genes are associated with the overall survival in patients with LIHC and LUAD. (A) The Kaplan-Meier Plotter demonstrated the associations between E2F1-8 and overall survival in patients with LIHC. The log-rank test was used to determine the overall survival P-value. (B) The Kaplan-Meier Plotter demonstrated the association between E2F1-8 and overall survival in patients with LUAD. LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; HR, hazard ratio.
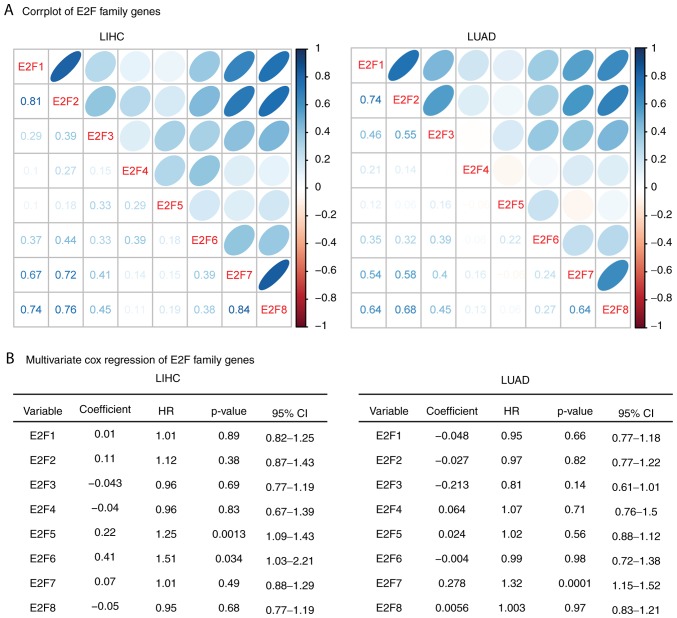
Correlation or independence of the E2F family genes is identified in patients with LIHC and LUAD
The results of the present study demonstrated the similar functions of the E2F family genes in overall survival in patients with LIHC and LUAD. Thus, the present study aimed to determine the connection between E2F family genes. The Spearman's correlation test demonstrated a high correlation between E2F1, E2F7 and E2F8 in LIHC and LUAD expression datasets (Fig. 4A). E2F2 was also highly correlated with E2F7 and E2F8 in LIHC and LUAD. Additionally, E2F7 and E2F8 were correlated with each other (Fig. 4A).
Figure 4.
Correlation or independence of the E2F family genes in patients with LIHC and LUAD. (A) Corrplots demonstrated the correlation between each E2F family gene in LIHC and LUAD. The numbers and the size of the circle represented the correlation coefficients. (B) Multivariate cox regression was used to test the association between E2F family genes in determining the overall survival of patients with LIHC and LUAD. LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; HR, hazard ratio; CI, confidence interval.
The present study also used multivariate cox regression analyses to reveal the connection that the E2F family genes have in determining the overall survival of patients with LIHC and LUAD. It was revealed that E2F5 and E2F6 were independent prognostic markers in patients with LIHC, and E2F7 was an independent prognostic marker in patients with LUAD (Fig. 4B). These results were consistent with the gene expression data, and E2F5 and E2F6 demonstrated little connection with other E2F family genes (Fig. 4A). E2F1-4, E2F7 and E2F8 genes were interconnected with each other, so those genes were not independent prognostic markers (Fig. 4B).
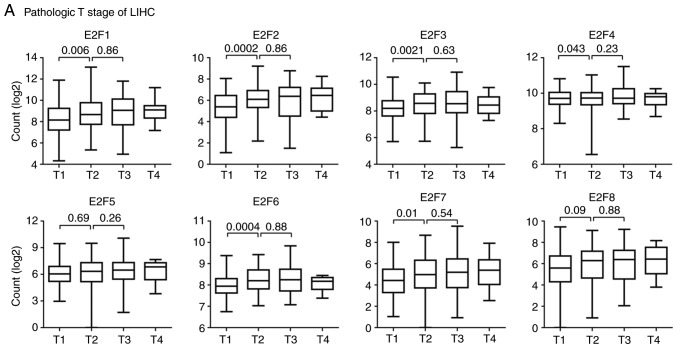
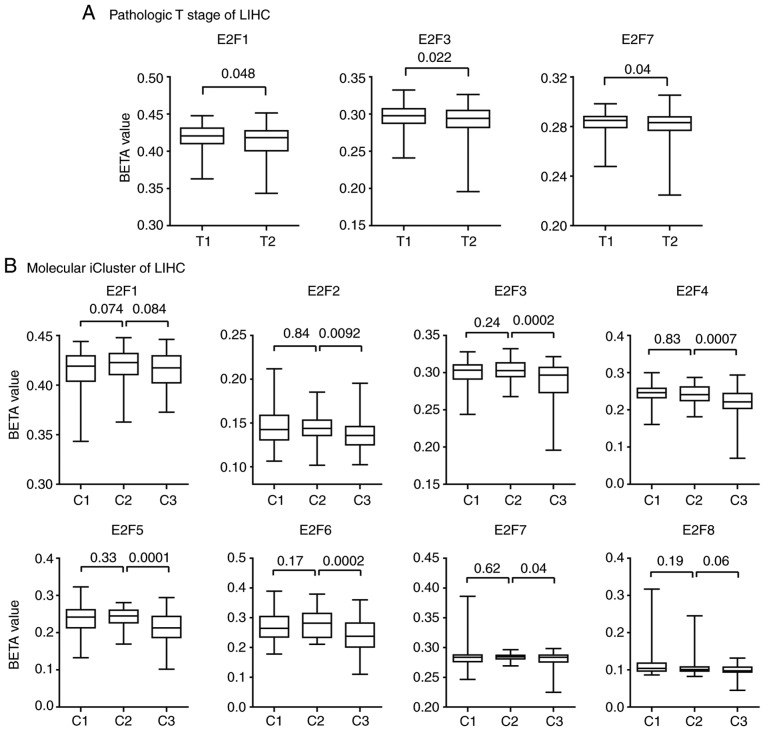
E2F family genes are downregulated in patients with T1 stage and iCluster2 LIHC subtypes
The Kaplan-Meier Plotter analysis suggested that the E2F family genes may play more important roles in LIHC than other tumor types (Fig. 3A). LIHC is a heterogeneous disease, reflected by differences in genomic or epigenetic aberrations, pathological stages and responses to therapies (35–37). Pathological stages are basic prognostic information to evaluate the malignant characteristics of the disease (38). The present study then tested the expression levels of E2F family genes in patients with different pathological stages of LIHC.
The majority of patients (98%) with LIHC in TCGA database were in the M0 or N0 stage, without distant or lymphatic metastasis. However, the expression levels of the E2F family genes in different pathological T stages were quite different. Compared with patients with T1 stage LIHC, E2F1-4, E2F6 and E2F7 were all highly expressed in patients with T2 stage LIHC (Fig. 5A). However, the expression levels of E2F5 and E2F8 in patients with T1 and T2 stage LIHC were not different. Furthermore, there were no differences observed between the expression level of the E2F family genes and patients with T2 stage or T3 stage LIHC (Fig. 5A).
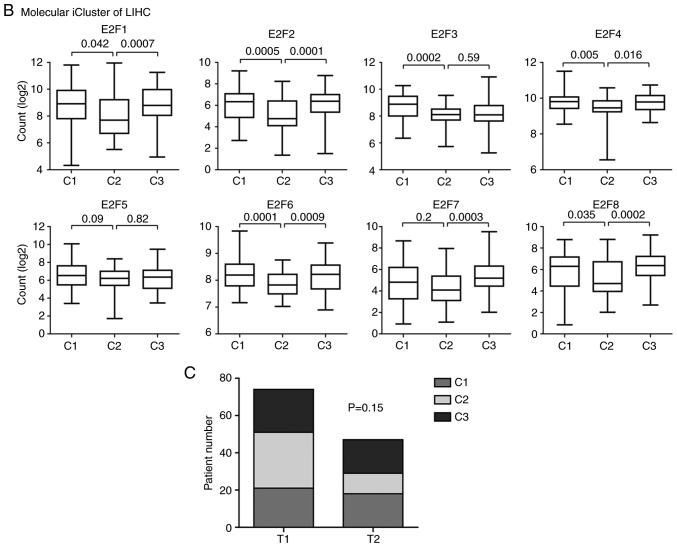
Figure 5.
E2F family genes are downregulated in patients with LIHC with T1 stage and the iCluster2 subtype. Box plots demonstrated the expression levels of the E2F family genes in different LIHC (A) pathological T stages and (B) iCluster. (C) The contingency graph demonstrated the distribution of T1 and T2 stage of patients with LIHC in each iCluster subtype. P-values were determined using the χ2 test. LIHC, liver hepatocellular carcinoma.
Besides the basic pathological classification of LIHC, patients with LIHC were divided into three iClusters based on genomic alterations, gene expression profiles and DNA methylation aberrations (24). Furthermore, patients with different iCluster subtype demonstrated different clinical outcomes. Patients with the iCluster1substype had a worse prognosis than patients with the iCluster2 and iCluster3 subtypes (24). The present study then assessed the expression levels of the E2F family genes in patients with different iCluster subtypes of LIHC. Compared with the iCluster1 or iCluster3 subtypes, the expression levels of E2F1-4, E2F6 and E2F7 were relatively lower in patients with iCluster2 subtype of LIHC (Fig. 5B). This low expression of the E2F family genes in iCluster2 was not due to the high proportion of patients with T1 stage LIHC (Fig. 5C).
The present study also assessed the expression levels of the E2F family genes in different pathological stages or molecular subtypes of patients with LUAD (25). However, no significantly different expression levels of E2F family genes were observed in different pathological stages (Fig. S2). E2F2 and E2F8 were highly expressed in patients with T2 stage LUAD compared with patients with T1 stage (Fig. S2).
Increased expression levels of the E2F family genes are induced by multiple levels of genomic or epigenetic aberrations
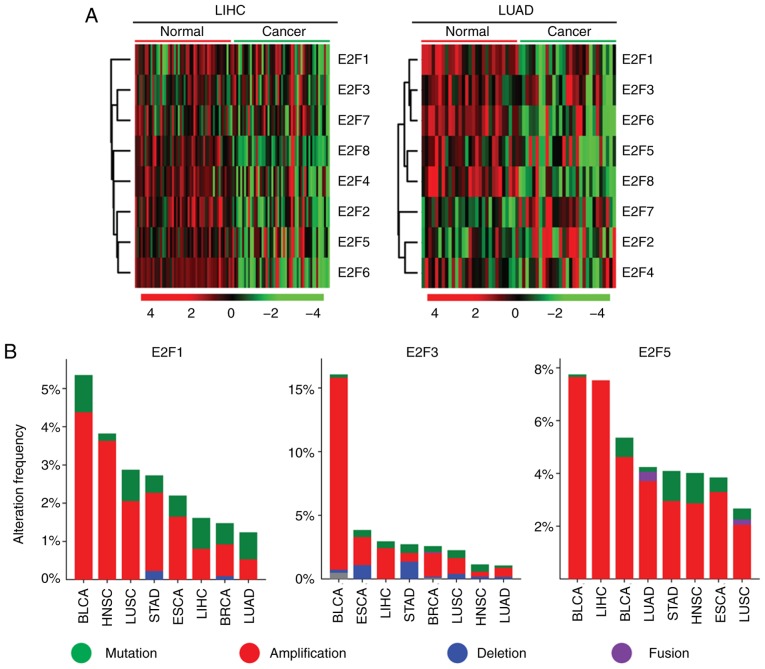
The present study aimed to determine the mechanisms that induced the activation of the E2F family genes in tumor development. The high expression levels of oncogenes are usually mediated by hypo-DNA methylation, DNA amplification and gene mutation (16). Compared with normal tissues, it was revealed that the E2F family genes exhibited hypo-DNA methylation patterns in LIHC and LUAD tumor tissues (Fig. 6A). Particularly LIHC, E2F2, E2F4, E2F5, E2F5 and E2F8 genes exhibited hypo-DNA methylation in tumor samples (Fig. 6A), and E2F1, E2F3 and E2F6 genes exhibited hypo-DNA methylation in LUAD (Fig. 6A). These observations suggested that DNA methylation was partially contributing to the activation of E2F family genes in the tumor cells.
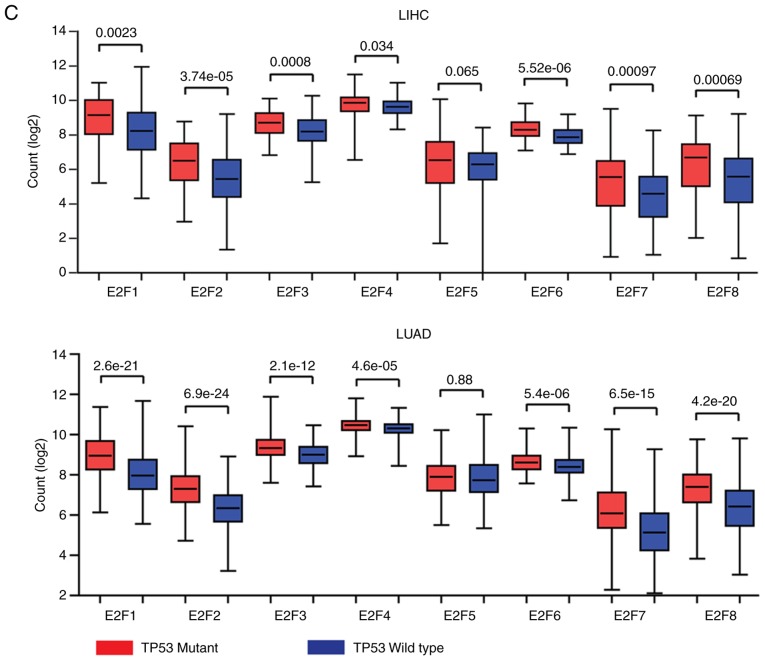
Figure 6.
Increased expression levels of the E2F family genes are induced by multiple levels of genomic or epigenetic aberrations. (A) Heatmaps demonstrated the methylation level (β value) of the E2F family genes in normal and tumor samples in patients with LIHC and LUAD. Hypermethylated (red), hypomethylated (green) and unchanged (black) genes were delineated. (B) Oncoprints demonstrated the alteration frequency of E2F1, E2F3 and E2F5 genes in patients with BLCA, HNSC, LUSC, STAD, ESCA, LIHC, BRCA and LUAD. (C) Box plots demonstrated the expression levels of the E2F family genes in patients with LIHC and LUAD. P-values indicated the differences between patients with TP53 mutant and TP53 wild-type cancer types. LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma.
Another factor determining the activation of E2F genes in tumor cells was genomic aberration, particularly DNA amplification. It was revealed that the E2F1 and E2F3 genes were present in higher proportions of DNA amplification in BLCA (Fig. 6B). BRCA, LIHC and BLCA tumor types also exhibited high levels of E2F5 amplification (Fig. 6B).
TP53 is a driver of mutations in several types of tumor (23,40,41). Loss of TP53 functions induces uncontrolled cell cycle progression, and resistance to cell apoptosis (42–44). The present study assessed whether TP53 regulated the expression levels of the E2F family genes. It was revealed that, except for E2F5, the other E2F family genes were all highly expressed in patients with TP53 mutant LIHC and LUAD, compared with those patients with TP53 wild-type LIHC and LUAD (Fig. 6C). Overall, the present study speculated that hypo-DNA methylation, DNA amplification and TP53 mutation were contributing to the high expression levels of E2F family genes in cancer cells.
E2F family genes exhibit hyper-DNA methylation patterns in patients with iCluster2 subtype LIHC
The present study demonstrated that the E2F family genes were downregulated in patients with pathological T1 stage and molecular iCluster2 subtypes of LIHC. However, the mechanisms underlying this phenotype were not clear. The present study analyzed the methylation patterns of the E2F family genes in patients with LIHC. It was revealed that the E2F family genes E2F1, E2F3 and E2F7 exhibited hyper-DNA methylation patterns in patients with T1 stage LIHC compared with those patients with T2 stage LIHC (Fig. 7A). No significantly different methylation intensities of other E2F family genes were observed between patients with pathological T1 and T2 LIHC.
Figure 7.
E2F family genes with hyper-DNA methylation patterns in patients with the iCluster2 subtype of LIHC. (A) Box plots demonstrated the methylation level (β value) of E2F1, E2F3 and E2F7 genes in patients with pathological T1 and T2 stage of LIHC. (B) Box plots demonstrated the methylation level of the E2F family genes in patients with different iCluster subtypes of LIHC. LIHC, liver hepatocellular carcinoma.
The methylation patterns of the E2F family genes in patients with different molecular iClusters of LIHC were also analyzed. Consistent with the low expression levels of the E2F family genes in patients with iCluster2 subtype of LIHC, the present study revealed that the methylation intensity of E2F2-7 in patients with LIHC was relatively higher, compared with patients with iCluster3 subtype LIHC. However, there was no statistically significant difference in methylation intensity in the E2F family genes between patients with iCluster1 and iCluster2 subtypes of LIHC (Fig. 7B).
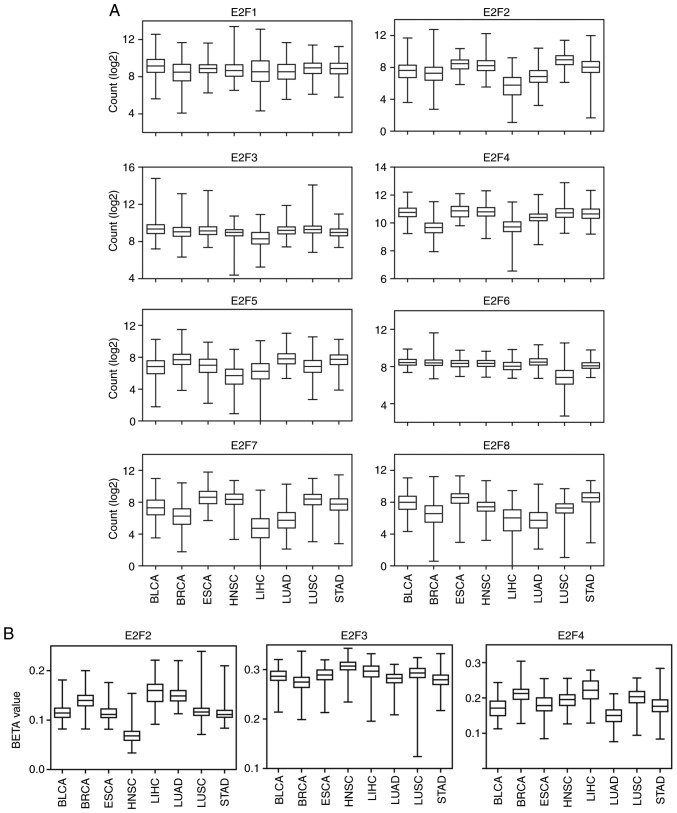
E2F family genes are expressed in particularly low levels in patients with LIHC
The present study compared the expression levels of E2F family genes in BLCA, BRCA, ESCA, HNSC, LIHC, LUAD, LUSC and STAD. It was revealed that, compared with other tumor types, E2F2-4, E2F7 and E2F8 were all expressed at relatively low levels in LIHC tumors (Fig. 8A). In addition, the methylation intensity of E2F2-4 was relatively higher in LIHC compared with other tumor types (Fig. 8B). However, the methylation level of E2F3 was higher in HNSC than in LIHC (Fig. 8B).
Figure 8.
E2F family genes are expressed in particularly low levels in patients with LIHC. (A) Box plots demonstrated the expression levels of the E2F family genes (log2 count) in different types of tumor. (B) Box plots demonstrated the methylation level (β value) of the E2F family genes in different types of tumor. LIHC, liver hepatocellular carcinoma.
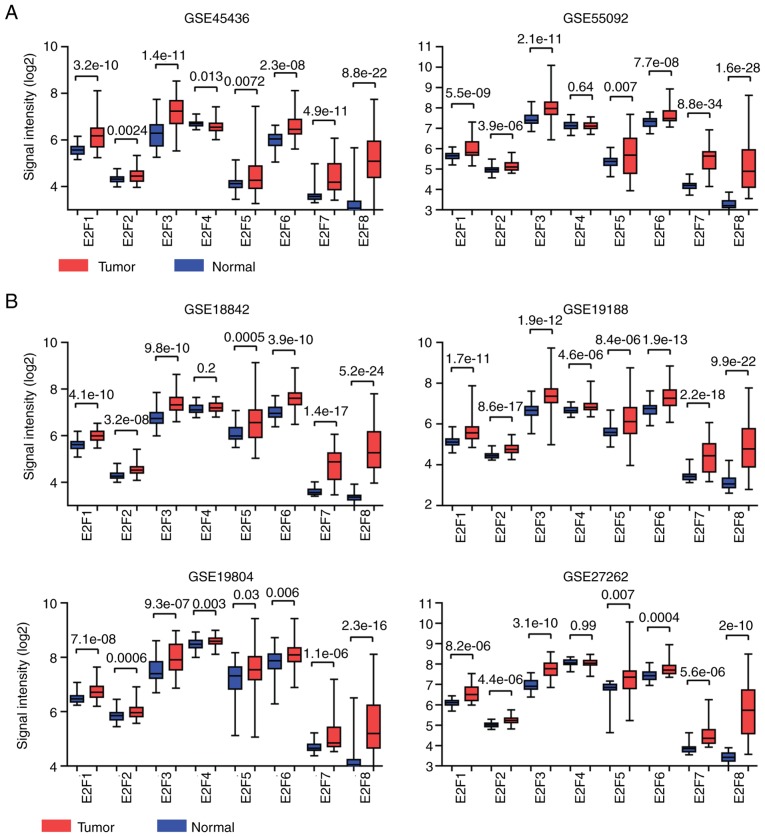
Expression and prognostic effects of the E2F family genes are validated from GEO datasets
From TCGA database, it was revealed that the majority of the E2F family genes were upregulated in several types of tumor tissue, and the upregulated E2F family genes were associated with worse prognosis in LIHC and LUAD tumor types. In order to further confirm these findings, the present study analyzed the functions of the E2F family genes from GEO datasets.
The GSE45436 and GSE55092 datasets included the expression profiles derived from normal liver and malignant liver tissues (27). The expression levels of the E2F family genes varied significantly between normal liver or malignant tissues. E2F3 was highly expressed, while the expression levels of E2F7 and E2F8 were relatively lower in liver tissues (Fig. 9A). However, compared with the normal liver tissues, the E2F family genes E2F1-8 were all highly expressed in liver cancer tissues (Fig. 9A). However, the expression of E2F4 between normal liver and cancerous liver tissues was not significantly different in the GSE55092 dataset (Fig. 9A).
Figure 9.
Expression levels and prognostic effects of the E2F family genes were validated from GEO datasets. (A) Box plots demonstrated the expression levels of the E2F family genes in the GSE45426 and GSE55092 datasets. P-values indicated the significance of different expression levels of the E2F family genes between normal liver tissues and cancerous liver tissues. (B) Box plots demonstrated the expression levels of the E2F family genes in the GSE18842, GSE19188, GSE19804 and GSE27262 datasets. P-values indicated the significance of different expression levels of the E2F family genes between normal lung tissues and cancerous lung tissues. (C) Association between E2F1-8 and overall survival in patients with lung cancer. The results were derived from combined multiple GEO datasets. GEO, Gene Expression Omnibus; HR, hazard ratio.
The expression levels of the E2F family genes between normal lung and malignant lung tissues were also analyzed using the GSE18842, GSE19188, GSE19804 and GSE27262 GEO datasets (28–31). Similar to liver tissues, E2F3 was highly expressed, while the expression levels of E2F7 and E2F8 were relatively lower in normal lung or malignant tissues (Fig. 9B). In addition, compared with the normal lung tissues, the E2F family genes were all highly expressed in lung cancer tissues, except E2F4 (Fig. 9B). These results were relatively consistent with the findings derived from TCGA database.
The Kaplan-Meier Plotter analysis also provided prognostic values of genes using combined GEO transcriptional data in non-small-cell lung cancer (45). Similar to TCGA data, high expression levels of the E2F family genes E2F1, E2F2, E2F4, E2F7 and E2F8 were all associated with low overall survival in patients with lung cancer, while, high expression levels of E2F3, E2F5 and E2F6 were associated with an improved prognosis in patients with LUAD (Fig. 9C).
Correlation between E2F family genes is validated in the GSE30219 lung cancer expression dataset
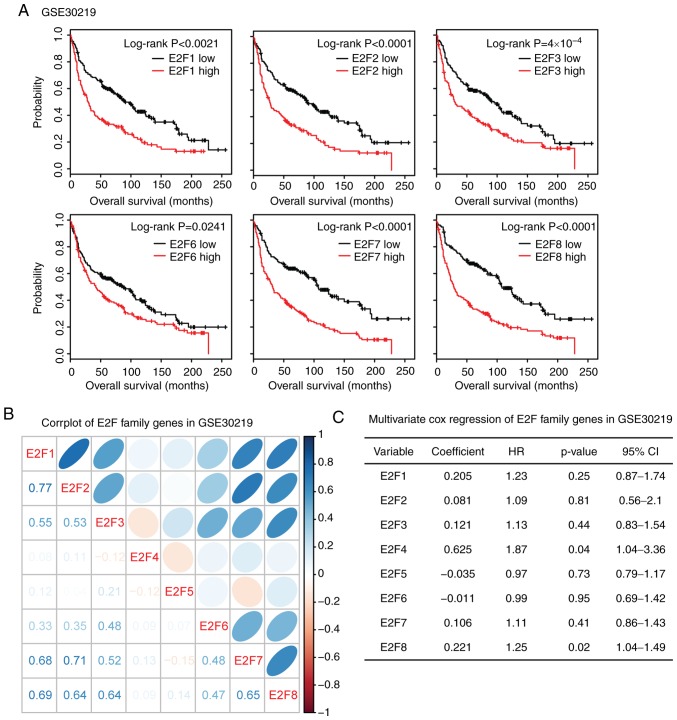
The present study validated the correlation of E2F family genes in the GSE30219 lung cancer expression dataset. This dataset was selected as it included both gene expression and overall survival time of patients with lung cancer (32). High expression levels of E2F1-3 and E2F6-8 were associated with poor overall survival (Fig. 10A). Results derived from this dataset suggested a strong correlation among E2F1, E2F7 and E2F8 (Fig. 10B). E2F2 was also strongly correlated with E2F7 and E2F8. In addition, E2F7 and E2F8 were correlated with each other (Fig. 10B). Multivariate cox regression analysis revealed those genes interconnected with each other in determining the overall survival of patients with LUAD. Only E2F4 and E2F8 were independent prognostic markers (Fig. 10C). All these results were quite similar to those derived from TCGA database.
Figure 10.
Correlation among E2F family genes was validated in the GSE30219 lung cancer expression dataset. (A) The Kaplan-Meier Plotter demonstrated the association between E2F1-3 and E2F6-8 and overall survival in patients with lung cancer. The results were derived from the GSE30219 dataset. (B) Corrplots demonstrated the correlation between each E2F transcription factor in the GSE30219 dataset. (C) Multivariate cox regression was used to test the association between the E2F family genes in determining the overall survival of patients with lung cancer.
Discussion
Using TCGA database, the present study analyzed the transcriptional profiles, DNA methylation characteristics, DNA amplification features and prognostic effects of the E2F family genes in several tumor types. Compared with the normal tissues, E2F family genes were highly expressed in multiple cancer tissues, including BLCA, BRCA, COAD, ESCA, HNSC, KIRC, KIRP, LIHC, LUAD, LUSC and STAD. Furthermore, the E2F transcription factors were highly enriched in the aforementioned tumors. The results of the present study suggested that the E2F family genes played important and universal roles in cancer development. Furthermore, since the E2F family genes were mostly associated with the cell cycle signaling pathway, the results supported previous observations that uncontrolled cell cycle progression was a hallmark of cancer (46,47).
The potential prognostic roles of E2F family genes in breast cancer (48), gastric cancer (49), liver cancer (50) and non-small-cell lung cancer (51) have been previously studied. For example, higher E2F1, E2F3, E2F5, E2F7 and E2F8 expression levels were significantly associated with lower overall survival in patients with breast cancer (48). E2F1, E2F3 and E2F4 were significantly associated with unfavorable outcomes in patients with gastric cancer (49). E2F1 and E2F2 upregulation was significantly correlated with poor prognosis in patients with non-small-cell lung cancer (51). However, in the present study, the E2F family genes had no or little prognostic effect in LUSC and STAD. On the contrary, the high expression levels of E2F family genes, E2F1-8, were all associated with unfavorable outcomes in patients with LIHC. In patients with LUAD, E2F1, E2F2, E2F4, E2F7 and E2F8, were significantly associated with unfavorable outcomes. The data from the present study suggested that the E2F family genes had more important functions in LIHC and LUAD than in other tumor types.
The expression levels of E2F family genes in tumor cells are regulated by several molecular mechanisms. In retinoblastoma (RB), RB protein was a critical regulator of E2F family genes (52). The present study demonstrated that, in LIHC and LUAD patients, the hypomethylation of the E2F family genes in tumor samples may explain the high expression levels of these genes in the tumor tissues. Moreover, in BLCA, the high expression levels of the E2F family genes may be due to DNA amplification. Furthermore, BRCA and LIHC tumor types exhibited high levels of E2F5 amplification. TP53 tumor suppressor is also a critical regulator of E2F family genes. E2F7 is a TP53 direct transcriptional target gene, connecting TP53 with RB protein (53). E2F family genes and TP53 bind the same promoter region to regulate downstream target genes (54). The present study demonstrated that, except for E2F5, the E2F family genes were all highly expressed in patients with TP53 mutant LIHC and LUAD, compared with patients with TP53 wild-type LIHC and LUAD.
Overall, the analysis in the present study provided a more in-depth understanding of the biological functions of the E2F family genes. However, the results require further clinical validation. The results of the present study suggested that the E2F family genes may serve as important biomarkers and therapeutic targets, particularly for patients with LIHC and LUAD.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Fujian Provincial Maternity and the Children's Hospital (grant nos. YCXB 18-10 and YCXM 19-04).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in TCGA (tcga.xenahubs.net) and GEO (www.ncbi.nlm.nih.gov/geo) repositories.
Authors' contributions
HW designed the study and wrote the manuscript. HW, XW and LX performed the data analysis. HC and JZ designed the study and supervised the work.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 2.Liu H, Tang X, Srivastava A, Pécot T, Daniel P, Hemmelgarn B, Reyes S, Fackler N, Bajwa A, Kladney R, et al. Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles. Nat Cell Biol. 2015;17:1036–1048. doi: 10.1038/ncb3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgunova E, Yin Y, Jolma A, Dave K, Schmierer B, Popov A, Eremina N, Nilsson L, Taipale J. Structural insights into the DNA-binding specificity of E2F family transcription factors. Nat Commun. 2015;6:10050. doi: 10.1038/ncomms10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893. doi: 10.1158/0008-5472.CAN-10-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benevolenskaya EV, Frolov MV. Emerging links between E2F control and mitochondrial function. Cancer Res. 2015;75:619–623. doi: 10.1158/0008-5472.CAN-14-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertoli C, Herlihy AE, Pennycook BR, Kriston-Vizi J, de Bruin RAM. Sustained E2F-dependent transcription is a key mechanism to prevent replication-stress-induced DNA damage. Cell Rep. 2016;15:1412–1422. doi: 10.1016/j.celrep.2016.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kent LN, Bae S, Tsai SY, Tang X, Srivastava A, Koivisto C, Martin CK, Ridolfi E, Miller GC, Zorko SM, et al. Dosage-Dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest. 2017;127:830–842. doi: 10.1172/JCI87583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarangelo A, Lo N, Teng R, Kim E, Le L, Watson D, Furth EE, Raman P, Ehmer U, Viatour P. Recruitment of pontin/reptin by E2f1 amplifies E2f transcriptional response during cancer progression. Nat Commun. 2015;6:10028. doi: 10.1038/ncomms10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent LN, Rakijas JB, Pandit SK, Westendorp B, Chen HZ, Huntington JT, Tang X, Bae S, Srivastava A, Senapati S, et al. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest. 2016;126:2955–2969. doi: 10.1172/JCI85506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park SA, Platt J, Lee JW, Lopez-Giraldez F, Herbst RS, Koo JS. E2F8 as a novel therapeutic target for lung cancer. J Natl Cancer Inst. 2015;18:107. doi: 10.1093/jnci/djv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara K, Yuwanita I, Hollern DP, Andrechek ER. Prediction and genetic demonstration of a role for activator E2Fs in myc-induced tumors. Cancer Res. 2011;71:1924–1932. doi: 10.1158/0008-5472.CAN-10-2386. [DOI] [PubMed] [Google Scholar]
- 13.Lan W, Bian B, Xia Y, Dou S, Gayet O, Bigonnet M, Santofimia-Castaño P, Cong M, Peng L, Dusetti N, Iovanna J. E2F signature is predictive for the pancreatic adenocarcinoma clinical outcome and sensitivity to E2F inhibitors, but not for the response to cytotoxic-based treatments. Sci Rep. 2018;8:8330. doi: 10.1038/s41598-018-26613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saenz-Ponce N, Pillay R, de Long LM, Kashyap T, Argueta C, Landesman Y, Hazar-Rethinam M, Boros S, Panizza B, Jacquemyn M, et al. Targeting the XPO1-dependent nuclear export of E2F7 reverses anthracycline resistance in head and neck squamous cell carcinomas. Sci Transl Med. 2018;27:447. doi: 10.1126/scitranslmed.aar7223. [DOI] [PubMed] [Google Scholar]
- 15.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun W, Bunn P, Jin C, Little P, Zhabotynsky V, Perou CM, Hayes DN, Chen M, Lin DY. The association between copy number aberration, DNA methylation and gene expression in tumor samples. Nucleic Acids Res. 2018;46:3009–3018. doi: 10.1093/nar/gky131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisenberger DJ. Characterizing DNA methylation alterations from the cancer genome atlas. J Clin Invest. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-Cancer landscape of aberrant DNA methylation across human tumors. Cell Rep. 2018;25:1066–1080. doi: 10.1016/j.celrep.2018.09.082. [DOI] [PubMed] [Google Scholar]
- 19.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, Cancer Genome Atlas Research Network The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutter C, Zenklusen JC. The cancer genome atlas: Creating lasting value beyond its data. Cell. 2018;173:283–285. doi: 10.1016/j.cell.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 21.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;35 doi: 10.1126/science.aan2507. eaan2507. [DOI] [PubMed] [Google Scholar]
- 22.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173:291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, Colaprico A, Wendl MC, Kim J, Reardon B, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network. Electronic address, corp-author. wheeler@bcm.edu and CancerGenome Atlas Research Network: Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang HW, Hsieh TH, Huang SY, Chau GY, Tung CY, Su CW, Wu JC. Forfeited hepatogenesis program and increased embryonic stem cell traits in young hepatocellular carcinoma (HCC) comparing to elderly HCC. BMC Genomics. 2013;14:736. doi: 10.1186/1471-2164-14-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melis M, Diaz G, Kleiner DE, Zamboni F, Kabat J, Lai J, Mogavero G, Tice A, Engle RE, Becker S, et al. Viral expression and molecular profiling in liver tissue versus microdissected hepatocytes in hepatitis B virus-associated hepatocellular carcinoma. J Transl Med. 2014;12:230. doi: 10.1186/s12967-014-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Fárez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 29.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fouret R, Laffaire J, Hofman P, Beau-Faller M, Mazieres J, Validire P, Girard P, Camilleri-Bröet S, Vaylet F, Leroy-Ladurie F, et al. A comparative and integrative approach identifies ATPase family, AAA domain containing 2 as a likely driver of cell proliferation in lung adenocarcinoma. Clin Cancer Res. 2012;18:5606–5616. doi: 10.1158/1078-0432.CCR-12-0505. [DOI] [PubMed] [Google Scholar]
- 31.Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC, Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103:1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY, Lantuejoul S, Hainaut P, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy A, Lanczky A, Menyhart O, Gyorffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8:9227. doi: 10.1038/s41598-018-29514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, et al. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416–423. doi: 10.1038/nm843. [DOI] [PubMed] [Google Scholar]
- 36.Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, Lim JY, Heo J, Kim YJ, et al. Sixty-five gene-based risk score classifier predicts overall survival in hepatocellular carcinoma. Hepatology. 2012;55:1443–1452. doi: 10.1002/hep.24813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kate RJ, Nadig R. Stage-Specific predictive models for breast cancer survivability. Int J Med Inform. 2017;97:304–311. doi: 10.1016/j.ijmedinf.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376:2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 40.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bertucci F, Ng CKY, Patsouris A, Droin N, Piscuoglio S, Carbuccia N, Soria JC, Dien AT, Adnani Y, Kamal M. Genomic characterization of metastatic breast cancers. Nature. 2019;569:560–564. doi: 10.1038/s41586-019-1056-z. [DOI] [PubMed] [Google Scholar]
- 42.Vousden KH, Lane DP. P53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 43.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 45.Gyorffy B, Surowiak P, Budczies J, Lanczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Huang J, Yang D, Xiang S, Sun J, Li H, Ren G. Expression patterns of E2F transcription factors and their potential prognostic roles in breast cancer. Oncol Lett. 2018;15:9216–9230. doi: 10.3892/ol.2018.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manicum T, Ni F, Ye Y, Fan X, Chen BC. Prognostic values of E2F mRNA expression in human gastric cancer. Biosci Rep. 2018;21:38. doi: 10.1042/BSR20181264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang YL, Ning G, Chen LB, Lian YF, Gu YR, Wang JL, Chen DM, Wei H, Huang YH. Promising diagnostic and prognostic value of E2Fs in human hepatocellular carcinoma. Cancer Manag Res. 2019;11:1725–1740. doi: 10.2147/CMAR.S182001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Z, Shi R, Yuan K, Wang Y. Expression and prognostic value of E2F activators in NSCLC and subtypes: A research based on bioinformatics analysis. Tumour Biol. 2016;37:14979–14987. doi: 10.1007/s13277-016-5389-z. [DOI] [PubMed] [Google Scholar]
- 52.Narita M, Nunez S, Heard E, Narita M, Lin AW, Hearn SA, Spector DL, Hannon GJ, Lowe SW. Rb-Mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/S0092-8674(03)00401-X. [DOI] [PubMed] [Google Scholar]
- 53.Aksoy O, Chicas A, Zeng T, Zhao Z, McCurrach M, Wang X, Lowe SW. The atypical E2F family member E2F7 couples the p53 and RB pathways during cellular senescence. Genes Dev. 2012;26:1546–1557. doi: 10.1101/gad.196238.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ran LK, Chen Y, Zhang ZZ, Tao NN, Ren JH, Zhou L, Tang H, Chen X, Chen K, Li WY, et al. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated X protein-dependent apoptotic pathway. Clin Cancer Res. 2016;22:3372–3382. doi: 10.1158/1078-0432.CCR-15-1638. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in TCGA (tcga.xenahubs.net) and GEO (www.ncbi.nlm.nih.gov/geo) repositories.