Abstract
Glioblastoma (GBM) is a lethal brain cancer with a median survival time of approximately 15 months following treatment. Common in vitro GBM models for drug screening are adherent and do not recapitulate the features of human GBM in vivo. Here we report the genomic characterization of nine patient derived, spheroid GBM cell lines that recapitulate human GBM characteristics in orthotopic xenograft models. Genomic sequencing revealed the spheroid lines contain alterations in GBM driver genes such as PTEN, CDKN2A, and NF1.
Two spheroid cell lines, JHH-136 and JHH-520, were utilized in a high-throughput drug screen for cell viability using a 1,912 member compound library. Drug mechanisms that were cytotoxic in both cell lines were Hsp-90 and proteasome inhibitors. JHH-136 was uniquely sensitive to Topoisomerase 1 inhibitors while JHH-520 was uniquely sensitive to Mek inhibitors. Drug combination screening revealed that PI3 Kinase inhibitors combined with Mek or proteasome inhibitors were synergistic. However, animal studies to test these drug combinations in vivo revealed that Mek inhibition alone was superior to the combination treatments. This data shows these GBM spheroid lines are amenable to high-throughput drug screening and that this dataset may deliver promising therapeutic leads for future GBM preclinical studies.
Keywords: glioblastoma, high-throughput screening, spheroid, drug combination screening
Introduction
Glioblastoma (GBM) is the most common primary malignant brain tumor in adults. The current standard of care for patients diagnosed with GBM is maximal safe tumor resection with optional implantation of BCNU polymer wafers (Gliadel) followed by combination temozolomide (Temodar) and radiation therapy1. Despite these treatments, the median survival time remains 14–18 months following diagnosis2–5. There is a pressing need for new GBM therapeutics to improve patient outcomes.
Many pharmacological agents have entered clinical trials for GBM; however only four drugs are currently FDA approved for GBM therapy. The failure of many GBM clinical trials is attributed to pre-clinical testing in cell and animal models that do not recapitulate features of human GBM, and therefore are poor predictors of how effective a drug will be in patients. Traditional adherent GBM cell lines, such as LN-229 and U-87MG, are grown in serum and do not resemble the characteristics of the primary tumor they were derived from when grown in vivo. Cells that produce spheriods when grown in serum free media with added growth factors, hereafter known as oncospheres, more closely mirror the genetic and phenotypic expression patterns from the original tumor6. Finally, oncosphere cell lines implanted intracranially in mice show histological characteristics of human GBM, including areas of necrosis and an invasive growth pattern7.
We have generated novel oncosphere cell lines, each derived from a human GBM or GBM variant that forms tumors in vivo with human GBM characteristics8. Here we report the genomic characterization of nine oncosphere cell lines. Two oncosphere lines were then used for a high throughput cell viability drug screen of 1,912 compounds and identified cytotoxic compounds were further validated using a non-high throughput format in additional cell lines. We found that certain compound mechanisms were effective in all cell lines, while others were active in some lines but not others. Pairwise drug combination screening of 30 compounds, giving 435 discrete drug combinations, revealed that PI3 Kinase inhibition combined synergistically with many different compounds to reduce viability of oncosphere cell lines. Finally, two drug combinations were tested in vivo to determine which could extend survival. This report is the largest screen of well-characterized compounds using GBM oncosphere lines and the first report of high throughput drug combination screening utilizing GBM oncospheres.
Materials and Methods
Compounds, antibodies and reagents
BIIB021 (#S1175) and TAK-733 (#S2617) were obtained from Selleck Chemicals (Houston, TX). Bardoxolone methyl (#1772) was obtained from Axon MedChem (Groningen, Netherlands). PD0325901 (P-9688) and GDC-0941 (G-9252) was obtained from LC Laboratories (Woburn, MA). Marizomib (SalA-100) was obtained from FIVEphoton Biochemicals (San Diego, CA). Neurocult NS-A Proliferation kit (#05751) and heparin (#07980) were obtained from StemCell Technologies (Vancouver, British Columbia). Basic-FGF (AF-100–18) and EGF (AF-100–15) were obtained from PeproTech (Rocky Hill, NJ). alamarBlue (DAL1100) was obtained from ThermoFisher Scientific (Waltham, MA). CellTiter-Glo (G7573) and Caspase-Glo 3/7 (G8092) were obtained from Promega (Madison, WI). NF-1 antibody (sc-67) and beta-actin antibody (sc-47778 HRP) were obtained from Santa Cruz Biotechnology (sc-67). PTEN antibody (#9559T) and goat anti-rabbit IgG (7074P2) secondary antibody were obtained from Cell Signaling Technology (Danvers, MA). Phosphokinase antibody array (ARY003B) was obtained from R&D Systems (Minneapolis, MN).
Cell Lines and Culture Conditions
Methods for the creation, establishment and validation of oncosphere cell lines has been previously published8. Br23C, JHU-0879, JHH-136, JHH-68, JHH-227, JHU-1016B, JHH-505 and JHH-520 were created by the Riggins and Gallia laboratories. All patients were consented and samples were de-identified upon collection. Samples with a JHU designation were collected using an additional research protocol, which allowed for collection of two vials of blood. HSR-GBM1 cells were a gift from Sara Piccirillo and Angelo Vescovi to the Riggins lab. All cell lines were grown as spheroids in Neurocult NS-A Proliferation media containing EGF, bFGF and heparin8. LN-229 (CRL-2611) and U-87MG (#HTB-14) were obtained from the America Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco’s Modified Eagle Medium (ThermoFisher Scientific, 11965–092) containing 10% fetal bovine serum (Hyclone) and 1% Penicillin/Streptomycin (ThermoFisher Scientific, 15140122). All cells were maintained in standard incubator conditions (37°C, 5% CO2, 95% humidity).
Exome Capture and Next Generation Sequencing
DNA was extracted from oncosphere cell lines using a DNeasy Blood and Tissue Kit (Qiagen, Venlo, The Netherlands). The STR profile of each sample was determined prior to sequencing to ensure each sample was independent. For JHU-0879, JHH-136, JHH-68, JHH-227, JHU-1016B, JHH-505, and JHH-520, DNA was extracted from cells that were less than 15 passages away from the passage number used for in vivo tumor establishment previously published8. Early passage cells were not available for Br23C and HSR-GBM1 so the earliest available was used. JHU-0879 and JHU-1016B samples had matched normal DNA which was extracted from whole blood using a DNeasy Blood and Tissue Kit (Qiagen). The amount of DNA in each sample was quantified using a Nanodrop Spectrophotometer (ThermoFisher Scientific). Library construction was completed at Johns Hopkins Microarray Core Facility (Baltimore, MD) and exome capture was completed at the J.Craig Venter Institute (Rockville, MD).
Methods for sequencing, reads mapping, variant identification, and assessment of copy number variation and have been previously published9. Briefly, genomic DNA was fragmented and 200–300 base pair segments were selected for sequencing. The exome capture was done using the SureSelect All Exon 50 Mb Target Enrichment kit (Agilent, Santa Clara, CA) and sequencing was done using Illumina HiSeq2000 platform. Image analysis and base calling were performed using Illumina’s Casava1.8.2 software.
Reads were mapped to reference genome (GRch37) using the Burrows-Wheeler Aligner (version 0.5.9). Regions that could benefit from realignment were identified using the GATK Realigner Target Creator (version 1.0.5506). GATK (version 1.5) was used to realign reads covering localized indels, recalibrate quality values, as well as locate, filter and annotate variants. JHU-1016B and JHU-0879 had whole blood available for sequencing so somatic mutations were eliminated from the final analysis. Somatic changes including point mutations and small indels were called based on comparison between somatic and tumor sequences. For samples with no somatic material, case versus controls were compared. Only missense mutations were assessed for functional impact by PhyloP, SIFT, Polyphen, and Mutation Taster 10 programs. For evaluation of copy number alterations, amplifications were defined as case vs control log2 ratio greater than eight, meaning greater than 4 copies of the gene were present.
Immunoblotting and Phosphokinase Profiling Arrays
Oncospheres were dissociated into single cells then pelleted by centrifugation at 300xg for 5 minutes. Media was discarded and pellets were washed using PBS and centrifuged at 300xg for 5 minutes. Following removal of PBS, an appropriate volume of ice cold RIPA buffer containing protease and phosphatase inhibitors was added to the pellet. Following a 30 minute incubation on ice, cells were centrifuged at 14000 RPM at 4°C for 15 minutes. Supernatant was collected and protein levels were quantified using a Pierce BCA assay kit (ThermoFisher Scientific). For PTEN, 20µg of denatured protein was loaded on a NuPAGE 4–12% Bis-Tris gel (Invitrogen, NP0322) and run at 100V for 75 minutes using 1X MES buffer. Proteins were transferred to a PVDF membrane at 30V for 90 minutes at 4°C. For NF1, 20µg of denatured protein was loaded on to a NuPAGE 3–8% Tris-acetate gel (Invitrogen, NP03552) and run at 100V for 75 minutes using 1X Tris Acetate buffer. Proteins were transferred to a PVDF membrane at 30V overnight at 4°C. Membranes were blocked in 5% nonfat dry milk in TBST (1X TBS/0.15% Tween20) mixture for 60 minutes at room temperature while rocking. PTEN antibody and NF1 antibody were added at a 1:1000 dilution in 5% nonfat dry milk in TBST and incubated overnight at 4°C with rocking. Membranes were washed three times with 1x TBST for 10 minute with rocking. Horseradish peroxidase (HRP) conjugated secondary anti-rabbit antibody was diluted 1:10,000 in 5% milk-TBST and incubated on each membrane for 60 minutes at room temperature with rocking. For phosphokinase arrays (R&D Systems), 190µg of protein was applied to the antibody array according to manufacturer instructions. Membrane proteins and antibody arrays were visualized using Pierce Chemiluminescence kit (Thermo). Following visualization of PTEN and NF1, membranes were incubated in Restore Western Blot Stripping Buffer (ThermoFisher, #21059) for 10 minutes at room temperature with rocking. Membranes were then blocked then washed as before followed by addition of beta-actin directly conjugated to HRP at a 1:10,000 dilution in 5% milk-TBST overnight at 4°C with rocking. Film capture was used for PTEN, NF1 and beta-actin while antibody arrays were visualized using a Bio-Rad Universal Hood II Gel Doc. Antibody arrays were quantitated using ImageQuant TL (GE Healthcare, Boston, MA) software.
Quantitative High Throughput (qHTS) Single Agent Drug Screening
The Mechanism Interrogation PlatE (MIPE) 4.0 is a collection of 1,912 small molecules that target signaling pathway components that are altered in many different cancers. All MIPE 4.0 compounds are plated in an 11 point dose response with a top concentration of 10mM and a 1:3 titration and has been used for screening previously11–13. High base solid bottom white Greiner plates (#789173) were used throughout. JHH-136 and JHH-520 spheroids were dissociated into single cells then plated at 500 cells per well in 5μL of complete NeuroCult media using a Multidrop liquid dispenser (Thermo). LN-229 and U-87MG cells were plated at 500 cells per well in 5µL of cell culture media. Immediately after plating, a Pintool dispenser (Kalypsys) was used to add 23nL of MIPE titrated compounds to columns 5 to 48 and control compounds to columns 1–4. The final top concentration of MIPE compounds is 47µM and a 1:3 titration. Column 1 contained media only, column 4 contained DMSO, and columns 2 and 3 contained the proteasome inhibitor bortezomib at a final concentration of 9.2μM. Plates were then incubated for 48 hours with low evaporation stainless steel cell culture Kalypsys lids in standard incubator conditions. For cell viability measurements, 3μL per well of CellTiter-Glo reagent (Promega) was dispensed using an Aspect Automation dispenser with solenoid valves (Lee Valves). After incubation for 15 minutes at room temperature, the luminescence signal was measured using a ViewLux CCD-based multi-label reader (Perkin Elmer, Waltham, MA). Cell viability values were normalized to the DMSO only control values as 100% activity, and media only controls as 0% activity. Dose response data for each compound was fit to a 4-parameter Hill curve using a grid-based algorithm developed in house. Using the fits, we determined the potency (AC50) and efficacy (% max response) and area under the curve (AUC) parameters for each compound tested. In addition to the curve fit parameters, we assigned each fit a “curve class reponse” (CRC), which is a heuristic classification scheme that allows one to readily identify the quality and nature of a dose response curve. Thus, a well-defined curve, with fully defined asymptotes, an R2 > 0.9 and an efficacy greater than 80% would be assigned a class of 1.1. Curves with a missing or asymptote and poorer efficacy would be considered a class 2 curve, and curves with activity at a single dose point would be considered class 3 (or inconclusive). Finally curves with no dose response would be classified as class 4 (inactive). See Inglese et al14 for a more detailed description of curve classes.
Single Agent Follow up Screening
BIIB021, Bardoxolone methyl, and TAK-733 were selected for hit verification. Cells were dissociated during the log growth phase and plated at 1000 cells per well in 178μL of complete Neurocult media into a clear bottom 96 well plate. Following an overnight incubation at standard incubator conditions, 2μL of each compound was added to columns 3 through 10 at the following final concentrations: 10μM, 5μM, 1μM, 0.5μM, 0.1μM, 0.05μM, 0.01μM, and 0.005μM. Columns 1 and 12 served as a no treatment control, Column 2 contained DMSO and Column 10 contained bortezomib at a final concentration of 2μM. Plates were stored at standard incubator conditions for the duration of the experiment. 20μL of Alamar Blue Reagent was added to each well and plate readings were taken at 0, 24, 48, 72, and 96 hours using a Perkin Elmer Wallac 1420 Multilabel Counter. For Br23C and JHH-227, plate readings were taken every 48 hours due to slow in vitro growth rates. Each plate contained 6 replicates and all values were normalized to the DMSO control. GraphPad Prism 5 was used to calculate IC50 values.
Drug Combination Screening
These methods have been described previously11. The initial drug combination screen was done in a 6x6 matrix format encompassing a grid of 36 wells, containing five concentrations of one compound in combination with five concentrations of a second compound in a 25 well grid, plus 10 additional wells of all five concentrations of single agent compounds and 1 well with DMSO only. 30 compounds were selected for pairwise matrix screening based on their single agent activity giving 435 discrete drug combinations. An appropriate volume of each compound was added to 384 well black Greiner plates (#788876). 10nL/well of each compound were acoustically dispensed in a 6x6 format into an empty 1536 well high base solid bottom white Greiner plates (#789173) using an ATS-100 (EDC Biosystems). Plates were then covered with stainless steel cell culture Kalypsys lids and stored in standard incubator conditions. The following day, JHH-136 cells were added to drug combination plates as in single agent screening at 1000 cells/well in 5 µL of complete Neurocult media. Plates were stored with stainless steel cell culture Kalypsys lids in standard incubator conditions for 48 hours and then 3 µL per well of CellTiter-Glo luminescent cell viability assay reagent (Promega) was added using an Aspect Automation dispenser with solenoid valves (Lee Valves). The plates were then incubated for 15 minutes at room temperature and the signal was captured using a ViewLux (Perkin Elmer). Follow up drug combination screening was done in a 10x10 matrix format encompassing a 100 well grid, meaning 9 concentrations of each compound was combined with 9 concentrations of a second compound in a 81 well matrix grid, plus 9 concentrations of each compound as a single agent and 1 well with DMSO only. Cell viability matrix screening was completed using a 10x10 format in duplicate for JHH-136 and JHH-520 using 46 discrete drug combinations using the same methods as above and results were aggregated using the mean activity. Apoptosis assays were done in JHH-136 using a 10x10 matrix format. Cells were incubated for 8 or 24 hours with compound, and then 3 µL of Caspase Glo 3/7 luminescent apoptosis assay reagent (Promega) was added to the plates. After a 30 minute incubation at room temperature, signal was captured using a ViewLux. As caspase activation time point may vary between cell lines and compounds, the apoptosis data was gated between DMSO control (caspase activation 0%) and the top 5% quantile (caspase activation 100%) of the entire assay.
Animal Drug Efficacy Studies
Animals were maintained using approved Institutional Animal Care and Use Committee (IACUC) protocols. 35 female athymic nude mice (NCI-Fredrick) aged 4–6 weeks were anesthetized with a ketamine/xylazine mixture and fixed into a stereotactic frame. 500,000 JHH-520 cells were intracranially implanted through a burr hole drilled 2mm lateral to the sagittal suture and 1mm anterior to the coronal suture at a depth of 3mm below the dura as previously described15. Surgical incisions were sealed with staples. Seven days post implantation, animals were randomized into six groups and drug treatment was initiated. Groups were as follows: Control (n=6), GDC-0941 alone (n=5), PD0325901 alone (n=5), Marizomib alone (n=5), GDC-0941 plus PD0325901 (n=7) and GDC-0941 plus Marizomib (n=7). Control animals received vehicle (0.5% CMC/0.2% Tween80 in water) by oral gavage. GDC-0941 was diluted in vehicle and administered at a dose of 75mg/kg by oral gavage. PD0325901 was diluted in vehicle and administered at a dose of 10mg/kg by oral gavage. Oral drugs were administered once daily, five days per week and continued until animals were sacrificed. Marizomib animals received 75µg/kg of drug diluted in 1X PBS administered by intravenous (i.v.) tail vein injection starting on day 7 post-implantation and continued twice weekly with a 3 day rest between doses. Marizomib dosing was stopped after 19 treatments on day 70 post implantation. Combination treatment groups received dual treatments at the same doses listed above within a 60 minute window. All animals were weighed thrice weekly for the duration of the experiment. Animals were sacrificed upon signs of intracranial tumor burden and de-brained. Brains were fixed in 10% formalin solution for further analysis. Kaplan-Meier curves and statistical tests were performed using GraphPad Prism.
Results
Each Oncosphere Cell Line Has a Distinct Alteration Profile
Each oncosphere cell line was derived from an individual patient tumor. Therefore we explored deleterious point mutations and copy number variation within each cell line. We assessed alterations in the glioblastoma candidate cancer (CAN) genes because mutations in these genes are drivers of glioblastoma cell growth16. Table 1 shows the CAN gene alterations present plus one additional oncogene, MYC. The full list of genomic alterations in all nine cell lines can be found in supplementary data. Genomic alterations were not found in IDH1, CDK4 or PIK3CA. CDKN2A alterations were the most common, present in seven of nine lines (77%). Oncosphere cell lines lacking alterations in CDKN2A contained alterations in RB1, a tumor suppressor gene within the same pathway as CDKN2A. Of note, we report only two lines with high level (> 10 copies) EGFR amplification, however, many cell lines contained low level (3–10 copies) EGFR amplification. We explored additional oncogenes and tumor suppressor genes implicated as drivers in other cancer types but found very few alterations (Supplemental Table 1).
Table 1.
Genomic alterations present in GBM candidate cancer (CAN) genes for nine oncosphere cell lines.
| HSR-GBM1 | JHH-520 | JHU-1016B | JHH-227 | JHH-136 | JHH-68 | JHU-0879^ | BR 23C | JHH-505* | |
|---|---|---|---|---|---|---|---|---|---|
| CDKN2A | WT/D74P fs*45 | Hom Del | Hom Del | Hom Del | Hom Del | Hom Del | Hom Del | ||
| TP53 | WT/L130I | H179D | G244S | C176F | |||||
| EGFR | 16 copies | 16 copies | |||||||
| PTEN | Del/C124W | Del/R378I | Hom Del | Hom Del | Del/K66E | Hom Del | |||
| NF1 | WT/A1676T | Hom Del | WT/L115T fs*42 | I526S | |||||
| RB1 | S671F fs*8 | Del/E837G fs*2 | |||||||
| PIK3R1 | WT/F156L, Q157del, E158del | ||||||||
| MYC | 18 copies | 14 copies |
= GBM with primitive neuroectodermal (PNET) features
= GBM with oligodendroglial component
Del = deleted copy of gene
WT= wild type copy of gene
Hom Del = homozygous deletion
Compared to previous sequencing studies utilizing GBM patient samples16–18, our oncosphere panel had 25% to 36% increase in mutations in PTEN and NF1 (Supplemental Table 2) which indicates that tumors with these mutations have a selective advantage for long term in vitro growth. The Br23C sample had a matching patient xenograft, Br23X, which was generated from the same tumor tissue. Br23X was sequenced in a previous study16 so we compared CAN gene mutations for Br23C with those previously reported for Br23X (Supplementary Table 3). We found that the mutations in PTEN, NF1, and TP53 were identical in xenograft and cell line, while mutations in RB1 and MYC were present only in the cell line. This data is evidence that the alterations within the CDKN2A/RB1 pathway provide a selective advantage for long term in vitro growth.
We then verified the presence or absence of PTEN and NF1 protein by western blot in eight oncosphere lines (Figure 1A and 1B). JHH-505 was excluded from analysis due to low quantity of protein. Three cell lines had PTEN protein present, HSR-GBM1, JHU-1016B and JHU-0879 while all others had no PTEN. Four cell lines had NF1 present, JHH-227, JHH-136, JHH-68 and JHU-0879, while all others had no NF1. Certain cell lines showed discrepancies between the mutation status and protein expression of PTEN and NF1. The genomic and protein expression data was combined to delineate which GBM driver signaling pathways were altered within each oncosphere line (Table 2). The most commonly mutated pathways were the CDKN2A/RB1/CDK4 pathway and the PTEN/PIK3CA/PIK3R1 pathway, with 7 of 8 cell lines containing an alteration of one gene/protein in each of these pathways.
Figure 1.
Protein expression profiling of PTEN and NF1 in oncosphere cell lines
A. Western blot of PTEN protein for eight oncosphere cell lines. Beta-actin was used as a loading control.
B. Western blot of NF1 protein for eight oncosphere cell lines. Beta-actin was used as a loading control.
C. Bar graph showing the quantitated intensity of dots on the phospho-kinase antibody array. Black bars are for JHH-136 and red is for JHH-520. Dot intensity was normalized to positive and negative controls included on each array. Only proteins that were significantly different between each cell line (p < 0.01) are shown here. Full dataset and array images are in Supplemental Figure 1.
Table 2.
Summary of genomic and protein analysis of GBM CAN signaling pathways
| HSR-GBM1 | JHH-520 | JHU-1016B | JHH-227 | JHH-136 | JHH-68 | JHU-0879 | BR 23C | |
|---|---|---|---|---|---|---|---|---|
| CDKN2A/RB1/CDK4 | CDKN2A | CDKN2A | CDKN2A | CDKN2A | CDKN2A | RB1 | RB1 | |
| PTEN/PIK3CA/PIK3R1 | PTEN | PTEN | PTEN | PTEN | PTEN | PTEN | PTEN | |
| NF1/RAS | NF1 | NF1 | NF1 | NF1 | ||||
| EGFR | EGFR | EGFR |
We selected two cell lines, JHH-136 and JHH-520, for phosphokinase profiling of 46 proteins to examine downstream kinase activation (Supplementary Figure 1). Of the 46 proteins examined, 17 proteins had significantly higher expression (p<0.001) in JHH-136 compared to JHH-520 (Figure 1C). Many of these proteins are directly phosphorylated by phosphoAkt, such as mTOR and CREB, evidence that JHH-136 has higher activated Akt levels than JHH-520. Hsp60 was the most highly expressed protein in both cell lines. Taken together, this data shows that our GBM oncosphere cell line panel contains cell lines with unique GBM driver pathway alteration profiles. This data paired with the in vivo characterization of these oncosphere cell lines8 makes them an excellent research and discovery tool for GBM therapeutics. We chose to use JHH-136 and JHH-520 in a qHTS screen in order to examine differential drug sensitivities and possible novel therapeutics.
Oncosphere Cell Lines Show Differential Sensitivity to Single Agent Compounds
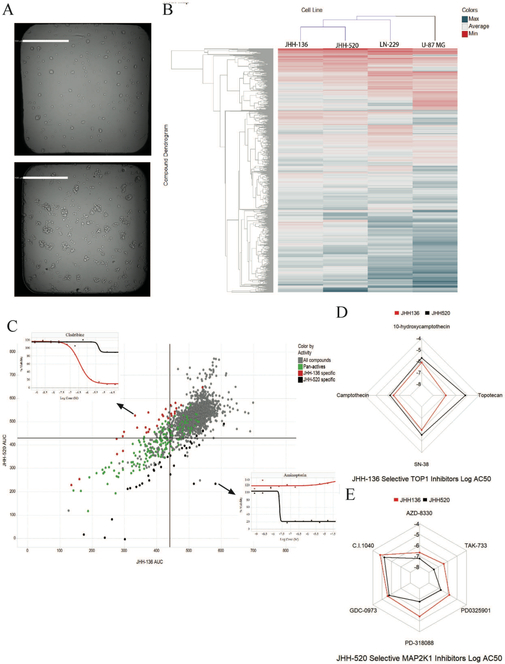
We first sought to compare drug responses between oncosphere cell lines and adherent GBM cell lines. JHH-136, JHH-520, LN-229 and U-87MG were screened for cell viability using the MIPE 4.0 compound library. All single agent screening data is available for download here [https://tripod.nih.gov/matrix-client/?p=121]. Figure 2A shows JHH-136 and JHH-520 spheroid morphology after 48 hours in a 1536 well plate. Each compound in the MIPE 4.0 library was annotated with gene target, mechanism of action, and cellular process information to allow for aggregation of data to search for novel biological insight.
Figure 2.
Single agent screening data for JHH-136 and JHH-520
A. A representative brightfield image of JHH-136 (top image) and JHH-520 (bottom image) spheroid cells in 1536 well plates 48 hours after plating. Scale bars in white are 500 µm in length.
B. Heatmap of compound AUC values in JHH-136, JHH-520, LN-229 and U-87MG with unbiased hierarchical clustering by compound and cell line. Color gradient is by AUC value with potent compounds in red and inactive compounds in blue.
C. Scatterplot of AUC values for each compound in JHH-136 on the x axis vs JHH-520 on the y axis. Each circle on the plot is one compound. Circles in green are active in both cell lines (pan-active), dots in red are active only in JHH-136 and dots in black are active only in JHH-520 and gray dots are the remaining compounds. Inset dose response curves are for the compounds that were highly selective for a single cell line. Cladribine was for JHH-136 and Aminopterin was for JHH-520.
D. Radar plot for Topo1 inhibitors which were enriched in the JHH-136 specific active compound list. Each point on the plot represents the Log AC50 value for that compound with each dot connected by a line. Red lines are for JHH-136 and black lines are for JHH-520.
E. Radar plot for MAP2K inhibitors which were enriched in the JHH-520 specific active compound list. Each point on the plot represents the Log AC50 value for that compound with each dot connected by a line. Red lines are for JHH-136 and black lines are for JHH-520.
An unbiased hierarchical clustering of AUC values for all four GBM cell lines screened is shown in Figure 2B. LN-229 and U-87MG cluster separately from both oncosphere lines indicating that cell growth conditions can affect drug response. To compare the oncosphere drug responses, a deltaAUC value was calculated for each compound by subtracting the AUC value in JHH-136 from JHH-520. This allowed for a more direct comparison of compound performance across each cell line. Pan-active compounds were defined as having a CRC of −1.1, −1.2, and −2.1 in both cell lines and a deltaAUC between −80 and 80 (Figure 2C, green circles), which yielded 160 compounds for the two oncosphere lines. Pan-actives were aggregated by gene target and a target enrichment analysis (using Fishers exact test) was done in order to compare pan-actives to the MIPE 4.0 library targets overall, and a false discovery rate (FDR) was used for p-value correction. Hsp90, HDAC1, PSMD1, CDK1, PIK3CA, and NFKB1 all were significantly overrepresented (adjusted p-value < .01). These targets are all directly involved in cell cycle regulation, global transcriptional regulation, and protein folding. The PIK3CA pathway was altered in both oncosphere cell lines screened which explains the overrepresentation of this target. Overall, these mechanisms are critical for growth of all GBM cells, therefore we hypothesized that compounds with these mechanisms would have similar activity across multiple GBM oncosphere cell lines, regardless of their mutation profile.
To validate that this hypothesis, we selected two pan-active compounds for testing across multiple oncosphere cell lines. The Hsp90 inhibitor, BIIB021, and Bardoxolone methyl, an Nf-kB inhibitor, were tested for viability against 6 oncosphere cell lines. Both compounds had similar sub-micromolar IC50 values and all dose response curves contained two asymptotes, as in the primary screen (Supplemental Figure 2). We also examined these two compounds in the high throughput screen against LN-229 and U-87MG and found them to be active similarly to the oncosphere cell lines (Supplementary Figure 2). This is evidence that compounds targeting Hsp90 or Nf-kB mechanistic classes may be good candidates for reducing GBM cell line viability regardless of genetic background.
Next, we examined actives specific to either JHH-136 or JHH-520 to determine if cell line genomic profiles correlated with drug sensitivity. To find highly specific actives for either cell line, we defined specific actives as compounds with a deltaAUC greater than 80 or less than −80 and having a maximum response (MAXR) lower than 60 in either cell line. Compounds with a negative deltaAUC were more active in JHH-136 than in JHH-520 and compounds with a positive deltaAUC were more active in JHH-520 than in JHH-136. Using these methods, 29 JHH-136 specific actives and 34 JHH-520 specific actives were identified (Figure 2C, red and black circles respectively), and target-wise differential analysis was completed for each group of compounds to determine targets for which a statistically significant difference in AUC between cell lines existed.
For JHH-136 selective actives, proteasome and topoisomerase 1 (TOP1) inhibitors had a significantly (p = 0.0012 and 0.0059 respectively) lower AUC in JHH-136 compared to JHH-520. The four compounds targeting TOP1 had a median deltaAUC of −125.404 ± 27.211 (Figure 2D). Of these four compounds, three of them are camptothecin analogs and the fourth is camptothecin’s active metabolite, SN-38, indicating that JHH-136 is highly sensitive to this structural class of compounds. The compound with the most negative deltaAUC was Cladribine (Figure 2C, inset), a nucleoside analog which targets Adenosine Deaminase (ADA) causing DNA replication arrest and DNA damage19. Given that both TOP1 and ADA are both critical to DNA replication, this data shows that JHH-136 is highly sensitive to compounds that interfere with DNA replication.
For JHH-520 selective actives, Mek (MAP2K1) inhibitors had a significantly (p = 0.0001) lower AUC in JHH-520 compared to JHH-136. The six Mek inhibitors had a median deltaAUC of 90.097 ± 8.247 (Figure 2E). Aminopterin (Figure 2H), an inhibitor of dihydrofolate reductase (DHFR), had the most positive deltaAUC (Figure 2C inset). Two more DHFR inhibitors were highly specific to JHH-520, as well as two inhibitors of HPRT1, which indicated that this cell line was highly sensitive to blockade of purine synthesis and related pathways. Given the lack of NF1 protein present in this cell line, the sensitivity to Mek inhibition is expected. Since NF1 alterations were present in some GBM oncosphere cell lines, we hypothesized that Mek inhibitors would have variable activity across GBM oncosphere cell lines.
To test this hypothesis, TAK-733, a JHH-520 selective active, was screened against 6 oncosphere cell lines (Supplemental Figure 2). Activity was highly variable across all cell lines tested with IC50 values ranging from 16 µM in HSR-GBM1 to 30 nM in JHH-520, and Br23C was unable to calculate an IC50 due to compound inactivity. Taken together, this data is evidence that GBM cell lines with varying mutational profiles are sensitive to compounds which directly target critical cellular processes such as cell cycle, transcription, protein folding and commonly mutated GBM pathway components such as PI3 Kinase. However certain classes of compounds have activity only in certain cell lines with a specific mutation profile, such as Mek inhibitors in NF1 altered cell lines.
PI3 Kinase inhibitors are synergistic in combination with Mek inhibitors
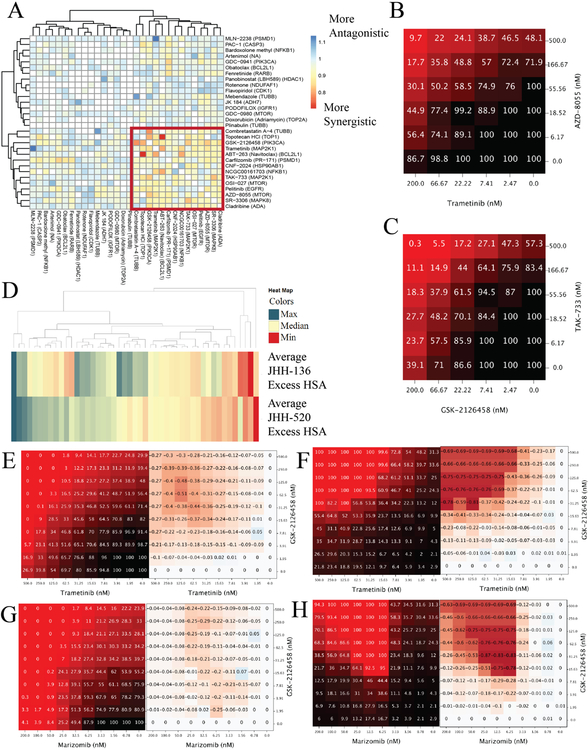
We next sought to determine if drug combination viability screening could reveal drug mechanisms that would be synergistic when combined. A high-throughput drug combination screening platform, which has been previously described11, was utilized for this purpose. 30 compounds were selected for drug combination screening based multiple criteria. Single agent performance as well as enriched targets were taken into account. Weakly active and inactive compounds were also included if they had targets which were enriched in single agent screens or had scientific evidence of blood brain barrier (BBB) penetration. All 30 compounds were combined individually with each other, resulting in 435 discrete drug combinations; these were all screened in a 6x6 matrix format for cell viability. The full list of compounds and single agent performance is listed in Supplementary Table 4. All drug combination screening data is available here [https://tripod.nih.gov/matrix-client/?p=121]. JHH-136 was used for the primary matrix screen and multiple synergy metrics were assessed for all combinations. Hierarchical clustering of each combination by Gamma synergy metric20 revealed 14 compounds, which were highly synergistic when combined with each other (Figure 3A). Within these 14 compounds, PI3 Kinase, mTOR and Mek inhibitors were present, specifically combinations involving either GSK-2126458 or AZD-8055 were all highly synergistic with Trametinib and TAK-733 (Figure 3B and 3C). Additional synergistic combinations involved CNF-2024 and Carfilzomib, a HSP90 inhibitor and proteasome inhibitor, respectively.
Figure 3.
Drug combination screening data
A. Heatmap of gamma synergy values for each compound in combination with all others tested in the 6x6 matrix screen. Color gradient is from red to blue with red being most synergistic and blue being most antagonistic. Unbiased hierarchical clustering of gamma synergy revealed a highly synergistic cluster of compounds, highlighted by the red box.
B. Heatmap of cell viability values of Trametinib, a MEK inhibitor, in combination with AZD-8055, an mTOR inhibitor. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values shown inside the grid are % viability and color gradient is from black to red with black being 100% viability and red being 0% viability.
C. Heatmap of cell viability of TAK-733, a MEK inhibitor, in combination with GSK-2126458, a PI3 Kinase/mTOR inhibitor. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values shown are % viability and color gradient is from black to red with black being 100% viability and red being 0% viability.
D. Heatmap of Excess HSA synergy value for 47 combinations in each neurosphere cell line. Color gradient is from red to blue with red being most synergistic and blue being most antagonistic.
E. Heatmap of cell viability values of GSK-2126458 in combination with Trametinib in the JHH-136 cell line. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values in left heatmap are % viable cells and values in right heatmap are calculated delta HSA synergy values.
F. Heatmap of 8 hour apoptosis induction of GSK-2126458 in combination with Trametinib in the JHH-136 cell line. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values in left heatmap are % apoptotic cells and values in right heatmap are calculated delta HSA synergy values.
G. Heatmap of cell viability GSK-2126458 in combination with Marizomib in the JHH-520 cell line. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values in left heatmap are % viable cells and values in right heatmap are calculated delta HSA synergy values.
H. Heatmap 8 hour apoptosis induction of GSK-2126458 in combination with Marizomib in the JHH-520 cell line. Values shown on the x and y axis is the concentration of compound in that row or column in nanomolar. Values in left heatmap are % apoptotic cells and values in right heatmap are calculated delta HSA synergy values.
We then selected 47 discrete combinations (Supplemental Table 5) for an expanded cell viability combination screen in a 10x10 format to allow for more precise determination of the synergistic window of each combination. Marizomib, a BBB penetrant proteasome inhibitor21, was added for this screen as well as other known BBB penetrant compounds. Combinations were tested for viability in JHH-136 and JHH-520 as well as apoptosis induction after 8 and 24 hours of drug exposure in JHH-136. Hierarchical clustering of the Excess Highest single agent (HSA) value for each drug combination (Figure 3D) in each cell line revealed that overall most combinations had similar synergy in both lines. The focused 10x10 drug combination cell viability data were rank ordered from most synergistic to most antagonistic using the Gamma synergy metric for each combination in each cell line. Self-crosses were excluded from analysis leaving 42 ranked combinations. Trametinib, a Mek inhibitor, combined with GSK-2126458 (PI3 Kinase/mTOR) (Figure 3E), AZD-8055 (mTOR), Navitoclax (Bcl-2/Bcl-xL), CNF-2024 (Hsp90) or Pelitinib (EGFR) were within the top 10 most synergistic combinations for both cell lines and also induced apoptosis after 8 hours of compound exposure (Figure 3F). This data is strong evidence that targeting Mek, PI3 Kinase, mTOR or Bcl-2 simultaneously causes a synergistic decrease in cell viability as well as induction of apoptosis as early as 8 hours after drug exposure. Proteasome inhibitors, such as Carfilzomib and Marizomib had excellent synergy when combined with GSK-2126458 (Figure 3G) and induced apoptosis at low nanomolar concentrations after 8 hours of drug exposure (Figure 3H). Overall, the drug combination studies showed trends in which mechanisms can synergize to decrease GBM cell growth and induce apoptosis.
GDC-0941 paired with PD0325901 and PD0325901 alone extend survival in vivo
Finally, we examined whether the in vitro data could be replicated in vivo. The JHH-520 cell line was an excellent candidate for in vivo experimentation. Animals implanted intracranially with this cell line develop highly invasive tumors with areas of necrosis and mitotic features, and implanted animals had median survival time (MST) of 82 days8. We selected two drug combinations for testing in vivo, a PI3 Kinase inhibitor paired with either a Mek inhibitor or a proteasome inhibitor.
GDC-0941 as a PI3 Kinase inhibitor was a candidate as it was tested in single agent screening. There are reports that GDC-0941 penetrates the BBB, however pharmacokinetic studies have not been reported that indicate maximal doses attainable in the brain22. Structurally, GDC-0941 and GSK-2126458, the compound tested in our combination studies are dissimilar; however, they have a common target of the p110 subunit of PI3 Kinase. Trametinib has minimal BBB penetration23, however, PD0325901 is a MEK inhibitor that is able to penetrate the BBB and has already been tested as a single agent in an intracranial GBM model24. Marizomib is reported to have better activity in GBM compared to Bortezomib25. Given this data, we elected to test GDC-0941 with PD0325901 and GDC-0941 with Marizomib in vivo using the JHH-520 xenograft model.
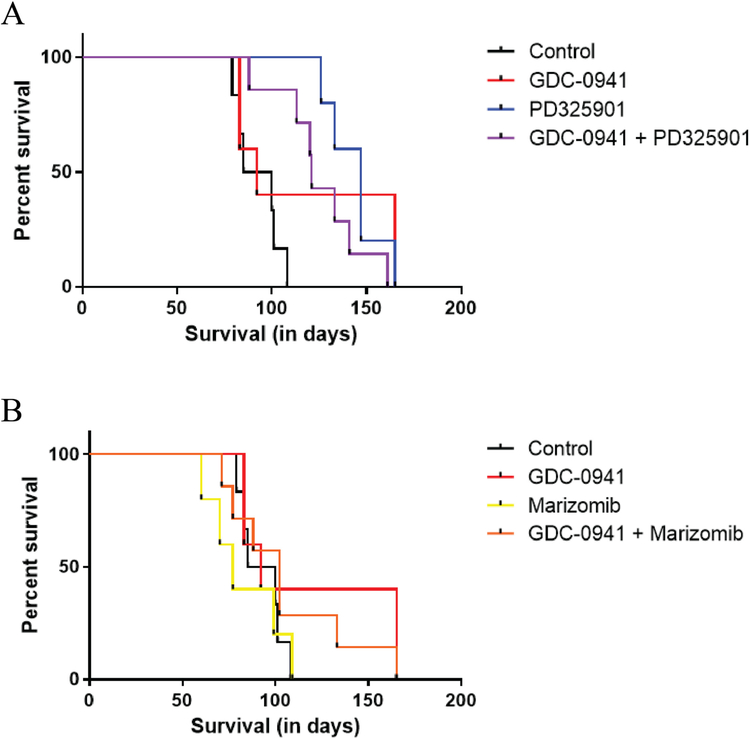
Control animals received vehicle only and treatment groups received either GDC-0941 alone, PD0325901 alone, Marizomib alone, GDC-0941 plus PD0325901, or GDC-0941 plus Marizomib. The control group had an MST of 92.5 days. The MST of GDC-0941 alone was 92 days, PD0325901 alone was 147 days and the combination of GDC-0941 plus PD0325901 was 121 days (Figure 4A). Only PD0325901 alone and combination provided a significant improvement in median survival time over the control group (p =0.0014 and p=0.0020, respectively). The MST of Marizomib alone was 77 days while the combination of Marizomib plus GDC-0941 had a MST of 102 days (Figure 4B) neither of which were a significant improvement over control groups (p = 0.2611 and p=0.5677, respectively). All Marizomib treatment groups had early toxicity issues which likely caused a lower MST than compared to the control group. Overall, the treatment groups with the longest survival were PD0325901 alone or GDC-0941 plus PD0325901 which is evidence that targeting PI3 Kinase and Mek simultaneously or Mek alone can extend survival in a GBM animal model.
Figure 4 –
Orthotopic xenograft drug combination survival data
A. Kaplan-Meier curves showing animal survival for control, GDC-0941 alone, PD0325901 alone and GDC-0941 plus PD0325901 combination groups.
B. Kaplan-Meier curves showing animal survival for control, GDC-0941 alone, Marizomib alone and GDC-0941 plus Marizomib combination groups.
Discussion
Each Oncosphere Cell Line is a Unique, Independent GBM Model
All oncosphere cell lines included within this study have been characterized in a previous publication8 which details clinical data, doubling times, and in vivo tumor formation with histological characteristics of human GBM. This study adds genomic mutation data to these well-characterized lines. Genetic alterations common in primary GBM tissues have been widely studied and genes that drive GBM formation and growth have been determined16–18. We found that our GBM oncosphere cell lines have alterations in GBM CAN genes at similar frequencies to studies using primary tissue. The most common alterations found were loss of CDKN2A and PTEN, the former results in uncontrolled cell growth and the latter causes overactive signaling through the PI3 Kinase/AKT pathway. Only two of nine oncosphere lines contained high level EGFR amplification, which is a lower frequency than reported in studies sequencing primary tumor samples. However for our study, we elected to consider cell lines with greater than 10 copies of EGFR as amplifications while previous studies utilized a lower threshold. Multiple oncosphere cell lines, such as JHH-136 and JHH-520, contained 3–4 copies of EGFR evidence that this alteration is prevalent in GBM tumors and cell lines.
NF1 and PTEN alterations were present at a higher frequency than reports sequencing primary tumor samples. Our oncosphere panel also had some differences in mutation frequency compared to commonly used monolayer GBM cell lines26, 27. These differences between GBM oncosphere cell line, primary GBM tissue, and monolayer GBM cell lines suggest a selection bias of oncosphere cell culture methods. However, evidence has shown that long term growth in serum-containing media can increase the frequency of certain mutations6, therefore it is challenging to compare mutation frequencies between monolayer GBM lines grown in serum-containing media for over 50 years28 to oncosphere cell lines with a short culture time in non-serum containing media. When protein expression was verified by western blot, we found a few inconsistencies with the sequencing data. These could be explained by alterations in antibody recognition sequence, epigenetic silencing mechanisms, mutations not considered in regulatory regions, or other unforeseen mutational affects resulting in a loss of protein expression.
Previous studies show that GBM oncospheres grown in serum free conditions better maintain the parental tumor genomic mutation and RNA expression profile than adherent cell lines grown with serum.6 Additionally, our GBM oncosphere cell lines can form intracranial tumors, which contain features of human GBM such as invasive growth and areas of necrosis8. U-87MG, LN-229 and other adherent GBM cell lines lack the ability to form tumors in vivo that show the pathological hallmarks of human GBM, such as invasive tumor growth, which makes them a poor model for pre-clinical studies used to inform clinical trials. Taken together, oncosphere cell lines are highly representative of the histology and disease progression of GBM patients, yet it remains unclear if they are a better model for pre-clinical studies. We hope that through widespread use of our GBM oncosphere cell lines, there can be pre-clinical studies done to inform successful clinical trials to improve treatment options for GBM patients.
Drug Efficacy can be dictated by the Characteristics of the Model Tested
Our qHTS study, using a focused collection of compounds with clinical relevance in cancer, revealed a number of different drug classes that were effective at stopping GBM cell growth. We utilized two oncosphere cell lines with two different mutation profiles for screening and found multiple targets, such as Hsp90 and proteasome inhibitors, that were enriched in the list of compounds that were active in both cell lines tested. Topoisomerase inhibitors were the only dominant drug class that was selectively active in JHH-136, indicating that targeting DNA replication and mitosis was cytotoxic to this cell line. Mek inhibitors were the dominant class of compounds that were active only in JHH-520, showing that targeting signaling pathways was effective for this cell line. This effect was likely due to the absence of NF1 protein in the JHH-520 cell line. The classes of compounds which were highly active in a single cell line could be caused by differences in protein expression or activated signaling pathways which were not assessed in this study. An additional observation was that some compounds, specifically inhibitors of Nicotinamide phosphoribosyltransferase or Mek, were inactive or weakly active in the adherent GBM cell lines tested, yet were highly active in one or both oncosphere cell lines. This indicates that screening studies using only adherent cell lines may be excluding efficacious compounds.
The drug combination studies revealed synergistic effects that were common across cell lines as well as some that were selective. PI3 Kinase inhibition paired with inhibition of Mek, proteasome or Bcl-2 was synergistic in both oncosphere cell lines. However, JHH-136 had multiple top ranked synergistic combinations involving Pelitinib, an EGFR inhibitor yet these combinations were additive in JHH-520. We believe these differences in single agent and combination drug response between cell lines are due to the differences in genomic mutation and protein expression profile of JHH-136 and JHH-520. Further exploration of the cellular mechanisms that explain these compound synergies will be necessary. In making this full screening dataset public, we hope others will utilize this data as a starting point for further preclinical investigation.
Translation of the in vitro results to in vivo experiments was not straightforward. PD0325901 when dosed as a single agent had the best MST. PD0325901 has already been tested as a single agent in an intracranial GBM model and showed a survival benefit24. The cause of the lack of synergy in vivo between GDC-0941 and PD0325901 is likely due to pharmacokinetic issues related to GDC-0941. A recent study testing Buparlisib in combination with PD0325901 showed the combination had a better MST than the single agent in subcutaneous glioma xenografts, however the same combination tested using orthotopic xenografts, PD0325901 was superior to the combination29, similar to the data presented here. This is evidence that more effort should be focused upon the development of a blood-brain barrier penetrant PI3 Kinase inhibitor. Marizomib, a proteasome inhibitor produced by a marine bacteria, Salinispora tropica, has conflicting reports about its ability cross the BBB21, 25 and has been tested in GBM in combination with HDAC inhibitors21. In our testing, treatment groups containing Marizomib did not show a survival benefit most likely due to toxicity issues and the complexity of the BBB penetration of the compounds at the doses tested. Taken together, our panel of GBM oncosphere cell lines are suitable for use in high throughput screening and drug combination studies however, there remains the issue of translating in vitro efficacy into in vivo efficacy.
Supplementary Material
Acknowledgments
We would like to thank Dr. Natalia J. Martinez and Steve Titus for the usage of the U-87MG and LN-229 cell lines, Jonathan M. Keller for qHTS screening assistance, Dr. Qi Zhao and Dr. Ewen Kirkness for DNA sequencing and data analysis, Dr. Aleksandra Michalowski for assistance with data analysis and Dr. Matthew Hall for critical reading of the manuscript. This work was funded by NIH Intramural Research Program at NCATS, the Virginia and D.K. Ludwig Fund for Cancer Research (GJR), and the Department of Neurosurgery at Johns Hopkins School of Medicine (GLG).
References
- 1.Nabors LB; Portnow J; Ammirati M; et al. Central Nervous System Cancers, Version 1.2015. J Natl Compr Canc Netw 2015, 13, 1191–202. [DOI] [PubMed] [Google Scholar]
- 2.Gallia GL; Brem S; Brem H. Local treatment of malignant brain tumors using implantable chemotherapeutic polymers. J Natl Compr Canc Netw 2005, 3, 721–8. [DOI] [PubMed] [Google Scholar]
- 3.Lai A; Tran A; Nghiemphu PL; et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol 2011, 29, 142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R; Mason WP; van den Bent MJ; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005, 352, 987–96. [DOI] [PubMed] [Google Scholar]
- 5.Walker MD; Green SB; Byar DP; et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N Engl J Med 1980, 303, 1323–9. [DOI] [PubMed] [Google Scholar]
- 6.Lee J; Kotliarova S; Kotliarov Y; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [DOI] [PubMed] [Google Scholar]
- 7.Galli R; Binda E; Orfanelli U; et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 2004, 64, 7011–21. [DOI] [PubMed] [Google Scholar]
- 8.Binder ZA; Wilson KM; Salmasi V; et al. Establishment and Biological Characterization of a Panel of Glioblastoma Multiforme (GBM) and GBM Variant Oncosphere Cell Lines. PLoS One 2016, 11, e0150271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borodovsky A; Meeker AK; Kirkness EF; et al. A model of a patient-derived IDH1 mutant anaplastic astrocytoma with alternative lengthening of telomeres. J Neurooncol 2015, 121, 479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwarz JM; Rodelsperger C; Schuelke M; et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 2010, 7, 575–6. [DOI] [PubMed] [Google Scholar]
- 11.Heske CM; Davis MI; Baumgart JT; et al. Matrix Screen Identifies Synergistic Combination of PARP Inhibitors and Nicotinamide Phosphoribosyltransferase (NAMPT) Inhibitors in Ewing Sarcoma. Clin Cancer Res 2017, 23, 7301–7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews Griner LA; Zhang X; Guha R; et al. Large-scale pharmacological profiling of 3D tumor models of cancer cells. Cell Death Dis 2016, 7, e2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen I; Mathews-Greiner L; Li D; et al. Transcriptomic profiling and quantitative high-throughput (qHTS) drug screening of CDH1 deficient hereditary diffuse gastric cancer (HDGC) cells identify treatment leads for familial gastric cancer. J Transl Med 2017, 15, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inglese J; Auld DS; Jadhav A; et al. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A 2006, 103, 11473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai RY; Staedtke V; Aprhys CM; et al. Antiparasitic mebendazole shows survival benefit in 2 preclinical models of glioblastoma multiforme. Neuro Oncol 2011, 13, 974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons DW; Jones S; Zhang X; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brennan CW; Verhaak RG; McKenna A; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van den Neste E; Cardoen S; Offner F; et al. Old and new insights into the mechanisms of action of two nucleoside analogs active in lymphoid malignancies: fludarabine and cladribine (review). Int J Oncol 2005, 27, 1113–24. [PubMed] [Google Scholar]
- 20.Cokol M; Chua HN; Tasan M; et al. Systematic exploration of synergistic drug pairs. Mol Syst Biol 2011, 7, 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manton CA; Johnson B; Singh M; et al. Induction of cell death by the novel proteasome inhibitor marizomib in glioblastoma in vitro and in vivo. Sci Rep 2016, 6, 18953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salphati L; Pang J; Plise EG; et al. Preclinical pharmacokinetics of the novel PI3K inhibitor GDC-0941 and prediction of its pharmacokinetics and efficacy in human. Xenobiotica 2011, 41, 1088–99. [DOI] [PubMed] [Google Scholar]
- 23.Gilmartin AG; Bleam MR; Groy A; et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res 2011, 17, 989–1000. [DOI] [PubMed] [Google Scholar]
- 24.See WL; Tan IL; Mukherjee J; et al. Sensitivity of glioblastomas to clinically available MEK inhibitors is defined by neurofibromin 1 deficiency. Cancer Res 2012, 72, 3350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson MJ; Blank JL; Bruzzese FJ; et al. Comparison of biochemical and biological effects of ML858 (salinosporamide A) and bortezomib. Mol Cancer Ther 2006, 5, 3052–61. [DOI] [PubMed] [Google Scholar]
- 26.Abaan OD; Polley EC; Davis SR; et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res 2013, 73, 4372–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil V; Pal J; Somasundaram K. Elucidating the cancer-specific genetic alteration spectrum of glioblastoma derived cell lines from whole exome and RNA sequencing. Oncotarget 2015, 6, 43452–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allen M; Bjerke M; Edlund H; et al. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med 2016, 8, 354re3. [DOI] [PubMed] [Google Scholar]
- 29.McNeill RS; Canoutas DA; Stuhlmiller TJ; et al. Combination therapy with potent PI3K and MAPK inhibitors overcomes adaptive kinome resistance to single agents in preclinical models of glioblastoma. Neuro Oncol 2017, 19, 1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.