Abstract
Chinese Hamster Ovary (CHO) cells, commonly used in the production of therapeutic proteins, are aneuploid. Their chromosomes bear structural abnormality and undergo changes in structure and number during cell proliferation. Some production cell lines are unstable and lose their productivity over time in the manufacturing process and during the product’s life cycle. To better understand the link between the genome structural changes and productivity stability, an Immunoglobulin G (IgG) producing cell line was successively single-cell cloned to obtain subclones that retained or lost productivity, and their genomic features were compared. Although each subclone started with a single karyotype, the progeny quickly diversified to a population with a distribution of chromosome numbers that is not distinctive between the parent and among subclones. Comparative genomic hybridization (CGH) analysis showed that the extent of copy variation of gene-coding regions among different subclones stayed at levels of a few percent. Genome regions that were prone to loss of copies, including one with a product transgene integration site, were identified in CGH. The loss of the transgene copy was accompanied by loss of transgene transcript level. Sequence analysis of the host cell and parental producing cell showed prominent structural variations within the regions prone to loss of copies. Taken together, we demonstrated the transient nature of clonal homogeneity in cell line development and the retention of a population distribution of chromosome numbers; we further demonstrated that structural variation in the transgene integration region caused cell line instability. Future cell line development may target the transgene into structurally stable regions.
Keywords: Chinese Hamster Ovary cells, CGH, karyotype, cell stability, genome instability
INTRODUCTION
Chinese hamster ovary (CHO) cells are industrial workhorses for the production of recombinant protein therapeutics, such as monoclonal antibodies and Fc-fusion proteins, which require proper folding and post-translational modifications for their biological activity (Bandyopadhyay et al., 2014; Wurm, 2013). The production cell line for these biologics is traditionally generated by random integration of the product transgene into the host CHO cell followed by transgene amplification and screening for high producing cell clones (Bandyopadhyay et al., 2017). In addition to a high productivity, the cell line must also sustain its productivity, not only during the manufacturing process but also throughout the product’s life cycle. To mitigate risks related to genetic changes in the producing cell line, single cell cloning is performed prior to the establishment of the cell stock to ensure the homogeneity of the starting cell population (EMA, 1998; FDA, 1997). This minimizes the probability that a subpopulation of cells overtakes the population, possibly causing changes in the productivity or product quality.
For normal diploid cells, such as many different types of stem cells, single cell cloning ensures the homogeneity of the ensuing cell population. However, aneuploid cell lines, including CHO cells, have abnormal chromosome number and structure. During proliferation, they continuously undergo genomic changes such as mutations, deletions, duplications, and other structural alterations due to errors in DNA replication and repair, and mistakes in chromosome segregation. As a result, these cells have a wide distribution of chromosome number, which has been shown in commonly used cell lines such as HEK293 (Stepanenko et al., 2015), MDCK (Gaush et al., 1966; Wunsch et al., 1995), and Vero cells (Bianchi & Ayres, 1971; Osada et al., 2014; Rhim et al., 1969). This heterogeneity in chromosome number and structure has also been demonstrated in CHO cells (Davies & Reff, 2001; Deaven & Petersen, 1973; Derouazi et al., 2006; Vcelar, Melcher, et al., 2018; Worton et al., 1977).
For a production CHO cell line, a large number of cell divisions are required to expand the cell population and have enough cells to fill a manufacturing bioreactor and to create enough cell banks to encompass a product’s life cycle. The subsequent accumulation of genome aberrations over time can lead to genetic and phenotypic heterogeneity among CHO cells, even those which are clonally derived (Frye et al., 2016). This heterogeneity can occur in the form of genomic and epigenomic variation (Feichtinger et al., 2016; Rouiller et al., 2015) or changes to cell phenotype or productivity (Kim et al., 1998; Ko et al., 2017).
There are a number of reported mechanisms leading to production instability (Barnes et al., 2003). These include loss of transgene copy number (Chusainow et al., 2009; Kim et al., 2011), promoter methylation (Osterlehner et al., 2011; Yang et al., 2010), and other epigenetic silencing mechanisms in the promoter region (Veith et al., 2016). In most studies, gene amplification was used to acquire multiple copies of transgene. How the loss of transgene copy number is affected by structural changes in the surrounding genome regions has not been elucidated.
Using Comparative Genome Hybridization (CGH), it has become possible to globally survey copy number changes in genomic loci within CHO cells (Vishwanathan et al., 2017). High throughput sequencing technologies have also greatly facilitated whole genome sequencing and the detection of structural changes in the genome. The combination of these tools with classical karyotyping and chromosome counting allows us to evaluate the effect of genome stability on phenotype stability in CHO cells. We chose to study productivity because it is the most important phenotype of an industrial cell line, and because it is readily measurable after single cell cloning.
We first eliminate the population heterogeneity of a recombinant DG44 cell line by single cell cloning to obtain subclones of high and low productivities, then allowed the heterogeneity to reestablish. The genomic heterogeneity of the populations derived from high and low producing subclones was investigated at the macroscopic level using karyotyping and chromosome counts, and at the microscopic gene and sequence levels using genome sequencing and CGH data. We also identified the primary transgene integration sites in the genome and examined local genomic alterations at the integration loci.
MATERIAL AND METHODS
Cell Lines and Culture Conditions
A recombinant CHO-DG44 cell line producing IgG (rDG_IgG) was used in this study. rDG_IgG and all derived clones were grown in T-flasks (Corning) in CHO-S-SFM-II Medium (Gibco). Cells were incubated at 37°C in a humidified incubator with 5% CO2. Single cell cloning was performed by limiting dilution at a seeding density of 0.2 cells per well.
Preparation of metaphase cells and Chromosome counting
Approximately 5 x 106 cells in exponential phase of growth were submitted to the Cytogenomics core at the University of Minnesota for metaphase spreads and G-banding analysis. Briefly, cells were cell cycle arrested by colcemid treatment, followed by incubation in 0.075M KCl at 37°C. Cells were fixed in a 3:1 solution of methanol:acetic acid, and then dropped onto slides. Slides were treated with a trypsin solution for improved banding. Wright’s stain was used for G-banding and visualization of chromosomes. Imaging of metaphase spreads was done on a Nikon Diaphot using a 100x oil objective. At least 70 chromosome spreads were counted per sample. Chromosome counting was done manually using ImageJ (NIH).
Genome Sequencing
Genomic DNA from H1 was extracted using DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA) and paired-end sequenced (101 bp) using Illumina Hi-seq 2000. Sequencing data for the DG44 genome was obtained from Lewis et al. (2013) (Accession number: SRS406582). Raw sequencing reads were quality-trimmed and were mapped to an annotated genome, UMN2.0 (Vishwanathan et al., 2016) using the gap-enabled aligner BWA mem (Li & Durbin, 2009) in paired-end mode. The SAMtools suite of algorithms (v.0.1.18) (Li et al., 2009) was used to process the resulting alignments. Reads with minimum paired-end mapping quality of 20 were used. Structural variant analysis was performed using DELLY (Rausch et al., 2012).
Comparative genomic hybridization (CGH)
A full description of comparative genomic hybridization (CGH) can be found in the Supplementary Information. Briefly, a CGH array based on Expressed Sequence Tags (ESTs) from the Chinese hamster genome was custom designed and manufactured by Agilent. Genomic DNA extracted from either CHO cell lines or Chinese Hamster liver tissue was labeled using the SureTag DNA Labeling Kit, followed by hybridization to the CGH array. Raw microarray hybridization intensity data was normalized by a LOWESS method. In order to identify segments of the genome with variation in copy number relative to liver, a circular binary segmentation-based algorithm was used called DNAcopy (Olshen et al., 2004; Venkatraman & Olshen, 2007). This algorithm uses statistical analysis to smooth out noisy CGH probe data and call discreet regions of the genome as having a particular copy number.
Integration Site Analysis & qPCR
A description of these methods can be found in the Supplementary Information.
RESULTS
Derivation of subclones
A CHO cell line (rDG_IgG) that has been extensively cultured in the laboratory (De Leon Gatti et al., 2007; Kantardjieff et al., 2010; Yee et al., 2008) was subcloned and IgG productivity was quantified (Fig. 1). The highest producing clone, H1, was isolated and expanded for a second round of subcloning 35 days after the first subcloning (~ 30 population doublings). The two subclones with highest and lowest IgG titer derived from H1 (designated as H1H2 and H1L2) were again expanded to obtain enough cells for further characterization. H1H2 was then subcloned for the third round 55 days after the second subcloning (~40 population doublings). The highest and lowest producing clones were again obtained and designated as H1H2H3 and H1H2L3 respectively (Fig. 1). The growth rates of the derived clones were all similar.
Figure 1.
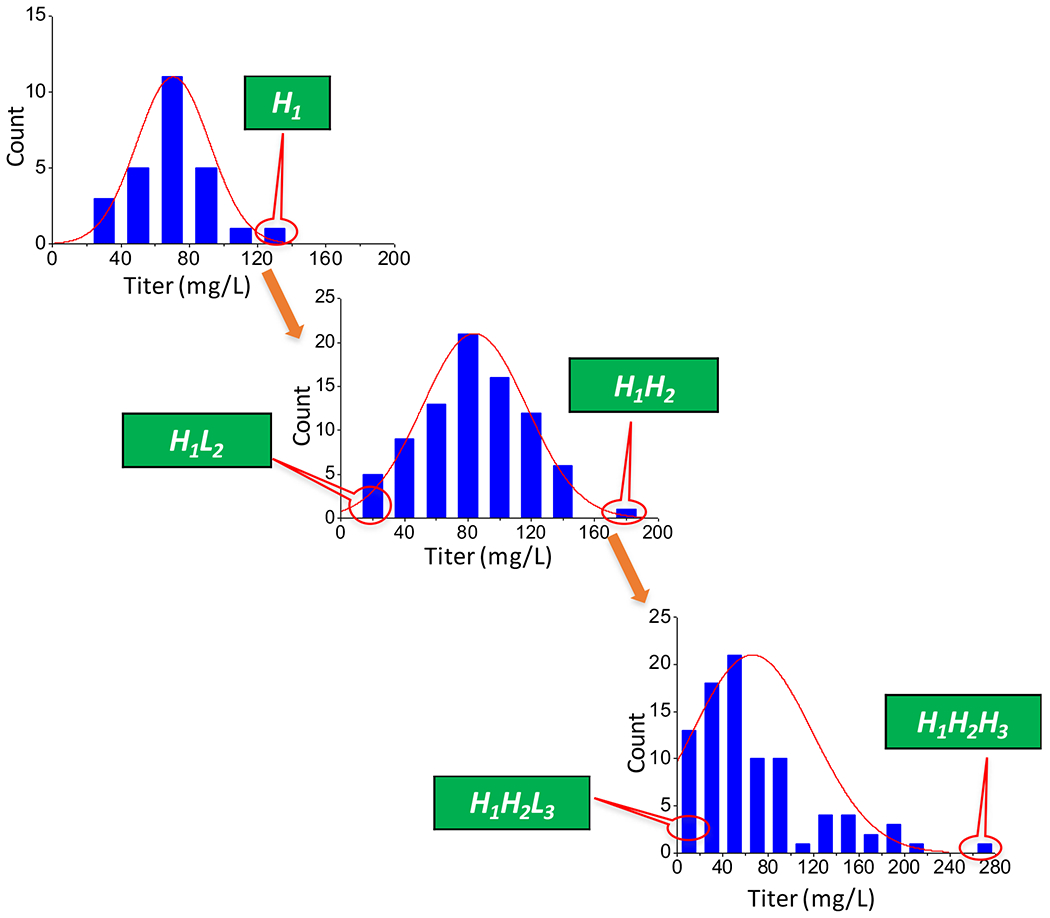
Derivation of cell lines through subcloning and the titer distribution.
Rapid rise of heterogeneity in karyotype and chromosome number
The inbred female Chinese hamster from which CHO cells were derived had 11 pairs of chromosomes (classically the chromosomes were numbered 1, 2, X, 4 until 11, skipping chromosome 3) (Deaven & Petersen, 1973). Figure 2A shows representative cell karyotypes from H1H2 and H1H2H3 based on the classical assignment of Chinese hamster chromosomes. Each metaphase spread exhibited a few normal appearing chromosomes as well as some abnormal ones. The copy number of each normal chromosome in the H1H2 and H1H2H3 cells analyzed is shown in Figure 2B. Each line represents the chromosome count for one metaphase spread analyzed. More than half of the cells retained a very similar distribution of normal chromosomes for both H1H2 and H1H2H3. Chromosomes 6 retained two copies in all cells. No cell was found to lose all copies of chromosome 1, while some trisomic cells were seen.
Figure 2.
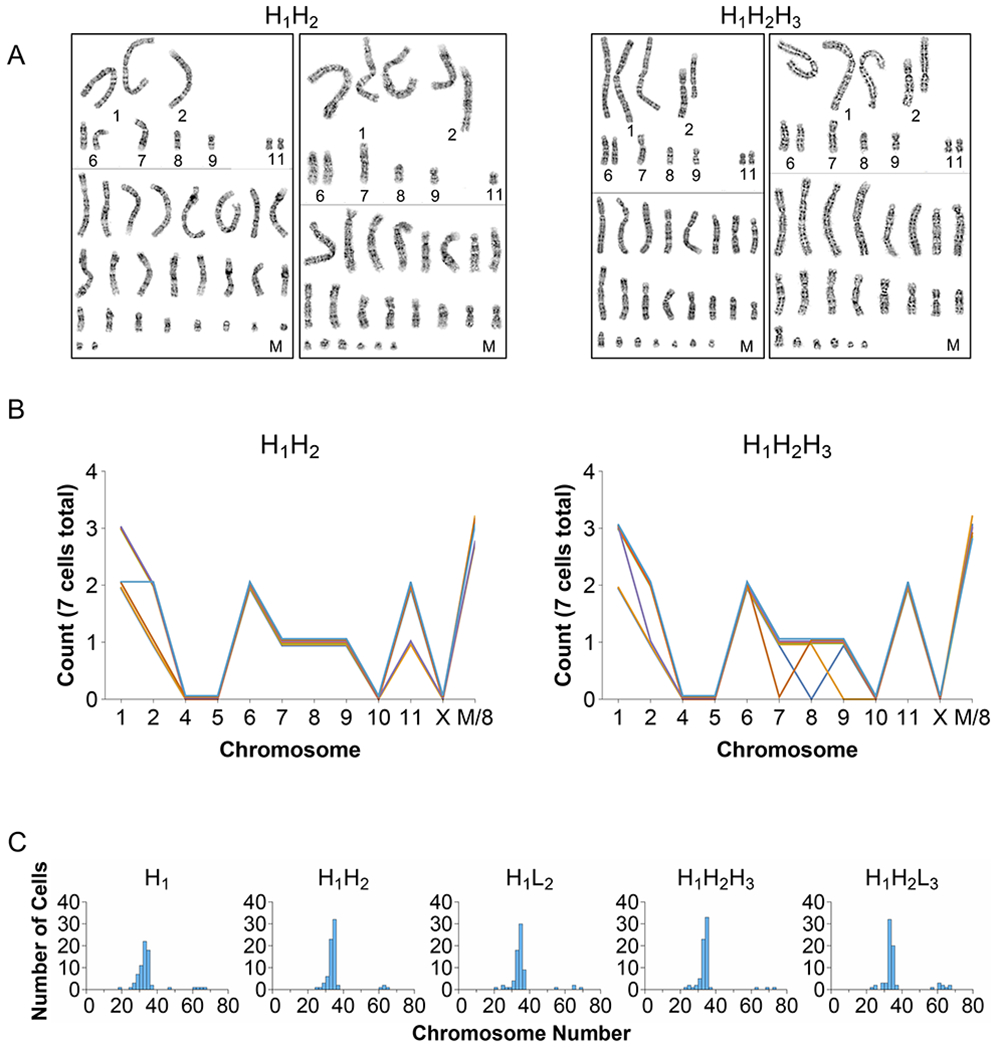
(A) Representative karyotype images from H1H2 and H1H2H3. (B) Distribution of all the chromosomes from the karyotypes. The marker chromosome numbers are divided by 8 for visualization purposes (M/8). (C) Chromosome number distributions for all the subclones obtained from ~70 metaphase spreads.
To survey a large number of cells, we utilized chromosome counting of metaphase spreads instead of karyotyping each chromosome. Chromosome numbers of approximately 70 cells were obtained for all five subclones. Representative images are shown in Supplementary Figure S1. All the cell lines showed a pseudo-triploid chromosome set of ~35 chromosomes with broad cell-to-cell variance in chromosome number within the population (Fig. 2C). A small number of cells in each cell line had a lower chromosome number (~15-25) while some had almost 70 chromosomes. The mode of chromosome number as well as the chromosome number distribution was largely similar among the five subclones. H1H2L3 may have a slightly higher number of cells with a very high number of chromosomes compared to its sister H1H2H3 and parent H1H2. Nevertheless, the data show that in the population doublings required to attain sufficient cells for systematic investigation, the once “clonal” population displayed a spread of chromosome numbers. Although the chromosome number and karyotype of each initial cell from subcloning was not known and were likely to be different from each other, the resulting populations had remarkably similar chromosome number distributions. The small sample size did not reveal any discernible difference in chromosome number in the phenotypically “abnormal” low-producing subclones.
Gain and loss of gene copy number among subclones
To assess the genomic changes at a microscopic scale, a comparative genomic hybridization (CGH) array (focused on transcript-coding regions) was used to detect gain or loss of genomic regions. Since the reference DNA was from a diploid genome, the normalized signal ratio of the sample to the reference for a probe reflects the gain or loss of abundance level of the corresponding gene. It should be noted that the genomic structure of the cell population was heterogeneous, and in a given genomic region, some cells may have no gain or loss of copy, while a subpopulation might have gained or lost copies. It is also possible that cells with multiple structural changes in the same gene region may appear to be distinct copy number change events, or that different degrees of copy number change might occur in a region that is prone to structural changes. Thus, the gene copy number data may not show discrete changes in copy numbers (e.g. one allele changed or both alleles changed) as in the case of a homogeneous population with a diploid genome.
Pairwise comparisons of CGH data for the subclones are shown in Figure 3. The dotted horizontal and vertical lines a, b mark one copy gain with respective to liver (log21.5 = 0.58 i.e. 3 copies instead of 2), while the lines c, d mark one copy loss with respect to liver (log20.5 = −1 i.e. 1 copy instead of 2 copies). 5,072 (3.02 %) probes had gained copy(-ies) in both H1 and H1H2 as compared to liver (upper yellow box, Fig. 3A). Similarly, 932 (0.55 %) probes lost copy(-ies) for both H1 and H1H2 (lower yellow box, Fig. 3A). Similar numbers of about 5000 for gain of copy(-ies) and 900 for loss of copy(-ies) were seen in the other comparisons. Despite the seemingly large change of chromosome number from normal diploidy, the genome showed a relatively small gain and loss of copies at the gene probe level compared to a diploid reference.
Figure 3.
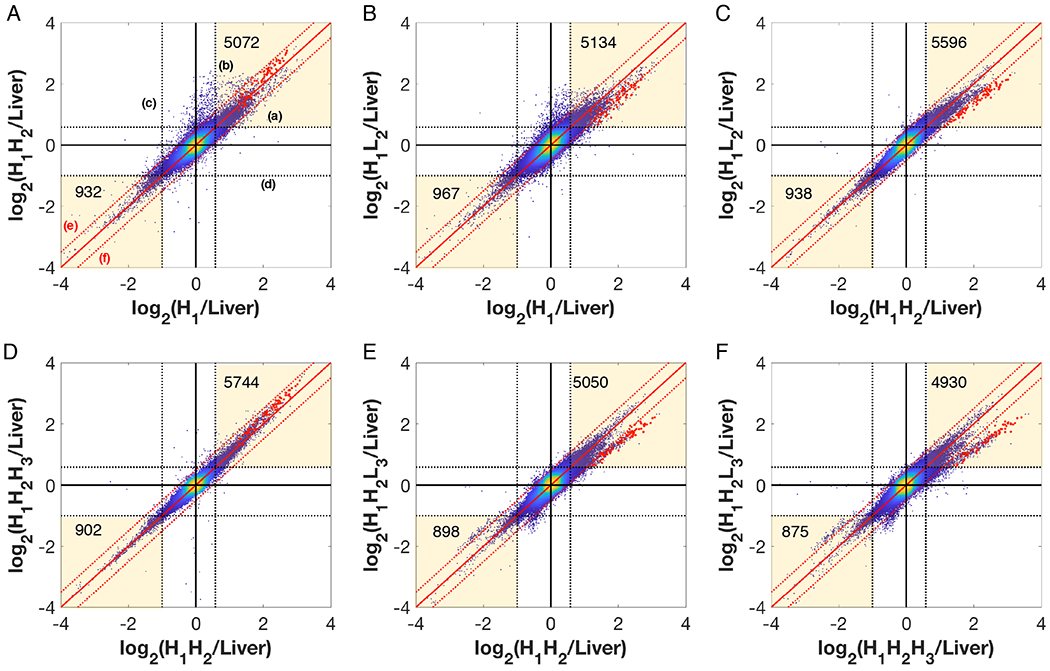
Pairwise comparison of log2(Sample/Liver) probe intensities for the 5 subclones. The dotted black lines (labeled a and b), both horizontal and vertical, are the bounds for one copy gain with respect to liver (log21.5 = 0.58) (i.e. 3 copies instead of 2). The dotted lines (labeled c and d) mark the bounds for loss of one copy with respect to liver (log20.5 = −1), i.e. 1 copy instead of 2 copies. The solid red line along the diagonal indicates the line x=y which implies both samples have the same copy number. The red dashed diagonal lines (labeled e and f) indicate the bounds for 1.5-fold difference in copy between the two cell lines. The red dots are the probes belonging to a genomic 4 Mbp region which show loss during high-to-low producer transition.
The 45° solid red lines (x=y) mark the probes for which the two samples have the same copy number relative to liver. The dotted red lines e, f indicate the bounds for 1.5-fold gain or loss from one cell line to the other. The vast majority of probes lie within the bounds of 1.5-fold change (region bounded by red dashed lines). 959 (0.57 %) probes (outside the upper red dashed line labeled e) show a higher copy number in H1H2 compared to its parent H1 (Fig. 3A), whereas 280 (0.17 %) probes show a lower copy number in H1 than H1H2 (outside the lower red dashed line labeled f). Similarly, 1199 probes showed a gain in H1L2 compared to its parent H1 and 753 probes showed a loss in H1L2 compared to H1 (Fig. 3B). H1H2H3 compared to H1H2 showed a very narrow spread of probe intensities (82 probes are higher and 35 probes are lower, Fig. 3D), indicating that these two samples are very similar at the gene copy abundance level, whereas H1H2L3 and its parent H1H2 show a higher spread, with 271 probes higher and 786 probes lower in H1H2L3, (Fig. 3E).
Figure 3C shows the comparison of two sublines derived from the same parent, H1H2 and H1L2, where 145 (0.09 %) probes showed an increased copy number in H1H2 over H1L2 while 87 (0.05 %) probes showed a lower copy number. H1H2L3 showed higher divergence from its sister clone H1H2H3 (276 probes ↑ and 1281 probes ↓, Fig. 3F). Interestingly, both low producers (H1L2 and H1H2L3) had ~100 probes with gained copy from the diploid reference show a decrease in copy number from their respective high producing sister clones (H1H2 and H1H2H3) (Fig. 3C and F).
CNV between high and low producing clones
Among the probes which showed consistent loss during high-to-low producer transitions, ~85 probes (Fig. 3, red dots) belonged to a 4 Mbp region within the genome, flanked by the genes Csmd3 and Rspo2 (Fig. 4A, yellow box). Each point represents the (log2) intensity ratio of the sample to diploid genome (liver) of a probe lined up according to its position in the genome region. The black line denotes the mean (log2) intensity ratio of a contiguous segment computed by DNAcopy. The region marked in yellow is amplified in the parent H1. The high producing clones, H1H2 and H1H2H3, retained the gain of copy number seen in the parent, but the low producers H1L2 and H1H2L3 showed a loss in copy number from their respective parents, H1 and H1H2. Importantly, the loss of the amplified segment happened in two independent transition events from a high producing parent to low producing clones.
Figure 4.
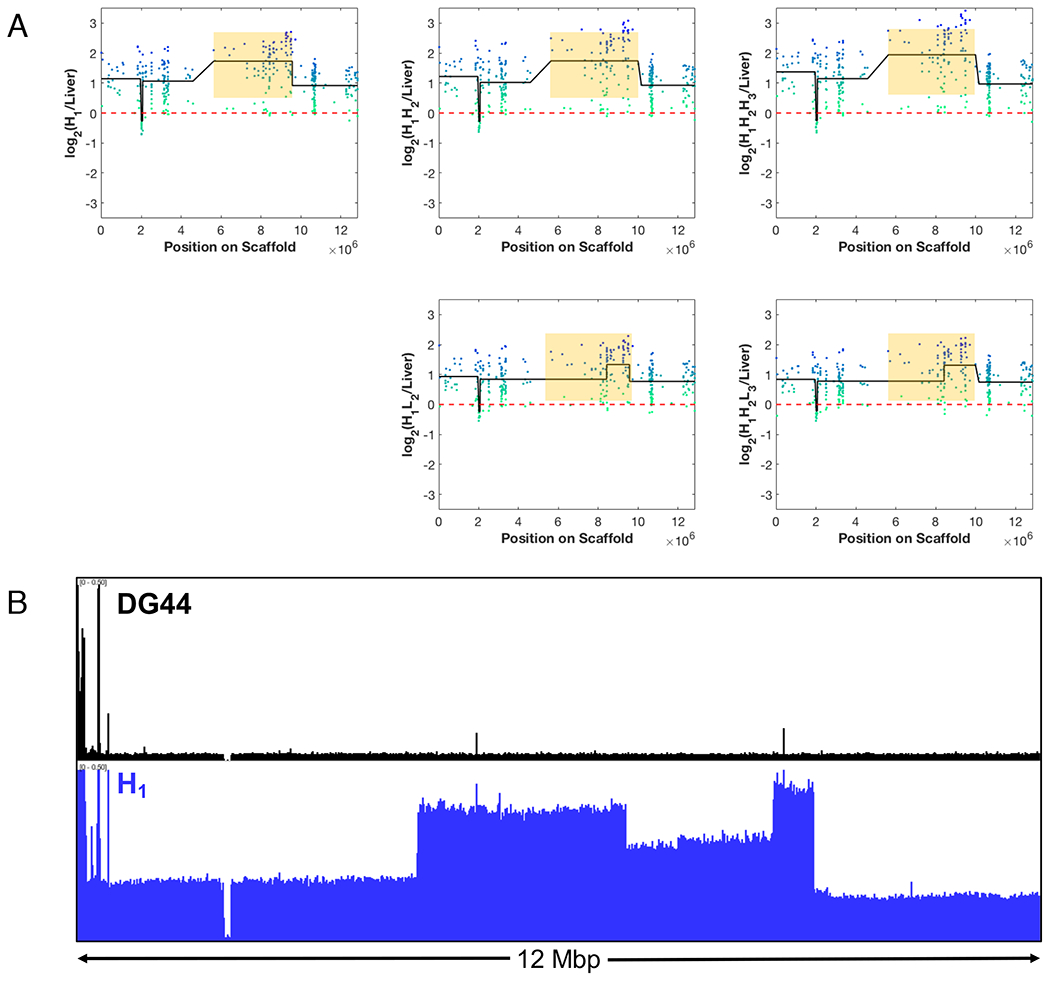
(A) Two independent losses of a 4 Mbp genomic region seen during high-to-low producer transition. The green-blue dots represent the log-ratio intensities for each probe in the region relative to liver. The black line is the mean intensity of the segments as identified by the DNAcopy. (B) Normalized sequencing read pileup (Reads per Million) for DG44 and H1. H1 shows amplification of the entire genomic region, correlating with the CGH data.
We further investigated this region using whole genome sequencing read pileups from H1 and the host cell line DG44 (Fig. 4B). The pileup of sequencing reads in this region for the host line DG44 showed almost a uniform sequence depth in the entire genomic region except a few highly amplified spikes at the left end. In contrast, the pileup for H1 showed increased depth in read pileups over the entire region. Notably, a nearly 4 Mbp region was further amplified to different levels in three segments. This is consistent with the DNAcopy call of ~4 Mbp segmental gain of copy from the CGH data. The CGH data lacked the spatial resolution to divide the segment further into different levels of amplification as the CGH microarray probes covered only the gene coding regions. Nevertheless, the sequence read pileup confirmed the CGH finding that the repeatedly lost 4 Mbp region was amplified in H1.
We investigated the stability of this region using the CGH data reported previously for other cell lines (Vishwanathan et al., 2017). In CHO-K1, almost the entire segment has a loss of ~1 copy from diploid (log20.5 = −1 i.e. 1 copy instead of 2 copies, Fig. S2A). The loss was retained by K1 derived producing cell lines rK1_IgG and rK1_2C10 (Fig. S2B and C) and was also seen in DXB11 (Fig. S2D). However, the log2 segment mean relative to diploid of the DXB11 derived cell line rDX_Fcf was between 0 and −1 (Fig. S2E). This might indicate a heterogeneous population in which some cells retained the loss while some others gained a copy relative to DXB11. Interestingly, the other DXB11 derived cell line, rDX_IgG_32, gained a copy relative to DXB11 and returned to the level of diploid cells (Fig. S2F). DG44 shows a normal ploidy state (Fig. S2G), consistent with the sequencing pileup data. In the DG44 derived lines, different segments of this region were amplified to varying extents as illustrate by the plots of H1L2 and H1H2L3 (Fig. 4A).
Overall, the data suggests that this region of genome is not only prone to change in copy number over a long stretch of segments as seen in the transition of high producing cells to low producing cells but has also been deleted or amplified in various CHO cell lines. This further suggests that some genome regions may be more prone to structural changes.
Identification of transgene integration sites
Having shown that the occurrence of low producing clones was accompanied by recurring loss of copies in some genome regions, we identified the transgene integration sites to determine possible gain or loss of copies in those regions. Three methods were employed to identify the transgene integration sites in the genome. First, biotinylated probes were used to capture the genome fragments containing sequences of different regions of the vector (Fig. S3). The isolated fragments were then sequenced using Illumina sequencing. Due to the short-read length of Illumina MiSeq (250bp), the confidence for mapping of split reads was low in some cases. From reads containing both vector and genome sequences, three integration sites were identified (Table S1). The read pileup showed a blunt boundary at the vector-genome junction and trailing read depth moving away from the integration site as expected (Fig. S4). Next, vector specific primers designed along the vector sequence were used to amplify DNA segments extending beyond the genome vector junction (Fig. S5). Amplified fragments were then separated on a gel (Fig. S6), extracted, and sequenced using Sanger sequencing. The longer genome sequences obtained gave higher confidence in the integration site identified. Using this method, we confirmed the previously determined integration sites and identified an additional site (Table S1). Further, DELLY was used on whole genome sequencing data of H1 to identify split reads that map to both the genome and the vector. Based on the paired-end and split reads spanning the integration junction, all but one of the integration sites found using the PCR-based method were identified, and a fifth site was also identified. The missing integration site within the intron of the Rc3h1 gene, as discussed later, had complex sequence rearrangements that made it difficult to identify. Combining the three methods we were able to identify the five transgene integration sites with high confidence (Table S1).
Loss of transgene in high-to-low producer transition
We next examined the CGH data on the integration sites identified for changes in copy number. The relative probe intensity at or near all integration loci was similar among all subclones except for the probe representing Rc3h1 (site #1 in Table S1). Rc3h1 had a much higher copy number than the diploid genome, suggesting that the locus had been amplified (Fig. 5A, marked in red circle). The copy number appeared similar in H1 and H1H2, but slightly higher in H1H2H3. However, the copy number was markedly reduced in low producing clones. Note that H1H2 and H1L2 were both derived from H1, while H1H2H3 and H1H2L3 were both derived from H1H2. The data thus showed that the Rc3h1 locus lost copy in two independent events associated with the emergence of low producers.
Figure 5.
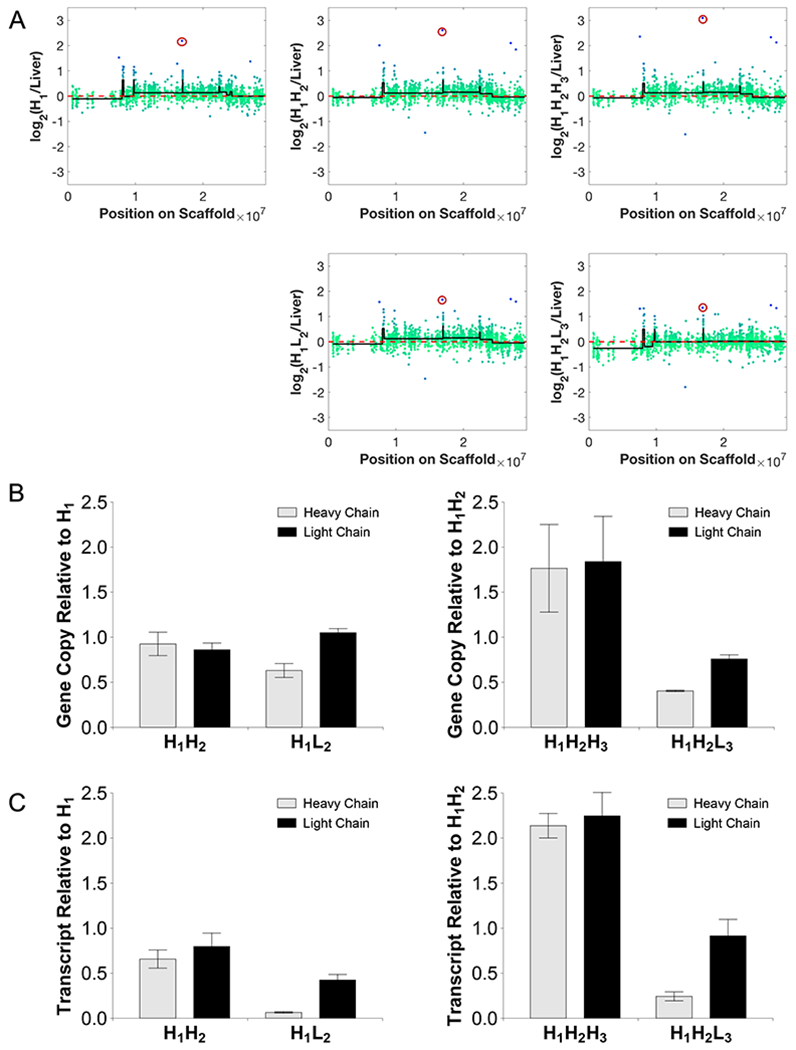
(A) Two independent losses of the gene probe for the integration site at Rc3h1 (circled in red) seen during high-to-low producer transition. The green-blue dots represent the log-ratio intensities for each probe in the region relative to liver. The black line is the mean intensity of the segments as identified by the DNAcopy. (B) Genomic qPCR results show loss of heavy chain correlating with the loss of the integration site. Results reported in terms of gDNA copy number fold change relative to the subclone parent. (C) Transcript expression as quantified by qRT-PCR shows loss of heavy and light chain transcript levels. Results reported in terms of transcript expression fold change relative to the subclone parent.
To examine whether the copy loss of Rc3h1 was accompanied by the loss in transgene copy, the copy numbers of the IgG heavy chain and light chain genes in the clones were quantified using qPCR (Fig. 5B). The high producing daughter cell line H1H2 retained the transgene copy number from its parent H1 whereas H1L2 showed a loss of IgG heavy chain copy compared to its parent H1. High producing daughter cell line H1H2H3 showed an increased copy number of IgG heavy chain compared to its parent H1H2, consistent with the increased probe intensity for Rc3h1, the integration site. The low producing daughter cell line H1H2L3 showed a loss in copy number of IgG heavy chain compared to its parent H1H2. IgG transcript expression from qRT-PCR also showed a similar trend where the low producing clones showed a 6 to 15-fold decrease in the expression of IgG heavy chain (Fig. 5C). These results imply that the loss of productivity is likely to be a consequence of the loss of an IgG heavy chain gene copy at the Rc3h1 locus and subsequent loss in transcript expression.
This integration site was further investigated using the sequencing reads pileup of H1 and the parental cell line DG44 (Fig. 6). Figure 6A shows a 29 Mbp genomic region where the integration site is marked with a red vertical line. Though the read pileups for DG44 and H1 look very similar across the entire region, the region marked in yellow shows a slight (~1.2-fold) amplification and the integration site shows about a 10-fold amplification. Zooming in further (Fig. 6B), we see the integration locus is in a smaller segment of 31 Kbp that was amplified to ~8 times from diploid. DELLY identified tandem duplications and inversions within this segment, as well a large deletion. No deletions, duplications, or inversions were present at this site in DG44; there is thus no indication that the region surrounding this site is structurally unstable or has a higher propensity for duplication/amplification in the host cell line. The structural changes incurred after vector integration and amplification may have contributed to the instability of the locus and the loss in transgene copy number. Further zooming into the integration locus shows the sharp vector-genome boundary (Fig. 6C).
Figure 6.
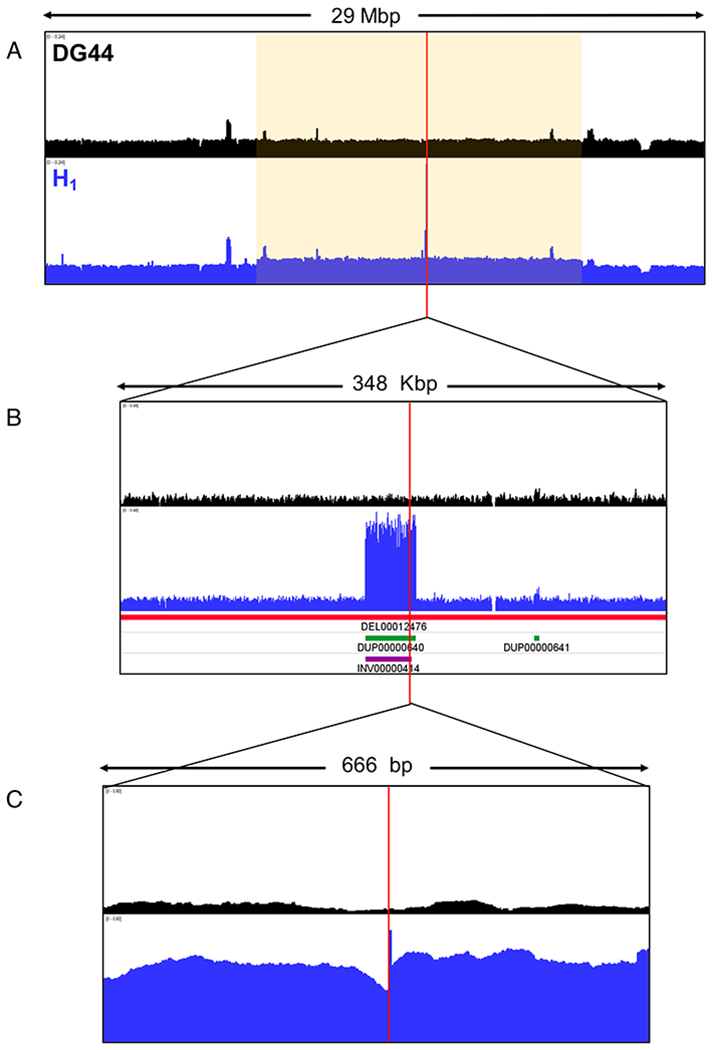
Normalized sequencing read pileup (Reads per Million) for DG44 and H1 in the integration region shown in the different magnifications. The red vertical line denotes the integration junction. The red horizontal bar indicates deletion, the green bar indicates tandem duplication, and the purple bar indicates inversion.
The sequence pileup around integration sites #2 and #3 (Table S1) are shown in Supplementary Figures S7A and S8A, along with the CGH data in the integration regions. Site 2 had an amplification with respect to DG44 (Fig. S7A), while site 3 did not show any amplification. The CGH data of the probes in the region did not show a significant difference in copy number among high and low producers (Fig. S7B and S8B).
DISCUSSION
The stability of recombinant CHO cell lines
Cell lines used in industrial production of biopharmaceuticals were initially clonal, originating from a single cell at some stage after the introduction of the transgene into the genome. This is to avoid the risk of a subpopulation within a heterogeneous population eventually out-growing the others and, as a result, altering the productivity or the product quality. It is now a regulatory requirement that production cell lines be initially phenotypically and genomically “clonal” (EMA, 1998; FDA, 1997). Recent studies have addressed the issue of the variability among single cell clones from clonally derived cell lines (Ko et al., 2017), and discussed the possibility for using cell pools for the production of non-clinical material (Munro et al., 2017).
Karyotyping of CHO host cells and assessment of chromosomal rearrangements has been demonstrated using various techniques such as G-banding, BAC-FISH (Bacterial Artificial Chromosome Fluorescence in Situ Hybridization), and chromosome painting (Baik & Lee, 2017; Cao et al., 2012b; Deaven & Petersen, 1973; Derouazi et al., 2006; Martinet et al., 2007; Vcelar, Jadhav, et al., 2018; Vcelar, Melcher, et al., 2018). DG44 was reported to have twenty chromosomes, with seven normal chromosomes, eleven chromosomes with rearrangements of normal chromosomes, and two uncharacterized marker chromosomes (Martinet et al., 2007). Other karyotyping studies on DG44 and CHO-K1 have found that chromosomes 1, 2, 4, 5, 9, and 10 (by classical nomenclature) are likely to have at least one intact copy (Cao et al., 2012a; Derouazi et al., 2006; Vcelar, Jadhav, et al., 2018; Vcelar, Melcher, et al., 2018). Our karyotyping data showed the presence of at least two copies of chromosomes 1 and 6, and at least one copy of chromosomes 2, 7, 8, 9, and 11. Comparing all of these studies, at least one intact copy of chromosomes 1, 2, and 9 seems to be stably present in all tested cell lines, and so these are promising targets for transgene integration.
Previous studies have also found a wide distribution of chromosome number in CHO cell lines, ranging from 15 to 50 chromosomes (Deaven & Petersen, 1973; Vcelar, Melcher, et al., 2018; Worton et al., 1977). Derouazi et al. (2006) found that the chromosome number for sixteen DG44-derived recombinant cell lines varied from 19 to 41. In our study of five cell lines also derived from DG44, we found a wide spread of chromosome numbers (20 - 70 chromosomes, Fig. 2C). Importantly, we demonstrated that this distribution is rapidly re-established by about 30 generations after starting from a single cell.
Clonal cells and population heterogeneity
By single cell cloning to ensure that all offspring cells originated from a particular karyotype and genome structure, we showed chromosomal reorganization and genomic structural changes occurred again to give a population with a wide distribution in karyotype and chromosome number. Interestingly, the redistribution of genomic organization was accompanied by relatively small levels of gain and loss of gene copies. The number of gene probes that showed gain or loss of copies from a diploid genome was about 4-5% in all the cell lines (Fig. 3). Further, in each subcloning, over the span of 30-40 population doublings, approximately 100 to 2000 probes (0.06 to 1.16 % of all probes) gained or lost copies from their immediate parent. However, the CGH microarray that we employed did not have a high spatial resolution. The Chinese hamster genome assembly and the sequencing analytical tools also await further enhancement. We thus refrain from determining the fraction of total genome regions that are subject to gain or loss of copies. Nevertheless, the CGH and sequencing data both indicate that the genes that changed copy number in CHO cell lines constitute only a minor fraction.
It is also interesting to note that the number of gene probes that showed a loss of copy is relatively small compared to those which gained copy. It is plausible that a cell with a more drastic loss of copies may have a growth disadvantage or be prone to death. Similarly, the cells that have an “extreme” karyotype (too many, too few, or too drastic of a change (e.g. losing both alleles of critical regions)) may not survive. Therefore, even though cells continue to undergo genomic structural changes, the population still maintains a relatively stable distribution of karyotype and gene copies. Taken together, the cell lines established by subcloning largely returned to a distribution of gene copy and chromosomal organization that is very similar to their parent. However, in this study we examined only the heterogeneity in the chromosome number, gene copy number, and the productivity phenotype. Even though only a small fraction of genes changed copy number in each pair of parent-subclone, extensive sequential subcloning will increase the probability of generating a new population that has a skewed property distribution compared to the ancestor cell line.
This thus raises the prospect that, in the “recloning” or re-purifying a cell line, one should focus on maintaining the property distribution of the population of the production cell line. For example, pooling a large number of cells that maintain the productivity and eliminating low producers from the population, rather than picking a colony that grew out from of single cell.
Productivity stability and transgene integration
Traditionally, a CHO production cell line is generated by integrating the product transgene into a random locus in the genome followed by the amplification of transgene copy number. The initial integration of the transgene after transfection typically occurs at one or a few loci (Derouazi et al., 2006; Kim & Lee, 1999; Zhou et al., 2010), and in the subsequent amplification the copy number increases over a wide range. The number of copies of transgene integrated is affected by the many factors, including the dose of plasmids and the extent of selective pressure applied. In some cases, a large number of copies were integrated even without amplification (Derouazi et al., 2006; Jiang et al., 2006; Yusufi et al., 2017). The amplification process typically amplifies not only the vector and transgene, but also the surrounding genomic region. Furthermore, breakage, ligation, and other structural changes may occur during amplification (Chusainow et al., 2009; Kim et al., 2001; Pallavicini et al., 1990). Loss of productivity in transgene amplified cell lines over long-term culture occurs frequently (Bailey et al., 2012; Chusainow et al., 2009; Kim et al., 2011). Hence, before a cell line is chosen to be the production cell line, extensive testing of its stability is commonly performed.
In this study, by identifying the transgene integration sites, we not only showed that transgene copy loss was likely the cause of the lost productivity, but also demonstrated that the lost copy in two independent isolations of low producing subclones was from the same integration site (Rc3h1 locus). The loss was accompanied by a decreased IgG heavy chain copy number and transcript level. Sequence analysis of the production parental line (H1) revealed extensive amplification and structural rearrangements in the Rc3h1 locus (tandem duplication, inversion, and deletion), possibly occurring in conjunction with the amplification process during cell line development. The data thus suggests that some amplified sequences are unstable and prone to repeated loss of copy number.
Intriguingly, from CGH data we found another segment of an amplified 4 Mbp region in the production cell line was also lost in both low producing subclones (Fig. 4A). From our archived CGH data this segment showed frequent gain or loss of copies in other CHO cell lines (Fig. S2). Although the segment did not harbor any integrated transgene, its loss appears to associate with the loss of the productivity or the loss of the transgene in Rc3h1 locus, as its copy number was lower only in the low producing subclones, but normal in the high producing subclones. It is possible that the region also harbors genes or epigenetic loci that confer traits related to productivity, although the region was not very transcriptionally active, and no notable gene was found. It is also possible the co-loss of copy number in this region and the Rc3h1 locus was related to other clonal events. It is plausible that the low producing sublines H1L2 and H1H2L3 arose from subclones that had DNA repair alterations that triggered copy loss in both Rc3h1 and the 4 Mbp region, and these may be linked to transgene copy change and loss of productivity. Regardless of the mechanism of the co-loss of the two regions, the data suggests that some genome regions are possibly prone to structural variation. Our results also demonstrate that by combining CGH and subcloning one can identify such genome regions that are more vulnerable to copy number loss. One can thus use these tools to help determine clone stability.
With the advances in genome engineering, transgene integration can now be targeted to a specific locus in the host cell genome, without resorting to the traditional selective pressure-based transgene amplification (Cristea et al., 2013; Inniss et al., 2017; Lee et al., 2016; Lee et al., 2015). In general, the site selected for targeted integration should be stable, and not prone to structural variation or copy number changes. Genome scale CGH and sequencing analysis of structural variants provide valuable information on the regions to be avoided. Additionally, the target genomic regions should be transcriptionally active and epigenetically accessible for gene expression. Recent work from our laboratory showed that a single copy of the transgene integrated into a genomic region of high chromatin accessibility and high transcriptional activity can have an expression level equivalent to or higher than a cell line with multiple copies (O’Brien et al. 2018, manuscript submitted for publication). The incorporation of genomic analysis and genome engineering into cell line development will allow this traditionally empirical operation to become a design-based process.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Takeshi Omasa for valuable discussion. We would also like to acknowledge Meghan McCann and Marina Raabe for their assistance with experiments and Conor O’Brien for his assistance with computational analysis. Computational resources were provided by the Minnesota Supercomputing Institute. SAO was supported in part by the NIGMS Biotechnology Training Program (T32GM008347- 22).
Footnotes
Publisher's Disclaimer: This is the peer reviewed version of the following article:
Bandyopadhyay, A. A.; O’Brien, S. A.; Zhao, L.; Fu, H.-Y.; Vishwanathan, N.; Hu, W.-S. Re-curring Genomic Structural Variation Leads to Clonal Instability and Loss of Productivity. Bio-technology and Bioengineering 2019, 116 (1), 41-53. https://doi.org/10.1002/bit.26823. which has been published in final form at https://doi.org/10.1002/bit.26823. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.
REFERENCES
- Baik JY, & Lee KH (2017). A framework to quantify karyotype variation associated with CHO cell line instability at a single-cell level. Biotechnol Bioeng, 114(5), 1045–1053. doi : 10.1002/bit.26231 [DOI] [PubMed] [Google Scholar]
- Bailey LA, Hatton D, Field R, & Dickson AJ (2012). Determination of Chinese hamster ovary cell line stability and recombinant antibody expression during long-term culture. Biotechnol Bioeng, 109(8), 2093–2103. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay A, Fu H-Y, Vishwanathan N, & Hu W-S (2014). Genomics and Systems Biotechnology in Biopharmaceutical Processing. Chemical Engineering Progress, 100. [Google Scholar]
- Bandyopadhyay AA, Khetan A, Malmberg LH, Zhou W, & Hu WS (2017). Advancement in bioprocess technology: parallels between microbial natural products and cell culture biologics. J Ind Microbiol Biotechnol, 44(4-5), 785–797. doi: 10.1007/s10295-017-1913-4 [DOI] [PubMed] [Google Scholar]
- Barnes LM, Bentley CM, & Dickson AJ (2003). Stability of protein production from recombinant mammalian cells. Biotechnol Bioeng, 81(6), 631–639. doi : 10.1002/bit.10517 [DOI] [PubMed] [Google Scholar]
- Bianchi NO, & Ayres J (1971). Heterochromatin location on chromosomes of normal and transformed cells from African green monkey (Cercopithecus aethiops). DNA denaturation-renaturation method. Exp Cell Res, 68(2), 253–258. [DOI] [PubMed] [Google Scholar]
- Cao Y, Kimura S, Itoi T, Honda K, Ohtake H, & Omasa T (2012a). Construction of BAC-based physical map and analysis of chromosome rearrangement in Chinese hamster ovary cell lines. Biotechnol Bioeng, 109(6), 1357–1367. doi: 10.1002/bit.24347 [DOI] [PubMed] [Google Scholar]
- Cao Y, Kimura S, Itoi T, Honda K, Ohtake H, & Omasa T (2012b). Fluorescence in situ hybridization using bacterial artificial chromosome (BAC) clones for the analysis of chromosome rearrangement in Chinese hamster ovary cells. Methods, 56(3), 418–423. doi: 10.1016/j.ymeth.2011.11.002 [DOI] [PubMed] [Google Scholar]
- Chusainow J, Yang Y, Yeo JHM, Toh P, Asvadi P, Wong NSC, & Yap MGS (2009). A study of monoclonal antibody producing CHO cell lines: What makes a stable high producer? Biotechnol Bioeng, 102(4), 1182–1196. doi: 10.1002/bit.22158 [DOI] [PubMed] [Google Scholar]
- Cristea S, Freyvert Y, Santiago Y, Holmes MC, Urnov FD, Gregory PD, & Cost GJ (2013). In vivo cleavage of transgene donors promotes nuclease-mediated targeted integration. Biotechnol Bioeng, 110(3), 871–880. doi: 10.1002/bit.24733 [DOI] [PubMed] [Google Scholar]
- Davies J, & Reff M (2001). Chromosome localization and gene-copy-number quantification of three random integrations in Chinese-hamster ovary cells and their amplified cell lines using fluorescence in situ hybridization. Biotechnol Appl Biochem, 33(Pt 2), 99–105. [DOI] [PubMed] [Google Scholar]
- De Leon Gatti M, Wlaschin KF, Nissom PM, Yap M, & Hu WS (2007). Comparative transcriptional analysis of mouse hybridoma and recombinant Chinese hamster ovary cells undergoing butyrate treatment. J Biosci Bioeng, 103(1), 82–91. doi: 10.1263/jbb.103.82 [DOI] [PubMed] [Google Scholar]
- Deaven LL, & Petersen DF (1973). The chromosomes of CHO, an aneuploid Chinese hamster cell line: G-band, C-band, and autoradiographic analyses. Chromosoma, 41(2), 129–144. doi: 10.1007/BF00319690 [DOI] [PubMed] [Google Scholar]
- Derouazi M, Martinet D, Schmutz NB, Flaction R, Wicht M, Bertschinger M, … Wurm FM (2006). Genetic characterization of CHO production host DG44 and derivative recombinant cell lines. Biochemical and Biophysical Research Communications, 340(4), 1069–1077. doi: 10.1016/j.bbrc.2005.12.111 [DOI] [PubMed] [Google Scholar]
- EMA. (1998). ICH Harmonized Tripartite Guideline Q5D Quality of biotechnological products: derivation and characterisation of cell substrates used for production of biotechnological/biological products. [PubMed]
- FDA. (1997). Points to consider in the manufacture and testing of monoclonal antibody products for human use (1997). U.S. Food and Drug Administration Center for Biologics Evaluation and Research. J Immunother, 20(3), 214–243. [DOI] [PubMed] [Google Scholar]
- Feichtinger J, Hernandez I, Fischer C, Hanscho M, Auer N, Hackl M, … Borth N (2016). Comprehensive genome and epigenome characterization of CHO cells in response to evolutionary pressures and over time. Biotechnol Bioeng, 113(10), 2241–2253. doi: 10.1002/bit.25990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye C, Deshpande R, Estes S, Francissen K, Joly J, Lubiniecki A, … Anderson K (2016). Industry view on the relative importance of “clonality” of biopharmaceutical-producing cell lines. Biologicals, 44(2), 117–122. doi: 10.1016/j.biologicals.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Gaush CR, Hard WL, & Smith TF (1966). Characterization of an established line of canine kidney cells (MDCK). Proc Soc Exp Biol Med, 122(3), 931–935. [DOI] [PubMed] [Google Scholar]
- Inniss MC, Bandara K, Jusiak B, Lu TK, Weiss R, Wroblewska L, & Zhang L (2017). A novel Bxb1 integrase RMCE system for high fidelity site-specific integration of mAb expression cassette in CHO Cells. Biotechnol Bioeng, 114(8), 1837–1846. doi: 10.1002/bit.26268 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Huang Y, & Sharfstein ST (2006). Regulation of recombinant monoclonal antibody production in chinese hamster ovary cells: a comparative study of gene copy number, mRNA level, and protein expression. Biotechnol Prog, 22(1), 313–318. doi: 10.1021/bp0501524 [DOI] [PubMed] [Google Scholar]
- Kantardjieff A, Jacob NM, Yee JC, Epstein E, Kok YJ, Philp R, … Hu WS (2010). Transcriptome and proteome analysis of Chinese hamster ovary cells under low temperature and butyrate treatment. J Biotechnol, 145(2), 143–159. doi: 10.1016/j.jbiotec.2009.09.008 [DOI] [PubMed] [Google Scholar]
- Kim M, O’Callaghan PM, Droms KA, & James DC (2011). A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng, 108(10), 2434–2446. doi: 10.1002/bit.23189 [DOI] [PubMed] [Google Scholar]
- Kim NS, Byun TH, & Lee GM (2001). Key determinants in the occurrence of clonal variation in humanized antibody expression of cho cells during dihydrofolate reductase mediated gene amplification. Biotechnol Prog, 17(1), 69–75. doi: 10.1021/bp000144h [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim NS, Ryu CJ, Hong HJ, & Lee GM (1998). Characterization of chimeric antibody producing CHO cells in the course of dihydrofolate reductase-mediated gene amplification and their stability in the absence of selective pressure. Biotechnol Bioeng, 58(1), 73–84. [PubMed] [Google Scholar]
- Kim SJ, & Lee GM (1999). Cytogenetic analysis of chimeric antibody-producing CHO cells in the course of dihydrofolate reductase-mediated gene amplification and their stability in the absence of selective pressure. Biotechnol Bioeng, 64(6), 741–749. [PubMed] [Google Scholar]
- Ko P, Misaghi S, Hu Z, Zhan D, Tsukuda J, Yim M, … Shen A (2017). Probing the importance of clonality: Single cell subcloning of clonally derived CHO cell lines yields widely diverse clones differing in growth, productivity, and product quality. Biotechnol Prog. doi: 10.1002/btpr.2594 [DOI] [PubMed] [Google Scholar]
- Lee JS, Grav LM, Pedersen LE, Lee GM, & Kildegaard HF (2016). Accelerated homology-directed targeted integration of transgenes in Chinese hamster ovary cells via CRISPR/Cas9 and fluorescent enrichment. Biotechnol Bioeng, 113(11), 2518–2523. doi: 10.1002/bit.26002 [DOI] [PubMed] [Google Scholar]
- Lee JS, Kallehauge TB, Pedersen LE, & Kildegaard HF (2015). Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Sci Rep, 5, 8572. doi: 10.1038/srep08572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NE, Liu X, Li Y, Nagarajan H, Yerganian G, O’Brien E, … Palsson BO (2013). Genomic landscapes of Chinese hamster ovary cell lines as revealed by the Cricetulus griseus draft genome. Nat Biotechnol, 31(8), 759–765. doi: 10.1038/nbt.2624 [DOI] [PubMed] [Google Scholar]
- Li H, & Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25(14), 1754–1760. doi: 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, … Durbin R (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics, 25(16), 2078–2079. doi : 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet D, Derouazi M, Besuchet N, Wicht M, Beckmann J, & Wurm FM (2007, 2007//). Karyotype of CHO DG44 cells. Paper presented at the Cell Technology for Cell Products, Dordrecht. [Google Scholar]
- Munro TP, Le K, Le H, Zhang L, Stevens J, Soice N, … Goudar CT (2017). Accelerating patient access to novel biologics using stable pool-derived product for non-clinical studies and single clone-derived product for clinical studies. Biotechnol Prog, 33(6), 1476–1482. doi: 10.1002/btpr.2572 [DOI] [PubMed] [Google Scholar]
- O’Brien SA, Lee K, Fu HY, Lee Z, Le TS, Stach CS, … Hu WS (2018). Single Copy Transgene Integration in a Transcriptionally Active Site for Recombinant Protein Synthesis. Manuscript submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, & Wigler M (2004). Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics, 5(4), 557–572. doi : 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- Osada N, Kohara A, Yamaji T, Hirayama N, Kasai F, Sekizuka T, … Hanada K (2014). The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res, 21(6), 673–683. doi : 10.1093/dnares/dsu029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlehner A, Simmeth S, & Gopfert U (2011). Promoter methylation and transgene copy numbers predict unstable protein production in recombinant Chinese hamster ovary cell lines. Biotechnol Bioeng, 108(11), 2670–2681. doi: 10.1002/bit.23216 [DOI] [PubMed] [Google Scholar]
- Pallavicini MG, DeTeresa PS, Rosette C, Gray JW, & Wurm FM (1990). Effects of methotrexate on transfected DNA stability in mammalian cells. Mol Cell Biol, 10(1), 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, & Korbel JO (2012). DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics, 28(18), i333–i339. doi: 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim JS, Schell K, Creasy B, & Case W (1969). Biological characteristics and viral susceptibility of an African green monkey kidney cell line (Vero). Proc Soc Exp Biol Med, 132(2), 670–678. doi: 10.3181/00379727-132-34285 [DOI] [PubMed] [Google Scholar]
- Rouiller Y, Kleuser B, Toso E, Palinksy W, Rossi M, Rossatto P, … Broly H (2015). Reciprocal Translocation Observed in End-of-Production Cells of a Commercial CHO-Based Process. PDA JPharm Sci Technol, 69(4), 540–552. doi: 10.5731/pdajpst.2015.01063 [DOI] [PubMed] [Google Scholar]
- Stepanenko A, Andreieva S, Korets K, Mykytenko D, Huleyuk N, Vassetzky Y, & Kavsan V (2015). Step-wise and punctuated genome evolution drive phenotype changes of tumor cells. MutatRes, 771, 56–69. doi: 10.1016/j.mrfmmm.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Vcelar S, Jadhav V, Melcher M, Auer N, Hrdina A, Sagmeister R, … Borth N (2018). Karyotype variation of CHO host cell lines over time in culture characterized by chromosome counting and chromosome painting. Biotechnol Bioeng, 115(1), 165–173. doi: 10.1002/bit.26453 [DOI] [PubMed] [Google Scholar]
- Vcelar S, Melcher M, Auer N, Hrdina A, Puklowski A, Leisch F, … Borth N (2018). Changes in Chromosome Counts and Patterns in CHO Cell Lines upon Generation of Recombinant Cell Lines and Subcloning. Biotechnol J, 13(3), e1700495. doi: 10.1002/biot.201700495 [DOI] [PubMed] [Google Scholar]
- Veith N, Ziehr H, MacLeod RA, & Reamon-Buettner SM (2016). Mechanisms underlying epigenetic and transcriptional heterogeneity in Chinese hamster ovary (CHO) cell lines. BMC Biotechnol, 16, 6. doi: 10.1186/s12896-016-0238-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatraman ES, & Olshen AB (2007). A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics, 23(6), 657–663. doi: 10.1093/bioinformatics/btl646 [DOI] [PubMed] [Google Scholar]
- Vishwanathan N, Bandyopadhyay A, Fu HY, Johnson KC, Springer NM, & Hu WS (2017). A comparative genomic hybridization approach to study gene copy number variations among Chinese hamster cell lines. Biotechnol Bioeng, 114(8), 1903–1908. doi : 10.1002/bit.26311 [DOI] [PubMed] [Google Scholar]
- Vishwanathan N, Bandyopadhyay AA, Fu HY, Sharma M, Johnson KC, Mudge J, … Hu WS (2016). Augmenting Chinese hamster genome assembly by identifying regions of high confidence. Biotechnol J, 11(9), 1151–1157. doi: 10.1002/biot.201500455 [DOI] [PubMed] [Google Scholar]
- Worton RG, Ho CC, & Duff C (1977). Chromosome stability in CHO cells. Somatic Cell Genetics, 3(1), 27–45. doi: 10.1007/BF01550985 [DOI] [PubMed] [Google Scholar]
- Wunsch S, Gekle M, Kersting U, Schuricht B, & Oberleithner H (1995). Phenotypically and karyotypically distinct Madin-Darby canine kidney cell clones respond differently to alkaline stress. J Cell Physiol, 164(1), 164–171. doi: 10.1002/jcp.1041640121 [DOI] [PubMed] [Google Scholar]
- Wurm F (2013). CHO Quasispecies—Implications for Manufacturing Processes. Processes, 1(3), 296–311. doi: 10.3390/pr1030296 [DOI] [Google Scholar]
- Yang Y, Mariati, Chusainow J, & Yap MG (2010). DNA methylation contributes to loss in productivity of monoclonal antibody-producing CHO cell lines. J Biotechnol, 147(3-4), 180–185. doi: 10.1016/j.jbiotec.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Yee JC, de Leon Gatti M, Philp RJ, Yap M, & Hu WS (2008). Genomic and proteomic exploration of CHO and hybridoma cells under sodium butyrate treatment. Biotechnol Bioeng, 99(5), 1186–1204. doi: 10.1002/bit.21665 [DOI] [PubMed] [Google Scholar]
- Yusufi FNK, Lakshmanan M, Ho YS, Loo BLW, Ariyaratne P, Yang Y, … Lee DY (2017). Mammalian Systems Biotechnology Reveals Global Cellular Adaptations in a Recombinant CHO Cell Line. Cell Syst, 4(5), 530–542 e536. doi: 10.1016/j.cels.2017.04.009 [DOI] [PubMed] [Google Scholar]
- Zhou H, Liu ZG, Sun ZW, Huang Y, & Yu WY (2010). Generation of stable cell lines by site-specific integration of transgenes into engineered Chinese hamster ovary strains using an FLP-FRT system. J Biotechnol, 147(2), 122–129. doi: 10.1016/j.jbiotec.2010.03.020 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


