Abstract
Toll-like receptors (TLRs) are a complex family of innate immune genes that are well characterized in mammals and birds but less well understood in nonavian sauropsids (reptiles). The advent of highly contiguous draft genomes of nonmodel organisms enables study of such gene families through analysis of synteny and sequence identity. Here, we analyze TLR genes from the genomes of 22 tetrapod species. Findings reveal a TLR8 gene expansion in crocodilians and turtles (TLR8B), and a second duplication (TLR8C) specifically within turtles, followed by pseudogenization of that gene in the nonfreshwater species (desert tortoise and green sea turtle). Additionally, the Mojave desert tortoise (Gopherus agassizii) has a stop codon in TLR8B (TLR8-1) that is polymorphic among conspecifics. Revised orthology further reveals a new TLR homolog, TLR21-like, which is exclusive to lizards, snakes, turtles, and crocodilians. These analyses were made possible by a new draft genome assembly of the desert tortoise (gopAga2.0), which used chromatin-based assembly to yield draft chromosomal scaffolds (L50 = 26 scaffolds, N50 = 28.36 Mb, longest scaffold = 107 Mb) and an enhanced de novo genome annotation with 25,469 genes. Our three-step approach to orthology curation and comparative analysis of TLR genes shows what new insights are possible using genome assemblies with chromosome-scale scaffolds that permit integration of synteny conservation data.
Keywords: immune system, orthology, reptile, reference genome, synteny, testudines
Introduction
Expansion of gene families through duplication creates the opportunity for the evolution of new functionalization, specialized subfunctionalization, or differences in gene expression (Ranz and Parsch 2012). Conversely, loss within a gene family may reflect reduced selective pressure to retain gene function sometime during a lineage’s evolutionary history (Albalat and Cañestro 2016). Within vertebrates, the immune response is a fundamental part of how an animal interacts with its environment. Toll-like receptors (TLRs) are a large family of signaling transmembrane proteins that trigger an innate immune response following pathogen exposure (Akira and Takeda 2004). Although somatic recombination produces genetic diversity in the adaptive immune system, genes in the innate immune system (e.g., TLRs) must evolve rapidly in other ways to keep pace with the evolution of pathogens. Although TLRs are well studied in endothermic mammals and birds (Hopkins and Sriskandan 2005; Velová et al. 2018), there is less known about TLR gene family evolution in nonavian sauropsids (nonavian reptiles; but see Kahn et al. 2019; Liu et al. 2019). The innate immune system of nonavian sauropsids remains poorly characterized overall (Zimmerman et al. 2010) due to the lack of experimental tools for functional immunology and seasonal changes in ectotherm physiology (Zapata et al. 1992; Sandmeier et al. 2016). Studying TLR evolution of nonavian sauropsids can provide insight into the adaptation of the innate immune response in this group, provide a broader amniote context to TLR evolution in birds and mammals, and shed light on gene evolution broadly.
Genome resources yield information about the gene repertoire of nonmodel species through comparative evolutionary analyses. These analyses depend in part on the contiguity of reference genomes. Here, we analyze two TLR subfamilies across 22 tetrapod genomes: the TLR7 subfamily (TLR7, TLR8, TLR9) that recognizes nucleotide-based pathogen-associated molecular patterns, and the more diverse TLR11 subfamily (TLR11, TLR12, TLR13, TLR21) that recognizes pathogen-associated molecular patterns in bacterial proteins, bacterial RNA, and CpG DNA viruses (Heil 2004; Keestra et al. 2010). To aid these analyses, we used new sequencing and scaffolding technology to yield chromosome-scale scaffolds and a de novo annotation for the threatened Mojave desert tortoise (Gopherus agassizii; genome build: gopAga2.0). We then employed a three-step integrative approach to manually curate TLR orthology assignments across 22 tetrapods. This study demonstrates that much can be learned about nonmodel organisms and well-studied gene families through a comparative genomic approach, which is broadly applicable to studies of other complex gene families.
Results
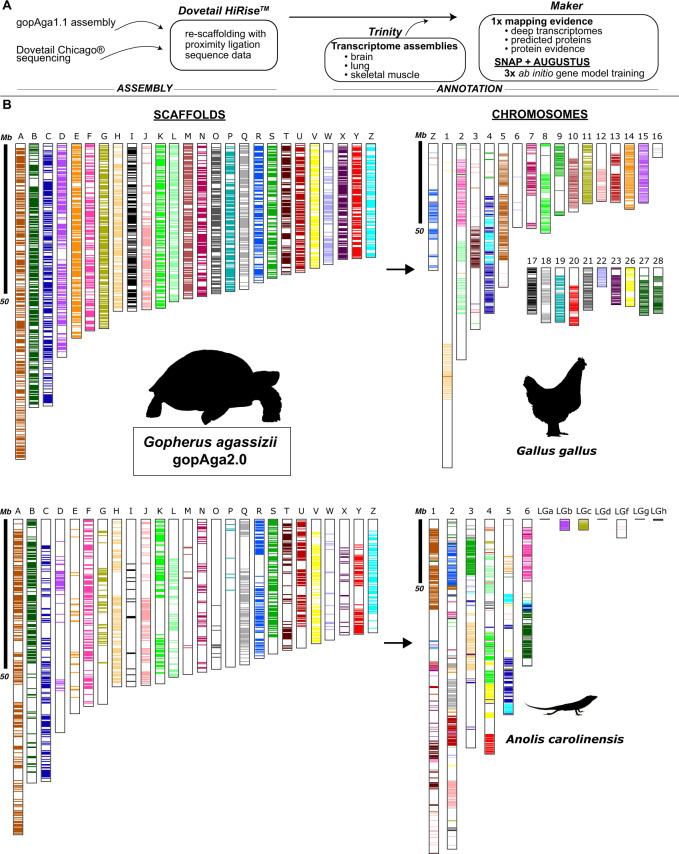
The final assembly length for the gopAga2.0 assembly is 2.34 Gb, with a longest scaffold length of 106.5 Mb, N50 of 28.36 Mb, and L50 of 26 scaffolds (table 1). The number of predicted haploid chromosomes is 26 (Olmo 1981), suggesting the new assembly contains chromosome-scale scaffolds. The final assembly length for gopAga2.0 is 2.34 Gb, with a longest scaffold length of 106.5 Mb, N50 of 28.36 Mb, and L50 of 26 scaffolds (table 1). The number of predicted haploid chromosomes is 26 (Olmo 1981), suggesting the new assembly contains chromosome-scale scaffolds. Bioinformatic chromosome painting reveals strong synteny conservation between chicken and desert tortoise (fig. 1B, top). As expected for more evolutionarily distant species, there is a decreased synteny conservation between desert tortoise and green anole, as fewer linkage blocks are identified and there is greater mixing of synteny blocks (mixed colors in fig. 1B, bottom).
Table 1.
Assembly and Gene Statistics across Genome Builds
| Assembly Statistics | |||||
|---|---|---|---|---|---|
| Total Length (Mb) | L50 (No. Scaffolds) | N50 (Mb) | Longest Scaffold (Mb) | ||
| gopAga1.0 | 2,399 | 2,592 | 0.25 | 2 | |
| gopAga2.0 | 2,338 | 26 | 28.36 | 107 | |
| Assembly BUSCO Statistics | Annotation Statistics | ||||
| Complete | Fragmented | Missing | Total Genes | Mean Gene Length (bp) | |
| gopAga1.0 | 92.7% | 4.6% | 2.7% | 20,172 | 19,062 |
| gopAga2.0 | 93.7% | 3.7% | 2.6% | 25,469 | 21,364 |
note.—Assembly BUSCO statistics are from the Tetrapoda gene set in the genome assemblies. Total genes and mean gene length are calculated based on the annotations for gopAga1.0 and gopAga2.0 (see supplementary table S2, Supplementary Material online).
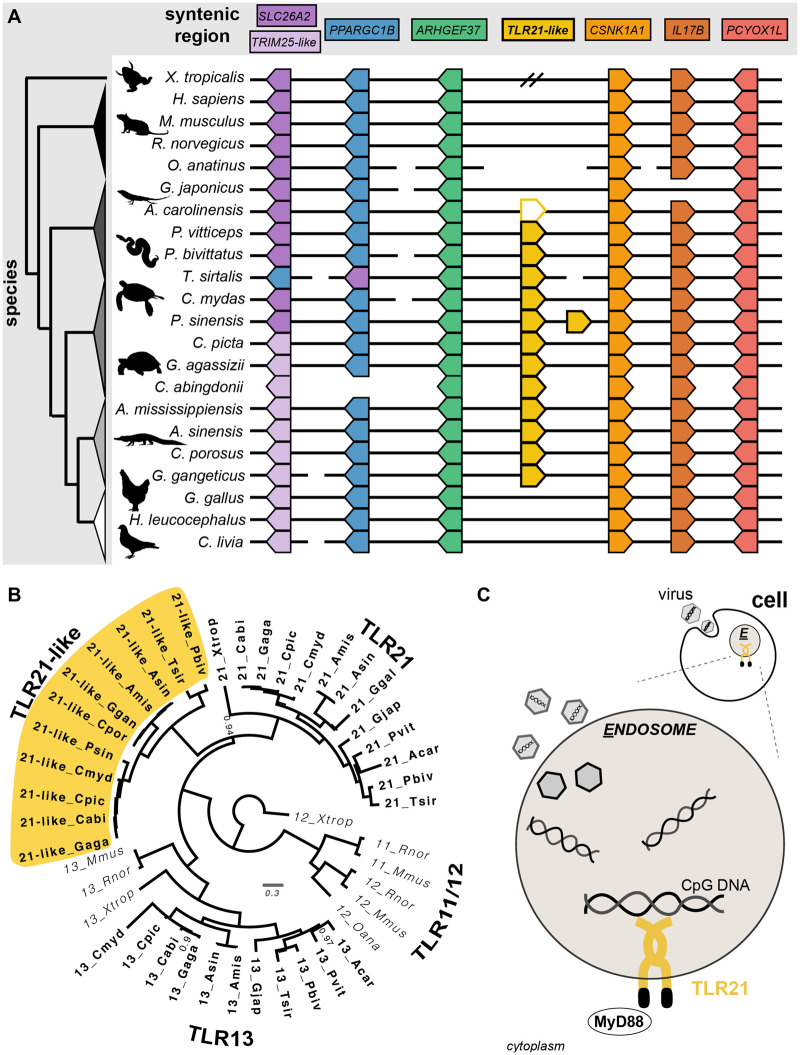
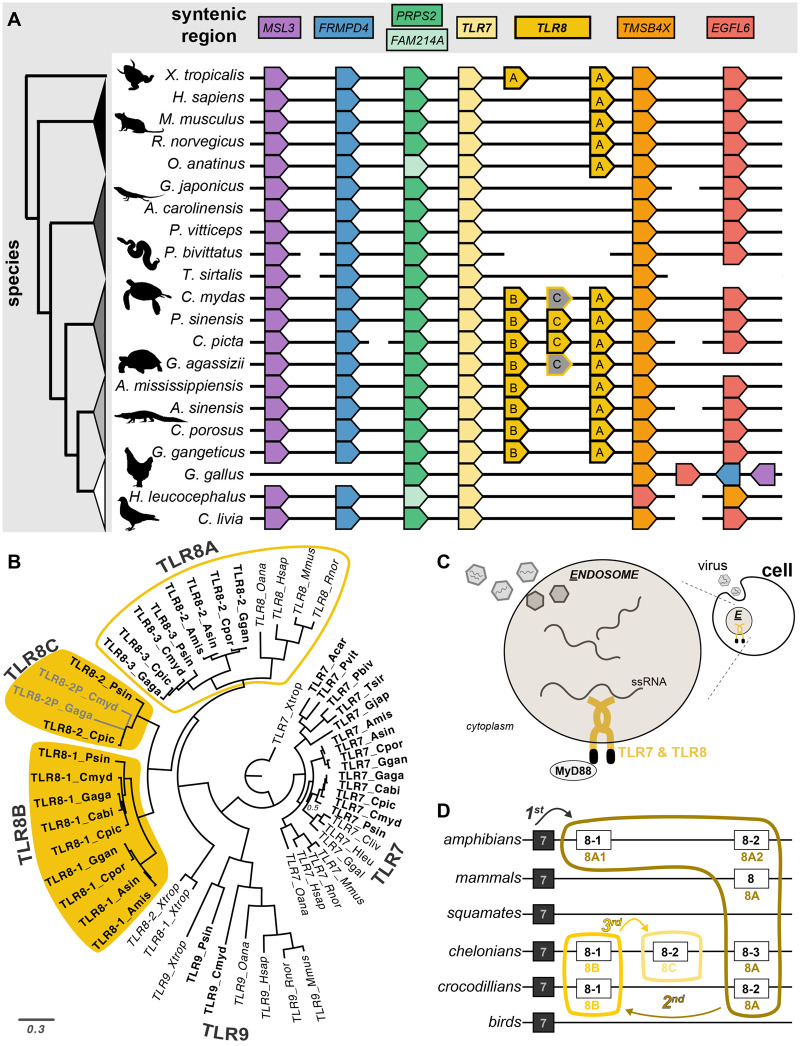
Regarding TLR evolution, we found that the TLR7 and TLR8 genes are tandemly arranged in a highly conserved syntenic region across tetrapod genomes (fig. 2A) and exhibit a large amount of gain/loss. The three-step orthology approach also revealed a new, misidentified TLR homolog exclusive to nonavian sauropsids, which we refer to as TLR21-like (fig. 3). For more details, see the Discussion section.
Fig. 3.
—TLR21-like gene discovered. (A) Syntenically conserved regions of TLR21-like (gold, middle) for 22 tetrapods. TLR21-like does not colocalize near TLR21 within the genome. Anolis carolinensis has LINGO1 (a leucine-rich repeat gene) instead of TLR21-like (yellow outline). Synteny information was not available for Celonoides abingdonii; therefore, only the present/absence of genes are shown (i.e., no black line). (B) Phylogenetic reconstruction of TLR11 gene subfamily including the TLR21-like gene discovered here (gold highlight). Posterior support values for nodes are 1 unless labeled. (C) Cartoon showing how TLR21 protein initiates the innate immune response by recognizing the CpG nucleotide sequence.
Discussion
TLR8 Gene Expansion
All species surveyed have one copy of TLR7, whereas the TLR8 gene varies among tetrapods. Our analyses confirm the duplication of TLR8 in turtles (Liu et al. 2019) and further reveals that TLR8 duplication (TLR8B) is also present in crocodilians. Mammals have only one copy of TLR8, and squamate reptiles and birds lack a TLR8 ortholog (fig. 2). In addition, we find that the third copy of TLR8 discovered in Chinese softshell turtle (Pelodiscus sinensis; Liu et al. 2019) is actually part of a second duplication event specific to testudines (TLR8C). The TLR8 protein recognizes single-stranded RNA viruses, suggesting this could produce subspecialization or expression differences of this function in crocodilians and especially turtles (fig. 2C; Heil 2004).
Fig. 2.
—TLR8 gene expansion in turtles. (A) Syntenically conserved region for TLR7 and TLR8 (gold, middle) for 21 tetrapods (Chelonoidis abingdonii is excluded because its TLR8 orthology cannot be determined from available data). Genes are labeled at the top and cladogram is shown on left. Black horizontal lines represent contiguous sequence. TLR8 is expanded once in testudines and crocodilian archosaurs (TLR8B) and a second time in testudines (TLR8C) followed by pseudogenization (gray genes). (B) Phylogenetic reconstruction of TLR7 gene subfamily; reptiles are bolded, gene expansions are highlighted in gold. Posterior support values for nodes are 1 unless labeled. (C) Cartoon summarizing how TLR7 and TLR8 proteins initiate the innate immune response by recognizing single-stranded RNA (ssRNA) viruses. (D) A summary of inferred gene duplication based on results in this article.
Within chelonians, the Pinta Island tortoise has two predicted TLR8 genes, but one consists of only 122 amino acids and contains no protein motifs. Gopherus agassizii has three TLR8 genes, one with a conserved open reading frame (TLR8-1/TLR8B), one that is pseudogenized based on accumulation of stop codons (TLR8-2/TLR8C, also pseudogenized in Chelonia mydas), and TLR8-3/TLR8A that is truncated to include only the N-terminal half but retains nine leucine-rich repeats (LRRs; the basis for TLR specificity and function), suggesting protein function is retained (supplementary fig. S2, Supplementary Material online). Predicted truncation of TLR8-3 occurs at the site of a derived TGA stop codon that—surprisingly—is polymorphic among individuals within G. agassizii (CGA/TGA; supplementary fig. S9, Supplementary Material online). The stop codon in the gopAga2.0 type specimen was validated by mapping available RNA-seq and genomic resequencing data of unrelated individuals (see Supplementary Material online). The CGA to TGA mutation can readily occur through deamination of cytosine to uracil and correction of uracil to thymine. A truncated TLR paralog has been observed in several fish species where there is both membrane-bound TLR5M gene and soluble TLR5S gene that lacks a TIR and transmembrane domain but retains 21 LRRs (reviewed in Rebl et al. 2010). These proteins were shown not only to exhibit different functions but also different tissue specificities. Tissue-specific expression patterns were also observed among the TLR8 paralogs in the Chinese softshell turtle (Liu et al. 2019).
Using the above results, we outline a new naming scheme based on the inferred evolutionary duplication history of TLR8. We suggest that TLR8A (TLR8-2, TLR8-3 sensu Liu et al. 2019) originated from TLR7 first because it is the paralog most shared across taxa (fig. 2D). TLR8B then duplicated from TLR8A within Archelosauria (testudines, crocodilians, birds) and was secondarily lost in birds. TLR8C then duplicated from TLR8B specifically within testudines, leading to pseudogenization within the two nonfreshwater species (green sea turtle and Mojave desert tortoise). The LRR motifs for TLR8-1 and TLR8-2 in the outgroup (western clawed frog) are nearly identical (supplementary fig. S7, Supplementary Material online) and differ from the other TLR8 motifs (supplementary fig. S2, Supplementary Material online), suggesting this duplication was separate. The duplication of TLR8 genes in nonavian sauropsids and the truncation in tortoises may reflect diversification of the innate immune response that warrants further study.
New TLR21-like Gene in Nonavian Sauropsids (Reptiles)
Many species have two genes labeled as TLR13. The first TLR13 is syntenically conserved across tetrapods (excluding birds and some crocodilians; supplementary fig. S5, Supplementary Material online). The second TLR13 gene is phylogenetically sister to, and reciprocally monophyletic with, TLR21 (fig. 3B) and occurs in a separate, highly conserved syntenic region (fig. 3A vs. supplementary fig. S5, Supplementary Material online vs. supplementary fig. S6, Supplementary Material online). Here, we refer to this gene as TLR21-like. The absence of a TLR21-like ortholog in mammals and birds, which include most traditional model systems, may explain why the sequence was not previously identified and reveals a potentially important component of the immune gene repertoire of nonavian sauropsids. The TLR21-like gene is exclusive to nonavian sauropsids (but absent in gecko) and has a highly conserved protein motif pattern (supplementary fig. S2B, Supplementary Material online), suggesting the duplication event could have occurred recently (e.g., by gene conversion) or that it arose on the sauropsid stem lineage, was strongly conserved, and was secondarily lost in birds and gecko. This second interpretation is likely given documented loss of TLR8, 9, and 13 in birds (Kahn et al. 2019). In Anolis carolinensis, the LINGO1 (LRR and lg domain-containing 1) gene is in the syntenic region of TLR21-like and contains five LRRs and could potentially be mislabeled or derived from TLR21-like. The Chinese softshell turtle has a tandem duplication of TLR21-like with an identical nucleotide sequence but separate gene model, suggesting a recent lineage-specific duplication (supplementary fig. S10, Supplementary Material online).
The TLR21 protein function is not well characterized, but involves recognizing CpG motifs of DNA viruses (fig. 3C; Keestra et al. 2010). In zebrafish, TLR21 is expressed in the early embryo and within the adult in organs of the innate immune system including intestine, spleen, and kidney (Yeh et al. 2013). Although TLR21 and TLR9 proteins have distinct CpG-DNA recognition sequences, they cooperatively mediate the pathogen-associated response. The duplication of a TLR21-like gene in nonavian sauropsids may have allowed for paralog specialization of tissue-specific expression and CpG-DNA recognition. Further characterization of this TLR21-like gene will be informative.
Conclusion
Comparative analyses of 22 tetrapod genomes revealed active TLR gene gain/loss across nonavian sauropsids, particularly the expansion of TLR8 in turtles, tortoises, and crocodilians and a novel TLR homolog exclusive to nonavian sauropsids, TLR21-like. The results from our three-step approach underscore the importance of integrating synteny, sequence similarity, and semantic searching to study gene family evolution. The syntenic analysis requires contiguous reference genomes and promises new insight into other biologically important gene families. Our findings were enabled by a revised draft of the Mojave Desert tortoise genome (gopAga2.0) that greatly improved assembly contiguity and the de novo annotation of 25,469 genes.
Materials and Methods
Genome Assembly and Annotation
For the gopAga2.0 assembly, the gopAga1.1 assembly (Webster, Dolby, et al. 2018) and skeletal muscle from the original individual were sent to Dovetail Genomics for chromatin-based long-range proximity ligation sequencing with the Chicago® method. These data were reanalyzed together with the gopAga1.0 paired-end and mate-pair Illumina HiSeq reads (SRX2367114–5, SRX2367341, SRX2367455, SRX2367692–3; Tollis et al. 2017) for rescaffolding using the HiRise software, which corrects for misjoins (Putnam et al. 2016; see Supplementary Material online).
We generated a de novo annotation for gopAga2.0 using genome-guided, tissue-specific transcriptomes generated in Trinity v2.5.1 (Grabherr et al. 2011), predicted proteins from the western painted turtle (NCBI PRJNA210179), and protein evidence from UniProtKB/Swiss-Prot database (fig. 1A). These data were mapped to the assembly using MAKER v3 (Campbell et al. 2015) followed by three rounds of ab initio gene model training with SNAP v2017-3-1 (Korf 2004) and AUGUSTUS v3.3.2 (Keller et al. 2011) within MAKER.
Fig. 1.
—Genome annotation and assembly. (A) Summary of methodology. (B) Results from chromosome painting of conserved synteny blocks shown between the Mojave Desert tortoise genome (gopAga2.0) and chicken (top right), and green anole lizard (bottom right). The following pairs of scaffolds (left) and chromosomes (right) between Mojave Desert tortoise and chicken are particularly conserved: B-27/28, D-15, E-14, G-11, I-17, O-21, W-22. Black bars show 50-Mb scale. Microchromosomes are labeled “LG”; there are no conserved synteny blocks for LGa, LGd, and LGg. Only homologous linkage blocks are colored.
Bioinformatic Chromosome Painting
To assess synteny conservation, we used bioinformatic chromosome “painting.” We downloaded EMBL genome files from Ensembl for green anole and chicken (Collins et al. 2004; Zhang et al. 2014). To process and reformat the gopAga2.0 genome file, we used a custom Perl script (Gfftofasta.pl modified from https://github.com/ISUgenomics/common_scripts) and subset proteins from the 26 most gene-rich scaffolds in the gopAga2.0 annotation (supplementary table S3, Supplementary Material online) using Biopython v1.73 (Cock et al. 2009). With conversion scripts in the CHROnicle package (v2015), we converted the three species files into SynChro-specific formatting. SynChro computed conserved synteny blocks with delta = 4, which is slightly conservative as it requires four consecutive genes to match across species to be considered a synteny block (the default is 2; Drillon et al. 2014).
TLR Evolution
To study TLR evolution, we used a three-step approach to curate orthology by: 1) searching by sequence similarity, 2) searching by semantic labels, and 3) assessing synteny conservation (supplementary fig. S1, Supplementary Material online). First, using human TLR protein as query sequences, or mouse/chicken sequences when human was unavailable (supplementary table S4, Supplementary Material online), we identified the top ten gene “hits” using BLASTp from the BLAST+ v2.9.0 (Camacho et al. 2009) for each of the seven TLR genes in 22 species: 14 nonavian sauropsids, 4 mammals, 3 birds, and the western clawed frog as outgroup (supplementary table S1, Supplementary Material online and Appendix). From the top ten RefSeq matches, we chose the top hit with an NP tag; if no hit had an NP tag then we used the top predicted hit (XP tag).
Second, we queried the NIH RefSeq database for the text string “Toll-like receptor” with the name of each study species and added to the list any unique gene models that had nonunique gene annotations (e.g., if two separate gene models were both labeled TLR21). Third, and most importantly, we used synteny conservation to verify orthology assignments of these candidate homologs. We visualized the syntenic region of each gene/gene model in every species using the Integrated Genomics Viewer (IGV, v2.3.47; Robinson et al. 2011), manually recorded gene IDs, gene order, and gene direction for five neighboring genes when available, and verified that each gene localized to the expected syntenic region. We excluded long noncoding RNAs, microRNAs, and unidentified loci. For sequences that fell outside the expected syntenic region, if that new syntenic region was the same in another species, it was retained (e.g., TLR21-like), if it fell in a syntenic region that was not recapitulated in other taxa, it was discarded (see Supplementary Material online). This three-step approach provides orthology assignments using complementary sources of evidence and allows for the discovery of novel genes that would be overlooked through sequence similarity alone.
We verified functional protein domains for some TLR sequences of interest using the Simple Modular Architecture Research Tool (SMART; Letunic and Bork 2018). Curated amino acid sequences were aligned using the Gonnet scoring matrix in ClustalW v2.1 (Larkin et al. 2007) with default settings (gap extension penalty = 0.20, gap opening penalty = 10). We visually inspected the amino acid alignments and removed short, highly variable regions that could not be unambiguously aligned. Original and manually curated alignments are available (see Supplementary Material online).
For tree reconstructions, we fit amino acid substitution models for the two subfamilies with ProtTest v3.2 (Darriba et al. 2011) and generated maximum likelihood and Bayesian inference trees on the CIPRES Science Gateway v3.3 (Miller et al. 2010) using the best-scoring models that were implemented in MrBayes (WAG, TLR7 family; VT, TLR11 family; supplementary table S5, Supplementary Material online). We generated maximum likelihood trees using 100 bootstrap iterations in RAxML-HPC v8.2.10 (Stamatakis 2014). In MrBayes v3.2.6 (Ronquist et al. 2012), we generated two runs of 10 million generations each, four chains per run and sampled every 10,000 generations. Runs reached convergence according to the potential scale reduction factor converging on 1; runs were aggregated using sump and sumt commands with the default 25% burn-in discarded. We visually compared topologies and found consistency across ML and BI trees. Markdown tutorials and scripts for all methods are provided (see Supplementary Material online).
Statement of Ethics Approval
Tortoise handling, transport, care, and tissue collection were approved by the Institutional Animal Care and Use Committee at Arizona State University (Protocol No. 13-1319 R). Acquisition and transport of the tortoise was also approved by the United States Fish and Wildlife Service (Recovery Subpermit No. FWSDTRO-1) and the Nevada Department of Wildlife (Export Permit No. S37016).
Supplementary Material
Acknowledgments
We thank C. Holt, M. Tollis, J. Cornelius, and C. Xu for assembly and annotation support. Dovetail Genomics Matching Grant (G.A.D. and K.K.); Fostering Postdoctoral Research in the Life Sciences seed grant from the School of Life Sciences at Arizona State University (G.A.D. and T.H.W.); US Geological Survey Cooperative Ecosystem Studies Units (CESU) award (G.A.D. and K.K.); Arizona Game and Fish Department Heritage (Grant No. I17001 to all authors); NIGMS (Grant No. R35GM124827 to M.A.W.); and funding from The College of Liberal Arts and Sciences at Arizona State University (K.K.). G.A.D. is a member of the Board of Directors for the Desert Tortoise Council, a nonprofit conservation organization that protects desert tortoises and their habitats. The DTC had no involvement in the design, implementation, or interpretation of this study.
Author Contributions
G.A.D., T.H.W., D.F.D., M.A.W., and K.K. contributed to genome assembly. G.A.D. and K.K. performed genome annotation. M.M., G.A.D., and K.K. performed TLR analyses. G.A.D. drafted and all authors edited the article.
Appendix
Additional genome references used that were not cited in the text because of space limits.
Data deposition: This project has been deposited at https://dataverse.harvard.edu/dataverse/gopAga2/. Tutorials for data-processing steps for SynChro and TLR gene family sequence curation and analysis are available at https://github.com/mmoral31/GopAga2.0-Genome.
Literature Cited
- Alföldi J, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477(7366):587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoe TA, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci U S A. 110(51):20645–20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DM, et al. 2009. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 7(5):e1000112–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges A, et al. 2015. High-coverage sequencing and annotated assembly of the genome of the Australian dragon lizard Pogona vitticeps. GigaScience 4(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, et al. 2010. The genome of the western clawed frog Xenopus tropicalis. Science 328(5978):633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, et al. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716. [DOI] [PubMed] [Google Scholar]
- Liu Y, et al. 2015. Gekko japonicus genome reveals evolution of adhesive toe pads and tail regeneration. Nat Commun. 6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML, et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428:493–521. [DOI] [PubMed] [Google Scholar]
- Perry BW, et al. 2018. Molecular adaptations for sensing and securing prey and insight into amniote genome diversity from the Garter snake genome. Genome Biol Evol. 10(8):2110–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, et al. 2013. Genomic diversity and evolution of the head crest in the rock pigeon. Science 339(6123):1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JA, et al. 2012. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 13(1):415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Q-H, et al. 2013. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23:1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453(7192):175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Literature Cited
- Akira S, Takeda K.. 2004. Toll-like receptor signalling. Nat Rev Immunol. 4(7):499–511. [DOI] [PubMed] [Google Scholar]
- Albalat R, Cañestro C.. 2016. Evolution by gene loss. Nat Rev Genet. 17(7):379–391. [DOI] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MS, Holt C, Moore B, Yandell M.. 2015. Genome annotation and curation using MAKER and MAKER-P. Curr Protoc Bioinformatics. 48:4.11.1–4.11.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, et al. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25(11):1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Lander ES, Rogers J, Waterston RH, Conso I.. 2004. Finishing the euchromatic sequence of the human genome. Nature 431:931–945. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27(8):1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drillon G, Carbone A, Fischer G.. 2014. SynChro: a fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS One 9(3):e92621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29(7):644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303(5663):1526–1529. [DOI] [PubMed] [Google Scholar]
- Hopkins PA, Sriskandan S.. 2005. Mammalian Toll-like receptors: to immunity and beyond. Clin Exp Immunol. 140(3):395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn I, et al. 2019. The vertebrate TLR supergene family evolved dynamically by gene gain/loss and positive selection revealing a host–pathogen arms race in birds. Diversity 11:1–25. [Google Scholar]
- Keestra AM, de Zoete MR, Bouwman LI, van Putten J.. 2010. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol. 185(1):460–467. [DOI] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S.. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27(6):757–763. [DOI] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2018. 20 Years of the SMART protein domain annotation resource. Nucleic Acids Res. 46(D1):D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Han Y, Chen S, Zhao H.. 2019. Genome-wide identification of Toll-like receptors in the Chinese soft-shelled turtle Pelodiscus sinensis and expression analysis responding to Aeromonas hydrophila infection. Fish Shellfish Immunol. 87:478–489. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T.. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. New Orleans (LA): Gateway Computing Environments Workshop (GCE). p. 1–8.
- Olmo E. 1981. Evolution of genome size and DNA base composition in reptiles. Genetica 57(1):39–50. [Google Scholar]
- Putnam NH, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 26(3):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, et al. 2018. Giant tortoise genomes provide insights into longevity and age-related disease. Nat Ecol Evol. 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Parsch J.. 2012. Newly evolved genes: moving from comparative genomics to functional studies in model systems: how important is genetic novelty for species adaptation and diversification? Bioessays 34(6):477–483. [DOI] [PubMed] [Google Scholar]
- Rebl A, Goldammer T, Seyfert HM.. 2010. Toll-like receptor signaling in bony fish. Vet Immunol Immunopathol. 134(3–4):139–150. [DOI] [PubMed] [Google Scholar]
- Robinson JT, et al. 2011. Integrative genomics viewer. Nat Biotechnol. 29(1):24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier FC, Horn KR, Tracy CR.. 2016. Temperature-independent, seasonal fluctuations in immune-function in a reptile, the Mojave desert tortoise (Gopherus agassizii). Can J Zool. 94(8):583–590. [Google Scholar]
- Shaffer HB, et al. 2013. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 14(3):R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollis M, et al. 2017. The Agassiz’s desert tortoise genome provides a resource for the conservation of a threatened species. PLoS One 12(5):e0177708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachana K, et al. 2011. Orthology prediction methods: a quality assessment using curated protein families. Bioessays 33(10):769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velová H, Gutowska-Ding MW, Burt DW, Vinkler M.. 2018. Toll-like receptor evolution in birds: gene duplication, pseudogenization, and diversifying selection. Mol Biol Evol. 35(9):2170–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2013. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat Genet. 45(6):701–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse RM, et al. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 35(3):543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster TH, Dolby GA, Sayres MAW, Kusumi K.. 2018. Improved draft of the Mojave Desert tortoise genome, Gopherus agassizii, version 1.1. PeerJ Preprints 6:e3266v4.
- Yeh DW, et al. 2013. Toll-like receptor 9 and 21 have different ligand recognition profiles and cooperatively mediate activity of CpG-oligodeoxynucleotides in zebrafish. Proc Natl Acad Sci U S A. 110(51):20711–20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata AG, Varas A, Torroba M.. 1992. Seasonal variations in the immune system of lower vertebrates. Immunol Today. 13(4):142–147. [DOI] [PubMed] [Google Scholar]
- Zhang G, et al. 2014. Comparative genomic data of the avian phylogenomics project. GigaScience 3(1):1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman LM, Vogel LA, Bowden RM.. 2010. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol. 213(5):661–671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.