Abstract
Neisseria spp. possess four genogroups of filamentous prophages, termed Nf1 to 4. A filamentous bacteriophage from the Nf1 genogroup termed meningococcal disease-associated phage (MDA φ) is associated with clonal complexes of Neisseria meningitidis that cause invasive meningococcal disease. Recently, we recovered an isolate of Neisseria gonorrhoeae (ExNg63) from a rare case of gonococcal meningitis, and found that it possessed a region with 90% similarity to Nf1 prophages, specifically, the meningococcal MDA φ. This led to the hypothesis that the Nf1 prophage may be more widely distributed amongst the genus Neisseria. An analysis of 92 reference genomes revealed the presence of intact Nf1 prophages in the commensal species, Neisseria lactamica and Neisseria cinerea in addition to the pathogen N. gonorrhoeae. In N. gonorrhoeae, Nf1 prophages had a restricted distribution but were present in all representatives of MLST ST1918. Of the 160 phage integration sites identified, only one common insertion site was found between one isolate of N. gonorrhoeae and N. meningitidis. There was an absence of any obvious conservation of the receptor for prophage entry, PilE, suggesting that the phage may have been obtained by natural transformation. An examination of the restriction modification systems and mutated mismatch repair systems with prophage presence suggested that there was no obvious preference for these hosts. A timed phylogeny inferred that N. meningitidis was the donor of the Nf1 prophages in N. lactamica and N. gonorrhoeae. Further work is required to determine whether Nf1 prophages are active and can act as accessory colonization factors in these species.
Keywords: meningococcal disease associated; MDA φ; Neisseria filamentous phage; Nf1; prophage; Inovirus; Neisseria gonorrhoeae, Neisseria lactamica, Neisseria cinerea
Introduction
Filamentous bacteriophages of the genus Inovirus are thin filamentous phage that contain a circular ssDNA genome (Day 2011). These phages mainly infect Gram-negative bacteria in which they replicate and shed phage particles from growing and dividing cells without killing the host. There are two groups of Inovirus: integrative “temperate” phages that can integrate their DNA within the host chromosome; and nonintegrative phages such as M13 (Rakonjac et al. 2011; Mai-Prochnow et al. 2015). Temperate filamentous phages, such as CTX φ of Vibrio cholerae and meningococcal disease-associated phage (MDA φ) of Neisseria meningitidis, infect the host cell through pili and then replicate in the cytoplasm, after which they integrate into the host genome to form a prophage (Campos et al. 2003; Meyer et al. 2016). Under induction conditions, the prophages release a replicative particle through rolling circle replication that is packaged into the virion and then released from the host cell using a self- or host-encoded secretin. In addition to morphological classification of Inovirus, the synteny of core genes and their sizes can be used to putatively identify the phages, as gene order tends to be the conserved feature rather than nucleotide sequence or protein sequence homology. Genes and genomes of Inovirus are organized in three modules based on function; replication genes, structural genes, and assembly and secretion genes (Mai-Prochnow et al. 2015).
The Neisseria genus includes pathogenic species such as N. meningitidis (causes invasive meningococcal disease) and Neisseria gonorrhoeae (causes gonorrhoea). Other members of the Neisseria genus such as Neisseria lactamica, Neisseria subflava, Neisseria cinerea, and Neisseria sicca are nonpathogenic species that colonize the human oral cavity, nasopharynx, and occasionally the genital tract. Several filamentous prophages have been identified in pathogenic Neisseria spp., and have been classified into four clades: Nf1, 2, 3, and 4. The filamentous prophage ecology is distinct between pathogenic members of the genus Neisseria. Meningococcal strains may carry either none, one, or up to four copies of Nf1 (MDA φ), as well as prophages Nf2 and Nf3. Gonococcal strains possess multiple copies of Nf4 filamentous prophages, as well as other truncated prophages, which have variable integration sites between strains (Kawai et al. 2005; Piekarowicz et al. 2006).
MDA φ (Nf1) was first described by Bille et al. (2005) who showed that it was associated with genetic lineages of N. meningitidis associated with invasive disease. The 8-kb MDA φ genome is highly conserved among N. meningitidis strains and has a similar genetic organization to other filamentous phages such as M13 or CTXφ (Bille et al. 2005; Meyer et al. 2016). The MDA φ genome encodes a functional filamentous phage that can infect other meningococcal strains through interaction with type IV pili, and is able to produce infectious virions which are released using host-encoded secretin PilQ (Meyer et al. 2016). MDA φ stabilizes intrabacterial interactions during microcolony formation by forming phage bundles that extend from the bacterial surface resulting in stable meningococcal mucosal colonization of the nasopharynx (Bille et al. 2017). Accordingly, improved carriage is believed to increase the incidence of bloodstream invasion associated with infection, while facilitating spread and persistence of the MDA φ carrying strain in the host population (Bille et al. 2017). Of the gonococcal Nf4 filamentous prophages only, Ngoφ6 has been shown to be a functional prophage which produces a circular single positive strand of DNA during bacterial culture (Piekarowicz et al. 2006). A constructed Ngoφ6 phagemid (designated pBS::Ngoφ6) is able to produce active phages that can infect and replicate in a number of Gram-negative bacteria, establishing that it has an unusually wide host range (Piekarowicz et al. 2014).
We recently isolated N. gonorrhoeae (ExNg63) from a rare case of gonococcal meningitis. During genome annotation, we identified a sequence with 90% sequence similarity to the Nf1 phage from N. meningitidis Z2491. We hypothesized that the possession of a Nf1 prophage in N. gonorrhoeae could be a factor associated with invasive disease in this species. This study was designed to examine the distribution, prevalence, and genetic diversity of filamentous prophages in the genus Neisseria with a focus on the prevalence of Nf1 prophage.
Materials and Methods
Bacterial Strains and Culture Conditions
Neisseria gonorrhoeae strain ExNg63 was isolated from cerebrospinal fluid of patient with meningitis in Australia in 2015. Isolates were stored in GC broth with 20% glycerol at −80 °C, were cultured under aerobic conditions with 5% CO2 at 37 °C on GC agar (Oxoid, Australia), and supplemented with 0.4% glucose, 0.01% glutamine, 0.2 mg/l of cocarboxylase, and 5 mg/l of iron (III) nitrate.
Whole-Genome Sequencing Assembly and Genomic Annotation
Genomic DNA extraction was performed using the DNeasy Blood and Tissue Kit (Qiagen, Germany). Genome sequencing ExNg63 was performed using the Illumina MiSeq platform (Illumina) with 2×300 base pair read lengths. The targeted sequencing depth was 120 with a minimum Phred quality score of 30. Reads were de novo assembled using SPAdes genome assembler version 9.0 (Bankevich et al. 2012). The quality of the assembled genome was assessed using the QUAST genome assembly evaluation tool (Gurevich et al. 2013). Sequencing and assembly quality statistics are as follows: number of contigs 97, total length: 2,152,527, GC (%) 52.50, N50: 61,914, N75: 36,955, L50: 9, and L75: 20. Bacterial Isolates Genome Sequence database (BIGSdb) genomics platform (PubMLST) tools—hosted on www. PubMLST.org/neisseria (last accessed 13 Feb 2020)—were used for annotation of the assembled isolate and for initial identification of the Nf1 genes (Jolley and Maiden 2010). ExNG63 has the PubMLST ID 46359.
PCR Amplification of the Entire Nf1 Prophage
The presence of Nf1 prophage in ExNg63 was detected during whole-genome sequencing and confirmed by Sanger sequencing of both strands of the 8-kb PCR amplified locus. The Nf1 genes were split across two contigs, to confirm that the entire Nf1 prophage was present and intact in the genome of ExNg63, a long-range PCR was preformed to detect the complete prophage region. The forward primer KAP823 (5′-TATATGATGCGCTCTATCAAAGGGGC-3′) upstream the RIF (NEIS0031) and the reverse primer KAP824 (5′CGCAGATGATATGTTGCCCGTCAAC-3′) downstream the IS110 transposase, conserved sequence specific to the Nf1 based on NCBI BLAST search, were used to amplify the entire Nf1 prophage using Phusion High-Fidelity DNA Polymerase (New England Biolabs). PCR conditions were as follows: 2 min at 98 °C followed by 35 amplification cycles of 10 s at 98 °C, 65 °C for 30 s, and then 72 °C for 10 min terminated with a final extension time of 10 min at 72 °C. Both forward and reverse strands of the 8-kb PCR product were sequenced using the Sanger method using BigDye Terminator v3.1 (Applied Biosystems, Australia) according to the manufacturer’s instructions at the Australian Genome Research Facility (Nedlands, Western Australia).
In Silico Data Sets
A summary of data sets and analysis performed in this article is shown in supplementary figure S5, Supplementary Material online. A total number of 91 genomes of Neisseria spp. strains with associated metadata were downloaded from NCBI and PubMLST databases. These include closed genomes of 44 N. meningitidis, 28 N. gonorrhoeae, 2 N. lactamica, 2 Neisseria species, N. weaveri, N. sicca, and Neisseria elongata subsp. glycolytica. In addition to 12 genomes in scaffolds or contigs of other Neisseria species where a closed genome of the species was not available were downloaded, these are as follows: Neisseria zoodegmatis, N. polysaccharea, N. mucosa, N. flavescens, Neisseria wadsworthii, Neisseria bacilliformis, Neisseria animaloris, N. dentiae, Neisseria canis, Neisseria arctica, N. subflava, N. shayeganii, and N. macacae. Accession numbers of strains are present in supplementary table S1, Supplementary Material online. The genomic sequence of a N. gonorrhoeae strain (ExNg63) (Neisseria PubMLST ID 46359) was also analyzed.
Protocols for the In Silico Detection of Filamentous Prophage in the Genomes of Neisseria
Intact filamentous prophage was defined as a set of genes that have the size and genetic organization similar to the open reading frames (ORFs) of Nf1–Nf3 in N. meningitidis strain Z2491 or Nf4 in N. gonorrhoeae strain FA1090.
NCBI BLAST was used to detect nucleotide sequence homology to Neisseria sp. filamentous phage. Geneious 7.1.5 software (https://www.geneious.com, last accessed February 13, 2020) was used to search for ORFs and analysis of sequences (Kearse et al. 2012). All stains were searched for the presence of homologs of genes encoding bacteriophage replication proteins, TspB gene and IS110 transposase. The sequence region from ORF1—encodes replication initiation factor—to the IS110 transposase gene with a total size of ∼8 kb was extracted and used for phylogenetic and bioinformatics analysis.
Phylogenetic Analysis and Population Structure of Filamentous Prophages
The whole-phage genomes were aligned using MAFFT alignment tool using the first base of the replication initiation factor gene as starting point. Maximum likelihood (ML) phylogenetic tree was constructed using MEGA7 with 500 bootstrap replicates (Kumar et al. 2016). hierBAPS was used to define genetic population groups of prophages using the aligned sequences (Cheng et al. 2013). Bayesian Evolutionary Analysis by Sampling Trees (BEAST) v 1.8.3 (Drummond et al. 2012) was used to infer time-measured phylogeny of a dated subset of 41 Nf1 sequences, these were extracted from 21 N. meningitidis, 3 N. gonorrhoeae, and a single N. lactamica genomes. The model used for the BEAST analysis started with a generalized time reversible model for substitutions between nucleotides at a given site, with a gamma distribution used to model variation across sites together with a proportion of invariant sites. A lognormal clock was assumed, together with a constant population size coalescent. From the conserved synteny of Nf1 prophage, it was inferred that the phage could be assessed as a single-phage species. Based upon this assumption, a BEAST analysis was performed using the date of isolation (tip dating) and not the substitution rate of the host genome. Easyfig: a genome comparison visualizer was used to compare different groups of the filamentous phage found in Neisseria genomes (Sullivan et al. 2011).
Distribution of Nf1 in the Genus Neisseria
The PubMLST database contains the whole-genome sequence of over 17,000 Neisseria sp. isolates. Supplementary table S3, Supplementary Material online, shows the frequency of Neisseria species in the database. To evaluate the presence of the Nf1 sequence in Neisseria sp. in a larger scale, the public database PubMLST was searched for the presence of Nf1-specific genes (ORF1, NEIS0031; ORF2, NEIS0030; ORF3, NEIS0029; ORF4, NEIS0028; and ORF5, NEIS0027) in all available Neisseria genomes (July 2019).
Because the remainder of the ORFs in Nf1 prophage has sequence similarity of over 95% with the sequence of Ngoφ9, these were excluded. The identification of Nf1 prophage-specific genes was performed using two methods. The first method relied on the PubMLST annotation to find genes that had above the 95% similarity to the reference genome from N. meningitidis FAM18. The second search relied on a BLAST search using the genome comparator tool implemented at the PubMLST database with parameters set at 85% for Min % identity and 80% for Min % alignment. The Nf1 positivity was defined as a genome containing at least four Nf1-specific genes with 85% minimum identity and 80% minimum alignment. To understand the distribution of Nf1 prophage in N. gonorrhoeae, a subset of 1,054 N. gonorrhoeae genomes available in PubMLST were analyzed using core genome multilocus-sequence typing for N. gonorrhoeae (cgMLST) to generate a core genome alignment. SNP sites were used to extract SNPs from the alignment, then MEGA7 was used to generate the NJ core genome SNP tree for N. gonorrhoeae with 100 bootstraps replicates. Population structure groups of the N. gonorrhoeae isolates were designated using hierBAPS tandem command line program implemented in BAPS v6.0 (Cheng et al. 2013). The cgMLST SNP tree then was colored by structure groups and plotted with the presence and absence of Nf1 sequence using the online tool, ITOL (Letunic and Bork 2007).
Assessing Mechanisms of Horizontal Gene Transfer in N. gonorrhoeae
Neisseria are naturally competent and are capable of DNA uptake from the environment. As possibly, the Nf1 sequence was acquired from N. meningitidis, mechanisms of horizontal gene transfer were analyzed using the genome comparator tool in PubMLST database for the 92 reference genomes including Restriction Modification system genes (R–M), CRISPR-associated endonuclease genes, pilin genes, and mismatch correction system (MMC) genes mutS (NEIS2138) and mutL (NEIS1378) (supplementary table S4, Supplementary Material online). REBASE a database for DNA restriction and modification: enzymes, genes, and genomes (Roberts et al. 2015) were used to identify R–M genes followed by annotation using PubMLST. In addition, Geneious 7.1.5 software (https://www.geneious.com, last accessed February 13, 2020) was used to search for the Nf1 targeted repeat (dRS3) flanking the Nf1 prophage in reference genomes and ExNg63 that are carrying the of Nf1 sequence.
Results
Nf Prophages Are Distributed across Both Pathogenic and Commensal Species of Neisseria
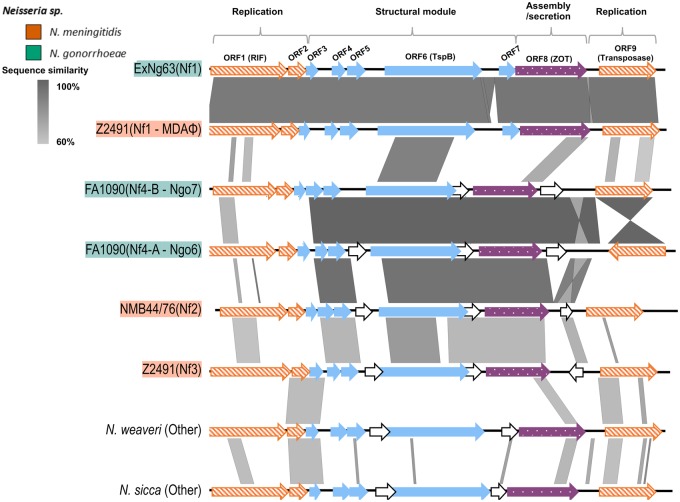
A gonococcal isolate was retrieved from a case of gonococcal meningitis in 2015 and was whole-genome sequenced. An initial annotation using the PubMLST database detected the presence of genes associated with an MDA φ. To determine if the putative phage locus was intact, it was amplified by PCR and completely sequenced using Sanger sequencing. The amplified region showed 90% similarity to the Nf1 of N. meningitidis Z2491 strain and was intact (fig. 1). Genes of highest sequence identity between the Nf1 prophages of ExNg63 and of strain Z2491 Nf1 were not related to a particular functional module even though the replication and structural genes showed high conservation. The gene with the lowest pairwise identity was the assembly gene (NEIS0023) at 87.8%, whereas the highest similarity was observed between the transposase coding gene (NMA1800) with 99% pairwise identity.
Fig. 1.
—Comparison of different groups of the filamentous prophages found in Neisseria genomes generated with Easyfig (Sullivan et al. 2011). Genes are colored based on function and organization of the Nf1 (MDAφ) presented by Bille et al. (2017). Nf1/Nf3 prophage of Z2491 and Ngoφ6/Ngoφ7 of FA1090 have been described previously (Kawai et al. 2005; Piekarowicz et al. 2006). Filamentous prophages in “other” category were too divergent to align. Replication genes are shown in orange (diagonal lines), genes encoding structural proteins are shown in blue (blocked), assembly and secretion gene is shown in purple, and genes in white (no fill) have no representative in Nf1. Vertical blocks between sequences indicate regions of shared similarity shaded according to BlastN.
To further examine the prevalence of Nf1 prophage in the genus of Neisseria, 92 annotated genomes of Neisseria spp., collected from the NCBI database and PubMLST, were investigated for the presence of complete filamentous prophages (supplementary table S1, Supplementary Material online). One hundred and sixty intact filamentous prophages were identified in 70 isolates (supplementary table S2, Supplementary Material online). All N. gonorrhoeae genomes (n = 29) in the data set had three to five complete prophage copies. Of the 44 closed genomes of N. meningitidis isolates, 34 isolates belonged to eight clonal complexes (cc) (cc4, cc5, cc8, cc11, cc32, cc41/44, cc269, and cc4821) and each isolate contained at least one copy of a filamentous prophage. The remaining ten closed meningococcal genomes from six cc (cc18, cc23, cc53, cc175, cc213, and cc4240/6688) did not carry any filamentous prophage. Eleven intact filamentous prophage genomes were identified in eight commensal Neisseria spp. These were three prophages in two N. lactamica isolates, two prophages in a single N. sicca isolate, and one phage from each of these species: Neisseria shayeganii, Neisseria weaveri, N. sp. KEM232 (identified as a novel species Neisseria chenwenguii by rMLST), N. subflava, Neisseria dentiae, Neisseria flavescens, and Neisseria macacae.
Phylogenetic and Population Structure Analyses of Neisseria Filamentous Prophages Reveal Five Main Clades
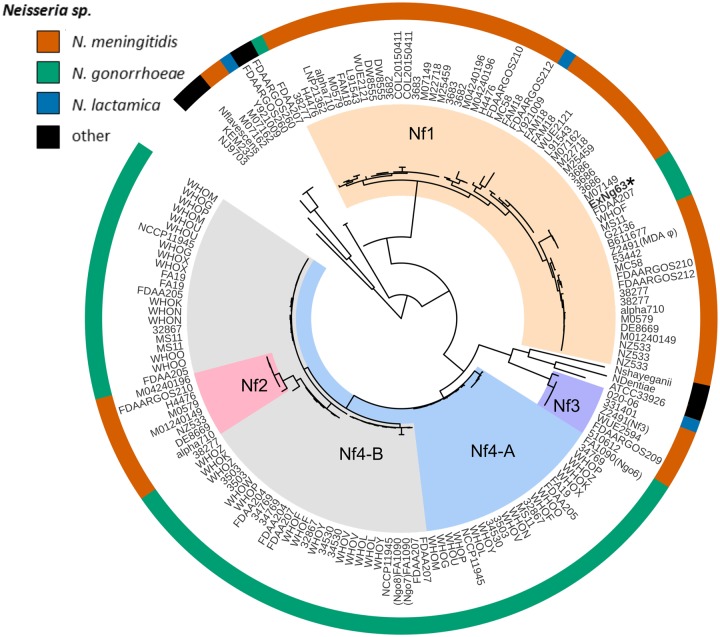
The ML phylogeny of 160 intact prophages is shown in figure 2. Five main population groups were identified using hierBAPS (fig. 2). The population groups were classified according to the Nf1–Nf4 genogroups previously defined by Kawai et al. (2005).
Fig. 2.
—Phylogeny and population groups of Neisseria filamentous (Nf) prophages. Whole-genome maximum likelihood phylogeny of 160 filamentous prophages found in Neisseria genomes highlighted with hierBAPS structure group (clade colors). Neisseria sp. are color coded (outer ring) as orange, Neisseria meningitidis; green, Neisseria gonorrhoeae; blue, Neisseria lactamica; and black, other Neisseria spp. The inner ring is the name of the strain. Because some isolates contain more than one copy of a prophage, the name of the strain can appear multiple times.
The Nf1 genogroup contained 50 filamentous phage genomes from 28 N. meningitidis isolates (one to four copies per isolate), including the meningococcal MDA φ of strain Z2491 (fig. 2). The 28 N. meningitidis isolates belonged to serogroups A, B, C, Y, and W and cc: cc4, cc8, cc11, cc32, cc41/44, cc269, and cc4821. This genogroup included four intact prophage genomes from N. gonorrhoeae including the Nf1 prophage from the gonococcal meningitis isolate ExNg63. Notably, one prophage from N. lactamica cc613 Y92-1009 (Pandey et al. 2017) clustered within genogroup Nf1.
Nf2 and Nf3 genogroups mainly contained N. meningitidis prophages. Even though previous studies have reported that the Nf2 prophage from N. meningitidis strain MC58 lacked the replication initiation factor gene (Kawai et al. 2005), many of the Nf2 prophages detected in this collection were intact. The Nf3 genogroup contained six prophages, of which five were from N. meningitidis, including the reported intact Nf3 prophage copy from N. meningitidis Z2491 and one prophage from N. lactamica cc640. All six N. meningitidis isolates carrying Nf2 prophage had at least one copy of Nf1 prophage, whereas five of the six meningococcal isolates that possessed Nf3 prophage did not possess Nf1 or Nf2 prophages.
The Nf4 genogroup, mostly observed in N. gonorrhoeae, was split into two subgroups termed Nf4-A and Nf4-B. Nf4-A contained prophages more similar to Ngoφ6 phage, whereas Nf4-B contained prophages related to Ngoφ7 and Ngoφ8 of N. gonorrhoeae strain FA1090. Ngoφ6 prophage has an inverted transposase gene when compared with Ngoφ7 and Ngoφ8 (Kawai et al. 2005; Piekarowicz et al. 2006). N. gonorrhoeae prophages from other isolates with an inverted transposase gene were present in both Nf4-A and Nf4-B. The prophages from the commensal Neisseria spp. were more divergent than the prophages from the pathogenic Neisseria spp. and did not form structure groups, but this may be the result of the small sample size. The Nf2 and Nf3 genogroups in N. meningitidis shared the same common ancestor as Nf4-A and Nf4-B which were predominantly present in N. gonorrhoeae (fig. 2).
Distribution of Nf1 Prophage in the Genus Neisseria
To determine the distribution of Nf1 in the genus Neisseria, all available whole-genome sequence data in the PubMLST Neisseria spp. database (as of July 2019 included 14,900 N. meningitidis, 4,415 N. gonorrhoeae, 330 N. lactamica, 29 Neisseria polysaccharea, 28 N. cinerea, 25 N. subflava, 15 Neisseria mucosa, 5 Neisseria bergeri, 4 Neisseria oralis and, 1 N. macacae) were searched for the presence of Nf1-specific genes annotated in the database as NEIS0031, NEIS0030, NEIS0029, NEIS0028, and NEIS0027 (supplementary table S3, Supplementary Material online). In the PubMLST database, genomes are annotated based on a cutoff value of 95% to the reference genome N. meningitidis strain FAM18. To allow detection of noncurated loci due to variable sequence similarity and alignment length, the genomes were BLAST searched using the genome comparator tool at the PubMLST for the Nf1-specific loci with 85% minimum allele identity and 80% minimum alignment. Nf1 positivity was defined as a genome containing at least four Nf1-specific genes.
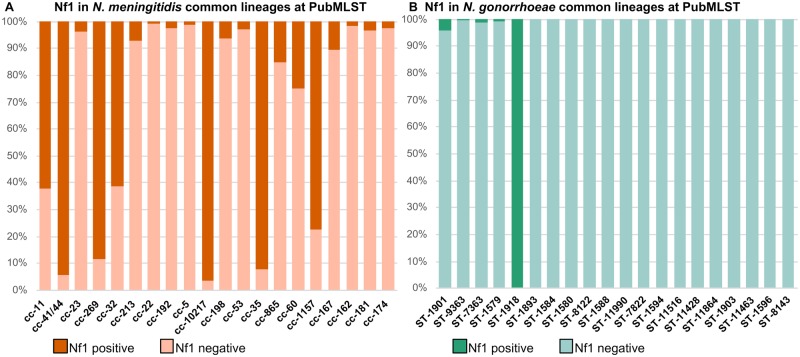
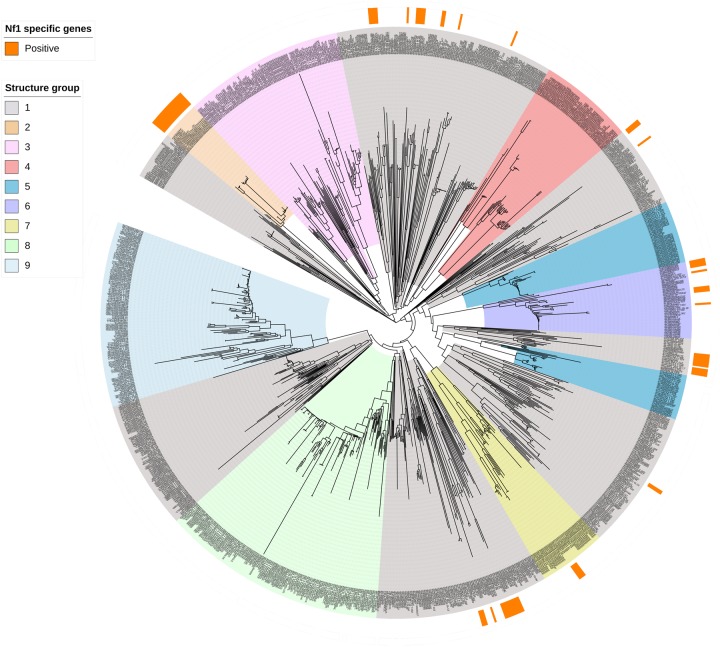
Specific genes of Nf1 prophage were present in 7,185 of 14,900 (48%) meningococcal unclosed genomes and were distributed widely across cc, including the rarely invasive cc53, cc192, and cc198 lineages (fig. 3A). Nf1-specific genes with similarity over the 95% cutoff value were detected in 6.7% (n = 296/4,415) of gonococcal genomes (fig. 3B). Of the eight records which reported that N. gonorrhoeae had been isolated from cases of invasive disease, only a single isolate possessed Nf1-specific genes. Where Nf1-specific genes were present, these had a restricted distribution among N. gonorrhoeae genetic lineages defined by four sequence types (STs); ST1918 (n = 152/152), ST1901 (n = 40/998), ST10932 (n = 17/17), and ST1590 (n = 15/29). To determine whether the restricted distribution of the Nf1 prophage corresponded to a particular structural group of N. gonorrhoeae, the association of Nf1-specific genes with a core genome MLST (cgMLST) SNP-based phylogeny of 1,054 gonococcal whole-genome sequences was performed (fig. 4). This phylogenetic tree showed that N. gonorrhoeae formed nine structure groups which is consistent with recent studies of N. gonorrhoeae evolution using Bayesian analysis (Ezewudo et al. 2015; Grad et al. 2016; Al Suwayyid et al. 2018; Lee et al. 2018; Sánchez-Busó et al. 2019; Wadsworth et al. 2019). Nf1 prophage was present in five of the nine gonococcal structure groups (fig. 4).
Fig. 3.
—The prevalence of Nf1-specific genes in Neisseria meningitidis (A) and Neisseria gonorrhoeae (B). The top 20 clonal complexes of N. meningitidis and the top 20 MLST sequence types of N. gonorrhoeae are shown in order of abundance in PubMLST.
Fig. 4.
—A SNP-based phylogeny of 1,054 Neisseria gonorrhoeae isolates from PubMLST database based on cgMLST scheme for N. gonorrhoeae. The outer ring (orange sections) indicates the presence of Nf1-specific genes in these isolates. The gonococcal clades are colored by the different structure groups identified by hierBAPS (Cheng et al. 2013).
The Nf1-specific genes were also detected in two human oropharynx commensal Neisseria species; N. lactamica and N. cinerea. Although the 330 N. lactamica genomes represented 46 genetic lineages, only the single lineage, ST3493, contained four or more Nf1-specific genes. This was found in all deposited genomes of this lineage (n = 227). In N. cinerea, two isolates (LNP26654, PubMLST ID 38984 and LNP28181, PubMLST ID 42061) possessed four or more Nf1 genes, corresponding to a carriage rate of 7% (n = 2/28) in available isolates. Identification of Nf1 genes in these commensal strains was not taken for further analysis as the remaining section of the phage was located on separate contigs (data not shown).
Integration Sites of the Nf1 Prophage in Intact Genomes Are Variable
Nf1 integration is catalyzed by an integrase which targets the repeat sequence dRS3 containing the CT conserved motif (supplementary fig. S1, Supplementary Material online) (Kawai et al. 2005; Meyer et al. 2016). dRS3 repeats are more abundant in N. meningitidis (700 per genome) than N. gonorrhoeae (200 per genome) (Marri et al. 2010). Because the Nf1 prophages were frequently at the ends of contigs and therefore unable to be closed in most genomes, we can only infer some limited associations with other factors using the library of 92 reference genomes (supplementary table S1, Supplementary Material online). Overall, the integration of Nf1 prophage is influenced by distribution and intensity of dRS3 (Nf1 subtype, ATTCCCRCCTRCGCGGRAAK) in the genome (Meyer et al. 2016) (supplementary fig. S1, Supplementary Material online).
Nf1 prophages in N. gonorrhoeae were integrated into different loci around the genome (supplementary fig. S1, Supplementary Material online). In the closed genomes of N. gonorrhoeae strains, MS11 and WHO-F, an Nf1 prophage was integrated into a pilS repeat region upstream of lpxC, whereas in strain FDAA207, Nf1 prophage was located at a different site between NEIS0638–NEIS0639. No similar Nf1 prophage integration sites were found in the 28 closed reference genomes of Nf1 prophage positive N. meningitidis genomes. In ExNg63, the single copy of Nf1 prophage was located between NEIS0276 and NEIS0288 and a similar integration site was found in N. meningitidis FAM18 (supplementary fig. S2, Supplementary Material online). Although the integration site was conserved between the two species, the alleles of NEIS0276 and NEIS0288 were unique to N. gonorrhoeae. The Nf1 prophage genes in the only reference genome of N. lactamica Y92-1009 were inserted into a pilS repeat region, albeit in a location associated with the locus encoding the transferrin-binding protein TbpB (supplementary fig. S2, Supplementary Material online). No similar insertion site for the Nf1 prophage was present in the 28 closed reference genomes of Nf1 positive N. meningitidis genomes.
Interrogation of Nf1 Receptor-Mediated and Homologous Recombination Mechanisms of Prophage Integration in N. gonorrhoeae
To further examine whether the presence of Nf1 prophage in N. gonorrhoeae was likely to be due to phage-mediated integration, we examined whether there was conservation of the phage receptor, PilE (major pilin subunit) in Nf1-positive gonococcal and meningococcal isolates compared with Nf1-negative gonococcal isolates (Meyer et al. 2016). A phylogenetic tree of the PilE amino acid sequences from 80 PilE positive Neisseria reference genomes in addition to ExNg63 did not demonstrate any clustering of PilE alleles with the presence of the Nf1 prophage in any particular species (supplementary fig. S3, Supplementary Material online).
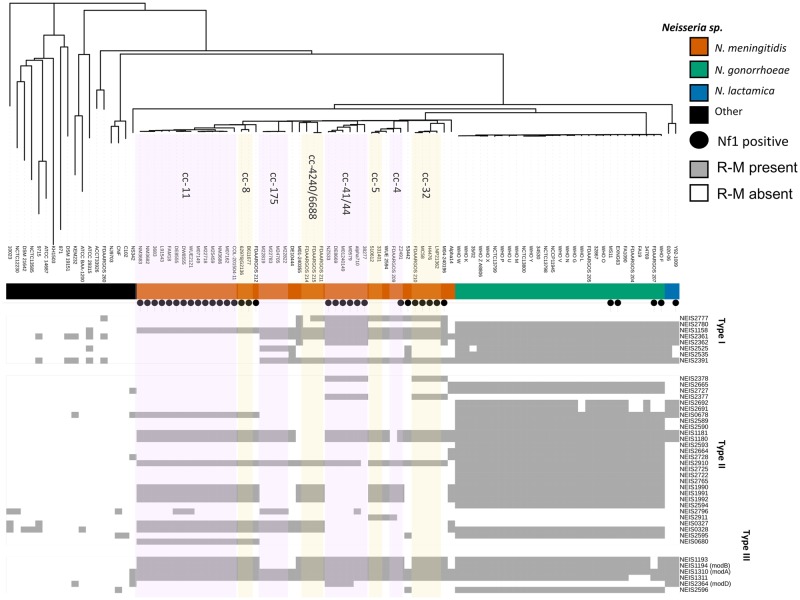
Apart from phage receptor-mediated entry into the bacteria host, the phages may have also been obtained by natural transformation and integrated into the host genome via homologous recombination. The distribution of Nf1 prophage across five of the nine structure groups of N. gonorrhoeae led to the hypothesis that the presence of the Nf1 prophage could be associated with the absence of restriction modification systems (R–M) that would normally restrict the entry of foreign DNA into these isolates (supplementary table S4, Supplementary Material online). Two R–M type I systems, 14 R–M type II systems, and 3 R–M type III systems were scanned in the 92 reference genomes and mapped to the core genome neighbor-joining phylogenetic tree in addition to the presence and absence of Nf1 prophage (fig. 5). Although the distribution of R–M systems is quite variable and associated with specific clonal complexes in N. meningitidis (Budroni et al. 2011), the distribution of the R–M systems in N. gonorrhoeae was reasonably invariant and showed no apparent association with MLST ST or the presence or absence of Nf1 prophage (fig. 5).
Fig. 5.
—Core genome neighbor-joining phylogeny of 92 reference Neisseria genomes from PubMLST database annotated with Neisseria spp., presence of the Nf1 prophage (black dot), and restriction modification systems (gray boxes). Neisseria spp. are color coded (outer ring) as orange, Neisseria meningitidis; green, Neisseria gonorrhoeae; blue, Neisseria lactamica; and black, other Neisseria spp. Neisseria meningitidis clonal complexes (cc) are highlighted in lavender and pink.
The frequency of homologous recombination with foreign DNA is influenced by whether the MMC is intact (Bucci et al. 1999; Davidsen et al. 2007) and increases when MMC genes mutS (NEIS2138) and mutL (NEIS1378) are inactive (Rotman and Seifert 2015). In the 92 reference genomes, 15 different mutated mutS alleles were identified; 8 alleles in N. meningitidis (7 of which were Nf1 positive), 2 in N. gonorrhoeae (1 of which was Nf1 positive, ExNg63), and 5 alleles in other Neisseria spp. isolates (none of which were Nf1 positive strains). The mutL analysis showed only two mutated alleles in Nf1 positive N. gonorrhoeae strains MS11 and N. sp. 10023. The mutS and mutL gene analysis in the 92 reference genomes showed that only two of the five gonococcal strains containing Nf1 prophage had frame shifted mut alleles: ExNg63 had a 207delT in mutS and MS11 had a mutL 1267delA. However, no association was found between mutS/mutL mutations and possession of Nf1 prophage in the collection of 92 Neisseria genomes (supplementary fig. S4, Supplementary Material online).
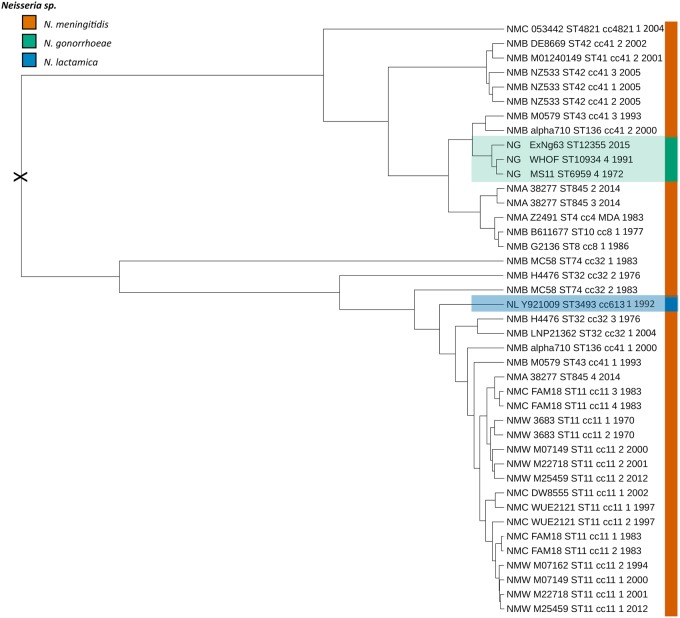
From the above analyses, it can be inferred that acquisition of the Nf1 prophage into N. gonorrhoeae and N. lactamica may have occurred via multiple mechanisms including infection and homologous recombination. To determine which Neisseria spp. was the likely source of the Nf1 prophage, a timed phylogeny using the prophage data set was conducted. A time-measured phylogeny (fig. 6) was inferred using Bayesian Evolutionary Analysis by Sampling Trees (BEAST) v 1.8.3 (Drummond et al. 2012), based on a dated subset of 41 Nf1 prophage sequences isolated between 1970 and 2015 that were extracted from 21 meningococcal, 3 gonococcal (including ExNg63), and a single N. lactamica whole-genome sequence (supplementary table S2, Supplementary Material online). From the conserved synteny of Nf1 prophage, it was inferred that the phage could be assessed as a single-phage species. Based upon this assumption, a BEAST analysis was performed using the date of isolation and not the substitution rate of the host genome. This revealed that the most likely ancestor of the Nf1 prophage-infecting N. gonorrhoeae and N. lactamica was N. meningitidis serogroup B cc-41/44 and N. meningitidis serogroup B cc-32, respectively (fig. 6).
Fig. 6.
—Time-measured phylogeny of a dated subset of 41 intact Nf1 prophage sequences. The most likely root of phylogeny is marked by an X. Tip labels show in the following order: serogroup for meningococcus (NMB and NMC) or gonococcus (NG), strain name, MLST sequence type (ST), clonal complex, filamentous phage copy (1–4), and year of isolation. NG serotypes A and B are based on porB subtypes porB1A and porB1B (Tam et al. 1982).
Discussion
We have described the identification and the distribution of filamentous bacteriophages in the genus of Neisseria with a focus on the Nf1 prophage genogroup corresponding to the MDA φ of N. meningitidis. Although prophages were detected in both pathogenic and commensal Neisseria spp., they were more abundant in N. meningitidis and N. gonorrhoeae. Phylogenetic and genomic population structure analysis categorized the Neisseria spp. filamentous prophages into five genogroups which corresponded to the four Nf1-4 subtypes described by Kawai et al. (2005). The Nf1 prophage was found in N. meningitidis, N. gonorrhoeae, N. lactamica, and N. cinerea, whereas Nf3 prophage was found in N. lactamica and N. meningitidis. In N. meningitidis, the pathogenic potential of MDA φ is dose-dependent and is related to the copy number of the phage in the genome (Omer et al. 2011). In the hyperinvasive lineages, isolates generally harbored more than one copy of the Nf1 prophage in different chromosomal locations (Meyer et al. 2016). In contrast, the closed genomes of N. gonorrhoeae and N. lactamica harbored only a single copy of the Nf1 prophage per host genome.
Nf2 prophages were found in eight N. meningitidis belonging to cc32, cc41/44, and cc269 and were co-resident with at least one copy of Nf1 prophage. To our knowledge, this is the first study to identify intact copies of Nf2 prophage in N. meningitidis. Nf3 prophage was not species-specific with prophages detected in five N. meningitidis belonging to cc4 and cc5 and in one N. lactamica strain 02006 of cc640. When an isolate possessed an Nf3 prophage, it was generally the only filamentous phage that was present (n = 5/6). The Nf4 genogroup could be split into two subgroups, Nf4-A and Nf4-B. All gonococcal isolates examined in this study have two to three intact copies of both Nf4 genogroup prophages. Novel species-specific filamentous pages that did not belong to any of the defined groups were also identified in six commensal Neisseria spp.: N. sicca, N. dentiae, N. shayeganii, N. macacae, and N. spp. KEM232 (N. chenwenguii). The maintenance of filamentous phages in Neisseria spp. may infer a role in colonization of the human host which has yet to be experimentally assessed.
Bille et al. (2005) has suggested the MDA φ could be exchanged by both receptor-mediated uptake into the bacterial host or by natural transformation of chromosomal DNA from lysed cells followed by homologous recombination into the recipient N. meningitidis host genome. We hypothesized that the former mechanism of receptor-mediated uptake might manifest in the conservation of the phage receptor, PilE, in Nf1-possessing N. gonorrhoeae isolates when compared with the Nf1 prophage negative isolates. However, the PilE protein alleles of N. gonorrhoeae possessing the Nf1 prophage did not cluster with the meningococcal alleles of PilE from Nf1 prophage positive isolates. This may reflect an ancestral acquisition of the Nf1 phage using a conserved receptor PilE allele which has since been overwritten by mutation and recombination. Of the three Nf1 prophage integration sites that are present in N. gonorrhoeae, only one was shown to be conserved (gonococcal strain ExNg63 and the meningococcal strain FAM18). As the flanking alleles in this locus were unique to all N. gonorrhoeae isolates, currently in the PubMLST database, this could be interpreted as either true phage-mediated integration or as natural transformation followed by homologous recombination that resulted in unique flanking allelles.
As there was no apparent conservation of the PilE receptor from Nf1 possessing N. lactamica, N. gonorrhoeae and N. meningitidis isolates, we examined whether the gonococcal isolates possessing Nf1 prophage had markers associated with increased natural transformation competency. R–M systems protect bacteria by restricting incoming foreign DNA such as phage DNA and we hypothesized the absence of an R–M system may promote phage infection. Although R–M systems are known to be associated with cc in N. meningitidis, R–M systems in N. gonorrhoeae were distributed across all the reference genomes of this species and no apparent association of the lack of an R–M system with possession of Nf1 phage was found. Methyl mismatch repair (MMR) systems are also known to act as a barrier to recombination between divergent sequences which occur during transduction with phages and recombination between partially divergent sequences. Inactivation of MMR systems dramatically increases recombination rates (Zahrt et al. 1994). Indeed, it has been shown that a N. meningitidis MutS mutator phenotype in Z5463 could have assisted an Nf1 duplication event after four passages in the descendant strain Z5463Pl (Omer et al. 2011). We found in the closed reference genomes of Nf1 positive isolates that N. gonorrhoeae strain MS11 and the unclosed genome of ExNg63 had a mutated mutL and mutS allele, respectively, whereas N. gonorrhoeae strains WHO-F and FDAA207 had intact alleles. However, in the 1,053 unclosed gonococcal genomes, the association of the mutator alleles with the presence of Nf1-specific genes was observed in only seven isolates of the 86 Nf1 positive gonococcal isolates in this collection, indicating that the co-occurrence of these elements was random. However, this analysis was confounded by the fragmentation of the unclosed genomes which resulted in only partial Nf1 identification in this library and as such, repeating this analysis with more closed reference genomes representing the diversity of N. gonorrhoeae is warranted.
Due to the restriction of the Nf1 phage in N. gonorrhoeae to a limited number of MLST STs, to one ST each in N. lactamica and N. cinerea, we hypothesized the Nf1 prophage was acquired by these species more recently than by N. meningitidis and, further, that N. meningitidis could have been the donor in this exchange. This was supported by BEAST time-measured phylogeny of a dated Nf1 subset, which suggests the most likely root of acquisition of Nf1 prophage into N. gonorrhoeae and N. lactamica was via N. meningitidis with the ancestral donor most likely related to cc41/44 and cc32, respectively. A second line of evidence that suggests Nf1 prophage acquisition was a late event in N. gonorrhoeae is the observation that the closed reference genomes of gonococcal isolates, FDAARGOS207 and WHO-F, had at least three copies of Nf4 phage. It has been shown in Escherichia coli that superinfection of the host by an incumbent coliphage effectively blocks a second superinfection by a second related coliphage due to the expression of gene III protein (NEIS0025) (Boeke et al. 1982). In this scenario, super infection with Nf1 prophage would be restricted due to this feature where related Nf4 prophages are already resident in the host strain (Kłyż and Piekarowicz 2018).
The role of Nf1 phage in N. gonorrhoeae and in commensal Neisseria spp. is unknown, but the elements identified in this study from the closed genomes were completely intact suggesting that they are maintained and thus functional. The presence of Nf1 prophage in N. gonorrhoeae is associated with some successful genetic lineages, most significantly the MLST ST1918 lineage that has persisted in United Kingdom for over 30 years (from 1982 to 2014) and caused an outbreak in Sheffield in the heterosexual population between 1995 and 2000 (Didelot et al. 2016). The gonococcal strain MS11 which has been used as a standard model for gonococcal colonization, has a single copy of the Nf1 prophage but the contribution of this phage in colonization studies has not been assessed (Higashi et al. 2007; Jerse et al. 2011). Of note, was the identification of the Nf1 prophage in the ST3493 N. lactamica Y92-1009 strain. This isolate was initially isolated in Northern Ireland in 1982 and has been successfully used as a probiotic to reduce the rate of N. meningitidis carriage (Deasy et al. 2015; Pandey et al. 2017). It will be useful to understand whether or not the possession of the Nf1 prophage in this strain is contributing to this competitive ability. It also remains to be seen whether the Nf1 prophage can be transferred to co-colonizing meningococcus strains and affect their virulence (Gbesemete et al. 2019).
This survey of intact filamentous phage elements in Neisseria spp. was limited to reference genomes in the PubMLST and NCBI databases. Due to the high conservation of the sequences of filamentous phage elements and the fact that they integrate into repetitive sequence regions, it is very difficult to retrieve contiguous regions surrounding prophage elements in unclosed genomes. Because of this issue, the incomplete phage Ngoφ9 was excluded from this analysis as it represents a truncated Nf1 prophage. Although Nf1-specific genes were identified in N. polysaccharea (M16 240183, PubMLST ID 42468 and LNP29576, PubMLST ID 58873), these were excluded from further analysis as they carried two or three Nf1-specific genes, respectively, and the contigs were not intact preventing further assessment. Future studies using a larger library of closed reference genomes of the genus of Neisseria would assist in further clarifying the diversity, distribution, and roles of filamentous phages in colonization of the human host by this genus.
To our knowledge, this is the first study that has reported the presence of Nf1 prophage in N. gonorrhoeae and the nonpathogenic species N. lactamica and N. cinerea. Nf1 prophage in N. meningitidis is an active phage that can enter a reproductive cycle, generate virions, infect naïve meningococcal strains and replicate within the meningococcal genome (Meyer et al. 2016). It has been shown to contribute to microcolony formation and act as an accessory colonization factor (Bille et al. 2017). The detection of Nf1 prophage elements in N. gonorrhoeae and human commensal Neisseria spp. suggests a wider role for this phage as a colonization factor for other Neisseria spp. Due to identification of Nf1 (MDA-like) prophage in Neisseria spp. besides N. meningitidis, we suggest the use of the term “MDA φ” for functional Nf1 phage that can replicate and produce virions and to use the “Nf1” term for intact prophage detected in Neisseria spp. genomes.
Supplementary Material
Acknowledgments
Collection of gonococcal isolates is facilitated by the National Neisseria Network in Australia. We thank Mr Christopher Mullally from the Marshall Center for Infectious Diseases at the University of Western Australia for his expertise in PubMLST Neisseria database analysis.
Literature Cited
- Al Suwayyid BA, et al. 2018. Genomic epidemiology and population structure of Neisseria gonorrhoeae from remote highly endemic Western Australian populations. BMC Genomics 19(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille E, et al. 2005. A chromosomally integrated bacteriophage in invasive meningococci. J Exp Med. 201(12):1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bille E, et al. 2017. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. 13(7):e1006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Model P, Zinder ND.. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol Gen Genet. 186(2):185–192. [DOI] [PubMed] [Google Scholar]
- Bucci C, et al. 1999. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell. 3(4):435–445. [DOI] [PubMed] [Google Scholar]
- Budroni S, et al. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 108(11):4494–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J, et al. 2003. Novel type of specialized transduction for CTX phi or its satellite phage RS1 mediated by filamentous phage VGJ phi in Vibrio cholerae. J Bacteriol. 185(24):7231–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, et al. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 30(5):1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen T, et al. 2007. Genetic interactions of DNA repair pathways in the pathogen Neisseria meningitidis. J Bacteriol. 189(15):5728–5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day L. 2011. Family inoviridae In: King AMQ, Adams MJ, Carstens EB, Leftkowitc EJ, editors. Virus taxonomy: classification and nomenclature of viruses: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego: Elsevier Academic Press; p. 375–384. [Google Scholar]
- Deasy AM, et al. 2015. Nasal inoculation of the commensal Neisseria lactamica inhibits carriage of Neisseria meningitidis by young adults: a Controlled Human Infection study. Clin Infect Dis. 60(10):1512–1520. [DOI] [PubMed] [Google Scholar]
- Didelot X, et al. 2016. Genomic analysis and comparison of two gonorrhea outbreaks. MBio 7(3):e00525–00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A.. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29(8):1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezewudo MN, et al. 2015. Population structure of Neisseria gonorrhoeae based on whole genome data and its relationship with antibiotic resistance. PeerJ 3:e806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gbesemete D, et al. 2019. Protocol for a controlled human infection with genetically modified Neisseria lactamica expressing the meningococcal vaccine antigen NadA: a potent new technique for experimental medicine. BMJ Open. 9(4):e026544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad YH, et al. 2016. Genomic epidemiology of gonococcal resistance to extended-spectrum cephalosporins, macrolides, and fluoroquinolones in the United States, 2000–2013. J Infect Dis. 214(10):1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G.. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29(8):1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi DL, et al. 2007. Dynamics of Neisseria gonorrhoeae attachment: microcolony development, cortical plaque formation, and cytoprotection. Infect Immun. 75(10):4743–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse A, et al. 2011. Estradiol-treated female mice as surrogate hosts for Neisseria gonorrhoeae genital tract infections. Front Microbiol. 2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC.. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11(1):595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Uchiyama I, Kobayashi I.. 2005. Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res. 12(6):389–401. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kłyż A, Piekarowicz A.. 2018. Phage proteins are expressed on the surface of Neisseria gonorrhoeae and are potential vaccine candidates. PLoS One 13(8):e0202437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RS, et al. 2018. Genomic epidemiology and antimicrobial resistance of Neisseria gonorrhoeae in New Zealand. J Antimicrob. 73(2):353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P.. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. [DOI] [PubMed] [Google Scholar]
- Mai-Prochnow A, et al. 2015. Big things in small packages: the genetics of filamentous phage and effects on fitness of their host. FEMS Microbiol Rev. 39(4):465–487. [DOI] [PubMed] [Google Scholar]
- Marri PR, et al. 2010. Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PLoS One 5(7):e11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J, et al. 2016. Characterization of MDAΦ, a temperate filamentous bacteriophage of Neisseria meningitidis. Microbiology 162(2):268–282. [DOI] [PubMed] [Google Scholar]
- Omer H, et al. 2011. Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLoS One 6(2):e17145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey AK, et al. 2017. Neisseria lactamica Y92–1009 complete genome sequence. Stand Genomic Sci. 12:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A, et al. 2014. Neisseria gonorrhoeae filamentous phage Ngoφ6 is capable of infecting a variety of Gram-negative bacteria. J Virol. 88(2):1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarowicz A, Majchrzak M, Klyz A, Adamczyk-Poplawska M.. 2006. Analysis of the filamentous bacteriophage genomes integrated into Neisseria gonorrhoeae FA 1090 chromosome. Pol J Microbiol. 55:251–260. [PubMed] [Google Scholar]
- Rakonjac J, et al. 2011. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr Issues Mol Biol. 13(2):51–76. [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, Macelis D.. 2015. REBASE – a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43(D1):D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman E, Seifert HS.. 2015. Neisseria gonorrhoeae MutS affects pilin antigenic variation through mismatch correction and not by pilE guanine quartet binding. J Bacteriol. 197(10):1828–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Busó L, et al. 2019. The impact of antimicrobials on gonococcal evolution. Nat Microbiol. 4(11):1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MJ, Petty NK, Beatson SA.. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27(7):1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam M, et al. 1982. Serological classification of Neisseria gonorrhoeae with monoclonal antibodies. Infect Immun. 36(3):1042–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth CB, Sater MR, Bhattacharyya RP, Grad YH.. 2019. Impact of population structure in the design of RNA-based diagnostics for antibiotic resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother. 63(8):e00549–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahrt TC, Mora GC, Maloy S.. 1994. Inactivation of mismatch repair overcomes the barrier to transduction between Salmonella typhimurium and Salmonella typhi. J Bacteriol. 176(5):1527–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.