Abstract
Primate brains vary dramatically in size and organization, but the genetic and developmental basis for these differences has been difficult to study due to lack of experimental models. Pluripotent stem cells and brain organoids provide a potential opportunity for comparative and functional studies of evolutionary differences, particularly during the early stages of neurogenesis. However, many challenges remain, including isolating stem cell lines from additional great ape individuals and species to capture the breadth of ape genetic diversity, improving the reproducibility of organoid models to study evolved differences in cell composition and combining multiple brain regions to capture connectivity relationships. Here, we describe strategies for identifying evolved developmental differences between humans and non-human primates and for isolating the underlying cellular and genetic mechanisms using comparative analyses, chimeric organoid culture, and genome engineering. In particular, we focus on how organoid models could ultimately be applied beyond studies of progenitor cell evolution to decode the origin of recent changes in cellular organization, connectivity patterns, myelination, synaptic development, and physiology that have been implicated in human cognition.
INTRODUCTION
Evolutionary changes have repeatedly transformed brain structure during the diversification of primates (Striedter, 2005). In particular, the human brain has expanded threefold since our divergence from a common ancestor with chimpanzee and over fifteen-fold since our divergence from old world monkeys (Herculano-Houzel, 2012). This expansion has been accompanied by changes in cortical microcircuits, an increased proportion of upper cortical layer neurons, increased connections between higher order association areas, more monosynaptic connections to motor neurons involved in dexterous movements, delayed myelination, and protracted periods of synaptic plasticity (Bianchi et al., 2013; Preuss, 2011; Sousa et al., 2017a; Striedter, 2005). Many of these differences in growth rate, brain size, and neuron number are apparent prior to birth (Sakai et al., 2013). Importantly, many of these recently evolved human traits may also be particularly vulnerable to dysfunction (Song et al., 2018; Varki, 2000; Van den Huevel et al., 2019). However, direct comparisons of gene expression, signaling pathway activation, and cell behavior during human, ape, and old world monkey brain development that could help to isolate the mechanisms underlying these recent evolutionary changes have been challenging due to the inaccessibility of developing ape brain tissue. Studying the cellular and genetic origin of these evolved differences requires an experimental system in which developmental processes can be compared and candidate sequence changes can be examined in a suitable genetic and cellular context.
Mouse models have enabled pioneering studies of how genes that changed in human evolution could influence brain development (Boyd et al., 2015; Charrier et al., 2012; Dennis et al., 2012; Florio et al., 2015). However, ancestors of mouse and human diverged roughly 90 million years ago. At the genomic level, this divergence makes it difficult to reconstruct many human-specific genetic changes because these changes often occur in regions with poor homology, and even when changes occur in homologous sequence, the broader genomic context of the region may differ. Similarly, at the cellular level, some cell types that are important for human brain development and function, such as outer subventricular zone radial glia, are rare or absent in the mouse brain (Fietz et al., 2010; Hansen et al., 2010; Wang et al., 2011). Finally, at the tissue level, the human brain is over 1000 times larger than the mouse brain, and special challenges associated with large brains, such as the formation of long-range connections, may be difficult to model in mouse. Therefore, despite the many strengths of mouse models in reproducibly capturing developmental patterning and cellular interactions, this model system alone may not be sufficient to identify evolved differences in developmental cell behavior or to identify the genetic and molecular changes that explain these differences.
Cultured cells from diverse species could offer a non-invasive window into the mechanisms underlying evolutionary differences provided they could be coaxed into undergoing similar developmental processes. Indeed, cultured skin cells and immune cells have revealed species differences between humans and other great apes (Barreiro et al., 2010; Lawlor et al., 1990). With the discovery that somatic cells from mouse and human could be reprogrammed to induced pluripotent stem cells (iPSCs) (Meissner et al., 2007; Takahashi et al., 2006; Yu et al., 2007), it became conceivable to imagine creating panels of iPSCs from diverse mammalian species that could be used to study how genetic changes underlie differences in developmental processes. This comparative iPSC approach has recently been applied to study human and chimpanzee iPSCs themselves (Gallego Romero et al., 2015; Marchetto et al., 2013; Wunderlich et al., 2014), iPSC-derived neural crest cells (Prescott et al., 2015), cardiomyocytes (Pavlovic et al., 2018), endoderm lineage (Blake et al., 2018), and forebrain lineage (Field et al., 2019; Mora-Bermúdez et al., 2016; Otani et al., 2016; Pollen et al., 2019; Kanton et al., 2019).
Organoid models, in particular, may further capture cell diversity and cellular interactions that normally occur during development. In principle, organoid models could help to bridge the gap between genome sequence differences and recently-evolved specializations of the human brain by providing an experimentally tractable system with the appropriate cellular and genetic background. Because experimental studies of development are otherwise inaccessible in apes, we know little about evolved differences in developmental cell behavior, as well as the tolerated variation in such processes within a species. Comparative analysis between species and chimeric co-culture in organoids could isolate these developmental differences and determine which differences are driven by cell intrinsic changes. Similarly, genome engineering in iPS-derived cell lines provides the opportunity to determine the extent to which genome sequence changes, including accelerated substitutions (Pollard et al., 2006), duplications (Sudmant et al., 2013), deletions (McLean et al., 2011), and insertions (Hellen and Kern, 2015; Kronenberg et al., 2018), contribute to specific evolved differences.
In this review, we describe the importance of expanding our representation of ape genetic diversity in a dish using iPSCs, the opportunity for harnessing organoid models to represent the diversity of cell types and communities, and general opportunities for improving the health and organization of organoid models. We then transition to discussing the first comparative studies of neural progenitor cell behavior and gene expression in human and ape organoids, and we close by examining how improvements in organoid models could enable mechanistic studies of evolutionary changes thought to be essential for human cognitive specializations.
REPRESENTING APE GENETIC DIVERSITY IN A DISH
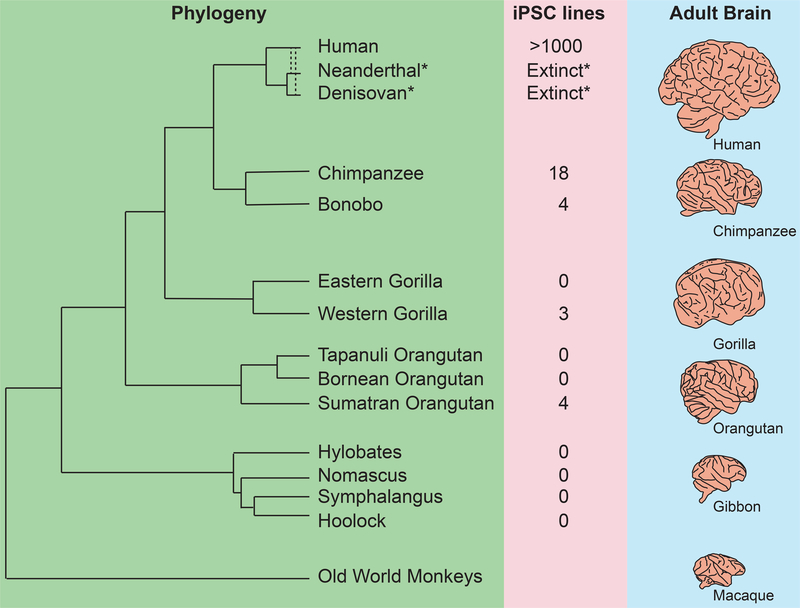
Collections of human and ape iPSC lines have the potential to represent divergence of developmental traits between species and variation within species. Characterizing these properties of divergence and variation is crucial for identifying adaptive changes in the human lineage, as well as for revealing both constrained processes and neutral variation tolerated during human and ape development. However, ape primary cells as a source for reprogramming are scarce and difficult to access and transfer given the endangered status of all non-human apes. In addition, although reprogramming of ape cells with protocols optimized for human cells is possible, that reprogramming fibroblasts from non-human primates is not as straightforward, likely due to species-specific differences in optimal culturing conditions (Nakai et al., 2018; Navara et al., 2018). Therefore, early studies that demonstrated proof of principle used only a few individuals and performed reprogramming with integrating retroviruses that may increase efficiency, but could have later consequences on cell behavior. Panels of integration free human and chimpanzee iPSC lines have now been generated and distributed (Gallego Romero et al., 2015), but there is limited representation of additional ape species (Figure 1; Table 1), and most studies have focused on divergence between human and chimpanzee.
Figure 1. Existing pluripotent stem cell lines sample a limited fraction of ape diversity.
Phylogeny describes the radiation of ape species since divergence from a common ancestor with old world monkeys, with gibbon and orangutan clades updated based on recent studies (Carbone et al., 2014; Nater et al., 2017). Second column indicates the approximate number of individuals currently reprogrammed across published papers. Note that although Neanderthal and Denisovan are extinct, a substantial portion of their genomes exists as introgressed alleles in human populations and is accessible to genetic mapping approaches (Vattathil and Akey, 2015). Third column shows approximate relative brain size across species based on the Comparative Mammalian Brain Collection (http://neurosciencelibrary.org)
Table 1.
Ape iPSC lines analyzed in recent studies, including newly generated lines:
| Publication | Number of Individuals | Reprogramming Method | Individuals |
|---|---|---|---|
| Marchetto et al., 2013 | 2 chimpanzee (new), 2 bonobo (new) | Lentiviral from fibroblast | PR00818, PR01209, AG05253B, PR01086 |
| Wunderlich et al., 2014 | 1 western gorilla (new), 1 bonobo (new), | Lentiviral from fibroblasts and endothelial cells | B1_i_1/2, G1_i_1/2a/2b/3a/3b, |
| Gallego Romero et al., 2015 | 7 chimpanzee (new) | Episomal from fibroblast | 3647, 3649, 3651, 4955, 8861, 40210, 40280 |
| Ramaswamy et al., 2015 | 2 Sumatran orangutan (new) | Retrovirus from fibroblasts | KB10460, KB10973 |
| Otani et al., 2016 | 2 chimpanzee | Lentiviral from fibroblast, | PR00818, PR01209, |
| Mora-Bermúdez et al 2016 | 2 chimpanzee (1 new), 1 Sumatran orangutan (new) | Lentiviral and Episomal from fibroblast and blood | PR00818, Sandra A, Toba |
| Ramsay et al., 2017 | 2 western gorilla (new), | Retrovirus from fibroblast | PR00053 and PR00075 |
| Blake et al., 2018 | 4 chimpanzee (1 new) | Episomal from fibroblast | 3647, 3649, 40300, 4955 |
| Pavlovic et al., 2018 | 10 chimpanzee (2 new) | Episomal from fibroblast | 3624, 3647, 3649, 3651, 4933, 4955, 8861, 40210, 40280, 40300 |
| Field et al., 2019 | 1 chimpanzee (new) 1 Sumatran orangutan (new) | Episomal from fibroblast | S008919 Josephine (11045–4593) |
| Pollen et al., 2019 | 8 chimpanzee (4 new) | Episomal from fibroblast | 3649, 4955, 8861, 40210, 40290, 3611, PR00226, PR00738 |
| Kanton et al., 2019 | 3 chimpanzee (1 new) 1 bonobo (new) | Retroviral and episomal from fibroblast and blood | PR00818, SandraA, JoC, Bokela |
Additional great ape stem cell lines would not only help to pinpoint key evolutionary specializations on the lineage leading to humans, but these lines could also help to interpret candidate disease-causing processes in humans. Humans underwent a population bottleneck to fewer than 10,000 individuals in the last 65,000 years, reducing the common genetic variation segregating in our population (Prado-Martinez et al., 2013). Recent studies have taken advantage of the greater extent of tolerated sequence variation naturally occurring in other primate species to efficiently predict de novo coding mutations that are likely to be pathogenic in humans (Sundaram et al., 2018). Using ape iPSC lines to characterize tolerated variation in gene regulation and developmental processes within primates species may further support interpretation of candidate human disease mechanisms, including those being studied using stem cells and organoid models. In addition, analyzing phenotypic divergence on additional branches of the great ape phylogeny will help to better resolve which changes on our own lineage are related to human traits and which are related to general patterns of divergence.
Finally, additional lines may support conservation for endangered primate species. Unfortunately, approximately 75% of non-human primate species’ populations are decreasing in size, and about 60% are under the risk of extinction in the near term (Estrada et al., 2017). Genome sequencing studies can assist conservation efforts to maximize genetic diversity retained in future generations. The extent to which repositories of cultured cells can also contribute to conservation efforts remains to be seen (Benirschke, 1984), but there are some promising avenues. Cell lines from apes have long been useful for understanding immunological differences (Lawlor et al., 1990), and recent studies of Zika virus in human and chimpanzee organoids suggest that organoids may be useful for studying species specific viral tropism and screening for protective drugs (Cugola et al., 2016). Similarly, pluripotent stem cells and organoids could support toxicology screens in other primates to identify compounds that may have species-specific influences on development (Walker et al., 2015). In the more distant future, iPSCs may also support the production of gametes for conservation purposes (Stanton et al., 2019), as proposed for the Northern rhinoceros (Ben-Nun et al., 2011; Hildebrandt et al., 2018). Thus, generating and distributing larger collections spanning many individuals from all ape species may support conservation efforts and represents an opportunity to learn a tremendous amount about our own specializations and vulnerabilities.
ORGANOID MODELS OF BRAIN DEVELOPMENT
Organoid models can capture cellular diversity, tissue organization, and aspects of function across diverse organ systems (Clevers, 2016). These models can be applied to characterizing how genetic diversity segregating within humans and diverging across species influences brain development. This section considers the status of brain organoid models and their fidelity to the developing brain.
Developmental Processes in Brain Organoids
Normal development occurs through natural processes of self-assembly and signaling between neighboring cells. Although these processes may be difficult to fully recapitulate in cell culture, pioneering studies demonstrated that mouse pluripotent stem cells aggregated together and exposed to minimal patterning cues could produce organoids containing structures resembling embryonic forebrain, optic cup, and pituitary epithelium (Eiraku et al., 2008; 2011; Suga et al., 2011; Watanabe et al., 2005). The forebrain organoids contained germinal zone-like structures, diverse neuronal cell types, and even spatial gradients (Eiraku et al., 2008), and the optic cup organoids further mimicked morphogenesis by invaginating in the absence of a lens to form neural retina and adjacent pigment epithelium (Eiraku et al., 2011). Subsequently, these protocols were adapted to human (Kadoshima et al., 2013; Lancaster et al., 2013) and extended to contain cells from many brain regions in individual whole brain organoids (Cederquist et al., 2019; Renner et al., 2017) or to resemble specific brain regions, including hippocampus and choroid plexus (Sakaguchi et al., 2015), spinal cord (Ogura et al., 2018), pituitary gland (Ozone et al., 2016), ventral telencephalon (Kim et al., 2019; Sloan et al., 2018; Xiang et al., 2017), thalamus (Xiang et al., 2019), hypothalamus and midbrain (Jo et al., 2016; Qian et al., 2018) and cerebellum (Muguruma et al., 2015).
Many studies have focused on cortical organoids and cortical-like regions that emerge in whole brain organoids. Because organoids are a relatively new model system, it is crucial to address what processes of normal development organoids recapitulate and what the opportunities are for improvement (Table 2). The appearance of a polarized neuroepithelium is one of the most striking features of cortical organoid models. Early in the patterning process, ventricular zone-like rosette structures containing radial glia spontaneously emerge in organoids. The arrangement of cells around these rosettes resembles the lateral ventricle of the embryonic telencephalon (Subramanian et al., 2017). Apical proteins localize to the inner lumen of these rosettes and radial glia-like cells undergo interkinetic nuclear migration dividing at the apical surface (Bershteyn et al., 2017; Lancaster et al., 2013). By 10 weeks of differentiation, radial glia also migrate away from the ventricle, express markers of outer subventricular zone radial glia (Qian et al., 2016), and undergo mitotic somal translocation (Bershteyn et al., 2017). These germinal zone structures and cell behaviors match observations of normal primate brain development (Hansen et al., 2010; Pollen et al., 2015), and have been used to study how patient genetic mutations influence cell behavior (Bershteyn et al., 2017; Blair et al., 2018; Iefremova et al., 2017).
Table 2.
Assessment of the fidelity of distinct features of cortical development in current organoid models.
| Category | Property | Correspondence in Organoids |
|---|---|---|
| Architecture | Ventricular and inner subventricular zones | Highly preserved |
| Outer subventricular zone | Preserved | |
| Intermediate zone and subplate | Absent, but cell types are present | |
| Cortical plate | Excitatory neuron compartment, but limited lamination | |
| Afferent and efferent projections | Limited, may require assembloids | |
| Cell types | Cell composition | Variable |
| Radial glia (ventricular and outer) | Preserved | |
| Intermediate progenitors | Preserved | |
| Excitatory neurons | Preserved, but subtypes require further characterization | |
| Inhibitory neurons | Preserved, but protocol dependent | |
| Astrocytes | Preserved | |
| Oligodendrocytes | Preserved, but protocol dependent | |
| Vasculature | Requires transplant or bioengineering | |
| Microglia | Likely requires transplant | |
| Cell type gene expression | Similar | |
| Developmental processes | Radial glia maturation | Present, but variable |
| Radial glia interkinetic nuclear migration | Preserved | |
| Radial glia mitotic somal translocation | Preserved | |
| Neuronal differentiation | Preserved | |
| Excitatory neuron migration | Present, requires more characterization | |
| Inhibitory neuron migration | Present, requires more characterization | |
| Dendritic spines | Present, requires more characterization | |
| Spontaneous activity | Present, requires long-term culture | |
| Myelination | Present, requires long-term culture | |
| Cell states | Cell cycle | Highly preserved |
| Gene co-expression networks | Highly preserved | |
| Glycolysis | Organoid-enriched | |
| ER Stress | Organoid-enriched | |
| Apoptosis | Organoid-enriched |
Neurons migrate from the ventricular zone-like structures of organoids outwards to the periphery. The outer zone of neurons in organoids has been described as a cortical plate-like region. Indeed neurons expressing multiple markers of deep and upper layer subtypes have been described in the periphery of cortical organoids at around 10 – 15 weeks (Qian et al., 2016). Nonetheless, the organization of neuronal migration and layer formation differs from primary tissue at comparable stages (Pollen et al., 2019). Organoid size is generally constrained to less than 5 mm in diameter, which is less than the typical span from the ventricle to the cortical plate during middle stages of neurogenesis (Pollen et al., 2019), and organoids often contain many rosettes, further limiting the migration distance of neurons. Within this compressed space, there is not a clearly demarcated intermediate zone or subplate, and markers of cortical layers often appear disorganized. In addition, non neuroectodermal-derived cells, including endothelial cells, pericytes, and microglia are missing as well subcortical targets of efferent connections and incoming afferent connections. Thus, tissue organization outside of the germinal zone and the representation of subcortical and non-neuroectodermal cell communities are active areas for improvement (Giandomenico et al., 2019).
Fidelity of Organoids to Normal Development
Single cell RNA sequencing of primary tissue samples and organoid models provides the opportunity to further evaluate the fidelity organoid models to developmental processes for the cell types that are present. Early single cell RNA sequencing studies indicated that the trajectory of neuronal differentiation observed in primary human samples (Pollen et al., 2014; Pollen et al., 2015) is well conserved in organoid models (Camp et al., 2015) and identified molecular signatures of outer radial glia-like cells (Bershteyn et al., 2017). More recently, several studies, across many additional individuals, indicate strong correlations of gene expression signatures between organoid and primary tissue and cell types (Pollen et al., 2019; Velasco et al., 2019; Yoon et al., 2019). For example, we found that the majority of gene co-expression modules present in primary samples independently emerge in organoid models, and that area-specific excitatory neurons appear spontaneously (Pollen et al., 2019). Nonetheless, several studies have observed variability in patterning across individuals (Osafune et al., 2008; Pollen et al., 2019; Quadrato et al., 2017). Some of the observed variability may be related to more permissive protocols (Velasco et al., 2019; Yoon et al., 2019), but this variability in composition does not appear to have a strong effect on gene expression in individual cell types. However, the observation that specific lines can be biased in their differentiation, regionalization, or patterning potential is an important consideration for comparative studies across species or of disease genotypes, particularly when phenotypes may involve differences in cell fate potential. Thus organoids already provide a useful model for examining how genetic changes influence gene expression in specific cell types, but the patterning of these types in reproducible combinations remains an active area of research (Velasco et al., 2019).
Importantly, strong differences in gene expression also exist between organoids and primary samples. Across three major protocols that vary in the extent of cortical patterning, organoid cells commonly exhibit increased glycolysis pathway gene expression and increased signatures of endoplasmic reticulum stress compared to primary cells of the same type (Pollen et al., 2019). These glycolysis signatures extend to newborn neurons that normally undergo oxidative phosphorylation (Zheng et al., 2016) and the signatures also include the unfolded protein response, but the influence of these ectopic pathways on neuronal maturation has not been examined in detail. Interestingly, the unfolded protein response is also induced in organoids after acute hypoxia (Paşca et al., 2019). Inhibiting the integrated stress response pathway by blocking EIF2A phosphorylation (Sidrauski et al., 2013) was discovered to be protective in hypoxic organoids, particularly in intermediate progenitor cells, which tend to be underrepresented in organoids relative to primary samples (Paşca et al., 2019). The same approach, or the use of other small molecules, could potentially be protective in organoids cultured at normoxia, and might bring gene expression and cell composition closer to normal development. Despite these ectopic stress pathways, it is often assumed that organoid cell types more faithfully adhere to normal development than cell types differentiated in monolayer. However, systematic comparisons of 2D and 3D approaches for the same lines by single cell RNA sequencing have not yet been performed. In addition, single cell RNA sequencing has been the main modality for comparison due to recent technological advances, but additional comparisons from chromatin accessibility (Amiri et al., 2018; Kanton et al., 2019) to microRNA (Nowakowski et al., 2018) to full isoform sequencing and proteomics (Bauernfeind et al., 2015) may be useful to further reveal aspects of organoid fidelity.
Overall, single cell RNA sequencing studies show that broad neuroectodermal derived cell types, gene co-expression networks, and trajectories spontaneously appear in organoids, but that patterning and metabolic differences may still lead to limitations. These limitations could be especially relevant to protein folding and neurodegenerative disorders in which the organoid stress response may already be sensitizing organoids (Halliday and Mallucci, 2015; Pollen et al., 2019). Thus, current forebrain organoid models recapitulate many features of early cortical germinal zones, but there are opportunities for histological and metabolic improvements.
GENERAL OPPORTUNITIES FOR IMPROVING ORGANOID MODELS
This section reviews general opportunities for improving organoid models. In particular, providing organoids with vasculature and microglia and incorporating a realistic extracellular matrix environment represent opportunities for improving tissue health and organization. In addition, both metabolism and the extracellular matrix represent properties that likely changed during primate brain expansion and diversification. Ultimately, a foundation of improved tissue health and organization will be required to further model many evolved traits and cellular interactions, as well as their dysfunction in disease.
Vascularization and microglia
Inclusion of non-neuroectodermal cell types may be crucial for improving organoid tissue quality. Vascularization, in particular, could improve the richness of developmental cell interactions and could potentially alleviate metabolic stress. In the developing brain, blood vessels invading from the pia play a role in both scaffolding and signaling in addition to supporting blood flow (Adams et al., 2015; Schmid et al., 2019). For example, vascular endothelial cells express proteins that can influence pyramidal neuron position, radial glia pial attachment, and astrocyte behavior (Segarra et al., 2018). However, generating vascularized tissues in a dish has been challenging.
Transplantation studies have long demonstrated that human pluripotent stem cell derived tissue can be vascularized in a mouse (Rossant and Papaioannou, 1984) and have recently highlighted that the transplant process reduces the cell death observed in cultured brain organoids (Mansour et al., 2018). Recent studies also indicate that it may be possible to mimic the developmental interplay between vasculature and neuroectoderm lineage cells by fusing brain organoids to blood vessel organoids (Wimmer et al., 2019), by seeding organoids with vascular cells (Pham et al., 2018; Schwartz et al., 2015), or by in situ generation of endothial cells via ETV2 overexpression within organoids (Cakir et al., 2019). These vascular cells may permit increased diffusion. However, without pumping oxygenated blood through organoid vessels, it remains unclear whether vasculature alone can alleviate the metabolic abnormalities of organoids. Alternatively, bioengineering approaches may support nutrient and oxygen delivery in organoids, using hydrogels (Grigoryan et al., 2019), subtractive fabrication (Sarig-Nadir et al., 2009) or carbohydrate glass as a sacrificial network scaffold (Miller et al., 2012a; Therriault et al., 2003). Of note, the human brain utilizes a large proportion of the body’s energy, and humans may have higher energetic demands than the other ape species (Pontzer et al., 2016; Preuss, 2011). Whether or not human-specific metabolic adaptations have recently evolved in brain cells has been difficult to study without access to primary tissue. Thus biologically and engineering inspired approaches to organoid vascularization could improve tissue health and organization and could potentially enable comparative studies of neurometabolism.
Microglia are also important for tissue homeostasis, acting not only as immune cells, but also contributing to the clearance of dead cells through phagocytosis, and the regulation of cell proliferation, differentiation, synapse formation, and pruning (Ginhoux and Prinz, 2015). Thus, microglia may respond to the increased apoptosis described in organoids by mitigating the necrotic core (Mansour et al., 2018), in addition to participating in normal developmental processes. However, microglia are derived from the yolk sac during embryonic development (Ginhoux and Garel, 2018), and have only rarely been observed in organoid protocols that pattern for neuroectoderm (Ormel et al., 2018). Three studies demonstrated that transplanted microglia migrate and integrate into brain organoids and physiologically respond to insults (Abud et al., 2017; Muffat et al., 2018; Song et al., 2019). Importantly, organoids containing transplanted microglia appeared to lack a necrotic core and displayed differential intracellular calcium signaling and increased response to pro-inflammatory stimuli compared to untransplanted organoids (Song et al., 2019). Together, these results suggest that brain organoids may provide the necessary microenvironment to support the long-term incorporation of other cerebral cell types, and hint that non-neuroectodermal cell types may improve tissue health and organization.
Extracellular Matrix and Bioprinting
Extracellular matrix (ECM) proteins play an important role in progenitor expansion and organization of developing brain circuits, providing both scaffolding and signaling sources (Pearlman and Sheppard, 1996). Fibronectin, for example, is present in the ventricular zone of the developing cortex and may promote cell division (Pearlman and Sheppard, 1996). Similarly, integrins play a key role in maintaining the high proliferative capacity of progenitors in the outer subventricular zone (Fietz et al., 2010). Indeed, primate outer subventricular zone radial glia strongly express a module of ECM genes that potentiate growth factor signaling, suggesting these cells may contribute to a self-sustaining progenitor niche away from the ventricle (Pollen et al., 2015). Organoid cells may endogenously produce important ECM genes that support self-organization. For example, organoid radial glia preserve expression of many ECM genes, and organoids contain a mesenchymal cell population that expresses DCN, BGN, OGN, and collagen genes found in meningeal fibroblasts (Pollen et al., 2019). However, the extent to which endogenous production of ECM proteins in organoids resembles that of primary tissue in protein types and localization has not been examined in detail.
Supplementing cultures with species- and tissue-specific ECM molecules could potentially improve the fidelity of organoid cell behavior and architecture. Many protocols already embed organoids in droplets of matrigel secreted by mouse sarcoma cells to induce extension of ventricular zones during early patterning (Lancaster et al., 2013). Whether the matrigel acts through scaffolding alone to induce ventricle expansion or also through other signaling pathways is unclear. Interestingly, the effects of ECM molecules may differ by species. For example, addition of HAPLN1, Lumican, and Collagen I, rapidly increases stiffness and induces folding in primary human organotypic cultures, but not in mouse or ferret cultures (Long et al., 2018). More generally, the encapsulation of neurons into gels derived from natural ECM molecules also leads to neurite extension and formation of active networks (Ma et al., 2004; Mooney et al., 2010). Distinct ECM molecules promote the survival of different cell types. Fibrinogen and thrombin gels, for example, promote the survival of neurons over glia cells (Mooney et al., 2010). Consistent with the importance of tissue-specific ECM signals, supplementing human brain organoids with porcine fetal brain, but not adult brain ECM extracts, results in higher baseline synchrony of neuronal activity, accelerated maturation, and long-term survival of projection neurons (Sood et al., 2019). New protocols that consider a combinatorial approach to ECM composition during aggregation and long-term culture could potentially improve the survival of neurons and the study of neuronal networks beyond modeling early circuit development. Similarly richer cell communities that include vasculature and meningeal fibroblasts may support further endogenous production of key ECM proteins.
Alternatively, the use of artificial hydrogels could provide scaffolding support and replace xenobiotic materials like matrigel with defined culture conditions. Such artificial hydrogels could potentially reduce cost and increase reproducibility, but may lack important bioactive molecules (Broguiere et al., 2019; Tsou et al., 2016). For example, polyethylene glycol hydrogels provide a particularly inexpensive and scalable platform for culture and can be augmented with bioactive components for reproducible organoid generation containing NPCs, microglia and endothelial cells (Barry et al., 2017a; Schwartz et al., 2015). As a proof of principle, encapsulation of primary neurons in polyethylene glycol and polysaccharide-based macroporous hydrogels showed a remarkably high level of neurite outgrowth and neuronal activity, despite the fact that the hydrogel scaffold lacked laminin peptides (Broguiere et al., 2019). Thus, ECM proteins likely play key roles in scaffolding and signaling during development that may shape cell fate potential and tissue organization, and there are encouraging signs that these can be harnessed endogenously and through engineering approaches to improve organoid models.
Finally, the proper cytoarchitecture of neuronal tissue is crucial for the correct function of the organism. For example, the reeler mouse, in which cortical layers are inverted, shows strong behavioral phenotypes, as well as impaired motor coordination, ataxia and tremors (D’Arcangelo et al., 1995; Salinger et al., 2003; Trommsdorff et al., 1999). While brain organoids recapitulate the sequential generation of deep layers followed by upper layer projection neurons, achieving proper cortical lamination has been challenging. 3D bioprinting could potentially enable engineering of cortical organization. In this approach, cells of interest are embedded into hydrogel filaments or other biomaterials and printed in a preprogrammed manner to obtain the desired cytoarchitecture (Grebenyuk and Ranga, 2019). Importantly, cell viability after bioprinting is high, reaching up 90% survival (Christensen et al., 2015). As a proof of principle, human neuronal stem cells have been bioprinted in bioink containing alginate, carboxymethyl-chitosan, and agarose (Gu et al., 2016). These cells form complex structures that upon in situ differentiation form synapses, neuronal networks and have spontaneous activity. Alternatively, the use of 2-photon driven polymerization of biocompatible materials has been used to directly induce the formation of thick, porous and complex scaffolds (Worthington et al., 2017). Neurons grown using this approach extend long processes through the pores and integrate as a system. Overall, there are many tradeoffs between guided self-assembly and engineering approaches, but combinations of the two may be crucial for further advances in tissue health and organization.
ORGANOID MODELS ENABLE COMPARATIVE STUDIES OF ORGANOGENESIS
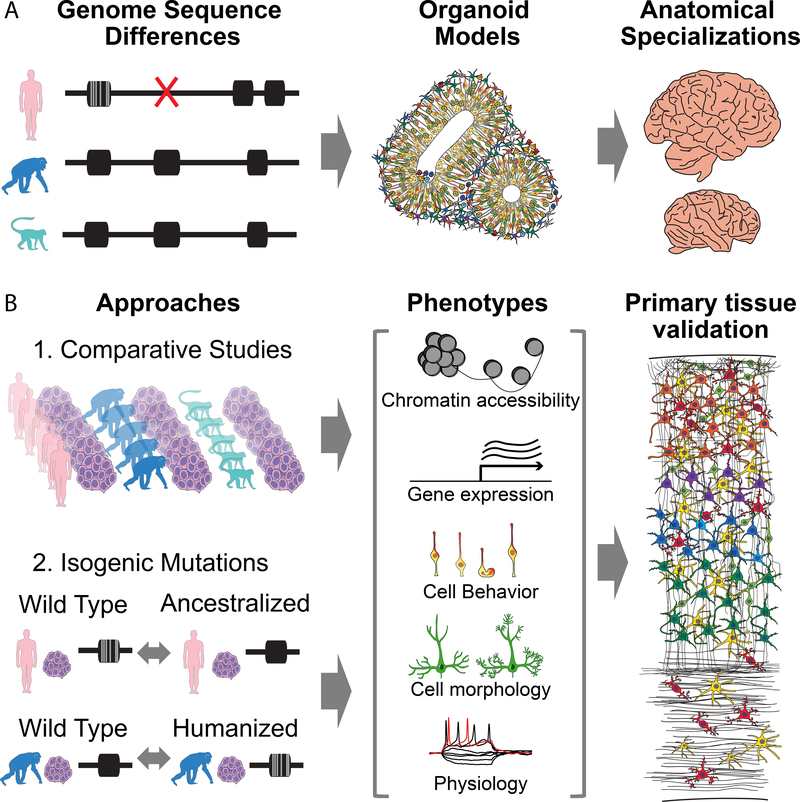
Comparative studies between human and primate brain organoids have only just begun, but could ultimately link causal genome sequence changes to changes in chromatin state, gene expression, and cell behavior underlying recent evolutionary changes (Figure 2A). This section outlines challenges facing comparative studies and provides two general approaches for determining the cellular origin of species differences and the causative genetic changes underlying developmental differences. Finally, this section highlights the first wave of comparative organoid studies that have focused on differences in progenitor cell behavior, gene expression and signaling pathway activation.
Figure 2. Organoid models help bridge the gap between human-specific genome sequence changes and evolved anatomical specializations of the human brain.
A) Human-specific genetic changes, including accelerated nucleotide substitutions, deletions, and duplications, as well as other insertions and inversions have been implicated in human evolution. Organoid models from humans and apes may provide the relevant cellular and genetic background for investigating the developmental consequences of these mutations. B) Two approaches for studying human-specific changes using organoids: First, comparative studies across sufficiently large numbers of biological replicates from human, chimpanzee, and other primate outgroups can reveal species differences during development that would otherwise be difficult to dissect. Second, re-creating evolutionary differences at individual loci in isogenic iPSC lines can directly dissect the function of individual mutations and may overcome some of the variability observed across iPSC lines. This can involve re-creating the ancestral state in human cells or examining the function of the human allele in a chimpanzee context. These iPSC lines can then be differentiated into organoid models or specific neural subtypes to dissect how the entirety of genetic differences across species or individual candidate loci influence phenotypes, including changes in chromatin state, gene expression, cell behavior, morphology, and physiology. Observed differences can then be anchored by validation in primary tissue from human and a non-human primate when possible.
Experimental Approaches and Challenges for Extending Comparative Studies
Performing comparative studies on the evolution of development in stem cell culture shares special challenges with disease modeling studies. In particular, there are many possible sources of variation in stem cell lines beyond genetic differences between source individuals and species, including cell culture derived mutations, epigenetic changes, possible differences in pluripotency state, and patterning differences between species (Carcamo-Orive et al., 2017). Therefore, ideal experimental designs could involve two paths (Figure 2B). In the first path, cross-species comparative studies should include a large number of individuals, while in the second path analysis of candidate mutations in isogenic lines can mitigate source of line to line variation. Both paths, to the extent possible, should anchor differences observed between species to changes also observed in primary brain tissue from human and non-human primates. Because ape primary samples are not available during early developmental stages, either developing macaque tissue or adult chimpanzee tissue has been used for validation in early studies. For example, two studies recently revealed that species differences in gene expression observed in iPSC-derived brain organoids or in cardiomyocytes across at least 8 individuals in each species recapitulate species-specific and tissue-specific differences observed in primary samples (Pavlovic et al., 2018; Pollen et al., 2019). Similarly, the duration of mitotic phases observed in organoids resembles that observed in primary human samples (Mora-Bermúdez et al., 2016). In addition to validating results in primary tissue, chimeric culture approaches, described further below, may be especially powerful to create a common garden environment that reduces batch effects and reveals cell intrinsic species differences (Otani et al., 2016).
Cross-species comparisons must consider several sources of biological variation that may be confounded with species differences in order to draw conclusions. Comparisons within species are generally safe in assuming that noise due to line-specific patterning or heterochrony is random and centered, but between species, these sources of variation can lead to systematic bias. This bias could involve systematic differences in patterning or maturation rate between species in a standardized protocol due to interspecies differences in the interaction between culture conditions and signaling pathways. These differences in patterning or maturation rates can then drive apparent differences in gene expression and other phenotypes that mainly relate to composition rather than to differences between cells of the same type (Barry et al., 2017b). Therefore, it is crucial to account for composition differences and systematic patterning biases in comparisons between species.
In principle, single cell RNA sequencing analyses enable comparison of molecularly defined cells of the same type between species. However, identifying cells of the same type between species through unbiased clustering approaches is challenging because species differences are reliably the major source of transcriptional variation, likely due to a combination of biological species differences, experimental confounds, and genome annotation artifacts. Recent analytical advances seek to overcome systematic sources of batch variation by enabling “alignment” and clustering of cells from multiple species in a shared low-dimensionality space (Haghverdi et al., 2018; Johansen and Quon 2019; Polański et al., 2019; Stuart et al., 2019; Welch et al., 2019). Nonetheless, these alignment methods involve heuristics, non-linear data transformations, distortions, and corrections that could over-fit putative orthologous cell types or depend on arbitrary user-defined parameters, without statistics describing the absolute or relative quality of the alignment output. Thus, in its current state, the field appears to need models that can “say no”. These models conceivably would provide an assessment of whether or not cells and clusters should be aligned in the first place by distinguishing and reporting variance due to technical artifacts, evolutionary changes, and underlying cell type homology. Current best practices should include evaluating how alignment results change between independent methods and evaluating whether co-clustered cells in joint analyses are indeed the most correlated groupings between independent analyses in each species. As a complementary approach, clustering-independent methods, such as comparing gene co-expression networks that independently arise between species (Pollen et al., 2019) may provide additional measures of homology.
Comparing gene expression between species is also challenging because of the superior genome assembly and transcriptome annotation in human, despite recent improvements in ape assemblies (Kronenberg et al., 2018). Approaches to overcome these biases include aligning reads only to orthologous exons, aligning reads to a consensus alignment between species with SNPs, indels and copy number variants masked, and aligning reads to the native genome of each species and mapping human annotations to other primates (Blekhman, 2012; Marchetto et al., 2013; Zhu et al., 2014, Pollen et al., 2019). Currently, each approach likely has strengths and weaknesses with respect to controlling for differences in gene models and read mapping rates between species versus capturing information from the greatest number of genes, including species-specific genes. Future studies may incorporate the best of each approach, considering consensus alignments for orthologous sequences, while also using higher quality de novo assembled genomes and improvements in gene annotations based on isoform sequencing across species and tissues to more fully account for molecular distinctions (Kronenberg et al., 2018). Overall, the sources of biological and technical variation that can confound species comparisons make validation of differences in available primary tissue samples particularly important.
As a second approach to reduce variation, human and chimpanzee iPSC lines may be directly modified to recreate ancestral or derived mutations respectively (Figure 2B). These modified lines can be compared directly to the unmodified (isogenic) version of the same line as a control to examine the necessity and sufficiency of human and chimpanzee mutations for particular phenotypes. For example, a recent study of human-specific NOTCH2NL duplications used CRISPR/CAS9 to ancestralize three human-specific functional paralogs of NOTCH2NL. Organoids with two homozygous and one heterozygous ancestral-like NOTCH2NL deletion showed premature neuronal differentiation, supporting a role for the NOTCH2NL duplicate genes in potentiating NOTCH signaling and maintaining proliferative divisions (Fiddes et al., 2018). Future studies may apply this isogenic approach more systematically to evaluate the quantitative contribution of candidate human-specific mutations to developmental cell behaviors. Together, both the comparative and the isogenic line approaches can now be applied across modalities from chromatin state, gene expression and proteomics, to cell behavior, morphology, and physiology, and these approaches will be particularly effective when anchored in species differences observed in primary tissue samples (Figure 2B).
Discovering Evolved Differences in Neural Progenitor Behavior
Studies of normal brain development have long predicted that changes in the proliferation of radial glia neural stem cells could underlie the evolutionary expansion of primate brains (Fish et al., 2008; Kriegstein et al., 2006; Rakic, 2009). Given the importance of radial glia in brain expansion and the preservation of radial glia cell behaviors and germinal zone structure in organoids, early studies have focused on this cell type. Comparisons of cell behavior in 2D, 3D, and chimeric co-culture suggest that human radial glia have an intrinsically greater proliferative capacity during deep layer neurogenesis than their macaque counterparts (Otani et al., 2016). The duration of cell cycle phases may also vary between species and could influence whether divisions are proliferative or neurogenic. Interestingly, time lapse imaging of cell cycle in neural progenitors of day 30 organoids revealed that the duration of metaphase, the portion of mitosis during which spindle fibers attach to chromosomes, lasts longer in human than in chimpanzee and orangutan neural stem cells (Mora-Bermúdez et al., 2016). However, by day 52, the duration of metaphase shrinks in human organoids to match that of other apes. Although these studies involved a small number of individuals from each species, the results hint that changes in human neural progenitor cell cycle behavior may influence early stages of cortical development when proliferative divisions are more common (Mora-Bermúdez et al., 2016).
Changes in signaling pathway activation could influence radial glia cell divisions and morphology. For example, human outer subventricular zone radial glia display increased activation of the LIFR-STAT3 self-renewal pathway compared to ventricular radial glia (Pollen et al., 2015). This signaling pathway is necessary for normal radial glia divisions in organotypic slice culture (Pollen et al., 2015) and activation of the pathway substantially amplifies the number of outer subventricular zone radial glia and the formation of upper layer neurons in organoids (Watanabe et al., 2017). Thus, this signaling pathway could contribute to the expansion of outer subventricular zone radial glia and their progeny among primates. Nonetheless, key genes in this pathway show conserved expression between human and macaque (Pollen et al., 2015). However, further analysis of gene expression patterns in human and chimpanzee radial glia across many individuals revealed divergent expression of genes involved with PI3K-AKT-mTOR signaling (Pollen et al., 2019). The mTOR pathway influences many aspects of cell growth, division, metabolism, and fate potential (Baser et al., 2019; Blair et al., 2018; Takei and Nawa, 2014) and displays enriched expression in outer subventricular zone radial glia compared to other cell types (Nowakowski et al., 2017). Immunohistochemistry and western blot analysis in organoids suggested that human radial glia undergo a quantitative increase in mTOR activation compared with chimpanzee radial glia that could be driven in part by increased expression of INSR and ITGB8. Analysis of primary tissue from human and macaque across stages of neurogenesis supported these findings and indicated that the changes in pathway activation may be derived on the human lineage (Pollen et al., 2019). Characterizing the upstream drivers and influence of these changes in pathway activation on radial glia cell behavior and morphology will require further study, but these early findings indicate that organoid models combined with analysis of primary tissue make it possible to compare the activation of major signaling pathways during neurogenesis.
Together, these early studies provide hints that human and ape organoid models can help us to decode cellular and molecular changes that recently evolved in the human lineage. While, current forebrain organoid models may be well suited to study how changes in radial glia of the germinal zone contribute to human brain expansion, possible improvements in organoid models discussed below may enable studies of richer phenotypes also associated with changes in human cognition.
IMPROVING ORGANOID MODELS TO STUDY TRAITS LINKED TO HUMAN COGNITION
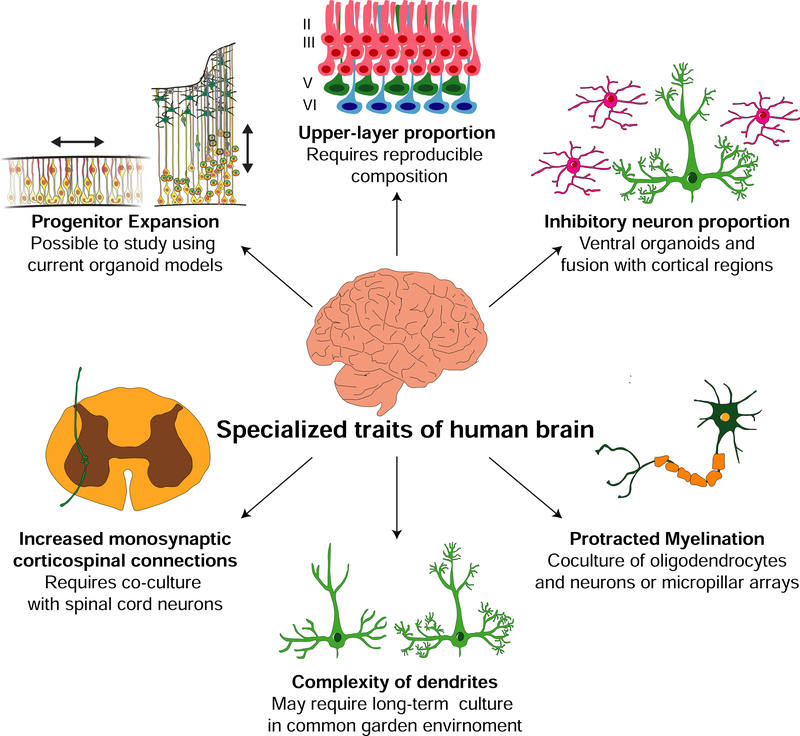
Careful histological comparisons (Bianchi et al., 2013; Raghanti et al., 2009) and transcriptomic studies in primary tissue (He et al., 2017; Konopka et al., 2012; Liu et al., 2012; Sousa et al., 2017b) have revealed several evolved postnatal differences between humans and other apes that influence cell composition, connectivity, and plasticity may be particularly important for human cognition beyond the quantitative increase in human brain size. However, studying the genetic and developmental sources of these evolved differences requires that organoid models mimic more sophisticated aspects of development beyond progenitor behavior in the germinal zone (Figure 3). In this section, we review promising opportunities in organoid modeling that could be applied to study specialized traits linked to cognition that recently changed in human evolution, including changes in cell type organization, connectivity relationships, protracted myelination, dendritic complexity, and physiology.
Figure 3. Reverse engineering the genetic and cellular basic of human cognitive specializations using organoid models will require methodological advances.
Comparative studies suggest several traits, including increased brain size, a reorganization of cell types, changes in connectivity patterns, protracted myelination and increased dendritic arborization contribute to human cognition. Current brain organoids recapitulate germinal zone structure and progenitor cell behaviors and have enabled comparative studies. On the other hand, studying the origin of the increased proportion of upper layer cortical excitatory neurons and cortical and thalamic inhibitory neurons will require reproducibility of cell fate potentials in organoid models across lines and species. In addition, studying traits like increased monosynaptic corticospinal connections and protracted myelination in humans depend on interactions between distinct cell types. These traits could be studied by co-culturing different cell types after differentiating them separately. Finally, the complexity of dendrites is a function of intrinsic properties of neurons, as well as connectivity with the surrounding cells. Studying differences across species therefore requires comparison of cells in the same background environment to determine whether the human increase in dendritic arborization and synaptogenesis are driven by cell-autonomous factors.
Cell type reorganization
Classical anatomical comparisons have suggested many evolutionary changes in cell type organization between primate and rodent brains. These differences may include changes in gene expression in homologous cell types, changes in the distribution of cell types, and possibly novel cell types in the adult brain. Comparative single cell genomic and spatial transcriptomic studies are poised to systematically reveal these quantitative and qualitative changes in available adult brain samples (Krienen et al., 2019). In parallel, organoids and in vitro models can help to evaluate the physiological relevance of species-specific changes in cell organization and to reveal the underlying developmental mechanisms, provided the cell types and brain regions can be modeled.
Upper layer cortical neurons provide connections across cortical areas. These cells have undergone changes in gene expression and abundance in primate evolution. For example, comparisons by in situ hybridization revealed that many markers specific to layer 5 cortical neurons in mouse are enriched in human upper layer cortical neurons (Zeng et al., 2012). Among these genes, the h-channel, HCN1, regulates subthreshold activity, and may contribute to the propagation of theta band frequency signals linked to learning and memory (Kalmbach et al., 2018; Rutishauser et al., 2010). Organoids containing mature upper layer neurons could support systematic studies on the functional consequences of these gene expression shifts. In addition to altered gene expression, the proportion of upper layer neurons has also expanded in humans (Aboitiz and Montiel, 2003; DeFelipe, 2011). Recent studies suggest that it is possible to compare fate potential and timing of neurogenesis between species (Kanton et al., 2019; Otani et al., 2016). Thus, the relative production of upper layer neurons and the cell intrinsic bias of radial glia clones to produce neurons of each subtype could be quantitatively examined in each species using in vitro models to identify the developmental basis for upper layer expansion, provided such protocols are reproducible across individuals.
As a subtle example of cell type reorganization, gene co-expression network analysis revealed increased expression of dopamine pathway related genes in human cortex and striatum compared to African apes (Sousa et al., 2017b). These findings support histological analyses suggesting changes in the distribution of tyrosine hydroxylase-expressing cells (Raghanti et al., 2009). The expression of tyrosine hydroxylase and DOPA decarboxylase does not confirm physiological dopamine production, but investigating the physiology of these cells in adult human and ape brains was impossible. Nevertheless, a subset of iPSC-derived inhibitory interneurons generated in a forebrain induction protocol resembled the cell types observed to have a divergent distribution in human forebrain. These in vitro-derived cells possessed the capacity to secrete dopamine, suggesting the observed change in cell organization could have important physiological consequences including a local source of dopamine in human cortex (Sousa et al., 2017b). Importantly, directly matching in vitro derived cells to primary cell types in cortex or striatum will be required to further establish the physiological relevance. Nonetheless, improvements in single cell RNA sequencing and impending comprehensive cell atlases across humans and apes should make it realistic to more precisely anchor in vitro-derived cell types and their measurable phenotypes to changes in organization and cell-type-specific gene expression whose consequences would otherwise be difficult to study in vivo.
More generally, the proportion of inhibitory interneurons has also changed substantially in cortical and thalamic evolution. However, studying these evolutionary changes using organoids may require modeling both the progenitors that give rise to inhibitory neurons and the behavior of interneurons in their ultimate destination. In rodents interneurons encompass ~15–20% of cortical neurons, while in primates they can represent over 40% of the neurons of the supragranular layers (DeFelipe, 2011). Similarly, interneurons represent about 5% of all neurons in the mouse thalamus compared to 30% in human thalamus (Arcelli et al., 1997). In both brain regions, the inhibitory neurons are not generated locally, but migrate in from a distinct brain region. In cortex, the inhibitory interneurons mainly migrate from the ventral telencephalon (Jones, 2009). In thalamus, less is known, but recent studies suggest the inhibitory neurons in sensory relay nuclei migrate from midbrain (Jager et al., 2016), while inhibitory neurons in higher order thalamic nuclei may also migrate from ventral telencephalon (Jager et al., 2019; Letinić and Kostović, 1997; Letinić and Rakic, 2001).
Cortical and thalamic organoids protocols mainly yield excitatory neurons (Pollen et al., 2019; Quadrato et al., 2017; Velasco et al., 2019; Xiang et al., 2019). However, new protocols enable production of organoids that resemble the medial ganglionic eminences, and fusion of these ventralized organoids with cortical organoids resulted in extensive interneuron migration towards the excitatory cells (Bagley et al., 2017; Birey et al., 2017; Xiang et al., 2017), resembling the in vivo context (Lim et al., 2018). Upon migration, the cells differentiated in all interneuron lineages, albeit at different than expected frequencies. For example, while parvalbumin-positive interneurons encompass ~40% of cortical interneurons (Rudy et al., 2010), they were observed at a low frequency and only after long culturing periods (>200 days in culture) (Bagley et al., 2017; Birey et al., 2017). This is in agreement with previous observations that parvalbumin-positive interneurons are rarely observed in vitro, and not observed in vivo until late stages of gestation (Maroof et al., 2013; Nicholas et al., 2013). Midbrain organoids have mainly been analyzed for dopaminergic neurons (Monzel et al., 2017; Qian et al., 2016) but do contain inhibitory neurons (Jo et al., 2016), although it remains unclear whether fusion with thalamic organoids would result in similar inhibitory neuron migration to thalamic organoids. Thus, region-specific organoids could enable comparative studies of evolutionary changes in inhibitory neuron production, but understanding the reorganization of these cell types in cortical and thalamic regions using organoids would still require improvements in modeling differentiation, maturation, and migration.
Overall, changes in cell type organization are challenging to model using organoids that lack the reproducibility of cell composition and tissue architecture of animal models. Nonetheless, organoids may enable modeling the physiological consequences of novel cell types or gene expression patterns whose role may be difficult to determine in comparisons of post-mortem primary tissue or in mouse model systems. Additionally, recent fusion experiments between organoid types hint that modeling migratory processes contributing to evolutionary changes in cell distribution may also now be tractable.
Increased subcortical and intracortical connectivity
Connectivity has also changed substantially in primate species. Cortical connections to motor neurons are thought to contribute to improved primate dexterity, while connections between cortical areas and through thalamus are thought to contribute to human cognition and behavioral flexibility (Rikhye et al., 2018; Striedter, 2005). Modeling evolutionary changes in these connections will require proper specification of source neurons and their targets. Although challenging, preliminary studies suggest organoids may permit modeling these important evolutionary differences.
Anterograde labeling experiments indicate that several large-brained primates have direct monosynaptic connections between corticospinal neurons and motor neurons of the spinal cord (Bortoff and Strick, 1993; Lemon, 2019; Lemon et al., 2002; Porter, 1987). These connections may contribute to the delicate and dexterous movements of primates (Bortoff and Strick, 1993; Porter, 1987). In particular, direct and possibly human-specific monosynaptic corticobulbar connections may contribute to human vocal dexterity and language production (Kuypers, 1958). However, we still do not know whether these connectivity differences are caused by intrinsic changes in the cortical projection neurons themselves, changes in target-derived factors, or changes in activity patterns that favor enlarged cortical networks in competition with subcortical nuclei (Herculano-Houzel, et al., 2016). Interestingly, rodents transiently develop cortico-motoneuronal connections (Maeda et al., 2016; Murabe et al., 2018; Ohno et al., 2004), and blocking Sema6D-PlexA1 signaling in mice prevents elimination of these transient cortico-motoneuron axons, leading to increased dexterity (Gu et al., 2017). Recent studies demonstrate that organoids grown in an liquid-air interface generate nerve tracks reminiscent of subcortical and intracortical projections (Giandomenico et al., 2019). Co-culture of these organoids with mouse spinal cord showed that these “organoid-fugal” axons can innervate spinal cord explants and elicit responses in adjacent muscles (Giandomenico et al., 2019). Similarly, recent studies found that organoids positioned either in two wells separated by a custom made chamber, or at two ends of an artificial hydrogel column, extended reciprocal axons to each other forming strong fasciculated bundles (Cullen et al., 2019; Kirihara et al., 2019). Strikingly, electrical stimulation of one organoid elicited a calcium response in the axonal fascicle and the distal organoid, suggesting the formation of complex networks (Cullen et al., 2019; Kirihara et al., 2019). Although the specificity of these connections has not been probed in detail, these studies hint that organoids may eventually enable comparative studies of cellular and genetic mechanisms contributing to subcortical and intracortical connectivity.
Importantly, connectivity patterns vary substantially across cortical areas. For example, human vocal dexterity may in particular depend on subcortical projection neurons from laryngeal motor cortex that specifically project through the corticobulbar tract to laryngeal motor neurons in the nucleus ambiguus (Simonyan, 2014). Similarly, connections between higher order association areas and between association areas and dorsal thalamus also have stereotyped areal patterns (Buckner and Krienen, 2013). Recent studies indicate that cortical area-specific excitatory neurons emerge prior to sensory experience and are further diversified in adult cortex (Cadwell et al., 2019; Nowakowski et al., 2017; Tasic et al., 2018). Thus, organoid based studies of connectivity evolution may require precise specification of appropriate cortical areas. Recently, we developed a classifier based on primary cell data and observed that excitatory projection neurons with area-specific molecular identities spontaneously emerge in human and ape organoids (Pollen et al., 2019). However, their organization within organoids and the fidelity of their connections has not yet been explored. In addition, we do not yet have a full catalog of area-specific signatures in primary tissue as early studies compared anterior and posterior cortical areas, and we do not know the extent to which molecular distinctions are arranged in discrete boundaries or gradients. Delivery of patterning molecules such as FGF8 to cortically specified organoids may permit further control of organoid arealization. Indeed, recent studies have directly engineered a patterning center-like structure to achieve dorsoventral gradients (Cederquist et al., 2019) and advances in microfluidics (Brassard and Lutolf, 2019) or in optical control of gene expression (Repina et al., 2019) could further support more reliable control over organoid arealization.
Properly specified areal organoids could then be examined for their connection ability to ex vivo mouse tissue (Giandomenico et al., 2019), to other region-specific organoids (e.g., medulla, spinal cord, dorsal thalamus, or higher order association areas) (Kirihara et al., 2019), or to in vivo targets through transplantation into mouse (Espuny-Camacho et al., 2013; Mansour et al., 2018; Mostajo-Radji and Pollen, 2018; Real et al., 2018; Stenevi et al., 1976). Early proof-of-principle studies using all three approaches indicate that mechanistic comparisons of connectivity development and evolution in humans and apes may soon be possible through control of areal, regional and subtype identity of pre- and postsynaptic neurons.
Protracted Myelination
Changes in myelination patterns may also be crucial for human cognition. The formation of a myelin sheath surrounding axons supports rapid communication of electrical signals across long distances (Rosenbluth, 1999). In the neocortex, myelin is gradually acquired throughout postnatal development following the activation of oligodendrocytes. Delayed myelination may support extended periods of plasticity (Fields, 2015; McGee et al., 2005), but may also contribute to multiple sclerosis susceptibility (Bove, 2018). In addition, disrupted myelination and white matter may contribute to other disorders including schizophrenia and bipolar disorder (Davis et al., 2003; Mighdoll, et al., 2015; Tønnesen et al., 2018; Van den Huevel et al., 2019). Importantly, the myelination process is particularly protracted in humans compared to other primates, continuing past age 25 (Miller et al., 2012b). In contrast, in chimpanzees, myelination density reaches its peak at the time of sexual maturation, around age 10 (Miller et al., 2012b). Similarly, at birth the human neocortex has almost no myelination while the chimpanzee cortex is ~20% myelinated (Miller et al., 2012b). These myelination differences could contribute to the accelerated divergence of lipid content reported in the human brain compared with other primates (Bozek et al., 2015).
Whether these species differences in myelination are controlled by general hormones, by neurons that signal to developing oligodendrocytes, or by cell intrinsic differences in oligodendrocytes themselves remains unknown. The heterogeneity of myelination across cortical layers and areas suggests the potential for local regulation. For example, humans contain a greater proportion of upper-layer excitatory neurons, which undergo myelination later than the deep cortical layers (Tomassy et al., 2014), and prefrontal cortex myelinates much later than primary visual cortex (Turner, 2019). Interestingly, pyramidal neurons may have the capacity to directly instruct their own myelination, as different neuronal subtypes have distinct myelination patterns (Micheva et al., 2016; Tomassy et al., 2014; Zonouzi et al., 2019), and there is limited molecular heterogeneity in oligodendrocytes across brain regions compared with neurons (Zeisel et al., 2018). In addition, when myelinated sheaths are ablated from individual axons, the axons ultimate remyelinate in their original patterns (Auer et al., 2018). Thus divergence in gene expression and composition of pyramidal neurons could potentially contribute to species-specific myelination timelines, but the source of these differences has been difficult to identify.
Comparing oligodendrocyte lineage cells or pyramidal neurons from a range of primate species in common garden environments could help to reveal the cellular source of species differences in myelination rates. For example, it would be interesting to examine whether human and chimpanzee neurons transplanted to a common garden environment in the mouse cortex undergo different rates of myelination instructed by species-specific differences in neuronal maturation and signaling. In addition, several protocols now exist to generate oligodendrocytes from embryonic stem cells, which could be used to examine cell intrinsic changes in oligodendrocyte maturation across species (Douvaras and Fossati, 2015; Numasawa-Kuroiwa et al., 2014; Stacpoole et al., 2013). These oligodendrocytes can myelinate neurons and also micropillar nanofiber arrays that mimic axons for increased experimental control (Mei et al., 2014).
Modeling myelination in organoids has been challenging, but recent studies demonstrated that the addition of specific growth factors, nutrients and hormones to organoid culturing media resulted in the expansion and differentiation of oligodendrocytes, and ultimately in vitro myelination in the organoid (Madhavan et al., 2018; Marton et al., 2019). In addition, the use of media with physiological levels of glucose accelerated the appearance of oligodendrocyte precursor cells by several weeks (Kim et al., 2019). Although these studies showed the progressive acquisition of myelin basic protein and the compaction of the myelin sheath, fine structural refinement including the presence of nodes of Ranvier was not reported in these organoids, even at 30 weeks in culture (Madhavan et al., 2018; Marton et al., 2019). The decades long timeline of human in vivo maturation, the long term nature of these in vitro experiments, and the uncertainty of the underlying cellular source of species differences makes future comparative myelination experiments challenging, However, the importance of the phenotype to cognition and to neurological disorders makes future studies worthwhile. Co-transplant studies of human and ape iPSC derived neural progenitors and oligodendrocyte precursors to mouse could provide an avenue for current studies as organoid models of these later stages continue to improve.
Increased Synaptogenesis and Dendritic Complexity
Rates of synaptic maturation, pruning, and overall synapse number also vary across primates. Comparative histological and transcriptomic studies across developmental stages suggest that cortical synapse number continues to increase in humans until age four to eight, but already peaks in the first year of chimpanzee and macaque postnatal development (Huttenlocher and Dabholkar, 1997; Liu et al., 2012; Petanjek et al., 2011; Rakic et al., 1986; Somel et al., 2009), while other studies also suggest an early acceleration of synaptogenesis in humans (Zhu et al., 2018). Continued developmental remodeling of synapses occurs well beyond adolescence in human prefrontal cortex, and the prolonged retention of early features of synaptic plasticity in humans, termed synaptic neoteny, has been implicated in learning and cognition (Petanjek et al., 2011). Synapse number varies dramatically across cortical areas within a species, with increased synaptic density and dendritic complexity in higher order association areas of cortex compared with primary processing areas (Bianchi et al., 2013; Elston et al., 2011; Hrvoj-Mihic et al., 2014). Despite substantial variation among cortical areas, comparisons between species of upper layer pyramidal neurons in prefrontal and temporal cortex, reveal that human excitatory neurons have larger dendritic arbors, longer dendritic segments and more dendritic spines than their chimpanzee and macaque counterparts in homologous areas (Bianchi et al., 2013; Deitcher et al., 2017; Elston et al., 2001; Mohan et al., 2015; Semendeferi et al., 2011; Teffer et al., 2013). This expansion is thought to increase the potential for integration of signals and abstraction of information in microcircuits of human prefrontal cortex, a key feature of human cognition. Interestingly, the increased dendritic complexity of human pyramidal neurons compared to rodent may also create additional neuronal compartments that perform non-linear transformations in individual neurons (Beaulieu-Laroche et al., 2018; Eyal et al., 2018; Tran-Van-Minh et al., 2015), but future work is required to examine whether this structure engenders richer computation and whether this compartmentalization also differs between humans and other primates.
Organoids and iPSC-derived pyramidal neurons may help to further reveal evolutionary changes in synaptic development and dendritic complexity and underlying mechanisms, (Hrvoj-Mihic et al., 2013; Hrvoj-Mihic et al., 2014). Indeed, a recent study indicates that molecular signatures related to early synaptogenesis and neuronal maturation appear later in human organoids than in chimpanzee and macaque organoids, suggesting that early timing differences following neurogenesis are preserved in vitro (Kanton et al., 2019). Nonetheless, neuronal maturation phenotypes are particularly challenging to study because synaptic development occurs over very long time periods, with many distinct phases from exuberant synapse formation through pruning and maintenance, and because the surrounding environment influences key aspects of development. To overcome these challenges, a recent study directly transplanted a mixture of human and chimpanzee iPSC-derived neural progenitor cells into the mouse cortex (Marchetto et al., 2019). The mouse cortical environment may provide connectivity partners crucial for normal synaptic development (Espuny-Camacho et al., 2013; Linaro et al., 2019; Mansour et al., 2018; Mostajo-Radji and Pollen, 2018; Real et al., 2018) and may reduce cell death compared to organoid models over long term experiments (Mansour et al 2018). Importantly transplanting a labeled mix of human and chimpanzee cells to the same cortical area enabled direct assessment of cell intrinsic differences in synaptic development. In this common garden environment, the human pyramidal neurons developed increased dendritic arborization and spine number, but this was visible only 8 – 19 weeks post-transplant. Interestingly, multielectrode arrays revealed that human in vitro cultures initially showed a lower mean firing rate, but ultimately acquired a higher firing rate after 6 weeks in culture (Marchetto et al., 2019). Together, these results are consistent with prevailing ideas about an extended duration of synaptic development in humans. Future studies can expand on this work by directly anchoring measurement of pyramidal neurons to area-specific cell types, by including additional individuals and stages of development, and by measuring phenotypes related to plasticity. The strategy of transplanting chimeric mixtures of human and ape cells into mouse pioneered in this study may be particularly beneficial for analyzing these synaptic phenotypes, but improvements in organoid maturation may also support further in vitro analyses.
Towards comparative neurophysiology and circuit activity
The computational function of the brain is an electrochemical property that arises from neuronal activity, cell composition, and connectivity relationships. If we hope to ultimately understand how differences between species’ cognition arise, we must examine the evolution of the basic circuitry of brain cells. Only comparative studies of in vivo physiology can currently address these questions at the circuit level. For example, recent single cell electrophysiology studies in the adult human and macaque brain suggest increased efficiency of information capacity in human anterior cingulate cortex neurons at the expense of robust coding (Pryluk et al., 2019). However, the capacity to perform in vivo human and primate studies at scale and to perform perturbations is limited. New tools, such as those supported by the BRAIN Initiative enabling further studies will ultimately be required to address these questions (Koroshetz et al., 2018).
Comparative ex vivo electrophysiology on surgical resections may also represent a powerful method for comparing physiological properties between human and macaque (Ting et al., 2018). For example, recordings of hippocampal slices from human and mouse suggest a possible divergence of spike-timing-dependent plasticity rules (Testa- Silva et al., 2010). Similarly, recordings in upper cortical layers indicate a functional role for HCN1 in dendritic conductance, with a possibly greater contribution of these channels to subthreshold synaptic integration and theta oscillations in human cortex (Kalmbach et al., 2018). However, these ex vivo resections are difficult to access and scale as an experimental model system, particularly at early stages of brain development. Organoids may help to fill these gaps, but the extent to which physiological properties of organoid neurons relate to normal physiological properties of developing brain remains unclear. Several studies have taken the first step of recording extracellular field potentials in brain organoids using multielectrode arrays, and one organoid study suggests a predictive relationship to oscillatory dynamics observed during normal brain development (Kawada et al., 2017; Mansour et al., 2018; Monzel et al., 2017; Quadrato et al., 2017; Trujillo et al., 2019). While organoid neural circuits will never share the receptive fields and precise connections that form in normal development, organoids offer the flexibility to selectively introduce cell types and to combine neurons from specific brain regions. This flexibility may allow organoid studies to complement in vivo and slice physiology by enabling reductionist studies of how cell type composition influences circuit properties outside of the normal space of circuits tolerated in vivo. In addition, organoids could enable studies of how homologous cell types from humans and apes perturb similar circuits in different ways. Speculatively, it may also be possible to construct novel architectures of neural circuits outside of the constraints of a normal brain (Hofman, 2014) that possess interesting properties and functions.
Both ex vivo brain slices and organoid cultures are particularly amenable to simultaneous recording and stimulation of large pools of neurons. One approach would involve viral delivery of cell-type-specific reporters that enable optical analysis of calcium signaling or voltage changes in specific cell types (Dimidschstein et al., 2016; Förster et al., 2017; Graybuck et al., 2019). Recently, genetically-encoded voltage indicators coupled with high speed cameras have been applied in vivo to support optogenetic manipulation and recording of neuronal activity in small pools of neurons (Adam et al., 2019; Piatkevich et al., 2019), and this approach could likely be extended to larger populations and timescales ex vivo. Following physiological analysis, reconstruction of the molecular identity of constituent cell types can be performed using spatial transcriptomic approaches or through interpretation of reporter gene expression. Ultimately, such longitudinal observation of circuit activity may reveal further species differences and could be used to study the cellular and molecular origin of these differences.
OUTLOOK
This review has considered human and primate-specific evolutionary differences whose cellular and genetic origin could conceivably be studied using organoid models. Indeed, several comparative studies have already revealed potential changes in progenitor behavior, signaling pathway activation, and gene expression. However, studying these and other traits will require many general improvements in organoid models including improved fidelity to primary tissue, more precise patterning of brain regions down the level of cortical areas or subcortical nuclei, and engineering communities of cell types that enable the study of neuronal migration and connectivity. It is remarkable how far unguided self-assembly can already go to mimic normal development in a dish, but incorporating engineering approaches such as delivering precise morphogen gradients and scaffolds could further add reproducibility to these processes. The possibility of chimeric culture of iPSC lines between species together in the same organoid or in a mouse environment may help to reduce experimental confounds and isolate cell intrinsic species differences across a range of molecular and cell behavior phenotypes. Meanwhile, genome engineering approaches now allow for systematic reconstruction of genetic changes in organoid models that could ultimately lead to a quantitative genetic understanding of evolved human specializations. In addition, organoid models offer the opportunity for high-dimensional phenotyping and longitudinal analysis of neurogenesis and neural circuit formation across genotypes and species that is only just beginning to be exploited. Multielectrode arrays provide one example of long-term analysis, but imaging of organoids that express fluorescent reporters of specific cell types and of processes such as neuronal activity provide a path to rich phenotyping of otherwise inaccessible species differences and to the evaluation of candidate causal mutations and their effect sizes. Thomas Henry Huxley and Richard Owen debated the uniqueness of the human brain and the origin of specialized features over 150 years ago (Gross, 1993; Huxley, 1863). Brain organoids from humans and great apes combined with genome engineering now make it possible to begin to dissect the cellular and genetic origin of these recently-evolved differences.
HIGHLIGHTS.
Brain organoids offer a window into the mechanisms underlying of evolution.
Organoids capture cell diversity and interactions that occur during development.
Organoid modeling could be applied to study specialized traits linked to cognition.
ACKNOWLEDGMENTS
We would like to thank Aparna Bhaduri, Bryan Pavlovic, Jenelle Wallace, Tomasz Nowakowski, Sofie Salama, Mircea Teodorescu and David Haussler for comments on this manuscript. This work was supported by a gift from the Schmidt Family Foundation, the National Institutes of Health (NIH) grant DP2OD027518 to A.A.P. and the UCSF Program for Breakthrough Biomedical Research (PBBR). M.A.M.-R. was supported by the NIH National Center for Advancing Translational Sciences, through UCSF-CTSI 5TL1TR001871-04.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aboitiz F, Montiel J (2003). One hundred million years of interhemispheric communication: the history of the corpus callosum. Brazilian Journal of Medical and Biological Research = Revista Brasileira De Pesquisas Medicas E Biologicas, 36(4), 409–420. [DOI] [PubMed] [Google Scholar]
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, et al. (2017). iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron, 94(2), 278–293.e9. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Y, Kim JJ, Lou S, Zhao Y, Xie ME, Brinks D, et al. (2019). Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature, 569(7756), 413–417. 10.1038/s41586-019-1166-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DL, Piserchia V, Economides JR, Horton JC (2015). Vascular Supply of the Cerebral Cortex is Specialized for Cell Layers but Not Columns. Cerebral Cortex (New York, N.Y. : 1991), 25(10), 3673–3681. 10.1093/cercor/bhu221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, et al. (2018). Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362(6420), eaat6720. doi: 10.1126/science.aat6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcelli P, Frassoni C, Regondi MC, De Biasi S, Spreafico R (1997). GABAergic neurons in mammalian thalamus: a marker of thalamic complexity? Brain Research Bulletin, 42(1), 27–37. [DOI] [PubMed] [Google Scholar]
- Auer F, Vagionitis S, Czopka T (2018). Evidence for Myelin Sheath Remodeling in the CNS Revealed by In Vivo Imaging. Current Biology, 28(4), 549–559.e3. 10.1016/j.cub.2018.01.017 [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, Reyzer ML, Caprioli RM, Ely JJ, Babbitt CC, Wray GA, et al. , High spatial resolution proteomic comparison of the brain in humans and chimpanzees. The Journal of Comparative Neurology 523, 2043–2061. 10.1002/cne.23777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley JA, Reumann D, Bian S, Lévi-Strauss J, Knoblich JA (2017). Fused cerebral organoids model interactions between brain regions. Nature Methods, 14(7), 743–751. 10.1038/nmeth.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Marioni JC, Blekhman R, Stephens M, Gilad Y (2010). Functional comparison of innate immune signaling pathways in primates. PLoS Genetics, 6(12), e1001249 10.1371/journal.pgen.1001249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C, Schmitz MT, Propson NE, Hou Z, Zhang J, Nguyen BK, et al. (2017a). Uniform neural tissue models produced on synthetic hydrogels using standard culture techniques. Experimental Biology and Medicine (Maywood, N.J.), 242(17), 1679–1689. 10.1177/1535370217715028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C, Schmitz MT, Jiang P, Schwartz MP, Duffin BM, Swanson S, et al. (2017b). Species-specific developmental timing is maintained by pluripotent stem cells ex utero. Developmental Biology, 423(2), 101–110. 10.1016/j.ydbio.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baser A, Skabkin M, Kleber S, Dang Y, Gülcüler Balta GS, Kalamakis G, et al. (2019). Onset of differentiation is post-transcriptionally controlled in adult neural stem cells. Nature, 566(7742), 100–104. 10.1038/s41586-019-0888-x [DOI] [PubMed] [Google Scholar]
- Beaulieu-Laroche L, Toloza EHS, van der Goes M-S, Lafourcade M, Barnagian D, Williams ZM, et al. (2018). Enhanced Dendritic Compartmentalization in Human Cortical Neurons. Cell, 175(3), 643–651.e14. 10.1016/j.cell.2018.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nun IF, Montague SC, Houck ML, Tran HT, Garitaonandia I, Leonardo TR, et al. (2011). Induced pluripotent stem cells from highly endangered species. Nature Methods 8, 829–831. 10.1038/nmeth.1706 [DOI] [PubMed] [Google Scholar]
- Benirschke K (1984). The frozen zoo concept. Zoo Biology, 3(4), 325–328. 10.1002/zoo.1430030405 [DOI] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR (2017). Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell, 20(4), 435–449.e4. 10.1016/j.stem.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi S, Stimpson CD, Bauernfeind AL, Schapiro SJ, Baze WB, McArthur MJ, et al. (2013). Dendritic morphology of pyramidal neurons in the chimpanzee neocortex: regional specializations and comparison to humans. Cerebral Cortex (New York, N.Y. : 1991), 23(10), 2429–2436. 10.1093/cercor/bhs239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature, 545(7652), 54–59. 10.1038/nature22330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Hockemeyer D, Bateup HS (2018). Genetically engineered human cortical spheroid models of tuberous sclerosis. Nature Medicine, 24(10), 1568–1578. 10.1038/s41591-018-0139-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake LE, Thomas SM, Blischak JD, Hsiao CJ, Chavarria C, Myrthil M, et al. (2018). A comparative study of endoderm differentiation in humans and chimpanzees. Genome Biology, 19(1), 162 10.1186/s13059-018-1490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R (2012). A database of orthologous exons in primates for comparative analysis of RNA-seq data. Available from Nature Precedings <http://hdl.handle.net/10101/npre.2012.7054.1> [Google Scholar]
- Bortoff GA, Strick PL (1993). Corticospinal terminations in two new-world primates: further evidence that corticomotoneuronal connections provide part of the neural substrate for manual dexterity. The Journal of Neuroscience : the Official Journal of the Society for Neuroscience, 13(12), 5105–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove RM (2018). Why monkeys do not get multiple sclerosis (spontaneously): An evolutionary approach. Evolution, Medicine, and Public Health, 2018(1), 43–59. 10.1093/emph/eoy002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd JL, Skove SL, Rouanet JP, Pilaz L-J, Bepler T, Gordân R, et al. (2015). Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Current Biology, 25(6), 772–779. 10.1016/j.cub.2015.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozek K, Wei Y, Yan Z, Liu X, Xiong J, Sugimoto M, et al. (2015). Organization and Evolution of Brain Lipidome Revealed by Large-Scale Analysis of Human, Chimpanzee, Macaque, and Mouse Tissues. Neuron, 85(4), 695–702. 10.1016/j.neuron.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Brassard JA, Lutolf MP (2019). Engineering Stem Cell Self-organization to Build Better Organoids. Cell Stem Cell, 24(6), 860–876. 10.1016/j.stem.2019.05.005 [DOI] [PubMed] [Google Scholar]
- Broguiere N, Husch A, Palazzolo G, Bradke F, Madduri S, Zenobi-Wong M (2019). Macroporous hydrogels derived from aqueous dynamic phase separation. Biomaterials, 200, 56–65. 10.1016/j.biomaterials.2019.01.047 [DOI] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM (2013). The evolution of distributed association networks in the human brain. Trends in Cognitive Sciences, 17(12): 648–665. 10.1016/j.tics.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Cadwell CR, Bhaduri A, Mostajo-Radji MA, Keefe MG, Nowakwoski TJ (2019). Development and Arealization of the Cerebral Cortex. Neuron, 103(6): 980–1004. 10.1016/j.neuron.2019.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakir B, Xiang Y, Tanaka Y, Kural MH, Parent M, Kang Y-J, et al. (2019). Engineering of human brain organoids with a functional vascular-like system. Nature Methods, 16: 1169–1175. 10.1038/s41592-019-0586-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Pnas, 112(51), 15672–15677. 10.1073/pnas.1520760112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Gnerre S, Veeramah KR, Lorente-Galdos B, Huddleston J, et al. (2014). Gibbon genome and the fast karyotype evolution of small apes. Nature, 513(7517), 195–201. 10.1038/nature13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, et al. (2017). Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Non-genetic Determinants of Heterogeneity. Cell Stem Cell, 20(4), 518–532.e9. 10.1016/j.stem.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, Studer L (2019). Specification of positional identity in forebrain organoids. Nature Biotechnology, 37(4), 436–444. 10.1038/s41587-019-0085-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, Kim J-E, Lambert N, de Marchena J, et al. (2012). Inhibition of SRGAP2 Function by Its Human-Specific Paralogs Induces Neoteny during Spine Maturation. Cell, 149(4), 923–935. 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Xu C, Chai W, Zhang Z, Fu J, Huang Y (2015). Freeform inkjet printing of cellular structures with bifurcations. Biotechnology and Bioengineering, 112(5), 1047–1055. 10.1002/bit.25501 [DOI] [PubMed] [Google Scholar]
- Clevers H (2016). Modeling Development and Disease with Organoids. Cell, 165(7), 1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JLM, Guimarães KP, et al. (2016). The Brazilian Zika virus strain causes birth defects in experimental models. Nature, 534(7606), 267–271. 10.1038/nature18296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen DK, Gordian-Velez WJ, Struzyna LA, Jgamadze D, Lim J, Wofford KL, et al. (2019). Bundled Three-Dimensional Human Axon Tracts Derived from Brain Organoids. iScience, 21, 57–67. 10.1016/j.isci.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T (1995). A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature, 374(6524), 719–723. 10.1038/374719a0 [DOI] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, et al. (2003). White matter changes in schizophrenia: evidence for myelin-related dysfunction. Archives of General Psychiatry, 60(5), 443–456. 10.1001/archpsyc.60.5.443 [DOI] [PubMed] [Google Scholar]
- DeFelipe J (2011). The evolution of the brain, the human nature of cortical circuits, and intellectual creativity. Frontiers in Neuroanatomy, 5, 29 10.3389/fnana.2011.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitcher Y, Eyal G, Kanari L, Verhoog MB, Atenekeng Kahou GA, Mansvelder HD, et al. (2017). Comprehensive Morpho-Electrotonic Analysis Shows 2 Distinct Classes of L2 and L3 Pyramidal Neurons in Human Temporal Cortex. Cerebral Cortex (New York, N.Y. : 1991), 27(11), 5398–5414. 10.1093/cercor/bhx226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, et al. (2012). Evolution of Human-Specific Neural SRGAP2 Genes by Incomplete Segmental Duplication. Cell, 149(4), 912–922. 10.1016/j.cell.2012.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi G-A, Guo L, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nature Publishing Group, 19(12), 1743–1749. 10.1038/nn.4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Fossati V (2015). Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nature Protocols, 10(8), 1143–1154. 10.1038/nprot.2015.075 [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, et al. (2011). Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature, 472(7341), 51–56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell, 3(5), 519–532. 10.1016/j.stem.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, DeFelipe J (2001). The pyramidal cell in cognition: a comparative study in human and monkey. Journal of Neuroscience, 21(17), RC163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Benavides-Piccione R, Elston A, Manger PR, DeFelipe J (2011). Pyramidal cells in prefrontal cortex of primates: marked differences in neuronal structure among species. Frontiers in Neuroanatomy, 5, 2 10.3389/fnana.2011.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, et al. (2013). Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron, 77(3), 440–456. 10.1016/j.neuron.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Estrada A, Garber PA, Rylands AB, Roos C, Fernandez-Duque E, Di Fiore A, et al. (2017). Impending extinction crisis of the world’s primates: Why primates matter. Science Advances, 3(1), e1600946 10.1126/sciadv.1600946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal G, Verhoog MB, Testa-Silva G, Deitcher Y, Benavides-Piccione R, DeFelipe J, et al. (2018). Human Cortical Pyramidal Neurons: From Spines to Spikes via Models. Frontiers in Cellular Neuroscience, 12, 181 10.3389/fncel.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL, et al. (2018). Human-Specific NOTCH2NL Genes Affect Notch Signaling and Cortical Neurogenesis. Cell, 173(6), 1356–1369.e22. 10.1016/j.cell.2018.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, et al. (2019). Structurally Conserved Primate LncRNAs Are Transiently Expressed during Human Cortical Differentiation and Influence Cell-Type-Specific Genes. Stem Cell Reports, 12(2), 245–257. 10.1016/j.stemcr.2018.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields RD (2015). A new mechanism of nervous system plasticity: activity-dependent myelination. Nature Reviews: Neuroscience, 16(12), 756–767. 10.1038/nrn4023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fietz SA, Kelava I, Vogt J, Wilsch-Bräuninger M, Stenzel D, Fish JL, et al. (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nature Neuroscience, 13(6), 690–699. 10.1038/nn.2553 [DOI] [PubMed] [Google Scholar]
- Fish JL, Dehay C, Kennedy H, Huttner WB (2008). Making bigger brains-the evolution of neural-progenitor-cell division. Journal of Cell Science, 121(Pt 17), 2783–2793. 10.1242/jcs.023465 [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, et al. (2015). Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science, 347(6229), 1465–1470. 10.1126/science.aaa1975 [DOI] [PubMed] [Google Scholar]
- Förster D, Dal Maschio M, Laurell E, Baier H (2017). An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nature Communications, 8(1), 116 10.1038/s41467-017-00160-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, et al. (2015). A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. eLife, 4, e07103 10.7554/eLife.07103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, et al. (2019). Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nature Publishing Group, 22(4), 669–679. 10.1038/s41593-019-0350-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Garel S (2018). The mysterious origins of microglia. Nature Publishing Group, 21(7), 897–899. 10.1038/s41593-018-0176-3 [DOI] [PubMed] [Google Scholar]
- Ginhoux F, Prinz M (2015). Origin of microglia: current concepts and past controversies. Cold Spring Harbor Perspectives in Biology, 7(8), a020537 10.1101/cshperspect.a020537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck LT, Sedeño-Cortés A, Nguyen TN, Walker M, Szelenyi E, Lenz G, et al. (2019). Prospective, brain-wide labeling of neuronal subclasses with enhancer-driven AAVs. bioRxiv, 525014 10.1101/525014 [DOI] [Google Scholar]
- Grebenyuk S, Ranga A (2019). Engineering Organoid Vascularization. Frontiers in Bioengineering and Biotechnology, 7, 39 10.3389/fbioe.2019.00039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, et al. (2019). Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science, 364(6439), 458–464. 10.1126/science.aav9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CGC (1993). Huxley versus Owen: the hippocampus minor and evolution. Trends in Neurosciences (Vol. 16, pp. 493–498). 10.1016/0166-2236(93)90190-W [DOI] [PubMed] [Google Scholar]
- Gu Q, Tomaskovic-Crook E, Lozano R, Chen Y, Kapsa RM, Zhou Q, et al. (2016). Functional 3D Neural Mini-Tissues from Printed Gel-Based Bioink and Human Neural Stem Cells. Advanced Healthcare Materials, 5(12), 1429–1438. 10.1002/adhm.201600095 [DOI] [PubMed] [Google Scholar]
- Gu Z, Kalambogias J, Yoshioka S, Han W, Li Z, Kawasawa YI, et al. (2017). Control of species-dependent cortico-motoneuronal connections underlying manual dexterity. Science, 357(6349), 400–404. 10.1126/science.aan3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L, Lun ATL, Morgan MD, & Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nature Biotechnology, 36(5), 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday M, Mallucci GR (2015). Review: Modulating the unfolded protein response to prevent neurodegeneration and enhance memory. Neuropathology and Applied Neurobiology, 41(4), 414–427. 10.1111/nan.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature, 464(7288), 554–561. 10.1038/nature08845 [DOI] [PubMed] [Google Scholar]
- He Z, Han D, Efimova O, Guijarro P, Yu Q, Oleksiak A, et al. (2017). Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nature Publishing Group, 20(6), 886–895. 10.1038/nn.4548 [DOI] [PubMed] [Google Scholar]
- Hellen EHB, Kern AD (2015). The role of DNA insertions in phenotypic differentiation between humans and other primates. Genome Biology and Evolution, 7(4), 1168–1178. 10.1093/gbe/evv012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S (2012). The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Pnas, 109, 10661–10668. 10.1073/pnas.1201895109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S, Kaas JH, de Oliveira-Souza R (2016). Corticalization of motor control in humans is a consequence of brain scaling in primate evolution. The Journal of Comparative Neurology 524(3), 448–455. 10.1002/cne.23792 [DOI] [PubMed] [Google Scholar]
- Hildebrandt TB, Hermes R, Colleoni S, Diecke S, Holtze S, Renfree MB, et al. (2018). Embryos and embryonic stem cells from the white rhinoceros. Nature Communications, 9(1), 2589 10.1038/s41467-018-04959-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman MA (2014). Evolution of the human brain: when bigger is better. Frontiers in Neuroanatomy, 8(14), 15 10.3389/fnana.2014.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvoj-Mihic B, Bienvenu T, Stefanacci L, Muotri AR, Semendeferi K (2013). Evolution, development, and plasticity of the human brain: from molecules to bones. Frontiers in Human Neuroscience, 7, 707 10.3389/fnhum.2013.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvoj-Mihic B, Marchetto MCN, Gage FH, Semendeferi K, Muotri AR (2014). Novel tools, classic techniques: evolutionary studies using primate pluripotent stem cells. Biological Psychiatry, 75(12), 929–935. 10.1016/j.biopsych.2013.08.007 [DOI] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS (1997). Regional differences in synaptogenesis in human cerebral cortex. The Journal of Comparative Neurology, 387(2), 167–178. [DOI] [PubMed] [Google Scholar]
- Huxley TH (1863). Evidence as to Man’s Place in Nature. London: Macmillan. [Google Scholar]
- Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, et al. (2017). An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Reports, 19(1), 50–59. 10.1016/j.celrep.2017.03.047 [DOI] [PubMed] [Google Scholar]
- Jager P, Calpin P, Durmishi X, Shimogori T, Delogu A (2019). Inhibitory interneurons distribute widely across the mouse thalamus and form ontogenetic spatial clusters. bioRxiv, 651745 10.1101/651745 [DOI] [Google Scholar]
- Jager P, Ye Z, Yu X, Zagoraiou L, Prekop H-T, Partanen J, et al. (2016). Tectal-derived interneurons contribute to phasic and tonic inhibition in the visual thalamus. Nature Communications, 7(1), 13579 10.1038/ncomms13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran H-D, Göke J, et al. (2016). Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell, 19(2), 248–257. 10.1016/j.stem.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen N, & Quon G (2019). scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biology, 20(1), 166–21. 10.1186/s13059-019-1766-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EG (2009). The origins of cortical interneurons: mouse versus monkey and human. Cerebral Cortex (New York, N.Y. : 1991), 19(9), 1953–1956. 10.1093/cercor/bhp088 [DOI] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Pnas, 110(50), 20284–20289. 10.1073/pnas.1315710110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanton S, Boyle MJ, He Z, Santel M, Weigert A, Sanchís-Calleja F, et al. (2019). Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422. doi: 10.1038/s41586-019-1654-9 [DOI] [PubMed] [Google Scholar]
- Kalmbach BE, Buchin A, Long B, Close J, Nandi A, Miller JA, et al. (2018). h-Channels Contribute to Divergent Intrinsic Membrane Properties of Supragranular Pyramidal Neurons in Human versus Mouse Cerebral Cortex. Neuron, 100(5), 1194–1208.e5. 10.1016/j.neuron.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawada J, Kaneda S, Kirihara T, Maroof A, Levi T, Eggan K, et al. (2017). Generation of a Motor Nerve Organoid with Human Stem Cell-Derived Neurons. Stem Cell Reports, 9(5), 1441–1449. 10.1016/j.stemcr.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Xu R, Padmashri R, Dunaevsky A, Liu Y, Dreyfus CF, Jiang P (2019). Pluripotent Stem Cell-Derived Cerebral Organoids Reveal Human Oligodendrogenesis with Dorsal and Ventral Origins. Stem Cell Reports, 12(5), 890–905. 10.1016/j.stemcr.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirihara T, Luo Z, Chow SYA, Misawa R, Kawada J, Shibata S, et al. (2019). A Human Induced Pluripotent Stem Cell-Derived Tissue Model of a Cerebral Tract Connecting Two Cortical Regions. iScience, 14, 301–311. 10.1016/j.isci.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, et al. (2012). Human-specific transcriptional networks in the brain. Neuron, 75(4), 601–617. 10.1016/j.neuron.2012.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koroshetz W, Gordon J, Adams A, Beckel-Mitchener A, Churchill J, Farber G, et al. (2018). The State of the NIH BRAIN Initiative. Journal of Neuroscience, 38(29), 6427–6438. 10.1523/JNEUROSCI.3174-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V (2006). Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nature Reviews: Neuroscience, 7(11), 883–890. 10.1038/nrn2008 [DOI] [PubMed] [Google Scholar]
- Krienen FM, Goldman M, Zhang Q, del Rosario R, Florio M, Machold R, et al. ,. Innovations in Primate Interneuron Repertoire. bioRxiv, 709501 10.1101/709501 [DOI] [Google Scholar]
- Kronenberg ZN, Fiddes IT, Gordon D, Murali S, Cantsilieris S, Meyerson OS, et al. (2018). High-resolution comparative analysis of great ape genomes. Science, 360(6393), eaar6343 10.1126/science.aar6343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers HG (1958). Corticobular connexions to the pons and lower brain-stem in man: an anatomical study. Brain, 81(3), 364–388. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature, 501(7467), 373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor DA, Warren E, Ward FE, Parham P (1990). Comparison of class I MHC alleles in humans and apes. Immunological Reviews, 113, 147–185. [DOI] [PubMed] [Google Scholar]
- Lemon R (2019). Recent advances in our understanding of the primate corticospinal system. F1000Research, 8, 274 10.12688/f1000research.17445.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon RN, Maier MA, Armand J, Kirkwood PA, Yang HW (2002). Functional differences in corticospinal projections from macaque primary motor cortex and supplementary motor area. Advances in Experimental Medicine and Biology, 508(Chapter 48), 425–434. 10.1007/978-1-4615-0713-0_48 [DOI] [PubMed] [Google Scholar]
- Letinić K, Kostović I (1997). Transient fetal structure, the gangliothalamic body, connects telencephalic germinal zone with all thalamic regions in the developing human brain. The Journal of Comparative Neurology, 384(3), 373–395. [DOI] [PubMed] [Google Scholar]
- Letinić K, Rakic P (2001). Telencephalic origin of human thalamic GABAergic neurons. Nature Neuroscience, 4(9), 931–936. 10.1038/nn0901-931 [DOI] [PubMed] [Google Scholar]
- Lim L, Mi D, Llorca A, Marín O (2018). Development and Functional Diversification of Cortical Interneurons. Neuron, 100(2), 294–313. 10.1016/j.neuron.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linaro D, Vermaercke B, Iwata R, Ramaswamy A, Libé-Philippot B, Boubakar L, et al. (2019). Xenotransplanted Human Cortical Neurons Reveal Species-Specific Development and Functional Integration into Mouse Visual Circuits. Neuron In Press. 10.1016/j.neuron.2019.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Somel M, Tang L, Yan Z, Jiang X, Guo S, et al. (2012). Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques. Genome Research, 22(4), 611–622. 10.1101/gr.127324.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long KR, Newland B, Florio M, Kalebic N, Langen B, Kolterer A, et al. (2018). Extracellular Matrix Components HAPLN1, Lumican, and Collagen I Cause Hyaluronic Acid-Dependent Folding of the Developing Human Neocortex. Neuron, 99(4), 702–719.e6. 10.1016/j.neuron.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Ma W, Fitzgerald W, Liu Q-Y, O’Shaughnessy TJ, Maric D, Lin HJ, et al. (2004). CNS stem and progenitor cell differentiation into functional neuronal circuits in three-dimensional collagen gels. Experimental Neurology, 190(2), 276–288. 10.1016/j.expneurol.2003.10.016 [DOI] [PubMed] [Google Scholar]
- Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, et al. (2018). Induction of myelinating oligodendrocytes in human cortical spheroids. Nature Methods, 15(9), 700–706. 10.1038/s41592-018-0081-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Fukuda S, Kameda H, Murabe N, Isoo N, Mizukami H, et al. (2016). Corticospinal axons make direct synaptic connections with spinal motoneurons innervating forearm muscles early during postnatal development in the rat. The Journal of Physiology, 594(1), 189–205. 10.1113/JP270885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, et al. (2018). An in vivo model of functional and vascularized human brain organoids. Nature Biotechnology, 36(5), 432–441. 10.1038/nbt.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, et al. (2013). Differential L1 regulation in pluripotent stem cells of humans and apes. Nature, 503(7477), 525–529. 10.1038/nature12686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Hrvoj-Mihic B, Kerman BE, Yu DX, Vadodaria KC, Linker SB, et al. (2019). Species-specific maturation profiles of human, chimpanzee and bonobo neural cells. eLife, 8, 1748 10.7554/eLife.37527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying S-W, Ganat YM, Merkle FT, et al. (2013). Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell, 12(5), 559–572. 10.1016/j.stem.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, et al. (2019). Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nature Publishing Group, 22(3), 484–491. 10.1038/s41593-018-0316-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee AW, Yang Y, Fischer QS, Daw NW, Strittmatter SM (2005). Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science, 309(5744), 2222–2226. 10.1126/science.1114362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, et al. (2011). Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature, 471(7337), 216–219. 10.1038/nature09774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Fancy SPJ, Shen Y-AA, Niu J, Zhao C, Presley B, et al. (2014). Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nature Medicine, 20(8), 954–960. 10.1038/nm.3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R (2007). Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nature Biotechnology, 25(10), 1177–1181. 10.1038/nbt1335 [DOI] [PubMed] [Google Scholar]
- Micheva KD, Wolman D, Mensh BD, Pax E, Buchanan J, Smith SJ, Bock DD (2016). A large fraction of neocortical myelin ensheathes axons of local inhibitory neurons. eLife, 5, 3347 10.7554/eLife.15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighdoll MI, Tao R, Kleinman JE, Hyde TM (2015). Myelin, myelin-related disorders, and psychosis. Schizophrenia Research, 161(1), 85–93. 10.1016/j.schres.2014.09.040 [DOI] [PubMed] [Google Scholar]
- Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, et al. (2012a). Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nature Materials, 11(9), 768–774. 10.1038/nmat3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, et al. (2012b). Prolonged myelination in human neocortical evolution. Pnas, 109(41), 16480–16485. 10.1073/pnas.1117943109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan H, Verhoog MB, Doreswamy KK, Eyal G, Aardse R, Lodder BN, et al. (2015). Dendritic and Axonal Architecture of Individual Pyramidal Neurons across Layers of Adult Human Neocortex. Cerebral Cortex (New York, N.Y. : 1991), 25(12), 4839–4853. 10.1093/cercor/bhv188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, et al. (2017). Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Reports, 8(5), 1144–1154. 10.1016/j.stemcr.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R, Tawil B, Mahoney M (2010). Specific fibrinogen and thrombin concentrations promote neuronal rather than glial growth when primary neural cells are seeded within plasma-derived fibrin gels. Tissue Engineering. Part A, 16(5), 1607–1619. 10.1089/ten.TEA.2009.0372 [DOI] [PubMed] [Google Scholar]
- Mora-Bermúdez F, Badsha F, Kanton S, Camp JG, Vernot B, Köhler K, et al. (2016). Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife, 5, 166 10.7554/eLife.18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostajo-Radji MA, Pollen AA (2018). Physiological Models of Human Neuronal Development and Disease. Neuron, 100(5), 1025–1027. [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Omer A, Durbin A, Bosch I, Bakiasi G, et al. (2018). Human induced pluripotent stem cell-derived glial cells and neural progenitors display divergent responses to Zika and dengue infections. Pnas, 115(27), 7117–7122. 10.1073/pnas.1719266115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y (2015). Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Reports, 10(4), 537–550. 10.1016/j.celrep.2014.12.051 [DOI] [PubMed] [Google Scholar]
- Murabe N, Mori T, Fukuda S, Isoo N, Ohno T, Mizukami H, et al. (2018). Higher primate-like direct corticomotoneuronal connections are transiently formed in a juvenile subprimate mammal. Scientific Reports, 8(1), 16536 10.1038/s41598-018-34961-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R, Ohnuki M, Kuroki K, Ito H, Hirai H, Kitajima R, et al. (2018). Derivation of induced pluripotent stem cells in Japanese macaque (Macaca fuscata). Sci Rep 8, 12187. doi: 10.1038/s41598-018-30734-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nater A, Mattle-Greminger MP, Nurcahyo A, Nowak MG, de Manuel M, Desai T, et al. (2017). Morphometric, Behavioral, and Genomic Evidence for a New Orangutan Species. Current Biology, 27(22), 3487–3498.e10. 10.1016/j.cub.2017.09.047 [DOI] [PubMed] [Google Scholar]
- Navara CS, Chaudhari S, McCarrey JR (2018). Optimization of culture conditions for the derivation and propagation of baboon (Papio anubis) induced pluripotent stem cells. PLoS ONE, 13(3): e0193195 10.1371/journal.pone.0193195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, et al. (2013). Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell, 12(5), 573–586. 10.1016/j.stem.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, et al. (2017). Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science, 358(6368), 1318–1323. 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Rani N, Golkaram M, Zhou HR, Alvarado B, Huch K, et al. (2018). Regulation of cell-type-specific transcriptomes by microRNA networks during human brain development. Nature Neuroscience 21(12):1784–1792. doi: 10.1038/s41593-018-0265-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numasawa-Kuroiwa Y, Okada Y, Shibata S, Kishi N, Akamatsu W, Shoji M, et al. (2014). Involvement of ER stress in dysmyelination of Pelizaeus-Merzbacher Disease with PLP1 missense mutations shown by iPSC-derived oligodendrocytes. Stem Cell Reports, 2(5), 648–661. 10.1016/j.stemcr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Sakaguchi H, Miyamoto S, Takahashi J (2018). Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development, 145(16), dev162214 10.1242/dev.162214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno T, Maeda H, Sakurai M (2004). Regionally specific distribution of corticospinal synapses because of activity-dependent synapse elimination in vitro. Journal of Neuroscience, 24(6), 1377–1384. 10.1523/JNEUROSCI.3903-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormel PR, Vieira de Sá R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, et al. (2018). Microglia innately develop within cerebral organoids. Nature Communications, 9(1), 4167 10.1038/s41467-018-06684-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, et al. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nature Biotechnology, 26(3), 313–315. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ (2016). 2D and 3D Stem Cell Models of Primate Cortical Development Identify Species-Specific Differences in Progenitor Behavior Contributing to Brain Size. Cell Stem Cell, 18(4), 467–480. 10.1016/j.stem.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, et al. (2016). Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nature Communications, 7(1), 10351 10.1038/ncomms10351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca AM, Park J-Y, Shin H-W, Qi Q, Revah O, Krasnoff R, et al. (2019). Human 3D cellular model of hypoxic brain injury of prematurity. Nature Medicine, 25(5), 784–791. 10.1038/s41591-019-0436-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic BJ, Blake LE, Roux J, Chavarria C, Gilad Y (2018). A Comparative Assessment of Human and Chimpanzee iPSC-derived Cardiomyocytes with Primary Heart Tissues. Scientific Reports, 8(1), 15312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlman AL, Sheppard AM (1996). Extracellular matrix in early cortical development. Progress in Brain Research, 108, 117–134. [PubMed] [Google Scholar]
- Petanjek Z, Judaš M, Šimic G, Rašin M-R, Uylings HBM, Rakic P, Kostović I (2011). Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Pnas, 108(32), 13281–13286. 10.1073/pnas.1105108108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham MT, Pollock KM, Rose MD, Cary WA, Stewart HR, Zhou P, et al. (2018). Generation of human vascularized brain organoids. Neuroreport, 29(7), 588–593. 10.1097/WNR.0000000000001014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Bensussen S, Tseng H-A, Shroff SN, Lopez-Huerta VG, Park D, et al. (2019). Population imaging of neural activity in awake behaving mice in multiple brain regions. bioRxiv, 616094 10.1101/616094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polański K, Park J-E, Young MD, Miao Z, Meyer KB, & Teichmann SA (2019). BBKNN: Fast Batch Alignment of Single Cell Transcriptomes. Bioinformatics, 36, 411 10.1093/bioinformatics/btz625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot M-A, Coppens S, Pedersen JS, et al. (2006). An RNA gene expressed during cortical development evolved rapidly in humans. Nature, 443(7108), 167–172. 10.1038/nature05113 [DOI] [PubMed] [Google Scholar]
- Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, et al. (2019). Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell, 176(4), 743–756.e17. 10.1016/j.cell.2019.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, et al. (2015). Molecular identity of human outer radial glia during cortical development. Cell, 163(1), 55–67. 10.1016/j.cell.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature Biotechnology, 32(10), 1053–1058. 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontzer H, Brown MH, Raichlen DA, Dunsworth H, Hare B, Walker K, et al. (2016). Metabolic acceleration and the evolution of human brain size and life history. Nature, 533(7603), 390–392. 10.1038/nature17654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R (1987). The Florey lecture, 1987. Corticomotoneuronal projections: synaptic events related to skilled movement. Proceedings of the Royal Society of London. Series B, Biological Sciences, 231(1263), 147–168. 10.1098/rspb.1987.0039 [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, et al. (2013). Great ape genetic diversity and population history. Nature, 499(7459), 471–475. 10.1038/nature12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, et al. (2015). Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell, 163(1), 68–83. 10.1016/j.cell.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss TM (2011). The human brain: rewired and running hot. Annals of the New York Academy of Sciences, 1225(S1), E182–E191. 10.1111/j.1749-6632.2011.06001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryluk R, Kfir Y, Gelbard-Sagiv H, Fried I, Paz R (2019). A Tradeoff in the Neural Code across Regions and Species. Cell, 176(3), 597–609.e18. 10.1016/j.cell.2018.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming G-L (2018). Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nature Protocols, 13(3), 565–580. 10.1038/nprot.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. (2016). Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell, 165(5), 1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature, 545(7652), 48–53. 10.1038/nature22047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghanti MA, Spocter MA, Stimpson CD, Erwin JM, Bonar CJ, Allman JM, et al. (2009). Species-specific distributions of tyrosine hydroxylase-immunoreactive neurons in the prefrontal cortex of anthropoid primates. Neuroscience, 158(4), 1551–1559. 10.1016/j.neuroscience.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P (2009). Evolution of the neocortex: a perspective from developmental biology. Nature Reviews: Neuroscience, 10(10), 724–735. 10.1038/nrn2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Eckenhoff MF, Zecevic N, Goldman-Rakic PS (1986). Concurrent overproduction of synapses in diverse regions of the primate cerebral cortex. Science, 232(4747), 232–235. 10.1126/science.3952506 [DOI] [PubMed] [Google Scholar]
- Ramaswamy K, Yik WY, Wang X-M, Oliphant EN, Lu W, Shibata D, et al. (2015). Derivation of induced pluripotent stem cells from orangutan skin fibroblasts. BMC Research Notes, 8(1), 577 10.1186/s13104-015-1567-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Marchetto MC, Caron M, Chen S-H, Busche S, Kwan T, et al. (2017). Conserved expression of transposon-derived non-coding transcripts in primate stem cells. BMC Genomics, 18(1), 214 10.1186/s12864-017-3568-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real R, Peter M, Trabalza A, Khan S, Smith MA, Dopp J, et al. (2018). In vivo modeling of human neuron dynamics and Down syndrome. Science, 362(6416), eaau1810 10.1126/science.aau1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M, Lancaster MA, Bian S, Choi H, Ku T, Peer A, et al. (2017). Self-organized developmental patterning and differentiation in cerebral organoids. The EMBO Journal, 36(10), 1316–1329. 10.15252/embj.201694700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repina NA, Bao X, Zimmermann JA, Joy DA, Kane RS, Schaffer DV (2019). Optogenetic control of Wnt signaling for modeling early embryogenic patterning with human pluripotent stem cells. bioRxiv, 665695 10.1101/665695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhye RV, Gilra A, Halassa MM (2018). Thalamic regulation of switching between cortical representations enables cognitive flexibility. Nature Publishing Group, 21(12), 1753–1763. 10.1038/s41593-018-0269-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbluth J (1999). A brief history of myelinated nerve fibers: one hundred and fifty years of controversy. Journal of Neurocytology, 28(4–5), 251–262. [DOI] [PubMed] [Google Scholar]
- Rossant J, Papaioannou VE (1984). The relationship between embryonic, embryonal carcinoma and embryo-derived stem cells. Cell Differentiation, 15(2–4), 155–161. [DOI] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J (2010). Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Developmental Neurobiology, 71(1), 45–61. 10.1002/dneu.20853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser U, Ross IB, Mamelak AN, Schuman EM (2010). Human memory strength is predicted by theta-frequency phase-locking of single neurons. Nature, 464(7290), 903–907. 10.1038/nature08860 [DOI] [PubMed] [Google Scholar]
- Sakaguchi H, Kadoshima T, Soen M, Narii N, Ishida Y, Ohgushi M, et al. (2015). Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nature Communications, 6(1), 8896 10.1038/ncomms9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Matsui M, Mikami A, Malkova L, Hamada Y, Tomonaga M, et al. (2013). Developmental patterns of chimpanzee cerebral tissues provide important clues for understanding the remarkable enlargement of the human brain. Proceedings. Biological Sciences, 280(1753), 20122398–20122398. 10.1098/rspb.2012.2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinger WL, Ladrow P, Wheeler C (2003). Behavioral phenotype of the reeler mutant mouse: effects of RELN gene dosage and social isolation. Behavioral Neuroscience, 117(6), 1257–1275. 10.1037/0735-7044.117.6.1257 [DOI] [PubMed] [Google Scholar]
- Sarig-Nadir O, Livnat N, Zajdman R, Shoham S, Seliktar D (2009). Laser photoablation of guidance microchannels into hydrogels directs cell growth in three dimensions. Biophysical Journal, 96(11), 4743–4752. 10.1016/j.bpj.2009.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F, Barrett MJP, Jenny P, Weber B (2019). Vascular density and distribution in neocortex. NeuroImage, 197, 792–805. 10.1016/j.neuroimage.2017.06.046 [DOI] [PubMed] [Google Scholar]
- Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Santos Costa V, et al. (2015). Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Pnas, 112(40), 12516–12521. 10.1073/pnas.1516645112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra M, Aburto MR, Cop F, Llaó-Cid C, Härtl R, Damm M, et al. (2018). Endothelial Dab1 signaling orchestrates neuro-glia-vessel communication in the central nervous system. Science, 361(6404), eaao2861 10.1126/science.aao2861 [DOI] [PubMed] [Google Scholar]
- Semendeferi K, Teffer K, Buxhoeveden DP, Park MS, Bludau S, Amunts K, et al. (2011). Spatial organization of neurons in the frontal pole sets humans apart from great apes. Cerebral Cortex (New York, N.Y. : 1991), 21(7), 1485–1497. 10.1093/cercor/bhq191 [DOI] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, et al. (2013). Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife, 2, e00498 10.7554/eLife.00498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonyan K (2014). The laryngeal motor cortex: its organization and connectivity. Current Opinion in Neurobiology, 28, 15–21. 10.1016/j.conb.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Andersen J, Paşca AM, Birey F, Paşca SP (2018). Generation and assembly of human brain region-specific three-dimensional cultures. Nature Protocols, 13(9), 2062–2085. 10.1038/s41596-018-0032-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, et al. (2009). Transcriptional neoteny in the human brain. Pnas, 106(14), 5743–5748. 10.1073/pnas.0900544106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JHT, Lowe CB, & Kingsley DM (2018). Characterization of a Human-Specific Tandem Repeat Associated with Bipolar Disorder and Schizophrenia. American Journal of Human Genetics, 103(3), 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Yuan X, Jones Z, Vied C, Miao Y, Marzano M, et al. (2019). Functionalization of Brain Region-specific Spheroids with Isogenic Microglia-like Cells. Scientific Reports 9, 11055 10.1038/s41598-019-47444-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood D, Cairns DM, Dabbi JM, Ramakrishnan C, Deisseroth K, Black LD, et al. (2019). Functional maturation of human neural stem cells in a 3D bioengineered brain model enriched with fetal brain-derived matrix. bioRxiv. 691907 10.1101/691907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Meyer KA, Santpere G, Gulden FO, Šestan N (2017a). Evolution of the Human Nervous System Function, Structure, and Development. Cell, 170(2), 226–247. 10.1016/j.cell.2017.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AMM, Zhu Y, Raghanti MA, Kitchen RR, Onorati M, Tebbenkamp ATN, et al. (2017b). Molecular and cellular reorganization of neural circuits in the human lineage. Science, 358(6366), 1027–1032. 10.1126/science.aan3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacpoole SRL, Spitzer S, Bilican B, Compston A, Karadottir R, Chandran S, Franklin RJM (2013). High yields of oligodendrocyte lineage cells from human embryonic stem cells at physiological oxygen tensions for evaluation of translational biology. Stem Cell Reports, 1(5), 437–450. 10.1016/j.stemcr.2013.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton MM, Tzatzalos E, Donne M, Kolundzic N, Helgason I, Ilic D (2019). Prospects for the Use of Induced Pluripotent Stem Cells in Animal Conservation and Environmental Protection. Stem Cells Translational Medicine 8(1), 7–13. 10.1002/sctm.18-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenevi U, Björklund A, Svendgaard NA (1976). Transplantation of central and peripheral monoamine neurons to the adult rat brain: techniques and conditions for survival. Brain Research, 114(1), 1–20. 10.1016/0006-8993(76)91003-9 [DOI] [PubMed] [Google Scholar]
- Striedter GF (2005). Principles of brain evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. (2019). Comprehensive Integration of Single-Cell Data. Cell, 177(7), 1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Bershteyn M, Paredes MF, Kriegstein AR (2017). Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nature Communications, 8(1), 14167 10.1038/ncomms14167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, et al. (2013). Evolution and diversity of copy number variation in the great ape lineage. Genome Research, 23(9), 1373–1382. 10.1101/gr.158543.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, et al. (2011). Selfformation of functional adenohypophysis in three-dimensional culture. Nature, 480(7375), 57–62. 10.1038/nature10637 [DOI] [PubMed] [Google Scholar]
- Sundaram L, Gao H, Padigepati SR, McRae JF, Li Y, Kosmicki JA, et al. (2018). Predicting the clinical impact of human mutation with deep neural networks. Nature Genetics, 50(8), 1161–1170. 10.1038/s41588-018-0167-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4), 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takei N, Nawa H (2014). mTOR signaling and its roles in normal and abnormal brain development. Frontiers in Molecular Neuroscience, 7, 28 10.3389/fnmol.2014.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teffer K, Buxhoeveden DP, Stimpson CD, Fobbs AJ, Schapiro SJ, Baze WB, et al. (2013). Developmental changes in the spatial organization of neurons in the neocortex of humans and common chimpanzees. The Journal of Comparative Neurology, 521(18), 4249–4259. 10.1002/cne.23412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa-Silva G, Verhoog MB, Goriounova NA, Loebel A, Hjorth J, Baayen JC, et al. (2010). Human synapses show a wide temporal window for spike-timing-dependent plasticity. Frontiers in Synaptic Neuroscience, 2, 12 10.3389/fnsyn.2010.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therriault D, White SR, Lewis JA (2003). Chaotic mixing in three-dimensional microvascular networks fabricated by direct-write assembly. Nature Materials, 2(4), 265–271. 10.1038/nmat863 [DOI] [PubMed] [Google Scholar]
- Ting JT, Kalmbach B, Chong P, de Frates R, Keene CD, Gwinn RP, et al. (2018). A robust ex vivo experimental platform for molecular-genetic dissection of adult human neocortical cell types and circuits. Scientific Reports, 8(1), 8407 10.1038/s41598-018-26803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassy GS, Berger DR, Chen H-H, Kasthuri N, Hayworth KJ, Vercelli A, et al. (2014). Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science, 344(6181), 319–324. 10.1126/science.1249766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Van-Minh A, Cazé RD, Abrahamsson T, Cathala L, Gutkin BS, DiGregorio DA (2015). Contribution of sublinear and supralinear dendritic integration to neuronal computations. Frontiers in Cellular Neuroscience, 9, 67 10.3389/fncel.2015.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, et al. (1999). Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell, 97(6), 689–701. 10.1016/s0092-8674(00)80782-5 [DOI] [PubMed] [Google Scholar]
- Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Chaim IA, Domissy A, Vandenberghe M, et al. (2019. 8). Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network Development. Cell Stem Cell, 25(4), 558–569. 10.1016/j.stem.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou Y-H, Khoneisser J, Huang P-C, Xu X (2016). Hydrogel as a bioactive material to regulate stem cell fate. Bioactive Materials, 1(1), 39–55. 10.1016/j.bioactmat.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner R (2019). Myelin and Modeling: Bootstrapping Cortical Microcircuits. Frontiers in Neural Circuits, 13, 34 10.3389/fncir.2019.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tønnesen S, Kaufmann T, Doan NT, Alnæs D, Córdova-Palomera A, Meer DVD, et al. (2018). White matter aberrations and age-related trajectories in patients with schizophrenia and bipolar disorder revealed by diffusion tensor imaging. Scientific Reports, 8(1), 14129 10.1038/s41598-018-32355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Heuvel MP, Scholtens LH, de Lange SC, Pijnenburg R, Cahn W, Van Haren NEM, et al. (2019). Evolutionary modifications in human brain connectivity associated with schizophrenia. Brain, 162, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A (2000). A chimpanzee genome project is a biomedical imperative. Genome Research, 10(8), 1065–1070. [DOI] [PubMed] [Google Scholar]
- Vattathil S, Akey JM (2015). Small Amounts of Archaic Admixture Provide Big Insights into Human History. Cell, 163(2), 281–284. 10.1016/j.cell.2015.09.042 [DOI] [PubMed] [Google Scholar]
- Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature, 570(7762), 523–527. 10.1038/s41586-019-1289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L, Baumgartner L, Keller KC, Ast J, Trettner S, Nieden, Zur NI (2015). Non-human primate and rodent embryonic stem cells are differentially sensitive to embryotoxic compounds. Toxicology Reports, 2, 165–174. 10.1016/j.toxrep.2014.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, LaMonica B, Kriegstein AR (2011). A new subtype of progenitor cell in the mouse embryonic neocortex. Nature Publishing Group, 14(5), 555–561. 10.1038/nn.2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Kamiya D, Nishiyama A, Katayama T, Nozaki S, Kawasaki H, et al. (2005). Directed differentiation of telencephalic precursors from embryonic stem cells. Nature Neuroscience, 8(3), 288–296. 10.1038/nn1402 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Buth JE, Vishlaghi N, la Torre-Ubieta, de L, Taxidis J, Khakh BS, et al. (2017). Self-Organized Cerebral Organoids with Human-Specific Features Predict Effective Drugs to Combat Zika Virus Infection. Cell Reports, 21(2), 517–532. 10.1016/j.celrep.2017.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, & Macosko EZ (2019). Single-Cell Multi-omic Integration Compares and Contrasts Features of Brain Cell Identity. Cell, 177(7), 1873–1887.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer RA, Leopoldi A, Aichinger M, Wick N, Hantusch B, Novatchkova M, et al. (2019). Human blood vessel organoids as a model of diabetic vasculopathy. Nature, 565(7740), 505–510. 10.1038/s41586-018-0858-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington KS, Wiley LA, Kaalberg EE, Collins MM, Mullins RF, Stone EM, Tucker BA (2017). Two-photon polymerization for production of human iPSC-derived retinal cell grafts. Acta Biomaterialia, 55, 385–395. 10.1016/j.actbio.2017.03.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich S, Kircher M, Vieth B, Haase A, Merkert S, Beier J, et al. (2014). Primate iPS cells as tools for evolutionary analyses. Stem Cell Research, 12(3), 622–629. 10.1016/j.scr.2014.02.001 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim K-Y, Sun P, et al. (2019). hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell, 24(3), 487–497.e7. 10.1016/j.stem.2018.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Tanaka Y, Patterson B, Kang Y-J, Govindaiah G, Roselaar N, et al. (2017). Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell, 21(3), 383–398.e7. 10.1016/j.stem.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S-J, Elahi LS, Paşca AM, Marton RM, Gordon A, Revah O, et al. (2019). Reliability of human cortical organoid generation. Nature Methods, 16(1), 75–78. 10.1038/s41592-018-0255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858), 1917–1920. 10.1126/science.1151526 [DOI] [PubMed] [Google Scholar]
- Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell, 174(4), 999–1014.e22. 10.1016/j.cell.2018.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Shen EH, Hohmann JG, Oh SW, Bernard A, Royall JJ, et al. (2012). Large-Scale Cellular-Resolution Gene Profiling in Human Neocortex Reveals Species-Specific Molecular Signatures. Cell, 149(2), 483–496. 10.1016/j.cell.2012.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, et al. (2016). Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. eLife, 5, 859 10.7554/eLife.13374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li M, Sousa AMM, Šestan N (2014). XSAnno: a framework for building ortholog models in cross-species transcriptome comparisons. BMC Genomics, 15(1), 343 10.1186/1471-2164-15-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, et al. (2018). Spatiotemporal transcriptomic divergence across human and macaque brain development. Science, 362(6420), eaat8077 10.1126/science.aat8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonouzi M, Berger D, Jokhi V, Kedaigle A, Lichtman J, Arlotta P (2019). Individual Oligodendrocytes Show Bias for Inhibitory Axons in the Neocortex. Cell Reports, 27(10), 2799–2808.e3. 10.1016/j.celrep.2019.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]