Abstract
BACKGROUND: Pancreatic ductal adenocarcinoma (PDAC) is a very lethal disease that can develop therapy resistance over time. The dense stroma in PDAC plays a critical role in tumor progression and resistance. How this stroma interacts with the tumor cells and how this is influenced by chemotherapy remain poorly understood. METHODS: The backbone of this study is the parallel transcriptome analysis of human tumor and mouse stroma in two molecular and clinical representative patient-derived tumor xenografts models. Mice (8 animals per group) were treated for 4 weeks with gemcitabine or control. We studied tumor growth, RNA expression in the stroma, tumor-associated macrophages (TAMs) with immunofluorescence, and cytokines in the serum. RESULTS: A method for parallel transcriptome analysis was optimized. We found that the tumor (differentiation, gene expression) determines the infiltration of macrophages into the stroma. In aggressive PDAC (epithelial-to-mesenchymal transition high), we find more M2 polarized TAMs and the activation of cytokines and growth factors (TNFα, TGFβ1, and IL6). There are increased stromal glycolysis, reduced fatty acid oxidation, and reduced mitochondrial oxidation (tricarboxylic acid cycle and oxidative phosphorylation). Treatment with gemcitabine results in a shift of innate immune cells, especially additional infiltration of protumoral M2 TAMs (P < .001) and metabolic reprogramming. CONCLUSIONS: Gemcitabine treatment of PDAC xenografts stimulates a protumoral macrophage phenotype, and this, in combination with a shift of the tumor cells to a mesenchymal phenotype that we reported previously, contributes to tumor progression and therapeutic resistance. Targeting M2-polarized TAMs may benefit PDAC patients at risk to become refractory to current anticancer regimens.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has a unique tumor microenvironment; it represents the most stroma-rich type of cancer. This stroma can comprise up to 90% of the tumor’s mass [1], and its components are very heterogeneous with multiple types of cells [stellate cells, endothelial cells, cancer-associated fibroblasts, lymphocytes, tumor-associated macrophages (TAMs), dendritic cells, and myeloid-derived suppressor cells (MDSCs)] and acellular components (physical dynamics of blood vessels, extracellular matrix, and soluble proteins) [[2], [3], [4]]. Physically, the dense stroma can block drug delivery by collapsing blood vessels and/or shielding the tumor cells from therapeutics, nutrients, and oxygen [5]. Its cellular components secrete cytokines and growth factors and, together with a hypoxic environment, drive tumor progression characterized by early invasion and metastasis [6,7]. Tumor cells and their neighboring stroma are engage in a two-way molecular communication that determines the tumor’s behavior and therapeutic resistance [1,8,9]. The importance of the stromal cells has recently been recognized but has not been investigated extensively [10,11]. To study these interactions will require biologically complex models such as genetically engineered mouse models or patient-derived xenograft (PDTX) models and techniques to discriminate the origin of the cells and their signals.
In the current study, the interaction between the stroma and tumor cells was investigated to explore the underlying mechanism of tumor progression and survival in two different human PDAC PDTX models with different disease biology. Tumor gene expression profiles of these cancer models were reported in our recent article [12]. The poorly differentiated and metastatic patient we investigated (EMT-high, PAC010) had a pronounced epithelial-to-mesenchymal transition (EMT) signature. In contrast, the moderately differentiated tumor model was from a localized tumor with an EMT-low profile (PAC006) [7,13].
In PDTX, the human stroma is replaced by murine stroma [14]. We used this as an opportunity to dissect the molecular characteristics of the stroma (mouse) and tumor cells (human). To be noted, in PDTX models, we can only investigate the tumor-infiltrating cells from the innate immune system as the host has a nonfunctional thymus whereby a large fraction of the stroma infiltrating cells are TAMs [15]. TAMs are a class of immune cells that are present in high numbers in the microenvironment of solid tumors and originate from bone marrow–derived blood monocytes or yolk sac progenitors. There is growing evidence that links increased recruitment of TAMs into the tumor tissue with that of tumor invasiveness, metastasis, immune escape, matrix remodeling, and therapeutic resistance [[16], [17], [18], [19], [20], [21]]. Conventionally, TAMs are classified as classical activated (M1) and nonclassical activated (M2) that reflect their distinct cellular function and metabolism. M1 macrophages are seen to have a proinflammatory and cytotoxic (antitumoral) function; M2 macrophages are anti-inflammatory (protumoral) and promote wound healing. This classification system might not fully reflect the actual plasticity of TAMs in the tumor microenvironment [[16], [17], [18], [19], [20], [21]].
By comparing the RNA expression of the stroma of EMT-high with that of EMT-low tumor model, we looked for specific pathways, regulatory molecules, and functions induced in the stroma by aggressive tumor cells, in particular related to the innate immune response. Subsequently, we investigated these markers in the stroma following gemcitabine treatment (widely used in the clinic) in our models that represent two distinct molecular and clinical presentations of PDAC. The present study combines basic biological understanding, unbiased molecular analysis of tumor and stroma, and a translational approach. A better understanding of TAMs in PDAC and the networks through which the tumor cells and TAMs communicate to promote tumor aggressiveness may contribute to new therapeutic targets that can augment current treatment.
Materials and Methods
Tumor Implantation, Treatment Procedure, Growth, and Sample Collection
The development of the PDTX models was recently reported [14]. For the current experiments, tumor tissues were implanted subcutaneously, and after the tumor had reached a volume of 100-200 mm3, the mice were randomly divided into groups of 8 mice. Group A (control) was treated with vehicle (0.9% NaCl), and group B (experimental) was treated for up to 28 days with intraperitoneal gemcitabine twice a week [gemcitabine 50 mg/kg (Hospira 38 mg/ml solution, Hospira Benelux, Belgium)]. The animal weight and tumor size were measured thrice a week, and the tumor volume was calculated [12]. Tumor tissue and serum were harvested as soon as the volume reached 1000 - 1500 mm3. At the time of harvesting, tissue samples were weighed, photographed, and stored for histological analysis and molecular profiling.
Animal care and all research procedures were executed in accordance with the applicable legal guidelines and under approval of the medical ethical committee for laboratory animals of the KU Leuven (P147/2012).
Histology and Immunohistochemistry
Immunohistochemistry was done on formalin-fixed, paraffin-embedded tumor tissue section to estimate human (tumor) and mouse (stroma) ratio using an antibody against human-specific Cytokeratin-pan antigen (Abcam, Cambridge, UK). In addition, immunofluorescence double staining was performed on formalin-fixed, paraffin-embedded tissue sections after antigen retrieval. As primary antibodies we used goat anti-mouse MMR/CD206 (R&D systems, AF2535-SP) or rat anti-mouse F4/80 antibody (Cl:A3-1 Bio-Rad, MCA497GA) monoclonal MHC Class II (I-A/I-E) (M5/114.15.2, eBioscience, 46-5321-82). Biotin-SP donkey anti-rat IgG (Jackson ImmunoResearch, 712-065-153) and donkey anti-Goat IgG (H+L) Alexa Fluor 568 (Invitrogen, A-11057) were used as secondary antibodies in combination with DAPI staining. Images were acquired on the Zeiss Axio Scan.Z1 using a ×20 objective and ZEN 2 software; images were processed using the software package QuPath (Version: 0.1.2) [22].
RNA Isolation, Next-Generation Sequencing, and Correct Classification of Reads per Organism in PDTX Samples
RNA was isolated with the RNeasy Kit (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. RNA sequencing and processing were performed by the VIB Nucleomics Core (www.nucleomics.be). In short, the poly-A–containing mRNA molecules were purified, converted into cDNA, and sequenced on an Illumina HiSeq 4000 full flow cell. Performance of Xenome [23] and in silico combined reference genome (ICRG) [24] methods for reads classification was compared using a simulated mix dataset (see supplementary file).
RNA sequencing data are available in Gene Expression Omnibus (NCBI) under number GSE118197.
Gene Expression Analysis
The data were analyzed using two complementary software suites: Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com) and Gene Set Enrichment Analysis (GSEA) (http://software.broadinstitute.org/gsea/index.jsp). The core analysis of IPA identifies the most significant biological functions and/or diseases and the potential upstream regulators (genes, RNA, and proteins). GSEA identified enrichment of hallmark gene sets as well as KEGG-pathways. Hierarchical clustering (based on KEGG defined pathways, https://www.genome.jp/kegg/pathway.html, or EMT signature [12]) of the mRNA expression of the individual samples was done using PermutMatrix program (Version 1.9.3 EN).
Enzyme-Linked Immunosorbent Assay
Serum levels of mouse interleulin-6 (IL6), interferon-γ (IFNγ), and transforming growth factor-1β (TGFβ1) were determined in duplicate by enzyme-linked immunosorbent assay (ThermoFischer scientific/Invitrogen, Waltham, MA).
Statistical Analysis
All statistics on quantitative reverse-transcription polymerase chain reaction (RT-qPCR) were performed using SPSS v23 (IBM). Statistical differences between groups were assessed with a Student's t test or the Mann-Whitney rank sum test when appropriate. For differences in gene expression assessed by RT-qPCR, ANOVA test with post hoc Tukey's procedure was used. A P value below .05 was considered statistically significant. For survival analysis, we performed a log-rank test using MedCalc software (version 19.0.6, MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2019).
Results
Design Parallel Transcriptome Analysis of Mouse Stroma Besides Human Tumor in a Single Sample
To test the hypothesis that tumor phenotype is linked to innate immune response, we analyzed the tumor (human) and stroma (mouse) of two PDTX-PDAC models by parallel transcriptome analysis in combination with immunofluorescence and protein analysis. Two tumor types from a previously established panel of PDAC-PDTX were selected (poorly differentiated PAC010 and the well/moderately differentiated tumor model PAC006) [12,14]. After engraftment in a group of immunocompromised mice, the tumor growth was monitored during treatment (Figure 1). To note, difference in growth rate for treated versus untreated PAC006 tumors demanded that we terminated the untreated animals 2 weeks in advance of the scheduled time. We cannot rule out that the moment of termination had an influence on our observations, either compensatory or enhancement; the expression analysis argues against this as we point out in the discussion.
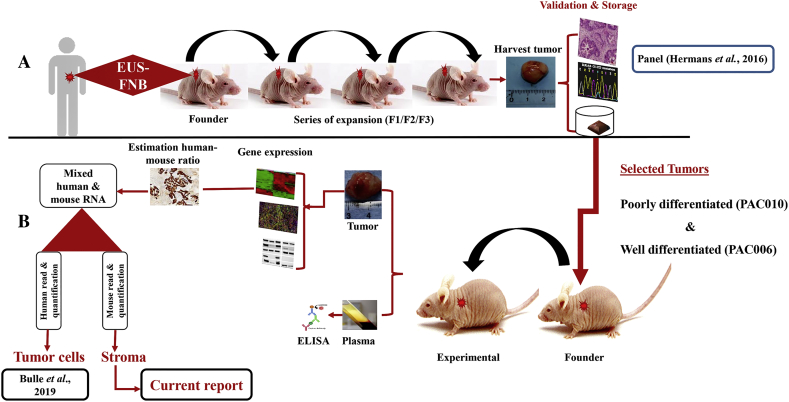
Figure 1.
Schematic representation of the study design: generation of patient-derived PDAC xenografts and their molecular characterization. (A) Establishment and validation design of patient-derived PDAC xenograft. (B) Molecular and histochemical characterization of the tumor cells and the stroma in models and in response to treatment.
The backbone of this study is the parallel transcriptome analysis of the human tumor and the mouse stroma. Sequenced reads from PDTX tissue samples are a mix of human and mouse reads that needed to be assigned to correct species prior to expression analysis. We investigated two approaches: 1) the Xenome classification [23] to discriminate human to mouse reads prior to mapping and 2) the In silico Combined human-mouse Reference Genome (ICRG) [24], where the reads are mapped to a combined genome and assigned to their species after mapping. To compare these approaches, we simulated different mix of reads from mouse and human skin fibroblasts sequenced datasets and compared their classification performances (see supplementary data). We found that Xenome discarded 1.2%-1.9% of the reads per sample due to ambiguity, cross-mapped reads, or unclassifiable reads. In our analysis, the ICRG and Xenome approaches on the experimental PDTX data showed similar results. Important to note, the ICRG is quite simple to implement compared to Xenome. We also investigated the effect of the read length (75 bp or 150 bp) and the use of single or paired-end reads, and the results show that the improvement is rather limited compared to increase in sequencing costs. We thus concluded that we can reliably quantify the gene expression for mixed species from a single sample using 75-bp single reads. To be noted, to perform the sequencing with enough coverage, we estimated on forehand the ratio of stroma (mouse) versus tumor (human) tissue using human specific cytokeratin staining (Supplementary Figure 2).
The separation of species-specific RNA reads resulted in two distinct transcriptomic profiles for each sample: one tumor profile based on human RNA sequences from the grafted tumor cells and one stromal profile from the mouse RNA sequences (Table 1). The comparison showed a big difference in gene expression between the tumors in the models (>4000 genes upregulated and >4000 genes downregulated).
Table 1.
Number of Differentially Expressed Genes Between Conditions
| Tumor |
Stroma |
|||
|---|---|---|---|---|
| Uncorr P Value < .001 |
Uncorr P Value < .001 |
|||
| log2 ratio < −1 | log2 ratio > +1 | log2 ratio < −1 | log2 ratio > +1 | |
| PAC010 vs PAC006 control | 4249 | 4033 | 587 | 848 |
| PAC006-GEM vs PAC006 | 708 | 1399 | 686 | 474 |
| PAC010-GEM vs PAC010 | 956 | 926 | 871 | 832 |
EMT Classification of PDAC Models Used in This Study
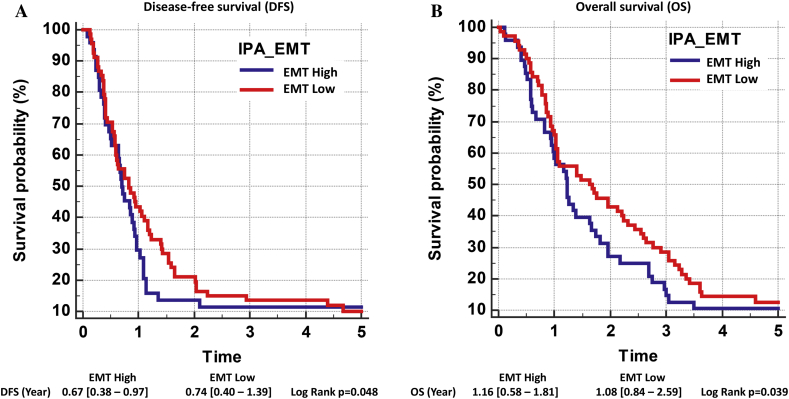
The growth of the tumors in the PDTX mice, their morphology, and their gene expression indicate two behaviorally different tumor models as were seen in the original patients. Analysis of the expression of a panel of EMT-related genes (defined by Ingenuity consortium, retrieved January 2019, www.ingenuity.com) [12] revealed higher expression of mesenchymal markers in aggressive cancer PAC010 (e.g., ZEB1, STAT3, VIM, and SNAI2) with a reduced expression of epithelial markers (e.g., CDH1 and CTNNB1) (EMT-high signature) compared to the moderately differentiated model PAC006 (EMT-low signature). In a clinical dataset of 118 PDAC patients [25], we performed hierarchical clustering using the reported PDAssigner 62 gene set for pancreatic cancer [26] and using the IPA-EMT gene set; both gene sets resulted in two clusters that showed good agreement (see Supplementary Figure 5). The patients clustered based on EMT signature into EMT-high versus EMT-low and showed significant differences in terms of survival (Figure 2).
Figure 2.
Survival analysis of human PDAC classified using IPA-EMT gene set. Hierarchical clustering was performed on 118 PDAC patients data retrieved from NCBI (GSE62165) using 55 EMT-associated genes [12]. H -settings: RNA expression = 2log values; dissimilarity = Pearson’s distance, HC = complete linkage, normalized rows = Z-score, seriation = multifragment heuristics. This signature could separate the patients into two groups (EMT-high patients and EMT-low patients). Using corresponding survival data and log-rank test, disease-free survival (DFS) and overall survival (OS) of these 118 patients were analyzed [25].
Identification of Top Enriched Processes in the Stroma for Aggressive PDAC
We performed a comprehensive gene expression analysis of the stroma components to identify critical regulatory molecules, pathways, cellular functions, and metabolic changes characteristic for an aggressive EMT-high PDAC. We found that between the stroma of EMT-high and EMT-low models (untreated), more than 1400 genes were differentially expressed (Table 1). Using GSEA, we could identify the Hallmark and Pathway gene sets that were most significantly different between both models (Table 2).
Table 2.
Top Hallmark Gene Sets Differentially Expressed in the Stroma Between EMT-High (PAC010) and EMT-Low (PAC006) Identified by GSEA. A: Top 10 positively enriched gene sets. B: Top 10 gene sets that were negatively enriched. NES, normalized enrichment score; FDR, false discovery rate. Size is the number of genes of the pathway.
| A | Gene Set (Positively Enriched) | Size | NES | FDR q-Val |
|---|---|---|---|---|
| 1 | TNFA signaling via NFKB | 180 | 2.14 | 0.000 |
| 2 | E2F targets | 195 | 2.14 | 0.000 |
| 3 | G2M checkpoint | 188 | 2.09 | 0.000 |
| 4 | Inflammatory response | 163 | 1.93 | 0.000 |
| 5 | IL6 JAK STAT3 signaling | 73 | 1.92 | 0.000 |
| 6 | Interferon alpha response | 86 | 1.91 | 0.000 |
| 7 | Unfolded protein response | 107 | 1.90 | 0.000 |
| 8 | MTORC1 signaling | 197 | 1.84 | 0.000 |
| 9 | Hypoxia | 159 | 1.81 | 0.001 |
| 10 | Glycolysis | 166 | 1.79 | 0.001 |
| B | Gene Set (Negatively Enriched) | Size | NES | FDR q-Val |
| 1 | Adipogenesis | 174 | −2.22 | 0.000 |
| 2 | Myogenesis | 122 | −2.20 | 0.000 |
| 3 | Bile acid metabolism | 74 | −1.98 | 0.000 |
| 4 | Oxidative phosphorylation | 185 | −1.92 | 0.000 |
| 5 | Hedgehog signaling | 28 | −1.84 | 0.002 |
| 6 | Estrogen response early | 151 | −1.61 | 0.021 |
| 7 | KRAS signaling DN | 80 | −1.61 | 0.018 |
| 8 | Fatty acid metabolism | 128 | −1.59 | 0.017 |
| 9 | Notch signaling | 27 | −1.59 | 0.017 |
| 10 | Estrogen response late | 147 | −1.53 | 0.025 |
In the aggressive tumor, there was a positive enrichment of the hallmark gene set for hypoxia (normalized enrichment score: NES +1.81) and several cell-cell signaling pathways (TNFα, IL6, etc.). Important differences were also found for gene sets representing metabolism (e.g., glycolysis NES +1.79, oxidative phosphorylation NES −1.92 and fatty acid metabolism NES −1.59).
Using IPA, we also identified the regulator molecules for pathways enriched in the EMT-high tumor model. The top regulators included intercellular signaling molecules (TGFβ1, IL1β, and TNFα) found in both the tumor cells [12] and the stromal compartment (Table 3, A).
Table 3.
IPA Analysis of Upstream Regulatory Molecules and Cellular Functions from Differentially Expressed Genes EMT-High Versus EMT-Low in the Stroma. A: Upstream regulator molecules identified by IPA. B: Functional annotation of processes for the cells in the stroma based on their underlying human tumor EMT-high (poorly differentiated, PAC010) versus EMT-low (well differentiated, PAC006) computed from differential gene expression of the mouse stroma.
| A | Upstream Regulator | Molecule Type | Corr. P Value | Activation Statea |
|---|---|---|---|---|
| IL1β | Cytokine | 4.56E-29 | Activated | |
| TNFα | Cytokine | 5.22E-29 | Activated | |
| TGFβ1 | Growth factor | 6.50E-29 | ||
| IFNγ | Cytokine | 7.78E-27 | Activated | |
| IL6 | Cytokine | 1.33E-16 | ||
| SMARCA4 | Transcrip. regulator | 3.60E-16 | ||
| IL10RA | Receptor | 4.03E-16 | Inhibited | |
| NFkβ (complex) | Complex | 6.55E-16 | Activated | |
| CEBPA | Transcrip. regulator | 2.01E-15 | ||
| IL13 | Cytokine | 2.65E-15 | ||
| B | Functions Annotation | Corr. P Value | ||
|---|---|---|---|---|
| Cell movement | 1.12E-41 | |||
| Migration of cells | 2.18E-40 | |||
| Leukocyte migration | 6.13E-33 | |||
| Inflammation of organ | 2.22E-28 | |||
| Cell movement of leukocytes | 5.79E-28 | |||
| Cellular infiltration | 1.84E-24 | |||
| Cellular infiltration by leukocytes | 4.47E-24 | |||
| Development of vasculature | 1.09E-23 | |||
| Cell movement of myeloid cells | 7.28E-23 | |||
| Cell movement of phagocytes | 1.52E-22 | |||
Activation state by IPA; blank = no prediction (see supplementary file).
These signaling molecules can support tumor progression and indicate a possible paracrine communication between the two entities. In addition, importantly, functional analysis of the stromal reaction revealed the increased recruitment of leukocyte (Table 3, B) in EMT-high PDAC, which was subsequently investigated by immunofluorescence staining (see below).
Stromal Reaction in Response to Gemcitabine Treatment in PDTX-PDAC
We investigate the effects of drug treatment on the different models. Gemcitabine treatment resulted in a high number of differentially expressed genes in the tumor and also in the stroma (Table 1). Using GSEA, we identified the hallmark gene sets significantly enriched after gemcitabine treatment (Supplementary Tables 1, A and B, and 2, A and B). We focused on the processes identified for aggressive PDAC (Table 2, Table 3): leukocytes/TAMs, related metabolic pathways, and cell-cell signaling.
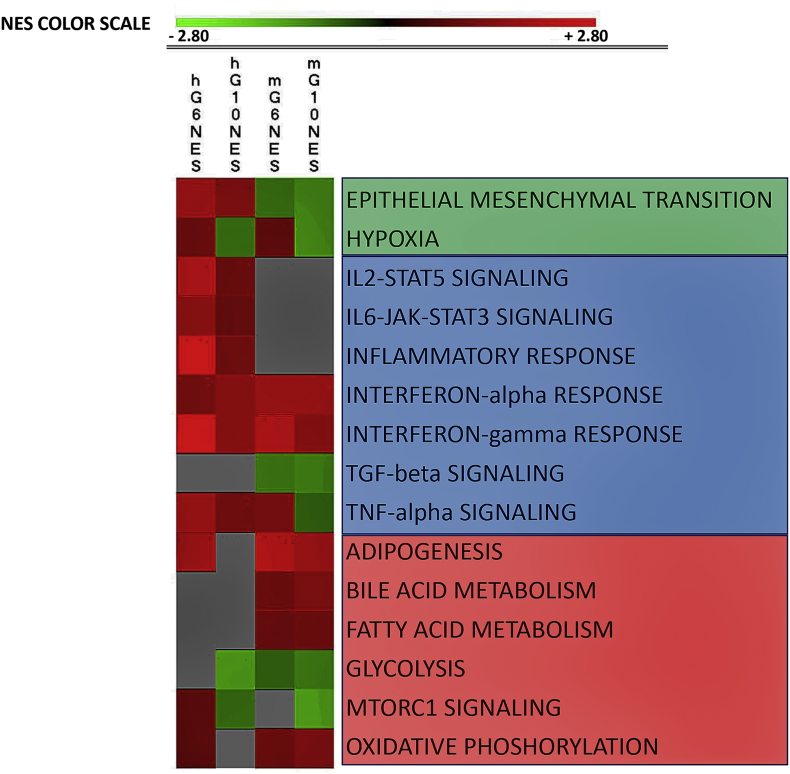
A graphic summary of how tumors/human (h) and stroma/mouse (m) responded to gemcitabine is presented in Figure 3. Two groups of related processes (cell-cell signaling and metabolism) are among the highest enriched gene sets. The individual processes were further analyzed through hierarchical clustering, IPA analysis, immunofluorescence, and/or protein determination in the serum.
Figure 3.
Graphic presentation of top enriched Hallmark gene sets in tumors and in the corresponding stroma as result of gemcitabine treatment. We analyzed the differential expressed genes for four conditions: (A) hG6NES: tumor PAC006 GEM-treated versus control, (B) hG10NES: tumor PAC010 GEM-treated versus control, (C) mG6NES: stroma PAC006 GEM-treated versus control, and (D) mG10NES: stroma PAC010 GEM-treated versus control. GSEA was used to identify Hallmark gene sets (for full list, see Supplementary Tables 1, A and B, and 2, A and B). The majority of the top gene sets could be functionally grouped into three main groups; they are presented using the normalized enrichment score (NES with P value < .05). The NES is indicated by a color; the intensity is scaled within each row so that the highest enrichment score corresponds to bright red and suppression to bright green. Gray: NES not significant (uncorrected P value > .05).
An IPA analysis was performed for the stroma when treated with gemcitabine (Table 4). This predicts that gemcitabine treatment inhibits TGFβ1, TNF, and IFNγ regulating signals in the stroma (Table 4, A).
Table 4.
IPA analysis using information from differentially expressed genes in the stroma of EMT-low (PAC006) and EMT-high (PAC010) in response to GEM treatment. Top upstream regulators (A) and associated cellular function (B) identified with their predicted activation state and P-value.
|
A Upstream Regulators | ||||||
|---|---|---|---|---|---|---|
| PAC010 vs PAC006 |
PAC6-GEM vs PAC006 |
PAC010-GEM vs PAC010 |
||||
| Act Statea | P-valueb | Act Statea | P-valueb | Act Statea | P-valueb | |
| TGFβ1 | 10−28 | INH | 10−34 | INH | 10−48 | |
| TNFα | Act | 10−28 | 10−16 | INH | 10−28 | |
| IL1β | Act | 10−28 | 10−10 | 10−21 | ||
| IFNγ | Act | 10−26 | 10−10 | INH | 10−30 | |
| IL-6 | 10−16 | 10−17 | 10−35 | |||
|
B Cellular Functions | |||
|---|---|---|---|
| PAC010 vs PAC006 |
PAC6-GEM vs PAC006 |
PAC010-GEM vs PAC010 |
|
| P-valueb | P-valueb | P-valueb | |
| Cell movement | 10−41 | 10−19 | 10−36 |
| Migration of cells | 10−40 | 10−18 | 10−34 |
| Leukocyte migration | 10−33 | 10−7 | 10−22 |
| Cell movement leukocytes | 10−28 | 10−6 | 10−20 |
| Cell infiltration leukocytes | 10−24 | 10−4 | 10−14 |
Activation state by IPA, INH = inhibited, ACT = activation and blank = no prediction
Corrected P-value. (see supplementary file).
We performed RT-qPCR and enzyme-linked immunosorbent assay to determine relative gene expression of selected cytokines and growth factor: mRNA (IL6, IL10, Hif1α, and TGFβ1) in the stroma and protein level (IL6, TGFβ1, and IFNγ) in circulation. The RT-qPCR confirmed the RNA sequencing. Gemcitabine treatment indicates a suppression of IL6 and TGFβ1 mRNA (Supplementary Table 3). At protein level, of the tested cytokines and growth factors, only IL6 yielded detectable serum levels in the untreated EMT-high model. This level was significantly suppressed following gemcitabine treatment [PAC010 = 234 (75-518) pg/ml vs PAC010-GEM treated = 22.5 (16-31) pg/ml; median (25%-75%); P = .029].
M2-like, CD206 posF4/80 posTAMs Are Increased in EMT-High Tumors
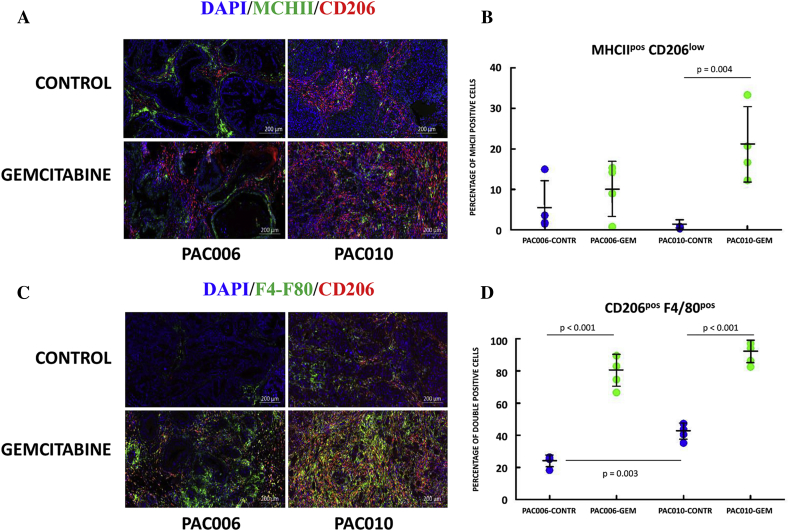
One of the major cell types recruited from the blood are macrophages that infiltrate the tumor, become TAMs, and can acquire in this microenvironment a protumoral or antitumoral phenotype under influence of cytokines, interleukins, and metabolic factors. Macrophages play a critical role in tumor progression. RNA-sequencing analysis revealed an association of infiltration of leukocytes and cytokine signaling (Table 3, Table 4). The effect of gemcitabine treatment on blood cell/leukocyte infiltration was more significant in the EMT-high model compared to EMT-low (Table 4, B). We performed histochemical analyses on formalin-fixed tumor slices using immunofluorescence dual staining for M2 and M1 polarized TAMs. We used mouse specific macrophage marker F4/80, along with CD206high, as indicator of M2 macrophages (Figure 4, C) and the combination of MHCIIhigh and CD206low as indicators of M1 macrophages (Figure 4, A). Gemcitabine treatment caused significantly (P < .001) increased expression of M2-like macrophages in both models (Figure 4, D). Between the models, our result also indicated statistically significantly higher expression of M2 macrophages in the EMT-high model (P = .003), suggesting a link with tumor aggressiveness (Figure 4, D). The expression of M1 markers between the models and for EMT-low in response to gemcitabine treatment was not statistically significant (Figure 4, B). However, in EMT-high, gemcitabine treatment resulted also in an increase of M1-like TAM (P = .004). These results were supported by heat map clustering of gene expression of TAMs markers [27]. The heat map revealed that, in EMT-low, the M1 phenotype was highly variable between the animals. Both M1 and M2 signatures could separate the gemcitabine-treated samples from untreated samples in the EMT-high model, supporting a treatment-induced immunological shift in TAMs (Supplementary Figure 6).
Figure 4.
Immunofluorescence staining of PDTX-PDAC tumor sections. Representative images for (A) DPI/MHCII/CD206 and (C) DPI/F4/80/CD206 from placebo- and gemcitabine-treated PDTX mice. Original magnification of histological images × 10; scale bar 200 μm. The graphs indicate percentage of cells stained for (B) M1-type MHCIIposCD206low macrophages or (D) double-positive M2-type CD206posF4/80pos macrophages. Sections were stained with DAPI, and percentage was calculated to the total number of cells using QuPath software. Each staining is representative for the analysis of tumor sections of four animals per group.
Hierarchical clustering of gene sets representing dendritic cells and MDSCs indicates more MDSCs in the untreated, EMT-high model (Supplementary Figure 6, D). Gemcitabine treatment results in increased antigen presentation (H2 genes and PD-L1 (CD274) are upregulated) (Supplementary Figure 6, C). We see an enrichment of MDSCs following treatment with gemcitabine, accompanied in the EMT-high model by increased expression of the immune checkpoint markers Lag3 (CD223) and Vista (Supplementary Figure 7, E-F).
The Metabolic Profile of EMT-High Tumors Favors M2-Polarized Macrophages
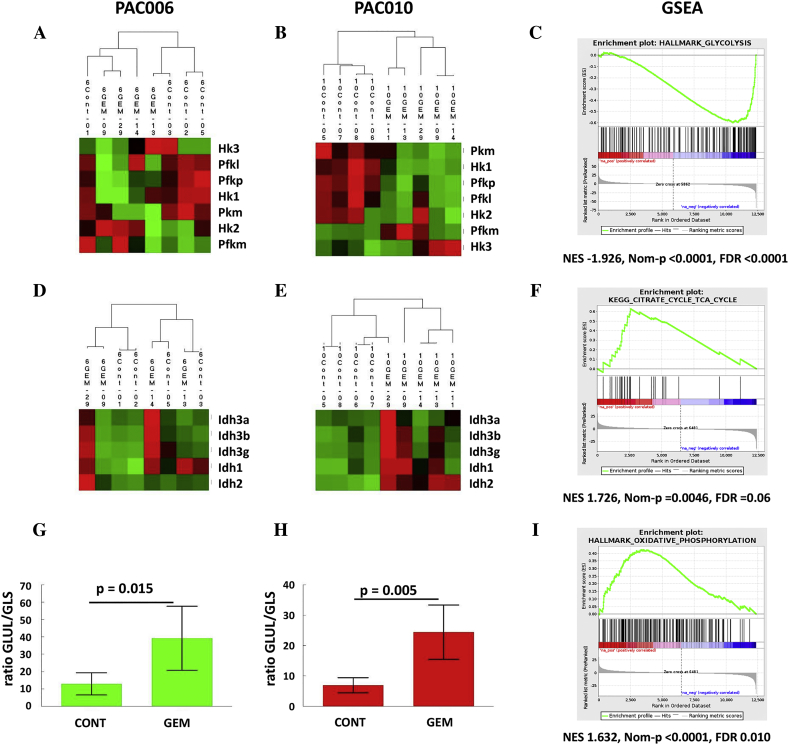
The functional phenotype of M1 and M2 macrophage subtypes is highly regulated at the transcriptional and metabolic level. Proinflammatory M1 macrophages consume glucose and rely heavily on glycolysis for ATP production. They also exhibit a defect in tricarboxylic acid (TCA) cycle. On the other hand, anti-inflammatory M2 macrophages have an intact TCA cycle and favor fatty acid oxidation as mechanism to produce ATP. We used RNA-seq expression data to explore change in glycolysis/gluconeogenesis (KEGG 00010), citrate cycle (TCA) (KEGG 00020), and oxidative phosphorylation (KEGG 00190). When we concentrate on the rate-limiting enzymes for glycolysis, we see by hierarchical clustering that gemcitabine treatment downregulated the majority of these enzymes (Figure 5, A and B). The same downregulation was found when the full gene set was examined (data not shown). This downregulation of glycolysis is supported by the GSEA (Figure 5, C).
Figure 5.
Changes in metabolic gene expression in PAC006 and PAC010 induced by gemcitabine. Hierarchical clustering of rate-limiting enzymes of glycolysis in (A) PAC006 and (B) PAC010. (C) Downregulation of glycolysis is confirmed by GSEA (e.g., PAC010). Key enzymes of TCA cycle are shown in (D) for PAC006 and (E) for PAC010. GSEA demonstrates enrichment of KEGG-TCA pathway (PAC006). The ratio of GLUL/GLS as marker for the use of glutamine to fuel the TCA cycle; (G) PAC006 and (H) PAC010. GSEA indicates a strong enrichment of (I) oxidative phosphorylation. The heat map shows the relative mRNA expression in glycolysis and TCA cycle of the mouse genes in the stroma in response to treatment of the xenografts. The red (high), black (middle), and green (low) colors indicate the relative expression intensity of each gene within a sample.
Discussion
A diagnosis of pancreatic cancer comes almost always with limited therapeutic perspectives and a bad prognosis for the patients. Several factors are considered important in this respect, one being the unique, stroma-rich microenvironment of a PDAC. The communication between the tumor and the stromal cells determines the evolution of the disease, but the underlying mechanism is only partly understood. To study the tumor-stroma interactions and to gain insight into possible therapeutic targets to improve treatment, we used two human tumor xenograft models of different clinical and morphological presentation. Gene expression analysis confirms the aggressive/poorly differentiated EMT-high model (PAC010) and the more moderate/differentiated tumor EMT-low model (PAC006). By comparing the gene expression of the stroma between these models, we looked for specific pathways, regulatory molecules, and functions characteristic for aggressive PDAC. Subsequently, we used these observations when we investigated the effect of gemcitabine treatment on the stroma.
In the untreated PDAC models, gene expression and functional analysis of the stroma indicated increased infiltration of leukocyte and macrophages. TAMs represent the most abundant leukocyte subpopulation in the stroma; they belong to the innate immune system [15] and can release signaling molecules facilitating cancer cell invasion, migration, angiogenesis, tumor progression, or metastasis [[28], [29], [30], [31], [32]]. In particular, TAMs secrete cytokines and growth factors that are involved in the induction of EMT in solid tumors [3,[33], [34], [35], [36]]. In PDTX, we could detect increased levels of IL6 in the serum of animals transplanted with EMT-high PDAC model in agreement with the gene expression analysis. Clinically, high serum levels of IL6 correlate with lower rates of survival and a higher chemoresistance [37]. The similarity of pathways found in the tumor [12] and in this study in the stroma suggests a possible paracrine communication between the two entities.
Within the tumor microenvironment, the TAMs come into contact with tumor-secreted factors that can polarize them toward M2 type macrophage [28,29,[38], [39], [40], [41], [42], [43]]. Immunofluorescence dual staining confirms a significantly higher presence of M2 polarized TAMs (CD206high on F4/80 mouse specific macrophage marker) in the EMT-high model. M2 macrophage infiltration is reported to be associated with a malignant tumor phenotype that correlates with poor prognosis [44,45].
Does the tumor-stromal interaction change when the tumor is treated with chemotherapy, and is such a change related to the phenotype of the PDAC? Gemcitabine-based combination chemotherapy is the standard therapy for most PDAC patients [46]. Resistance to gemcitabine and other drugs remains a major cause of therapeutic failure. Recently, we showed that gemcitabine treatment significantly induced the expression of mesenchymal markers in the residual tumor cells, indicating enrichment of metastatic and therapeutic resistance phenotype [12]. Other studies have also associated drug resistance of tumor cells with the acquisition of EMT phenotype [47,48]. To what extent the stroma under chemotherapy influences the survival is unknown. Gemcitabine treatment does not shift the microenvironment to a stroma as found for an aggressive PDAC (see GSEA or IPA) within the time frame we studied the PDTX. In contrast, cytokines like TNFα that are activated in EMT-high PDAC are inhibited by gemcitabine. However, in the present study, we clearly demonstrate that gemcitabine treatment induces an increased infiltration of M2 polarized TAMs in stroma of PDAC xenografts. In the more aggressive/EMT-high PAC010, following gemcitabine treatment, both TAM subtypes (M1 and M2) have increased. Interestingly, TAMs can contribute to EMT by exhibiting both pro- and anti-inflammation characteristics [49,50]. We further explored the metabolic pathways predominantly used in the stroma following gemcitabine treatment. The results support an increased M2 population. We found reduced glycolysis and upregulation of mitochondrial oxidative metabolism (TCA cycle and oxidative phosphorylation). Previous reports define them as metabolically M2 polarized TAMs [51,52]. In addition, upregulation of fatty acid oxidation [53] and increased GLUL/GLS ratio (indicating enhanced fueling of the TCA cycle through glutamine [27]) were identified. Unfortunately, in our study, we did not find a clear association between M2 macrophage infiltration and cytokines signaling (upstream regulators) as previously was reported (M1: commonly expresses higher levels of IL12, IL23, TNFα, and IL6 and M2: commonly expresses higher levels of IL10 and TGFβ1) [54]. The plasticity of the TAMs in the stroma allows also for cells with shared features of both M1 and M2, and the phenotype of TAMs can switch during different stages of tumor progression; these observations could explain this discrepancy [[55], [56], [57]]. There is some controversy in literature on how MDSCs react to gemcitabine treatment. Galluzi et al. report a reduction following treatment [58], while in the study by Plate et al. in patients with pancreatic cancer [59], after an initial reduction of the major MDSC subset (BDCA/CD1b), they found a rebound to levels equal or above baseline. In the present study, gene expression indicates that the MDSCs have increased and exhibit a stronger immune suppressive phenotype following gemcitabine treatment.
In a recent paper, it was reported that increased M2 TAMs in the PDAC stroma are associated with decreased survival under gemcitabine treatment. As a mechanism, they proposed that macrophage-released pyrimidines inhibit efficacy of gemcitabine therapy in pancreatic cancer [60]. This might be another mechanism by which M2 TAMs and tumor-stroma interactions have an impact on the patient’s prognosis. These observations as well as ours make M2 an attractive therapeutic target, especially in metastatic and therapy resistant PDAC [[61], [62], [63]]. Currently, a number of preclinical and clinical trials have been completed or are ongoing, targeting TAMs to treat different tumor types including pancreatic cancer (e.g., NCT03662412, NCT03184870, NCT01921699). These strategies aim to block monocyte recruitment [64], switch TAMs phenotype from M2 to M1 [65,66], deplete resident TAMs [67,68], or neutralize TAMs products [62,69].
Conclusion
Our PDTX models, representing two different clinically relevant phenotypes of PDAC, will contribute to a better understanding of tumor-stroma biology. When we take the expression analysis together of both PAC006 and PAC010, we think the effects in the stroma are firstly the result of the tumor EMT phenotype and the tumor-stroma interactions. Gemcitabine treatment results in a shift in the stroma to a predominantly immunosuppressive environment with M2 TAMs and MDSCs. This suggests a dynamic in the stroma that supports a mesenchymal phenotype of the tumor cells [12], influx of macrophages, and suppression of inflammation in the context of gemcitabine treatment. This molecular analysis identifies relevant pathways and molecular targets for new therapy development. Targeting M2-polarized TAMs may benefit PDAC patients at risk to become refractory to current anticancer regimens.
Conflict of Interest
All authors declare no potential conflicts of interest.
Funding Support
C. V. holds a mandate as Senior Clinical Investigator of the Research Foundation-Flanders (Belgium) (FWO). This study was partly supported by a research grant from “Kom op tegen Kanker” Belgium and VUYLSTEKE-FLIPTS FONDS LEVERKANKER.
Acknowledgements
The authors wish to thank Dr. E. Hermans and Profs. S. van der Merwe and F. Amant and the members of TRACE [TRACE: a patient-derived tumor xenograft platform (PDX) in translational cancer research at KU Leuven, Belgium] for their excellent work in the development of the EUS-PDTX models. We also want to thank the VIB Nucleomics Core facility for bioinformatics support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tranon.2020.01.004.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Luo G, Long J, Zhang B, Liu C, Xu J, Ni Q, Yu X. Stroma and pancreatic ductal adenocarcinoma: an interaction loop. Biochimica Biophysica Acta. 2012;1826:170–178. doi: 10.1016/j.bbcan.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Feig C, Gopinathan A, Gopinathan A, Neesse A, Chan DS, Cook N, Tuveson DA. The pancreas cancer microenvironment. Clin Cancer Res. 2012;18(16):4266–4276. doi: 10.1158/1078-0432.CCR-11-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonde AK, Tischler V, Kumar S, Soltermann A, Schwendener RA. Intratumoral macrophages contribute to epithelial-mesenchymal transition in solid tumors. BMC Cancer. 2012;12 doi: 10.1186/1471-2407-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada S, Masamune A, Shimosegawa T. Alteration of pancreatic cancer cell functions by tumor-stromal cell interaction. Front Physiol. 2013;4:318. doi: 10.3389/fphys.2013.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kota J, Hancock J, Kwon J, Korc M. Pancreatic cancer: stroma and its current and emerging targeted therapies. Cancer Lett. 2017;391:38–49. doi: 10.1016/j.canlet.2016.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Sluka P, Davis ID. Cell mates: paracrine and stromal targets for prostate cancer therapy. Nature Reviews Urology. 2013;10:441–451. doi: 10.1038/nrurol.2013.146. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Bosch N, Judith Vinaixa J, Pilar Navarro P. Immune evasion in pancreatic cancer: from mechanisms to therapy. Cancers. 2018;10(1):6. doi: 10.3390/cancers10010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bussard KM, Mutkus L, Stumpf K, Gomez-Manzano C, Marini FC. Tumor-associated stromal cells as key contributors to the tumor microenvironment. Breast Cancer Res. 2016;18(1) doi: 10.1186/s13058-016-0740-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu YS, Chung I, Wong WF, Masamune A, Sim MS, Looi CY. Paracrine IL-6 signaling mediates the effects of pancreatic stellate cells on epithelial-mesenchymal transition via Stat3/Nrf2 pathway in pancreatic cancer cells. Biochim Biophys Acta. 2017;1861(2):296–306. doi: 10.1016/j.bbagen.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Bulle AS, Dekervel J, Van der Merwe S, Van Cutsem E, Verslype C, van Pelt J. Relevance of the stroma in pancreatic ductal adenocarcinoma and its challenges for translational research. Journal of cancer treatment and diagnosis. J Cancer Treat Diagn Mini Review Art. 2017;1:1–15. [Google Scholar]
- 11.Netea-Maier RT, Smit JWA, Netea MG. Metabolic changes in tumor cells and tumor-associated macrophages: a mutual relationship. Cancer Lett. 2018;413:102–109. doi: 10.1016/j.canlet.2017.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Bulle A, Dekervel J, Libbrecht L, Deschuttere L, Lambrecht D, Van Cutsem E, Chris Verslype C, van Pelt J. Gemcitabine treatment induces epithelial-to-mesenchymal transition in patient-derived pancreatic ductal adenocarcinoama xenografts. Am J Transl Res 2019. 2019;11(2):765–779. PMID: 30899378. [PMC free article] [PubMed] [Google Scholar]
- 13.Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H, Boettcher M, Mougiakakos D, Reichardt W, Bronsert P. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19(5):518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 14.Hermans E, Van der Merwe SW, Depreeuw J, Dekervel J, Radaelli E, Roskams T, van Pelt J, Topal B, Verslype C, Prenen H. Successful application of endoscopic ultrasound guided fine needle biopsy to establish pancreatic patient-derived tumor xenografts: a pilot study. Endoscopy. 2016;48:1016–1022. doi: 10.1055/s-0042-113597. [DOI] [PubMed] [Google Scholar]
- 15.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Advanced Drug Delivery Reviews. 2016;99 Pt B:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 16.Wӧrmann SM, Diakopoulos KN, Lesina M, Algül H. The immune network in pancreatic cancer development and progression. Oncogene. 2014;33:2956–2967. doi: 10.1038/onc.2013.257. [DOI] [PubMed] [Google Scholar]
- 17.Mills CD. Anatomy of a discovery: M1 and M2 macrophages. Frontiers in Immunology. 2012;6(2121):1–12. doi: 10.3389/fimmu.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J Innate Immun. 2014;6(6):716–726. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aras S, Zaidi MR. TAMeless traitors: macrophages in cancer progression and metastasis. Br J Cancer. 2017;117(11):1583–1591. doi: 10.1038/bjc.2017.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goswami KK, Ghosh T, Ghosh S, Sarkar M, Bose A, Baral R. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cellular Immunology. 2017;316:1–10. doi: 10.1016/j.cellimm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Chimal-Ramírez GK, Espinoza-Sánchez NA, Chávez-Sánchez L, Arriaga-Pizano L, Fuentes-Pananá EM. Monocyte differentiation towards protumor activity does not correlate with M1 or M2 phenotypes. J Immunol Res. 2016;2016:6031486. doi: 10.1155/2016/6031486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, McQuaid S, Gray RT, Murray LJ, Coleman HG. QuPath: open source software for digital pathology image analysis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, Beresford-Smith B. Xenome—a tool for classifying reads from xenograft samples. Bioinformatics. 2012;28(12):i172–i178. doi: 10.1093/bioinformatics/bts236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callari M, Batra AS, Batra RN, Sammut SJ, Greenwood W, Clifford H, Hercus C, Chin SF, Bruna A, Rueda OM. Computational approach to discriminate human and mouse sequences in patient-derived tumour xenografts. BMC Genomics. 2018;19 doi: 10.1186/s12864-017-4414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janky R, Binda MM, Allemeersch J, Van den Broeck A, Govaere O, Swinnen JV, Roskams T, Aerts S, Topal B. Prognostic relevance of molecular subtypes and master regulators in pancreatic ductal adenocarcinoma. BMC Cancer. 2016;12(16):632. doi: 10.1186/s12885-016-2540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, Cooc J, Weinkle J, Kim GE, Jakkula L. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17(4):500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korf H, du Plessis J, van Pelt J, De Groote S, Cassiman D, Verbeke L, Ghesquière B, Fendt SM, Bird MJ, Talebi A. Inhibition of glutamine synthetase in monocytes from patients with acute-on-chronic liver failure resuscitates their antibacterial and inflammatory capacity. Gut. 2019;68:1872–1883. doi: 10.1136/gutjnl-2018-316888. [DOI] [PubMed] [Google Scholar]
- 28.Pollard JW. Tumor-educated macrophages promote tumour progression and metastasis. Nature Reviews Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 29.Mantovani A. Cancer Inflammation by remote control. Nature. 2005;435(7043):752–753. doi: 10.1038/435752a. 15944689 [DOI] [PubMed] [Google Scholar]
- 30.Meng F, Li W, Li C, Gao Z, Guo K, Song S. CCL18 promotes epithelial-mesenchymal transition, invasion and migration of pancreatic cancer cells in pancreatic ductal adenocarcinoma. Int. J. Oncol. 2015;46:1109–1120. doi: 10.3892/ijo.2014.2794. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Yu X, Gao L, Zhao Y, Lai J, Lu D, Bao R, Jia B, Zhong L, Wang F. Noninvasive imaging of CD206-positive M2 macrophages as an early biomarker for post chemotherapy tumor relapse and lymph node metastasis. Theranostics. 2017;7(17):4276–4288. doi: 10.7150/thno.20999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ye H, Zhou Q, Zheng S, Li G, Lin Q, Wei L, Fu Z, Zhang B, Liu Y, Li Z. Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(5):453. doi: 10.1038/s41419-018-0486-0. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C, Nitschke AM, Xiong W, Zhang Q, Tang Y, Bloch M, Elliott S, Zhu Y, Bazzone L, Yu D. Proteomic analysis of tumor necrosis factor-alpha resistant human breast cancer cells reveals a MEK5/Erk5-mediated epithelial-mesenchymal transition phenotype. Breast Cancer Res. 2008;10(6) doi: 10.1186/bcr2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen DP, Ning WR, Li XF, Wei Y, Lao XM, Wang JC, Wu Y, Zheng L. Peritumoral monocytes induce cancer cell autophagy to facilitate the progression of human hepatocellular carcinoma. Autophagy. 2018;14(8):1335–1346. doi: 10.1080/15548627.2018.1474994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CY, Xu JY, Shi XY, Huang W, Ruan TY, Xie P, Ding JL. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest. 2013;93(7):844–854. doi: 10.1038/labinvest.2013.69. [DOI] [PubMed] [Google Scholar]
- 36.Gao S, Hu J, Wu X, Liang Z. PMA treated THP-1-derived-IL-6 promotes EMT of SW48 through STAT3/ERK-dependent activation of Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2018;108:618–624. doi: 10.1016/j.biopha.2018.09.067. [DOI] [PubMed] [Google Scholar]
- 37.Holmer R, Goumas FA, Waetzig GH, Rose-John S, Kalthoff H. Interleukin-6: a villain in the drama of pancreatic cancer development and progression. Hepatobiliary Pancreat Dis Int. 2014;13(4):371–380. doi: 10.1016/s1499-3872(14)60259-9. 25100121 [DOI] [PubMed] [Google Scholar]
- 38.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr. Opin. Immunol. 2010;22:231–237. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 41.Sugimoto M, Mitsunaga S, Yoshikawa K, Kato Y, Gotohda N, Takahashi S, Konishi M, Ikeda M, Kojima M, Ochiai A. Prognostic impact of M2 macrophages at neural invasion in patients with invasive ductal carcinoma of the pancreas. Eur. J. Cancer. 2014;50:1900–1908. doi: 10.1016/j.ejca.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Liou GY, Döppler H, Necela B, Edenfield B, Zhang L, Dawson DW, Storz P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5(1):52–63. doi: 10.1158/2159-8290.CD-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Yan W, Mathew E, Kane KT, Brannon A, 3rd, Adoumie M, Vinta A, Crawford HC, Pasca di Magliano M. Epithelial-myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife. 2017;6 doi: 10.7554/eLife.27388. pii: e27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4:376. 23145206 [PMC free article] [PubMed] [Google Scholar]
- 45.De Palma M, Lewis CE. Macrophage regulation of tumor responses to anticancer therapies. Cancer Cell. 2013;23:277–286. doi: 10.1016/j.ccr.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J, Scheithauer W, Siena S, Tabernero J, Teixeira L. nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. JNCI J Natl Cancer Inst. 2015;107(2) doi: 10.1093/jnci/dju413. pii: dju413. [DOI] [PubMed] [Google Scholar]
- 47.Banyard JL, Bielenberg DR. The role of EMT and MET in cancer dissemination. Connect Tissue Res. 2015;56(5):403–413. doi: 10.3109/03008207.2015.1060970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. EMT-program is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helm O, Held-Feindt J, Grage-Griebenow E, Reiling N, Ungefroren H, Vogel I, Krüger U, Becker T, Ebsen M, Röcken C. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int J Cancer. 2014;135(4):843–861. doi: 10.1002/ijc.28736. [DOI] [PubMed] [Google Scholar]
- 50.Karnevi E, Andersson R, Rosendahl AH. Tumour-educated macrophages display a mixed polarisation and enhance pancreatic cancer cell invasion. Immunology and cell biology. 2014;92:543–552. doi: 10.1038/icb.2014.22. [DOI] [PubMed] [Google Scholar]
- 51.Mills EL, O’Neill LA. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. Eur. J. Immunol. 2016;46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- 52.Van den Bossche J, Saraber DL. Metabolic regulation of macrophages in tissues. Cellular Immunology. 2018;330:54–59. doi: 10.1016/j.cellimm.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 53.Namgaladze D, Brüne B. Fatty acid oxidation is dispensable for human macrophage IL-4–induced polarization. Biochim Biophys Acta. 2014;1841(9):1329–1335. doi: 10.1016/j.bbalip.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 54.Habtezion A, Edderkaoui M, Pandol SJ. Macrophages and Pancreatic ductal adenocarcinoma. Cancer Lett. 2016;381(1):211–216. doi: 10.1016/j.canlet.2015.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer research. 2006;66:11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 56.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends in immunology. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galluzzi L, Senovilla L, Zitvogel L, Kroemer G. The secret ally: immunostimulation by anticancer drugs. Nat Rev Drug Discov. 2012;11:215–233. doi: 10.1038/nrd3626. [DOI] [PubMed] [Google Scholar]
- 59.Plate JM, Plate AE, Shott S, Bograd S, Harris JE. Effect of gemcitabine on immune cells in subjects with adenocarcinoma of the pancreas. Cancer Immunol Immunother. 2005;54:915–925. doi: 10.1007/s00262-004-0638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halbrook CJ, Pontious C, Kovalenko I, Lapienyte L, Dreyer S, Lee HJ, Thurston G, Zhang Y, Lazarus J, Sajjakulnukit P. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 2019;29:1390–1399. doi: 10.1016/j.cmet.2019.02.001. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Q, Li Y, Niu Z, Zong Y, Wang M, Yao L, Lu Z, Liao Q, Zhao Y. Atorvastatin (Lipitor) attenuates the effects of aspirin on pancreatic cancerogenesis and the chemotherapeutic efficacy of gemcitabine on pancreatic cancer by promoting M2 polarized tumor associated macrophages. J Exp Clin Cancer Res. 2016;16(35):33. doi: 10.1186/s13046-016-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yao L, Wang M, Niu Z, Liu Q, Gao X, Zhou L, Liao Q, Zhao Y. Interleukin-27 inhibits malignant behaviors of pancreatic cancer cells by targeting M2 polarized tumor associated macrophages. Cytokine. 2017;89:194–200. doi: 10.1016/j.cyto.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Prenen H, Mazzone M. Tumor-associated macrophages: a short compendium. Cell Mol Life Sci. 2019;76:1447–1458. doi: 10.1007/s00018-018-2997-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pienta KJ, Machiels JP, Schrijvers D, Alekseev B, Shkolnik M, Crabb SJ, Li S, Seetharam S, Puchalski TA, Takimoto C. Phase 2 study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 (CCL2), in metastatic castration-resistant prostate cancer. Investig New Drugs. 2013;31:760–768. doi: 10.1007/s10637-012-9869-8. [DOI] [PubMed] [Google Scholar]
- 65.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–3446. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- 66.Palmieri EM, Menga A, Martín-Pérez R, Quinto A, Riera-Domingo C, De Tullio G, Hooper DC, Lamers WH, Ghesquière B, McVicar DW. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 2017;20(7):1654–1666. doi: 10.1016/j.celrep.2017.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Germano G, Frapolli R, Belgiovine C, Anselmo A, Pesce S, Liguori M, Erba E, Uboldi S, Zucchetti M, Pasqualini F. Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell. 2013;11(2):249–262. doi: 10.1016/j.ccr.2013.01.008. 23. [DOI] [PubMed] [Google Scholar]
- 68.Piaggio F, Kondylis V, Pastorino F, Di Paolo D, Perri P, Cossu I, Schorn F, Marinaccio C, Murgia D, Daga A. A novel liposomal clodronate depletes tumor-associated macrophages in primary and metastatic melanoma: anti-angiogenic and anti-tumor effects. J Control Release. 2016;223:165–177. doi: 10.1016/j.jconrel.2015.12.037. [DOI] [PubMed] [Google Scholar]
- 69.Angevin E, Tabernero J, Elez E, Cohen SJ, Bahleda R, van Laethem JL, Ottensmeier C, Lopez-Martin JA, Clive S, Joly F. A phase I/II, multiple-dose, dose-escalation study of siltuximab, an anti-interleukin-6 monoclonal antibody, in patients with advanced solid tumors. Clin Cancer Res. 2014;20:2192–2204. doi: 10.1158/1078-0432.CCR-13-2200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material