Abstract
The use of polymeric nanoparticles (NPs) as therapeutics has been steadily increasing over past decades. In vivo imaging of NPs is necessary to advance the therapeutic performance. 19F Magnetic Resonance Imaging (19F MRI) offers multiple advantages for in vivo imaging. However, design of a probe for both biodistribution and degradation has not been realized yet. We developed polymeric NPs loaded with two fluorocarbons as promising imaging tools to monitor NP biodistribution and degradation by 19F MRI.
These 200 nm NPs consist of poly(lactic-co-glycolic acid) (PLGA) loaded with perfluoro-15-crown-5 ether (PFCE) and PERFECTA. PERFECTA/PFCE-PLGA NPs have a fractal sphere structure, in which both fluorocarbons are distributed in the polymeric matrix of the fractal building blocks, which differs from PFCE-PLGA NPs and is unique for fluorocarbon-loaded colloids. This structure leads to changes of magnetic resonance properties of both fluorocarbons after hydrolysis of NPs.
PERFECTA/PFCE-PLGA NPs are colloidally stable in serum and biocompatible. Both fluorocarbons show a single resonance in 19F MRI that can be imaged separately using different excitation pulses. In the future, these findings may be used for biodistribution and degradation studies of NPs by 19F MRI in vivo using “two color” labeling leading to improvement of drug delivery agents.
Keywords: Fractal nanoparticles, Fluorocarbons, PLGA, SANS, 19F MRI, Degradation
1. Introduction
Polymeric nanotherapeutics have been developed for several decades, as they promise to improve the delivery of drugs and reduce side effects, and thus can have a strong impact on a treatment of various diseases [1–3]. Nevertheless, current drug delivery systems are still hampered by several drawbacks, to mention a few: low accumulation at the target site, insufficient control of drug release, and rapid blood clearance [3,4]. Monitoring biodistribution and degradation of NPs in vivo is essential to overcome these drawbacks, which has led to increased interest in theranostic NPs that additionally encapsulate an imaging agent [5–7]. Labeling NPs for monitoring both biodistribution and degradation in vivo without being limited by penetration depth remains a challenge to this day. Here, we developed polymeric NPs labeled with two different fluorocarbons, which hold a huge potential to act as a biodistribution and degradation probe with quantitative 19F MRI, due to their structural properties.
“Hot spot” fluorine MRI, which uses the magnetic moment of 19F nucleus, in addition to proton, has gained a lot of attention for clinical imaging over past years [8,9]. The signal from the 19F nucleus is directly detected, in contrast to SPIO or gadolinium-based labels, which are indirectly observed by modulation of the proton signal. The 19F nucleus has a unique combination of characteristics for imaging: (1) it has no endogenous background signal [10], (2) it displays a broad chemical shift for simultaneous “multi-color” in vivo tracking of different compounds [11–14], and, finally, (3) the obtained signal is linearly correlated to the amount of fluorine atoms, enabling quantitative imaging [15]. These properties make 19F MRI suitable for in vivo tracking of the biodistribution of labeled cell therapies [15–19], drug carriers [20,21], and molecular imaging agents [22]. None of the other imaging modalities combine all of these features.
Typically, liquid perfluorocarbons are used as a label for 19F MRI [8,10]. Perfluorocarbons contain high amount of fluorine nuclei and display balanced relaxation times that yield high signal intensity. Perfluoro-15-crown-5 ether (PFCE) is an ideal candidate for imaging, as it contains 20 chemically-equivalent fluorine nuclei that result in a single resonance signal, avoiding chemical shift artifacts. More recently, a new single-resonance probe PERFECTA, which is a crystalline solid at physiological temperature and contains 36 equivalent fluorine nuclei, was introduced [23].
Despite advantages for imaging, stabilizing perfluorocarbons in an aqueous solution is challenging, due to their hydrophobic and lipophobic character [24]. Typical imaging probes for 19F MRI are nanoemulsions stabilized with phospholipids or core-shell capsules with a polymeric shell and a core of a liquid perfluorocarbon [8,10,25,26]. Recently, we described PFCE-poly(lactic-co-glycolic acid) (PLGA) NPs that have a fractal multicore structure which contains multiple PFCE domains within one polymeric particle [27–29]. These NPs have been successfully used as cell tracking agents with 19F MRI. Moreover, they are cleared several times faster in vivo compared to core-shell systems, which is an advantage for biomedical applications [30].
For drug delivery applications tracking not only biodistribution, but also degradation of NPs is important. There are only few potential 19F labels, which could be used for in vivo tracking of NP break-down. However, these labels are unsuitable to monitor the biodistribution of the intact label [31,32]. In fact, there are major differences between tracking agents for NP biodistribution and NP degradation. Probes for NP degradation have an extremely short transverse relaxation time T2 to completely suppress the signal of 19F nuclei in the intact material. The suppression of the signal can be achieved either by sterically blocking fluorine atoms [31] or by coupling paramagnetic metal chelates, such as gadolinium [32,33], with the fluorinated compound. Thus, the 19F MRI signal can be detected only after degradation of the nanomaterial, hampering in vivo tracking of the intact nanomaterial. Simultaneous tracking of biodistribution and degradation of NPs have not been realized so far.
We hypothesized that combining two fluorinated molecules within the same particle could enable design of a probe for both biodistribution and degradation monitoring, given that both fluorocarbons separate from each other upon degradation. Due to miscibility gap between the perfluorocarbon and aqueous phase, it is unlikely that the two perfluorocarbons that are encapsulated within a core-shell structure will diffuse apart after degradation of the shell. However, for fractal structure this behavior was not clear. Therefore, the goals of this work were to develop nanoparticles loaded with two different fluorocarbons to study their colloidal properties, with focus on the structure and fluorocarbon interface, and, finally, to investigate their magnetic resonance properties upon hydrolysis, providing a proof of principle for our concept.
We decided to focus on PFCE and PERFECTA [23] as both molecules display a single resonance signal that would be advantageous for imaging allowing the use of common sequences. As a polymer for encapsulation we used PLGA, as this is a biodegradable and biocompatible polymer, which finds a broad use in drug delivery and controlled release applications [34]. We have developed the production method and performed intensive structural characterization. These data showed that nanoparticles have structural properties that potentially enable tracking of biodistribution and degradation, confirming the hypothesis. Moreover, nanoparticles were shown to be biocompatible and could be imaged with 19F MRI using a conventional sequence within a short time demonstrating that they will be suitable for in vivo imaging.
2. Materials and methods
2.1. Materials
Water was purified with a purification system from Merck Millipore. The following chemicals were used as received: poly(d,l-lactide-co-glycolide) (PLGA) Resomer® RG 502H, lactide:glycolide molar ratio 50:50 (Evonik Industries AG, Essen, Germany). 1,1,1,3,3,3-hexafluoro-propan-2-ol, ≥99% (HFIP) and poly(vinyl alcohol) (PVA, Mw 9000–10000 g/mol, 80% hydrolyzed, Prod. Nr. 360627, MKBF2590V) from Sigma Aldrich, Germany; dichloromethane, Ph. Eur. (DCM) (Merck, Darmstadt, Germany). Perfluoro-15-crown-5-ether (PFCE), 99%, was purchased from Exfluor, Round Rock USA. PERFECTA was synthesized as described previously [23]. For cell culture X-VIVO 15 medium (Lonza, Belgium) supplemented with 2% human serum was used. Phosphate-buffer saline (PBS) (Braun, Germany) was used to resuspend NPs for cell experiments and for cell washing.
2.1.1. Preparation of the NPs
For the synthesis of the NPs loaded with PERFECTA and PFCE, PERFECTA (100 mg, 0.1 mmol), PFCE (0.9 mL, 1.6 g, 2.8 mmol), and PLGA (100 mg, Mw = 7–17 kDa) were dissolved in HFIP (3 mL). The resulting solution was rapidly added with a glass pipette to an aqueous solution of poly(vinyl alcohol) (25.5 g, 1.96 wt-%) in a round bottom flask while sonication was started. The entire mixture was sonicated in ice-water bath for 3 min at an amplitude of 40% (3.2 mm microtip, Branson digital sonifier s250). After sonication the resulting emulsion was transferred into a beaker and stirred overnight at room temperature to evaporate the solvent (the evaporation of solvent seemed faster in a beaker).
The particles were isolated by centrifugation at 16,087g (11000 rpm) for 35 min in 50 mL centrifugation tubes and resuspended in 25 g of water followed by another centrifugation step. After washing, particles were resuspended in 4 mL of water, frozen with liquid N2 and freeze-dried. The resulting product was a white powder with a yield of typically 100 mg. Typical encapsulation yield of PERFECTA was 8.2 ± 1.7 wt-% and of PFCE 15 ± 4.7 wt-% (n = 3)
PFCE-PLGA NPs were prepared using a miniemulsion formulation method as described previously [27,28]: PLGA (100 mg) was dissolved in 3 mL dichloromethane followed by addition of perfluoro-15-crown-5 ether (0.9 mL). The resulting double phase liquid was rapidly mixed by pipetting it up and down and added with a glass pipette to solution of poly(vinyl alcohol) (25.5 g, 1.96 wt-%) in a round bottom flask while sonication was started. As PFCE does not mix with PLGA in DCM, PFCE and PLGA/DCM were pipetted up and down during the addition to ensure that both phases are added simultaneously. The entire mixture was sonicated in ice-water bath for 3 min at an amplitude of 40% (3.2 mm microtip, Branson digital sonifier s250).
After sonication, dichloromethane was evaporated at 4 °C overnight under stirring to achieve solidification of the particles. The particles were isolated by centrifugation at 16,087g for 35 min in 50 mL centrifugation tubes and resuspended in 25 g of water. The washing step was repeated three times with sonication in ultrasonic bath after second washing (sonication bath, Diagenode Bioruptor). After washing, particles were resuspended in 4 mL of water, frozen with liquid N2 and freeze-dried. The freeze-dried particles were collected as a white powder with a typical yield of 100 mg.
2.2. Characterization methods
CryoSEM was measured at JEOL 6330 Cryo Field Emission Scanning Electron Microscope (FESEM). The samples (8 μL) were pipetted in 2 rivets, which were then placed together. Next, the samples were frozen in liquid nitrogen slush and placed in an Oxford Alto 2500 cryo station with a cryo-transfer device. There the top rivet was broken and the sample was heated to −95 °C for 5 min, followed by a coating of 60/40 Au/Pd and transfer to the Cryo-SEM.
Multiangle DLS and Static LS were measured at ALV compact goniometer system equipped with ALV7004 correlator, ALV/LSE-5004 Goniometer, ALV / Dual High QE APD detector unit with fiber splitting device a set-up of 2 off detection system and a Uniphase Model 1145P He-Ne Laser. The laser wavelength and power were 632.8 nm and 22 mW, respectively. The temperature was controlled by a Julabo CF41 thermostatic bath. Water for dilutions was filtered with 0.45 μm hydrophilic filters. The concentration of samples was 0.01 mg/mL. For size determination under physiological conditions, NPs were added to RPMI or RPMI with 5 vol-% FBS, which were filtered through Millex-LCR filter, 0.45 μm pore size. The measurements were repeated at least three times.
DLS was measured at scattering angles θ = 30–150° in 10° steps. Data analysis was done with HDRC software, which was kindly provided by Prof. Manfred Schmidt, University of Mainz, Germany. The apparent hydrodynamic radii at different angles were obtained from a biexponential fitting of autocorrelation function, as it provides a good approximation of the radius for particles with polydisperse, monomodal size distribution. The absolute inverse z-averages of hydrodynamic radii were obtained by extrapolating the apparent radii q → 0. The second cumulant values μ2 were obtained from cumulant fitting at θ = 90°
Size determination under physiological conditions was done using the multicomponent analysis with HDRC software. Here, the autocorrelation function of nanoparticle-protein mixture was spited in a triexponential fit function for FBS (known component) and biexponential fit function for NPs (unknown component) [35]. The fitting coefficients of FBS were previously obtained from triexponential fitting of serum without NPs.
SLS for determination of radius of gyration Rg was measured at θ = 30–60° in 3° steps. This measurement range was selected, as it is needed for Guinier analysis (q × Rg ≤ 1).
The radius of gyration was determined from the angular dependence of the Rayleigh ratio according to the Guinier equation. The Rayleigh ratio Rθ was obtained as follows:
With a refractive index of solvent nwater = 1.333, refractive index of reference ntoluene = 1.494 and Rayleigh ratio of toluene Rtoluene = 1. 02 × 10−3 m−1 calculated as described by Wu [36].
Nuclear magnetic resonance spectroscopy was measured on Bruker Avance III 400 MHz spectrometer equipped with BBFO probe at 298 K (quantitative measurements and NOESY), or at Bruker Avance III 500 MHz spectrometer equipped with Prodigy BBO cryoprobe (relaxation measurements). Particles (typically 5–8 mg) were dissolved in 600 μL D2O. A 90° pulse was calibrated prior measurements.
2D Nuclear Overhauser Enhancement Spectroscopy (NOESY) was performed using a spectral width spanning from (−102) to (−62) ppm for 19F (377 MHz) using 2048 (F2) × 128 (F1) points. 96 scans per increment were acquired using an interscan relaxation delay of 2.0 s and a mixing time for dipole-dipole relaxation of 250 ms.
Longitudinal relaxation times T1 were measured using an inverse recovery sequence and transverse relaxation times T2 using the CPMG sequence. Relaxation times were measured with spectral width spanning from typically (−101) to (−62) ppm. At least 12 data points were acquired, which were then used for data fitting. An interscan delay was set to 5 × T1, which was estimated using a 1D version of the inversion recovery sequence and noting the null crossing point.
Quantitative 19F NMR was for determination of fluorocarbon content was measured in D2O as a solvent using known amount of NPs with trifluoroacetic acid (typically 1 μL) or sodium triflate (typically 1 mg) as a reference for quantification. An interscan relaxation delay was 20 s with TFA or 30 s with sodium triflate. The number of scans was between 8 and 32 depending on concentration of fluorinated compound.
Data evaluation was done with MestreNova 10.0 from Mestrelab (quantification, relaxation times) or with TopSpin 3.5 from Bruker (NOESY).
19F and 13C solid state NMR were measured on a Varian 400 MHz solid state NMR system using a 3.2 mm T3 design probe resonant for 19F at 376.272 MHz and 13C at 100.569 MHz. Magic angle spinning was employed at 16 kHz with 90° pulses of 2 μs for 19F and 3.75 μs for 13C. Ramped cross polarization from 19F to 13C was set-up on PTFE (Teflon). The Hartmann-Hahn matching condition was achieved with 78 kHz rf-field strength on 19F and 62 kHz on 13C with the difference the MAS frequency. Spinal decoupling with a pulse length of 7.2 μs and 7°phase was used to decouple 19F from 13C.
XRD was measured on a Panalytical Empyrean in reflection mode with fine-focus sealed tube, and PIXcel3D detector, using Cu Kα radiation. The scan range was from 2 to 50° 2-theta, with a step size of 0.013°.
Small Angle Neutron Scattering (SANS) was measured at D11 small angle diffractometer at ILL Grenoble and at Sans2d at ISIS spallation neutron source UK [37]. The description of settings of sans2d diffractometer is given in detail in supplementary information.
SANS measurements were done on D11 refractometer using 3 sample-detector distances of 1.5 m, 8 m and 39 m, with corresponding collimation distances of 8 m, 8 m and 40.5 m. The wave-length used was 6 Å, with a FWHM of 9%. Cylindrical Hellma cells were placed in a thermostated sample changer, the temperature was kept constant at 25 °C. A neutron beam of 13 mm in diameter was employed.
Scattering intensities were recorded using a two-dimensional position-sensitive 3He detector with a pixel size of 7.5 × 7.5 mm2 and an active area of 96 × 96 cm2. A dark current was measured and subtracted from all data. Solvents were measured and subtracted, too, taking into account the transmission values of all samples and solvents as obtained by a dedicated measurement. The data were first corrected pixel by pixel and in a second step radially regrouped in order to obtain a one-dimensional scattering curve in the form Intensity vs. Q. Data were put on an absolute scale making use of the secondary calibration standard H2O (cross-calibrated against h/d polymer blends), which has been measured in a 1 mm Hellma cell. The differential scattering cross section for H2O with a thickness of 1 mm has been determined to 0.983 1/cm for D11 at 6 Å neutron wavelength and 25 °C. Data were reduced using ILL’s standard software package LAMP [38,39].
Data analysis was done using the NIST SANS package, version 7.50, for Igor Pro (Wavemetrics) [40].
2.2.1. Cell culture and labeling
Peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy individuals after informed consent, using Ficoll density centrifugation (Lymphoprep, STEMCELL Technologies, Vancouver, Canada). Adherent monocytes were cultured in X-VIVO 15 medium supplemented with 2% human serum, and in the presence of interleukin-4 (300 U/mL) and granulocytemonocyte colony stimulating factor (450 U/mL) to obtain immature dendritic cells (DCs). Day 3 cells were harvested and labeled with NPs (resuspended in PBS right before the labeling) at concentration 2 mg of NPs/ 1×106 cells.
2.2.2. Cell viability
To investigate the influence of labelling on cell viability we performed an MTT assay. For this, 1×106 DCs were incubated in the presence of NPs for 72 h. Here, both PFCE-PLGA-NPs and PERFECTA/PFCE-PLGA-NPs samples were tested, including positive and negative control. After incubation, the excess of the label was removed by gentle washing with PBS. Next, cells were collected and placed in a 96-well flat-bottom plate, washed two time with PBS (100 μL/well) with centrifugation of 2 min between each wash. Next, 60 μL X-VIVO medium with 10 μL MTT (3-(4,5-dimethylthia zol-2-yl)-2,5-diphenyltetrazolium bromide) (concentration 4 mg/mL) was added to each well, followed by 1 h incubation at 37 °C. After incubation, the plate was centrifuged for 2 min and 100 μL of lysis buffer (isopropanol, 10% SDS, 2 N HCL, deionized water) was added to each well, and the plate was incubated for 15 min in dark at room temperature. Before the measurement, samples were resuspended to remove any precipitate of crystals. The plate was measured with iMark™ microplate reader (Bio-Rad, the Netherlands) at 595 nm. Cell not loaded with NPs were used as negative control and cells treated with DMSO were used as a positive control.
2.2.3. 19F magnetic resonance imaging
19F MRI was performed on a pre-clinical 11.7 T MRI scanner (Biospec 117/16, 500 MHz, Bruker, Ettlingen, Germany). PERFECTA/PFCE-PLGA NPs were homogeneously dissolved at a concentration of 10 mg/mL, the PFCE-only NPs and PERFECTA emulsion were diluted to contain a corresponding amount of fluorine atoms. The phantoms were imaged with a selective excitation 3D RARE-sequence with the following parameters: TR = 1500 ms, TE = 6.62 ms, RARE-factor 44, matrix size 64x44, FOV 32 × 22 mm, 16 slices with a thickness of 2 mm, 5 averages for a scan time of 2 min. To selectively excite, a transmission bandwidth of 1000 Hz was used, PERFECTA was excited at 470,753,521 Hz and PFCE at 470,744,387 Hz. To determine concentration limit of the probe we scanned the phantoms at various voxel sizes. The keep clinical applicability and in vivo relevance in mind, the scan time and reference voxel size were chosen accordingly. For scan time we used 8 min per excitation frequency to make a total of 16 min for both PFCs. For voxel size we aimed for 1 × 1 × 2 mm, as this is a sufficient resolution in our experience. Detection limit was determined to be a voxel size of 0.33 × 0.33 × 2 mm within the 8 min, this is a voxel size 10-fold smaller than the eventual in vivo voxel size. Therefore, a 1/10th concentration of the phantom concentration is the concentration limit.
3. Results and discussion
3.1. Synthesis and characterization of the NPs
NPs loaded with liquid fluorocarbons can be typically produced with a miniemulsion formulation method using non-fluorinated solvents [28]. PERFECTA, however, is a solid, which is poorly soluble in most non-fluorinated organic solvents. Our usual miniemulsion formulation method for PFCE-PLGA-NPs with dichloromethane as a solvent did not yield a sufficient encapsulation of PERFECTA. Therefore, we used 1,1,1,3,3,3-hexafluoro-pro pan-2-ol (HFIP), which is suitable to dissolve PLGA, PERFECTA, and PFCE. The solution of PLGA, PERFECTA, and PFCE in HFIP was added to aqueous solution of poly(vinyl alcohol) (PVA) as a nonionic surfactant and emulsified using sonication to provide the energy that is required for encapsulation of fluorocarbons [28]. As HFIP is miscible with water, the final process is a combination of miniemulsion and nanoprecipitation technique.
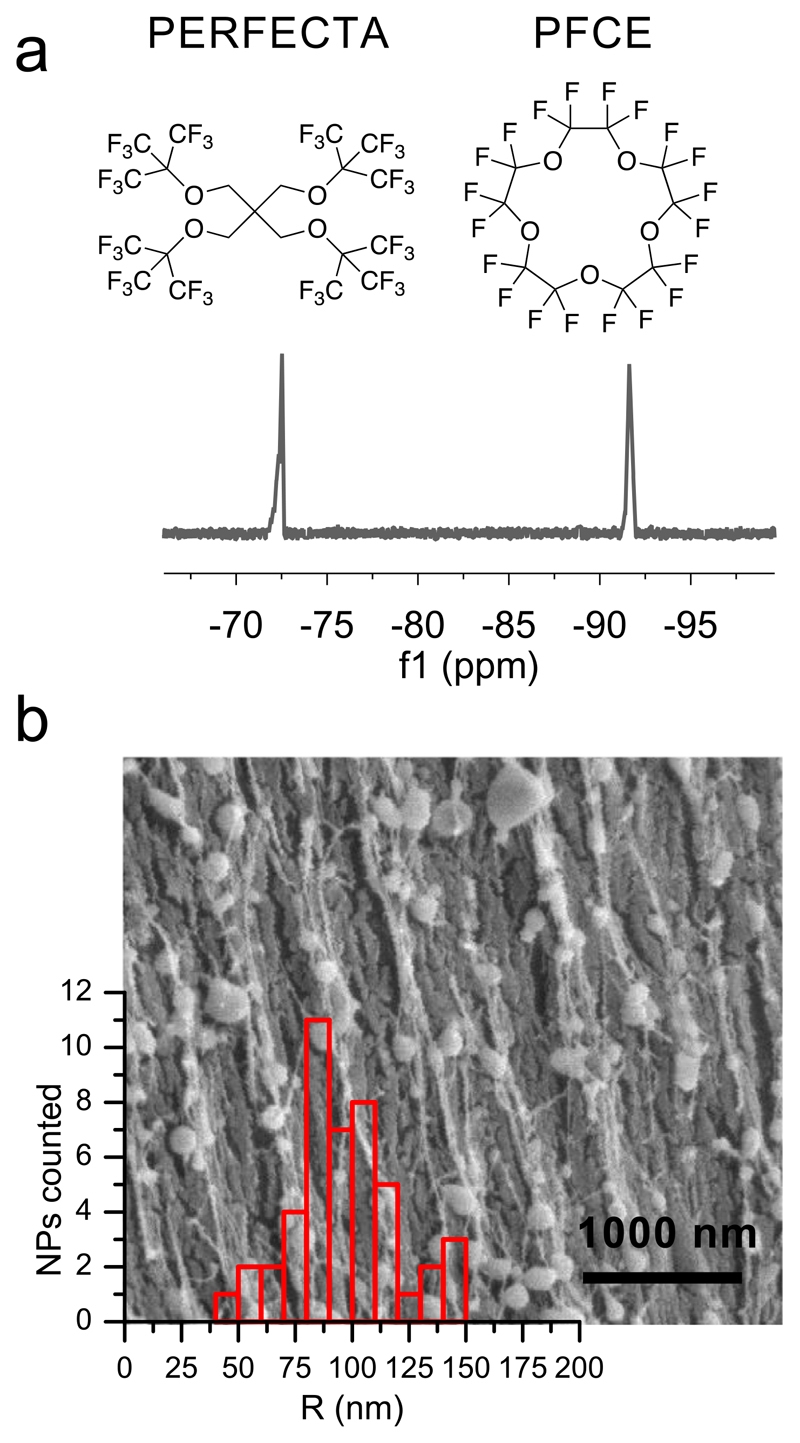
19F NMR spectroscopy on purified, freeze-dried NPs after redispersion in water demonstrated that both fluorocarbons were encapsulated in the NPs (Fig. 1a). The content of PERFECTA in the final NPs was 8.7 wt-% and of PFCE 10 wt-%, with some batch-to-batch variation (compare materials and methods), as shown by NMR spectroscopy. Cryogenic Scanning Electron Microscopy (cryo-SEM, Fig. 1b) demonstrated that the mean radius of the NPs was 96 ± 23 nm, indicating a certain polydispersity which is typical of PLGA NPs [34]. The average hydrodynamic radius of NPs was 164 ± 7 nm (second cumulant μ2 (90°) = 0.2) as determined by multi-angle Dynamic Light Scattering (scattering angle θ = 30–150°, compare Fig. S1). The radius of gyration Rg was 167 nm, as revealed by Guinier analysis of Static Light Scattering (SLS, θ = 30–60°, Fig. S1). The difference between multi-angle DLS and cryoSEM results from higher sensitivity of light scattering to bigger particles at small scattering angles [41]. Overall, the size distribution was comparable to PFCE-PLGA-NPs [28]. The zeta-potential of NPs was –5 mV ± 0.4 mV (in 5 mM sodium chloride), which is typical for colloids that are sterically stabilized with a non-ionic surfactant, such as PVA.
Fig. 1.
Characterization of PERFECTA/PFCE-PLGA NPs. (a) 19F NMR demonstrates that both PFCE and PERFECTA were encapsulated in the NPs (471 MHz in D2O). (b) CryoSEM shows that the majority of NPs have a mean radius of 96 ± 23 nm (47 NPs measured; cNP = 10 mg/mL in water). However, small numbers of larger and smaller particles could be detected. Scale bar 1000 nm.
While this method works well for obtaining PLGA NPs only loaded with PFCE, it was not possible to obtain PLGA-NPs loaded only with PERFECTA that had a narrow size distribution (compare supplementary information, section 1). Thus, the presence of the two fluorocarbons seem to be necessary to formulate PVA-stabilized PLGA NPs loaded with PERFECTA.
3.2. Structural characterization of the NPs
For biomedical applications, physicochemical properties of NPs, in particular the internal structure of NPs is important, as it can affect drug loading and/or the mechanism of degradation. Therefore, we performed an in-depth characterization of PERFECTA/PFCE-PLGA-NPs with different analytical techniques, to determine the arrangement of fluorocarbons inside the polymeric matrix.
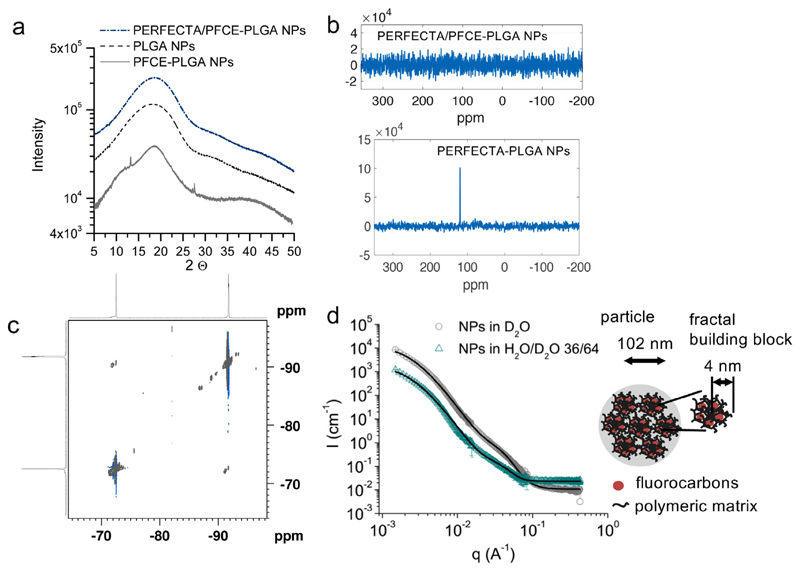
As pure PERFECTA is a crystalline solid at room or physiological temperature [23], we first measured powder X-ray diffraction (PXRD) to examine the crystallinity. PXRD patterns of PERFECTA/PFCE-PLGA-NPs were similar to PXRD patterns of PFCE-PLGA-NPs suggesting an amorphous structure of the NPs (Fig. 2a). As no crystalline peaks typical for PERFECTA [8] were detected, PXRD may indicates that PERFECTA loses its crystallinity upon encapsulation in PLGA matrix. Furthermore, cross polarization measurements between 19F and 13C nuclei by solid state NMR spectroscopy did not show any cross-polarization signal between the fluorocarbons (Fig. 2b, upper spectrum, corresponding 13C-spectra Fig. S2). As a control for these measurements we used PERFECTA-PLGA-NPs (compare section 1 in SI for synthesis and characterization). In contrast to PERFECTA/PFCE-PLGA-NPs, PERFECTA-PLGA-NPs displayed cross polarization between 19F and quaternary 13C nucleus of PERFECTA-molecule (Fig. 2b, lower spectrum). The absence of cross polarization signal in PERFECTA/PFCE-PLGA-NPs indicates that PERFECTA molecules have higher mobility within the polymeric matrix compared to PERFECTA-PLGA-NPs. Thus, PERFECTA molecules in PERFECTA/PFCE-PLGA-NPs show a liquid-like behavior, indicating that PERFECTA is in different environment and possibly close to PFCE.
Fig. 2.
(a) Powder X ray diffraction of PFCE/PERFECTA-PLGA-NPs compared to PFCE-PLGA-NPs and PLGA-NPs. No crystalline peaks could be detected, indicating that PERFECTA is mixed with PFCE and polymeric matrix. (b) 13C solid state NMR spectra obtained by cross-polarization from 19F. (c) In 19F-19F NOESY measurements PFCE (−92 ppm) and PERFECTA (−73 ppm) display NOE effect (cross-peak between both compounds), demonstrating that PFCE and PERFECTA are close to each other. 376.5 MHz, in D2O. (d) SANS scattering patterns of PERFECTA/PFCE-PLGA-NPs in D2O and H2O/D2O 36/64 (match conditions for PFCE) with simultaneous fit of the particle form factor with a fractal sphere model. SANS reveals that PERFECTA/PFCE-PLGA-NPs have a fractal structure with spherical building blocks that contain PFCE, PERFECTA, and polymer. c (NP) = 10 mg/mL.
To further study the interactions between both fluorocarbons in PERFECTA/PFCE-PLGA-NPs, we measured 19F-19F Nuclear Over-hauser Enhancement Spectroscopy (NOESY) on NPs in solution. NOESY detects cross-space magnetization transfer between different nuclei, when the distance between these nuclei is smaller than 6 Å. An NOE was observed between 19F nuclei of PFCE and PERFECTA, demonstrating that PFCE and PERFECTA molecules must be close to each other (Fig. 2c). This technique showed that F atoms of the molecules are close within the NPs, but not being a quantitative measurement, it is not possible to evaluate if all atoms or just some of them interact and to which extent.
Finally, we performed Small Angle Neutron Scattering (SANS) experiments at different solvent contrasts (100% deuterium oxide, water/deuterium oxide ratio 36/64 and 61/39 (v:v)) to describe the local structure of PERFECTA/PFCE-PLGA-NPs. In 100% deuterium oxide, all components of the NPs contribute to the scattering contrast. At a ratio of 36/64 (v:v) PFCE is matched and it does not contribute to the scattering (Fig. 2d, Table S2, Fig. S4 for results from a different beamline). For data analysis we performed model fitting of the absolute scattering intensity- the particle form factor, using a simultaneous fitting approach to reduce the number of fitted parameters.
Fluorocarbon-loaded colloids typically have a core-shell structure [8]. However, we have previously shown by SANS analysis that PFCE-PLGA-NPs have a fractal multi-core structure that consists of a fractal assembly of core-shell building blocks (NIST-model FractalPolyCore) [27]. Nevertheless, the form factor of PERFECTA/PFCE-PLGA-NPs could not be described by either of these models. In contrast, a fractal sphere model developed by Teixeira provided a good fit of the scattering curves (Fig. 2d, Table 1) [42]. This model describes a NP as a fractal assembly of spherical building blocks formed of PFCE, PERFECTA, and PLGA. The final particles are composed of spherical fractal building blocks of 4 nm, and have a radius of gyration Rg of 102 nm, as derived from the correlation length. A schematic representation of the interior structure of the PERFECTA/PFCE-PLGA-NPs is shown in Fig. 2 [42]. In PERFECTA/PFCE-PLGA-NPs, the encapsulated fluorocarbons are homogenously distributed between PLGA chains forming mixed spherical fractal building blocks. This arrangement is different from PFCE-PLGA-NPs, in which PFCE forms fractal building blocks composed of a fluorocarbon core surrounded by a PLGA shell (compare also Fig. S3 for differences between fractal sphere and fractal core-shell, NIST-models FractalPolySphere or FractalPolyCore). We expect that both fluorocarbons form very small clusters between polymeric chains. The resulting Rg of 102 nm is close to the value of the mean radius of 96 nm obtained from cryo-SEM analysis, confirming the results of model fitting.
Table 1.
Simultaneous fitting of SANS patterns of PERFECTA/PFCE-PLGA NPs in D2O and H2O/D2O 36:64 (v:v) using a fractal sphere model.i.
| Parameter | D2O | H2O/D2O (36/64) |
|---|---|---|
| Vol. fraction | 0.0072 | 0.0072 |
| Radius of fractal block (nm) | 4.1 | 4.1 |
| Polydispersity of fractal block | 0.36 | 0.30 |
| Fractal dimension | 3.05 | 3.05 |
| Correlation length (nm) | 41 | 41 |
| SLD sphere (Å−2) | 4.74 × 10−6 | 3.15 × 10−6 |
| SLD solvent (Å−2) | 6.36 ×10−6 | 3.87 ×10−6 |
| Bkg | 0.011 | 0.023 |
| Chi2/Npts | 3.5 | |
| Rg of NPs (nm) | 102 |
Volume fraction and SLD of solvent were fixed; block radius, fractal dimension and correlation length were fitted simultaneously.
Based on the content of PFCE and PERFECTA obtained from NMR spectroscopy, the expected scattering length density (SLD) of a fractal building block can be estimated to be 2.6 × 10−6 Å−2. In both solvents, the SLD obtained from the model fit is between the calculated SLD of particles and the SLD of solvent, indicating that the NPs might be hydrated with a solvent (Table 1).
NMR relaxation time measurements confirmed the structural difference between PERFECTA/PFCE-PLGA-NPs and PFCE-PLGA-NPs (Table 2). Both longitudinal (T1) and transverse (T2) relaxation times of PFCE were shorter in PERFECTA/PFCE-PLGA-NPs than in pure PFCE-PLGA-NPs. The effect is especially pronounced for T2 that is more sensitive to the environment of the nucleus than T1. The shortening of relaxation times is typical for molecules in confined environment, due to their lower mobility [43]. According to the SANS model, in PERFECTA/PFCE-PLGA-NPs, both fluorocarbons are distributed between the polymeric chains. In contrast, in PFCE-PLGA-NPs, PFCE forms several domains within the nanoparticle and has therefore, higher mobility. Additionally, the presence of PERFECTA can further contribute to the reduction of T2.
Table 2.
Relaxation times of PERFECTA/PFCE NPs before and after hydrolysis compared to PFCE NPs.i
| NPs | PFC | T1 [ms] | T2 [ms] |
|---|---|---|---|
| PERFECTA/PFCE-PLGA-NPs | PERFECTA | 542 | 145 |
| PFCE | 620 | 273 | |
| PFCE-PLGA-NPs NPs after hydrolysis |
PFCE | 794 | 492 |
| PERFECTA/PFCE-PLGA | PERFECTA | 520 | 207 |
| PFCE | 684 | 372 | |
| PFCE-PLGA | PFCE | 775 | 339 |
NPs in D2O, 471 MHz.
Usually, fluorocarbon-loaded colloids form core-shell structures with a core of liquid fluorocarbon, due to poor miscibility of fluorocarbons with both aqueous and organic emulsion phases. The only examples where the internal structure was different include PFCE-PLGA-NPs reported previously by our group [27] and multicore micelles formed by thermosensitive polymers with covalently-attached fluorocarbon and hydrocarbon side chains [44,45]. At last, the driving force for the formation of domains was still the tendency of fluorocarbons to segregate from hydrocarbon side chains. However, here the driving force for the formation of fractal structure seems to be different.
Possible explanation for the formation of such a fractal sphere structure composed of mixed PERFECTA, PFCE, and PLGA building blocks, in which all components are distributed homogenously, could be related to the use of HFIP as a solvent. In fact, HFIP allows dissolution of both fluorocarbons and PLGA and, being also miscible with water, might diffuse into the aqueous phase upon addition of organic HFIP solution to aqueous surfactant solution promoting the formation of PERFECTA/PFCE-PLGA building blocks arranged in a fractal spherical structure.
Both PERCECTA and PFCE are non-miscible with PLGA, and thus the formation of fractal structure seems not to be thermodynamically favored. Thus, kinetic effects, such as diffusion of solvent, are likely to play a role in the structure formation. Further effects could also influence the formation of the NPs. Particularly, we observed that the impurities in the surfactant, such as poly(propylene glycol) or shorter PVA chains, may generally affect the encapsulation of fluorocarbons, independent of the solvent or the method used.
3.3. 19F magnetic resonance properties alter after degradation of NPs
Finally, we wanted to test if both fluorocarbons can separate from each other upon the breakdown of the polymeric matrix. In the body, PLGA is usually metabolized to lactic and glycolic acid, which then enter into the tricarboxylic acid cycle [34]. After PLGA degradation, the surroundings of both fluorocarbons will change, leading to altered magnetic resonance properties. To study the behavior of both fluorocarbons after degradation, we hydrolyzed PERFECTA/PFCE-PLGA-NPs with aqueous sodium hydroxide solution. The solution of NPs turned from milky to transparent within few minutes after the addition of sodium hydroxide, indicating the success of hydrolysis. In 1H NMR spectra, signals from lactic and glycolic acid monomers could be detected (Fig. S5). In contrast, before the hydrolysis, no PLGA signals were observed in proton spectrum due to the reduced mobility of polymeric chains in the particles (Fig. S5). The choice of using alkaline conditions is related to the 1H NMR measurement after hydrolysis, for avoiding additional proton signals from water or organic acids in the same region as PLGA or PVA protons. Despite NPs would be hydrolyzed in acidic environment within the cell, acidic and alkaline hydrolysis are substitution reactions leading to the same final product.
To further investigate the effect of degradation on the 19F signal, we measured longitudinal and transverse relaxation times, T1 and T2, of both fluorocarbons before and after hydrolysis. Additionally, we compared the results with PFCE-PLGA-NPs (Table 2). After the hydrolysis of PERFECTA/PFCE-PLGA-NPs, T2 of both fluorocarbons increased, indicating that the fluorocarbons became more dynamic after the degradation of PLGA-matrix. In contrast, T2 decreased after degradation in PFCE-PLGA-NPs. Before hydrolysis PFCE-PLGA-NPs displayed longer transverse relaxation T2 compared to PERFECTA/PFCE-PLGA-NPs, with a difference of around 200 ms, which might be attributed to a different internal structure of both types of colloids. After hydrolysis, the difference between T2 of PFCE in PFCE-PLGA-NPs and PERFECTA/PFCE-NPs is only 43 ms. These changes in T2 may suggest that the surrounding environment of PFCE is similar after hydrolysis in both types of NPs.
After hydrolysis we could not observe any visible phase separation of fluorocarbons. In contrast, when a similar amount of PFCE was added to water, the phase separation could be observed by eye. DLS of hydrolyzed PERFECTA/PFCE-PLGA-NPs measured at a scattering angle of 173° showed the radius of 89 nm (PDI = 0.22), indicating that some colloidal structures were still present in the solution. However, the scattering intensity of this solution was lower than that of non-hydrolyzed NPs. The results were obtained without attenuation, while for measurements of intact particles of same concentration 36% attenuation were used (both set automatically by DLS machine). The size determination of hydrolyzed PFCE-PLGA-NPs was not possible due to high polydispersity of the obtained solution; the aggregates were around 200–300 nm. This behavior suggests that both fluorocarbons are still stabilized by the remaining surfactant after hydrolysis of PLGA matrix.
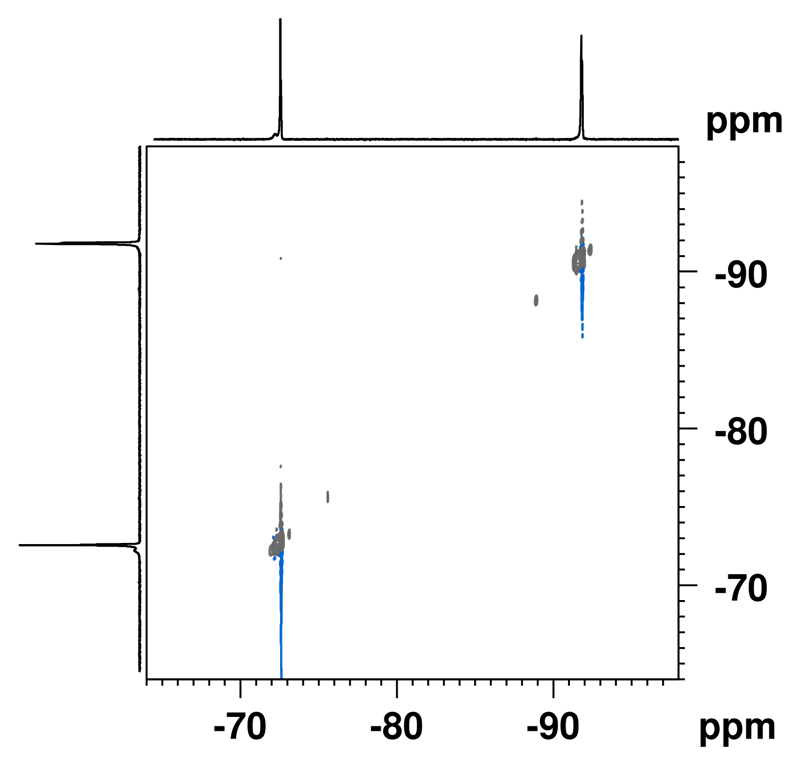
To further obtain information about the environment of both fluorocarbons after hydrolysis, we performed NOESY experiments. In contrast to intact particles, we could not detect sufficient NOE between PFCE and PERFECTA anymore (Fig. 3). However, it was difficult to distinguish between possible NOE-peak and noise after the hydrolysis. NOE is typically observed when the distance between two interacting nuclei is less than 6 Å. Hence, these observations suggest that after the hydrolysis the distance between both fluorocarbons increased. In case PERFECTA was dissolved in PFCE within the fractal structure of intact particles, the separation of both fluorocarbons after hydrolysis would be thermodynamically not favored. Thus, this behavior indicates that in intact NPs, PERFECTA is not completely dissolved in PFCE, but both molecules are still associated with each other to produce detectable NOE.
Fig. 3.
NOE-spectrum of PERFECTA/PFCE-PLGA-NPs after hydrolysis with sodium hydroxide. The absence of NOE-signal indicates the increased distance between both fluorocarbons after degradation of NPs. 400 MHz (1H), 376.5 MHz (19F), in D2O.
Overall the magnetic resonance properties of PERFECTA/PFCE-PLGA-NPs changed after hydrolysis. A simulation of complex in vivo conditions is difficult within an NMR tube. Therefore, here we used a simple method to study the magnetic properties of NPs after the degradation. The complexity of in vivo environment, such as physiological fluids and tissue, interactions with different biomolecules, as well as differences in oxygen content can affect the MR properties of fluorocarbons in vivo. Nevertheless, the change of the magnetic resonance properties provides the proof of our concept showing that the PERFECTA/PFCE-system is promising for tracking not just biodistribution but also degradation of NPs in vivo. Especially, we expect that the different structure of the two used fluorocarbons and the fact that PFCE is a liquid and PERFECTA a solid at physiological temperature, should lead to differences in clearance route of both fluorocarbons after degradation, resulting in different biodistribution. Hence, we expect that combining 19F MRI and relaxation time measurements could provide information on degradation of NPs and the environment of fluorocarbons, and thus holds potential to be used to monitor the biodistribution and degradation of drug carriers in later studies.
3.4. PERFECTA/PFCE-PLGA NPs are suitable as 19F MR imaging agents
The main intended application of these NPs is in vivo imaging agents for by 19F MRI. Therefore, we investigated their stability in biological milieus, performed cytotoxicity tests and 19F MRI.
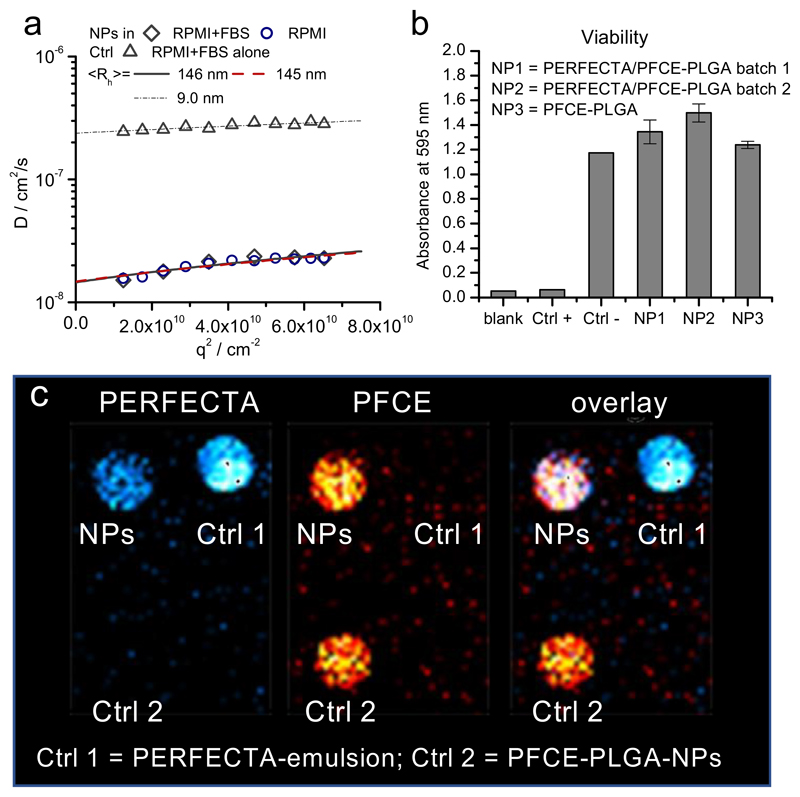
To be used in vivo, NPs need to be colloidally stable in physiological milieu. Therefore, we measured multi-angle DLS between 50° and 150° in cell culture medium with and without fetal bovine serum (FBS, Fig. 4a, for different scale compare also Fig. S6). For data analysis in serum, we used a multicomponent analysis developed by Rausch et al. to separate the scattering background of serum proteins from the scattering of the particles [35]. The angular dependency of resulting diffusion coefficients of NPs in serum-free and serum-containing medium, and of serum in medium without NPs is shown in Fig. 4a. The hydrodynamic radii, which were obtained from extrapolating q → 0, were 145 ± 7 nm in serum-free medium and 146 ± 16 nm in serum-containing medium. Thus, the NPs were colloidally stable and did not agglomerate in presence of serum, indicating only minor adsorption of biomolecules. As NPs are stabilized with PVA as a non-ionic surfactant, we expect that PVA provides sterical stabilization of nanomaterials and reduces the protein corona formation, as already described for PVA-stabilized colloids in the literature [46].
Fig. 4.
(a) Characterization of NPs in serum-containing cell culture medium RPMI 1640 using multi-angle light scattering. The angular dependency of diffusion coefficient D is shown, q = scattering vector that is related to scattering angle. The inverse z-average of radii were calculated using Stockes-Einstein equation from D at q = 0° that was obtained from linear extrapolation q → 0. No changes in size of NPs could be detected in serum-containing medium compared to serum-free medium. c(NP) = 0.01 mg mL−1. The measurements were done in triplicate. (b) PERFECTA/PFCE-PLGA-NPs do not affect cell viability, 1×106 cells were incubated in the presence of 2 mg of particles for 72 h (for each condition), followed by the viability analysis with MTT assay. NP1 and NP2 are two different batches of PERFECTA/PFCE-PLGA NPs. Each batch of NPs was tested as a triplicate (c) 19F MRI image of PERFECTA/PFCE-PLGA NPs (upper row, left, c = 10 mg/mL) with PERFECTA-emulsion (crtl 1; upper row, right, same PERFECTA-loading as NPs) and PFCE-PLGA-NPs (ctrl 2, lower row, 10 mg/mL). Left: selective excitation of PERFECTA (cyan), middle: selective excitation of PFCE, right: composite. RARE-sequence, transmission bandwidth 1 kHz, receiver bandwidth 15 kHz.
To investigate the NP effect on cell viability, dendritic cells (DCs) were incubated for 72 h with PFCE/PERFECTA-PLGA-NPs and PFCE-PLGA-NPs at a concentration usually used for cell labeling and then tested by MTT assay. PFCE-PLGA-NPs have now been approved for a clinical trial (clinicaltrials.gov ID NCT02574377) [11,28,29]. There was no significant difference on cell viability between PFCE-PLGA-NPs and PERFECTA/PFCE-PLGA-NPs (Fig. 4b) showing that both types of NPs are not cytotoxic. Thus, we expect that PERFECTA/PFCE-PLGA-NPs also should not display any in vivo toxicity and be suitable for biomedical applications.
Finally, we performed 19F MR imaging of the NP dispersions (Fig. 4c). As a control we used PFCE-PLGA-NPs and PERFECTA emulsion [23] to show a reference with selective imaging of only one of the molecules. Both PFCE and PERFECTA contain high amount of magnetically identical fluorine atoms and therefore, show singlet signals in their 19F NMR spectra. Importantly, the signals of both fluorocarbons are separated by 20 ppm. This large difference in the resonance frequency between the two molecules can be used for selective excitation. Signals from the controls, which were placed in the same imaging phantom, confirmed that we were detecting only one of the fluorocarbons (Fig. 4c, left panel imaging only of PERFECTA, middle only PFCE, right overlay of both fluorocarbons). As we used a conventional sequence for imaging liquid fluorocarbon emulsions (RARE sequence), we expect it should be possible to image the PERFECTA/PFCE-PLGA-NPs in vivo with signal-to-noise ratios comparable to PFCE-PLGA-NPs and a concentration limit of 1 mg/mL.
4. Conclusion and outlook
19F MRI is a powerful technique in the development of nanomedicine. However, 19F MRI probes are used just to monitor biodistribution of 19F label. Monitoring both biodistribution and degradation of nanoparticles have not been realized thus far. Our goal was to show that the encapsulation of two fluorocarbons within the same particle could be used for imaging of biodistribution and degradation of polymeric drug carriers. Therefore, we developed a fractal nanoscale “two color” MRI probe, loaded with PFCE and PERFECTA. As understanding the colloidal properties is essential for later application of this material, we performed extensive characterization with the emphasis on the internal structure. With 2D 19F NMR and SANS we showed that these NPs have a fractal structure and not a core-shell structure, which is typical for fluorocarbon-loaded colloids [8,10,26]. Thus, our particle can be described as an assembly of spherical building blocks, each of them containing PFCE, PERFECTA, and PLGA. Furthermore, we detected 19F-19F NOE between PFCE and PERFECTA, indicating their close proximity. After the hydrolysis of NPs, we observed changes in 19F relaxation times of both fluorocarbons. Moreover, we could not detect a sufficient NOE anymore, indicating that the distance between the two fluorocarbons had increased. These results provide the proof of our concept that both fluorocarbons can separate from each other after a breakdown of NPs. PFCE and PERFECTA both show single magnetic resonances and could be imaged separately using a selective excitation with imaging parameters suitable for in vivo studies. Given this property, imaging the biodistribution of PFCE and PERFECTA with different colors combined with relaxation time measurements may enable tracking both biodistribution and degradation of PLGA NPs in vivo.
PLGA used in our study is one of the best-established polymers for synthetic therapeutic NPs for clinical and preclinical use [34]. We expect, that NPs described in this work can be tailored to various applications of PLGA or other polymeric NPs, enabling imaging of both biodistribution and degradation of particles. Various techniques for multispectral 19F MRI were developed in the recent years [14,47]. Therefore, we expect that this work could have implications for in vivo biodistribution and drug delivery and release studies in the future.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcis.2019.12.083.
Acknowledgement
The authors kindly acknowledge Geert-Jan Janssen (RU) for help with cryoSEM, Jan van Hest (RU) and Daniella Wilson (RU) for access to chemistry facilities at Radboud University, Manfred Schmidt and Georg Conrad (Gutenberg University of Mainz) for providing the HDRC software for multi-angle DLS analysis, David Hess and PSCM labs for ALV light scattering access at ILL, Sarah Rogers for her support during measurements at ISIS neutron and muon source, Oya Tagit (RUMC), Carl Figdor (RUMC) and I. Jolanda M. de Vries (RUMC) for discussions.
Funding sources
This work was supported by ERC-2014-StG-336454-CoNQUeST. FBB and PM are thankful for funding to Regione Lombardia (Fondo per lo Sviluppo e la Coesione – FAS 2007–2013) and the project 2015-0023-TRANS-ALS is also acknowledged. SANS was supported by the Institut Laue-Langevin (ILL) (doi: https://doi.org//10.5291/ILL-DATA.9-10-1478). Experiments at the ISIS Pulsed Neutron and Muon Source were supported by a beamtime allocation from the Science and Technology Facilities Council. OK acknowledges Erasmus + staff travel grant for support of travel to ISIS neutron source.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Qiu H, Min YZ, Rodgers Z, Zhang LZ, Wang AZ. Nanomedicine approaches to improve cancer immunotherapy. WIRES Nanomed Nanobi. 2017;9(5) doi: 10.1002/wnan.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Hartshorn CM, Bradbury MS, Lanza GM, Nel AE, Rao J, Wang AZ, Wiesner UB, Yang L, Grodzinski P. Nanotechnology strategies to advance outcomes in clinical cancer care. ACS Nano. 2018;12(1):24–43. doi: 10.1021/acsnano.7b05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rosenblum D, Joshi N, Tao W, Karp JM, Peer D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03705-y. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shi JJ, Kantoff PW, Wooster R, Farokhzad OC. Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer. 2017;17(1):20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Janib SM, Moses AS, MacKay JA. Imaging and drug delivery using theranostic nanoparticles. Adv Drug Delivery Rev. 2010;62(11):1052–1063. doi: 10.1016/j.addr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang HL, Li XX, Tse BWC, Yang HT, Thorling CA, Liu YX, Touraud M, Chouane JB, Liu X, Roberts MS, Liang XW. Indocyanine green-incorporating nanoparticles for cancer theranostics. Theranostics. 2018;8(5):1227–1242. doi: 10.7150/thno.22872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kang H, Mintri S, Menon AV, Lee HY, Choi HS, Kim J. Pharmacokinetics, pharmacodynamics and toxicology of theranostic nanoparticles. Nanoscale. 2015;7(45):18848–18862. doi: 10.1039/c5nr05264e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tirotta I, Dichiarante V, Pigliacelli C, Cavallo G, Terraneo G, Bombelli FB, Metrangolo P, Resnati G. F-19 magnetic resonance imaging (MRI): from design of materials to clinical applications. Chem Rev (Washington, DC, U S) 2015;115(2):1106–1129. doi: 10.1021/cr500286d. [DOI] [PubMed] [Google Scholar]
- [9].Holland GN, Bottomley PA, Hinshaw WS. F-19 Magnetic-resonance imaging. J Magn Reson. 1977;28(1):133–136. [Google Scholar]
- [10].Ruiz-Cabello J, Barnett BP, Bottomley PA, Bulte JWM. Fluorine (F-19) MRS and MRI in biomedicine. NMR Biomed. 2011;24(2):114–129. doi: 10.1002/nbm.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Srinivas M, Tel J, Schreibelt G, Bonetto F, Cruz LJ, Amiri H, Heerschap A, Figdor CG, De Vries IJM. PLGA-encapsulated perfluorocarbon nanoparticles for simultaneous visualization of distinct cell populations by F-19 MRI. Nanomedicine. 2015;10(15):2339–2348. doi: 10.2217/NNM.15.76. [DOI] [PubMed] [Google Scholar]
- [12].Akazawa K, Sugihara F, Nakamura T, Matsushita H, Mukai H, Akimoto R, Minoshima M, Mizukami S, Kikuchi K. Perfluorocarbon-based 19F MRI nanoprobes for in vivo multicolor imaging. Angew Chem Int Ed. 2018;57(51):16742–16747. doi: 10.1002/anie.201810363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chirizzi C, Battista DD, Tirotta I, Metrangolo P, Comi G, Bombelli FB, Chaabane L. Multispectral MRI with dual fluorinated probes to track mononuclear cell activity in mice. Radiology. 2019;291(2):351–357. doi: 10.1148/radiol.2019181073. [DOI] [PubMed] [Google Scholar]
- [14].Jacoby C, Temme S, Mayenfels F, Benoit N, Krafft MP, Schubert R, Schrader J, Flögel U. Probing different perfluorocarbons for in vivo inflammation imaging by 19F MRI: image reconstruction, biological half-lives and sensitivity. NMR Biomed. 2014;27(3):261–271. doi: 10.1002/nbm.3059. [DOI] [PubMed] [Google Scholar]
- [15].Ahrens ET, Zhong J. In vivo MRI cell tracking using perfluorocarbon probes and fluorine-19 detection. NMR Biomed. 2013;26(7):860–871. doi: 10.1002/nbm.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moonshi SS, Zhang C, Peng H, Puttick S, Rose S, Fisk NM, Bhakoo K, Stringer BW, Qiao GG, Gurr PA, Whittaker AK. A unique F-19 MRI agent for the tracking of non phagocytic cells in vivo. Nanoscale. 2018;10(17):8226–8239. doi: 10.1039/c8nr00703a. [DOI] [PubMed] [Google Scholar]
- [17].Boehm-Sturm P, Mengler L, Wecker S, Hoehn M, Kallur T. In vivo tracking of human neural stem cells with F-19 magnetic resonance imaging. PLoS One. 2011;6(12):e29040. doi: 10.1371/journal.pone.0029040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ruiz-Cabello J, Walczak P, Kedziorek DA, Chacko VP, Schmieder AH, Wickline SA, Lanza GM, Bulte JWM. In vivo “hot spot” MR imaging of neural stem cells using fluorinated nanoparticles. Magn Reson Med. 2008;60(6):1506–1511. doi: 10.1002/mrm.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ahrens ET, Flores R, Xu HY, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat Biotechnol. 2005;23(8):983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- [20].Myerson J, He L, Lanza G, Tollefsen D, Wickline S. Thrombin-inhibiting perfluorocarbon nanoparticles provide a novel strategy for the treatment and magnetic resonance imaging of acute thrombosis. J Thromb Haemost. 2011;9(7):1292–1300. doi: 10.1111/j.1538-7836.2011.04339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bo SW, Yuan YP, Chen YP, Yang ZG, Chen SZ, Zhou X, Jiang ZX. In vivo drug tracking with F-19 MRI at therapeutic dose. Chem Commun (Cambridge, U K) 2018;54(31):3875–3878. doi: 10.1039/c7cc09898g. [DOI] [PubMed] [Google Scholar]
- [22].Southworth R, Kaneda M, Chen JJ, Zhang L, Zhang HY, Yang XX, Razavi R, Lanza G, Wickline SA. Renal vascular inflammation induced by Western diet in ApoE-null mice quantified by F-19 NMR of VCAM-1 targeted nanobeacons. Nanomed-Nanotechnol Biol Med. 2009;5(3):359–367. doi: 10.1016/j.nano.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tirotta I, Mastropietro A, Cordiglieri C, Gazzera L, Baggi F, Baselli G, Bruzzone MG, Zucca I, Cavallo G, Terraneo G, Bombelli FB, et al. A Superfluorinated molecular probe for highly sensitive in vivo(19)F-MRI. J Am Chem Soc. 2014;136(24):8524–8527. doi: 10.1021/ja503270n. [DOI] [PubMed] [Google Scholar]
- [24].Riess JG. Fluorous micro-and nanophases with a biomedical perspective. Tetrahedron. 2002;58(20):4113–4131. [Google Scholar]
- [25].Boissenot T, Fattal E, Bordat A, Houvenagel S, Valette J, Chacun H, Gueutin C, Tsapis N. Paclitaxel-loaded PEGylated nanocapsules of perfluorooctyl bromide as theranostic agents. Eur J Pharmaceut Biopharmaceut. 2016;108:136–144. doi: 10.1016/j.ejpb.2016.08.017. [DOI] [PubMed] [Google Scholar]
- [26].Pisani E, Tsapis N, Galaz B, Santin M, Berti R, Taulier N, Kurtisovski E, Lucidarme O, Ourevitch M, Doan BT, Beloeil JC, et al. Perfluorooctyl bromide polymeric capsules as dual contrast agents for ultrasonography and magnetic resonance. Imaging. 2008;18(19):2963–2971. [Google Scholar]
- [27].Koshkina O, Lajoinie G, Bombelli FB, Swider E, Cruz LJ, White P, Schweins R, Dolen Y, Dinther EV, Riessen NKV, Rogers SE, et al. Multicore liquid perfluorocarbon-loaded multimodal nanoparticles for stable ultrasound and 19F MRI applied to in vivo cell tracking. Adv Funct Mater. 2019;29(19) doi: 10.1002/adfm.201806485. 1806485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Swider E, Staal AHJ, van Riessen NK, Jacobs L, White PB, Fokkink R, Janssen GJ, van Dinther E, Figdor CG, de Vries IJM, Koshkina O, et al. Design of triphasic poly(lactic-co-glycolic acid) nanoparticles containing a perfluorocarbon phase for biomedical applications. RSC Adv. 2018;8(12):6460–6470. doi: 10.1039/c7ra13062g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Srinivas M, Cruz LJ, Bonetto F, Heerschap A, Figdor CG, de Vries IJM. Customizable, multi-functional fluorocarbon nanoparticles for quantitative in vivo imaging using F-19 MRI and optical imaging. Biomaterials. 2010;31(27):7070–7077. doi: 10.1016/j.biomaterials.2010.05.069. [DOI] [PubMed] [Google Scholar]
- [30].Staal AHJ, Becker K, Tagit O, Riessen NKv, Koshkina O, Veltien A, Bouvain P, Cortenbach K, Scheenen T, Flögel U, Temme S, et al. Single resonance, self-disassembling 19F magnetic resonance imaging nanoparticles for clinical applications with fast local and systemic clearance. 2019 (in submission) [Google Scholar]
- [31].Takaoka Y, Sakamoto T, Tsukiji S, Narazaki M, Matsuda T, Tochio H, Shirakawa M, Hamachi I. Self-assembling nanoprobes that display off/on F-19 nuclear magnetic resonance signals for protein detection and imaging. Nature Chem. 2009;1(7):557–561. doi: 10.1038/nchem.365. [DOI] [PubMed] [Google Scholar]
- [32].Mizukami S, Takikawa R, Sugihara F, Hori Y, Tochio H, Walchli M, Shirakawa M, Kikuchi K. Paramagnetic relaxation-based F-19 MRI probe to detect protease activity. J Am Chem Soc. 2008;130(3):794–795. doi: 10.1021/ja077058z. [DOI] [PubMed] [Google Scholar]
- [33].Keliris A, Mamedov I, Hagberg GE, Logothetis NK, Scheffler K, Engelmann J. A smart 19F and 1H MRI probe with self-immolative linker as a versatile tool for detection of enzymes. Contrast Media Mol Imaging. 2012;7(5):478–483. doi: 10.1002/cmmi.1470. [DOI] [PubMed] [Google Scholar]
- [34].Swider E, Koshkina O, Tel J, Cruz LJ, de Vries IJM, Srinivas M. Customizing poly(lactic-co-glycolic acid) particles for biomedical applications. Acta Biomater. 2018;73:38–51. doi: 10.1016/j.actbio.2018.04.006. [DOI] [PubMed] [Google Scholar]
- [35].Rausch K, Reuter A, Fischer K, Schmidt M. Evaluation of nanoparticle aggregation in human blood serum. Biomacromolecules. 2010;11(11):2836–2839. doi: 10.1021/bm100971q. [DOI] [PubMed] [Google Scholar]
- [36].Wu H. Correlations between the Rayleigh ratio and the wavelength for toluene and benzene. Chem Phys. 2010;367(1):44–47. [Google Scholar]
- [37].Lindner P, Schweins R. The D11 small-angle scattering instrument: a new benchmark for SANS. Neutron News. 2010;21(2):15–18. [Google Scholar]
- [38].Richard D, Ferrand M, Kearley GL. Analysis and visualisation of neutron-scattering data. J Neutron Res. 1996;4(1–4):33–39. [Google Scholar]
- [39].Lindner P. Neutrons, X-rays and Light: Scattering Methods Applied to Soft Condensed Matter. Elsevier Science Ltd; 2002. [Google Scholar]
- [40].Kline SR. Reduction and analysis of SANS and USANS data using IGOR Pro. J Appl Crystallogr. 2006;39:895–900. [Google Scholar]
- [41].Fischer K, Schmidt M. Pitfalls and novel applications of particle sizing by dynamic light scattering. Biomaterials. 2016;98:79–91. doi: 10.1016/j.biomaterials.2016.05.003. [DOI] [PubMed] [Google Scholar]
- [42].Teixeira J. Small-angle scattering by fractal systems. J Appl Crystallogr. 1988;21:781–785. [Google Scholar]
- [43].Korb JP. Multiscale nuclear magnetic relaxation dispersion of complex liquids in bulk and confinement. Prog Nucl Magn Reson Spectrosc. 2018;104:12–55. doi: 10.1016/j.pnmrs.2017.11.001. [DOI] [PubMed] [Google Scholar]
- [44].Kujawa P, Goh CCE, Calvet D, Winnik FM. Do fluorocarbon, hydrocarbon, and polycyclic aromatic groups intermingle? Solution properties of pyrene-labeled bis(fluorocarbon/hydrocarbon)-modified poly(N-isopropylacrylamide) Macromolecules. 2001;34(18):6387–6395. [Google Scholar]
- [45].Kujawa P, Liu RCW, Winnik FM. Do fluorocarbon, hydrocarbon, and polycyclic aromatic groups intermingle? A study of the interactions in water between fluorocarbon- and hydrocarbon-modified poly(N-isopropylacrylamides) J Phys Chem B. 2002;106(21):5578–5585. [Google Scholar]
- [46].Pillai GJ, Greeshma MM, Menon D. Impact of poly(lactic-co-glycolic acid) nanoparticle surface charge on protein, cellular and haematological interactions. Colloid Surf B. 2015;136:1058–1066. doi: 10.1016/j.colsurfb.2015.10.047. [DOI] [PubMed] [Google Scholar]
- [47].Schoormans J, Calcagno C, Daal MRR, Wüst RCI, Faries C, Maier A, Teunissen AJP, Naidu S, Sanchez-Gaytan BL, Nederveen AJ, Fayad ZA, et al. An iterative sparse deconvolution method for simultaneous multicolor 19F-MRI of multiple contrast agents. Magn Reson Med. 2020;83(1):228–239. doi: 10.1002/mrm.27926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.