Abstract
Type I interferons (IFNs), which activate many IFN-stimulated genes (ISGs), are known to regulate tumorigenesis. However, little is known regarding how various ISGs coordinate with one another in developing anti-tumor effects. Here we report that the ISG, UBA7, is a tumor suppressor in breast cancer. UBA7 encodes an enzyme that catalyzes the covalent conjugation of the ubiquitin-like protein product of another ISG (ISG15) to cellular proteins in a process known as “ISGylation”. ISGylation of other ISGs, including STAT1 and STAT2, synergistically facilitates production of chemokine-receptor ligands to attract cytotoxic T cells. These gene activation events are further linked to clustering and nuclear relocalization of STAT½ within IFN-induced PML bodies. Importantly, this coordinated ISG–ISGylation network plays a central role in suppressing murine breast cancer growth and metastasis, which parallels improved survival in breast cancer patients. These findings reveal a cooperative IFN-inducible gene network in orchestrating a tumor suppressive microenvironment.
Keywords: Type I interferon, interferon stimulated gene (ISG), ISGylation, PML phase separation, CXCL9/10, anti-tumor immunity
Introduction
The cellular and noncellular components of the tumoral niche create a specific tumor microenvironment (TME), where cells continuously sense danger and damage signaling by extracellular and intracellular pattern recognition receptors (PRRs) to balance the acts of the host immune system (1). One key event is the production of type I interferons (IFNs) in response to the activation of specific PRRs, such as cyclic GMP-AMP (cGAMP) synthase (cGAS), which plays a central role in regulating TME (2). IFNs activate the Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT) pathway, resulting in the induction of hundreds of IFN-stimulated genes (ISGs) (3), many of which remain uncharacterized (4). In general, it is thought that each of these ISGs may activate a pathway and multiple activated pathways collectively contribute to the overall anti-tumor effect. However, little is known regarding which ISGs are key drivers to the development of the anti-tumor effect and how these ISGs might coordinate with one another.
Type I IFNs have been well established to orchestrate both innate and adaptive immune response in cancer (2,5,6). This is accomplished by mobilizing diverse cell types in the host immune system, including natural killer cells, dendritic cells, tumor-associated macrophages (TAMs), B cells and T cells (5–7). A critical step in gaining anti-tumor immunity is to increase the traffic of activated effector T cells, such as cytotoxic CD8+ T cells, to infiltrate into the tumor bed to recognize and kill target cancer cells (2,7). While playing a central role in regulating T cell priming and homing in the TME (2), excessive IFN signaling is also known to modulate the TME to disable the anti-tumor function of effector T cells (8). These complex biological responses likely result from pleiotropic actions of ISGs and various feedback control mechanisms, which appear to be highly dependent on specific tumor contexts (4,9). Therefore, an important goal in enhancing anti-tumor immune response is to identify key ISG-mediated pathways that favor the development of an anti-tumor microenvironment, while minimizing the impact of those with adverse effects.
One such ISG, UBA7, encodes an ubiquitin-like modifier activating enzyme (UBA7, also known as UBE1L) that catalyzes the covalent conjugation of the ubiquitin-like protein product of another ISG (ISG15) to additional ISGs as well as other cellular proteins in a process termed “protein ISGylation” (10). ISG15 is conjugated to a host of proteins in response to propagate the effects of IFN stimulation and is known to play a role in anti-viral immunity, but the cellular consequences of UBA7-dependent ISGylation in the setting of cancer cells is not completely understood. Here, we uncover a regulatory network in which multiple UBA7-targeted ISGs act in a concerted fashion to activate key chemokines and attract cytotoxic T cells to the tumor bed. These findings reveal a synergistic ISG-mediated cellular program to achieve effective anti-tumor response in the host.
Results
Evidence for the ISGylation-Activating Enzyme UBA7 as a Tumor Suppressor
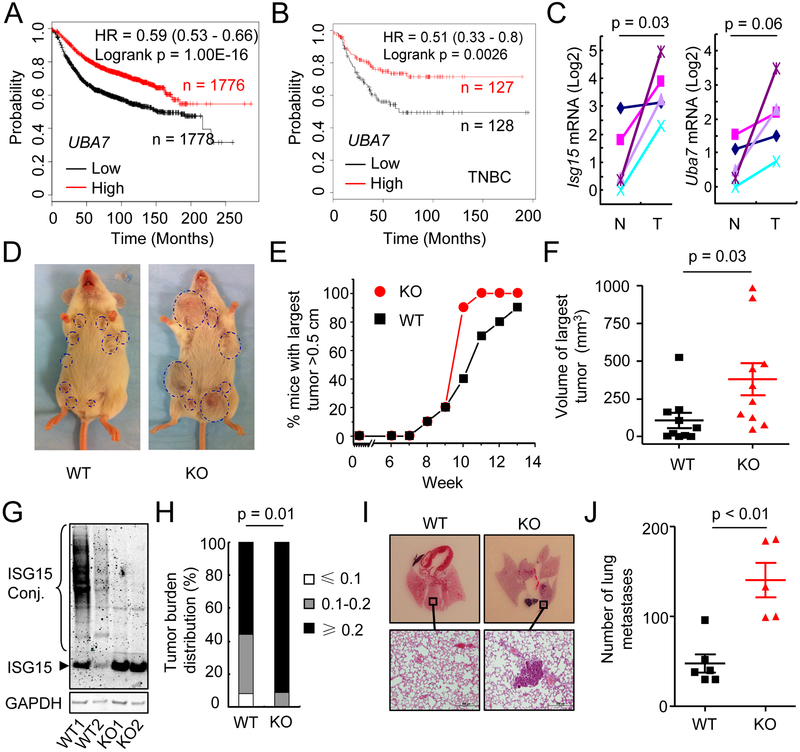
Type I IFNs have been well established to have immune stimulatory functions against malignancies, including breast cancer. To identify ISGs that contribute to IFN’s effect in preventing breast cancer progression, we examined the prognostic value of 81 individual ISGs showing correlated expression with ISG15, and another ISG, MX1, since both of them are highly induced by IFN (Figures S1A and S1B). We found that the expression of a subset of ISGs was significantly correlated with favorable relapse free survival (RFS) (Figure S1B and Table S1). Interestingly, one of the top ISGs positively correlated with patient survival is UBA7 (Figures 1A and S1B). This is in contrast to the lack of positive prognostic value for MX1 (Table S1), supporting the idea that not all ISGs positively contribute to anti-tumor immunity. Interestingly, ISG15, encoding for the ubiquitin-like donor for UBA7-mediated ISGylation, was also known to be highly elevated in breast cancer (11). Unlike UBA7, however, the induction of ISG15 was negatively correlated with patient survival (Table S1). This likely reflects the multifaceted function of ISG15, as free ISG15 is also responsible for attenuating IFN signaling by stabilizing the deISGylation protease USP18 (12). Importantly, the expression of UBA7 was associated with favorable RFS in triple negative breast cancer (Figure 1B). The association of UBA7 expression with patient survival was also validated in the TCGA dataset of breast cancer patients (Figure S1C). Collectively, these results strongly suggest that UBA7 may function as a critical tumor suppressive ISG in breast cancer.
Figure 1. Evidence for the ISGylation-activating enzyme UBA7 as a tumor suppressor.
A, Kaplan-Meier curves showing the correlation between Relapse Free Survive (RFS) and UBA7 expression in breast cancer (Affymetrix ID 203281_at UBA7).
B, Kaplan-Meier curves showing the correlation between RFS and UBA7 expression in triple negative breast cancer (TNBC) (Affymetrix ID 203281_at UBA7).
C, Expression of Isg15 and Uba7 in tumors compared with their adjacent normal tissues from PyVmT WT mice at 10 weeks of age (n = 5). p, paired t-test.
D, Representative photograph of PyVmT/WT and PyVmT/Uba7 KO mice at 10 weeks of age.
E, Percentage of mice with largest tumor diameter above 0.5 cm (n = 10/group).
F, Tumor volume of largest tumors in PyVmT/WT and PyVmT/Uba7 KO mice at 10 weeks of age (n = 10/group). p, student’s t-test.
G, Representative level of ISG15 and protein ISGylation in tumor tissue from PyVmT/WT and PyVmT/Uba7 KO mice.
H, Tumor burden (total tumor weight/body weight) distribution of PyVmT/WT (n = 25) and PyVmT/Uba7 KO (n = 23) mice at the endpoint. p, Fisher’s exact test.
I&J, Number of spontaneous lung metastasis in PyVmT/WT and PyVmT/Uba7 KO mice. Serial lung sections were stained with H&E (n = 5–6). p, student’s t-test.
We next modeled the tumor suppressive function of UBA7 in mice to establish its functional relevance. Previous studies have demonstrated that enhanced IFN signaling significantly changes the tumor microenvironment by increasing T lymphocyte infiltration, thereby causing significant shrinkage of tumors in the mouse mammary tumor virus (MMTV)-polyomavirus middle tumor antigen (PyVmT) breast cancer model (13,14). We therefore utilized this well-established mouse model to study the function of UBA7 in breast cancer. In line with a published RNA-seq dataset (15), we detected increased expression of both Uba7 and Isg15 in tumors compared to matched controls at week 10 (Figure 1C), indicating augmented IFN signaling during tumor progression. Knockout (KO) of Uba7 in mice did not appear to affect normal mammary development (Figure S2). Although both wild-type (WT) and Uba7-deleted (KO) MMTV-PyVmT mice developed palpable tumors around week 8, we found that tumors grew faster in KO mice compared to WT mice from week 9 to the endpoint when the mice were euthanized, as indicated by the number of mice that developed tumors over 0.5 cm (Figures 1D and 1E). At week 10, for instance, 90% of KO mice developed large tumors over 0.5 cm compared to 40% of WT mice (Figure 1E) and the average volume of the largest tumors in KO mice was ~380 mm3 compared to ~ 100 mm3 in WT mice (Figure 1F). Given that UBA7 is the activating enzyme for protein ISGylation, we detected both free ISG15 and ISG15-conjugated proteins in tumor lysates from WT mice at this stage, indicative of active protein ISGylation; while only free ISG15 was detected in tumor lysates from KO mice (Figure 1G). This agrees that UBA7 is a major activating enzyme responsible for tumor-induced protein ISGylation. We also monitored total tumor burden on these animals by quantifying total tumor weight normalized to body weight at the endpoint and found that KO mice had more relatively large tumors (those with tumor burden over 0.2) than WT mice (Figure 1H). In addition, KO mice also showed increased incidence of lung metastasis (Figures 1I–J). Collectively, our data on Uba7 KO mice revealed a tumor suppressive function of protein ISGylation. To further support this finding, we examined Usp18C61A/C61A knock-in (KI) mice in which ISGylation was increased by inactivating the deconjugating enzyme Usp18 (16) and found similar suppression to breast tumor growth (Figures S3A-C). Together, these results demonstrated that protein ISGylation was tumor suppressive in MMTV-PyVmT breast cancer model, which supports UBA7 as a critical tumor suppressive ISG in human breast cancer.
Protein ISGylation Stimulates Intratumoral Infiltration of T Lymphocytes
To understand how protein ISGylation suppressed tumor growth, we analyzed genes that were co-expressed with UBA7 from the TCGA breast cancer dataset. Excluding the expected association of classical ISGs, we found that >600 human genes showed co-expression with UBA7 (Figure S4A, R≥0.30). Ingenuity pathway analysis (IPA) revealed that top five canonical pathways are all related to T cells (Figure S4B). Interestingly, chemokine signaling pathway is also revealed by the analysis and CXCR3, which is mostly expressed on activated T lymphocytes, showed the highest correlation with UBA7 expression among all examined chemokine and chemokine receptors (Figure S4C). Therefore, we hypothesized that the tumor suppressive effect of UBA7 might be further reinforced by T lymphocytes in the TME.
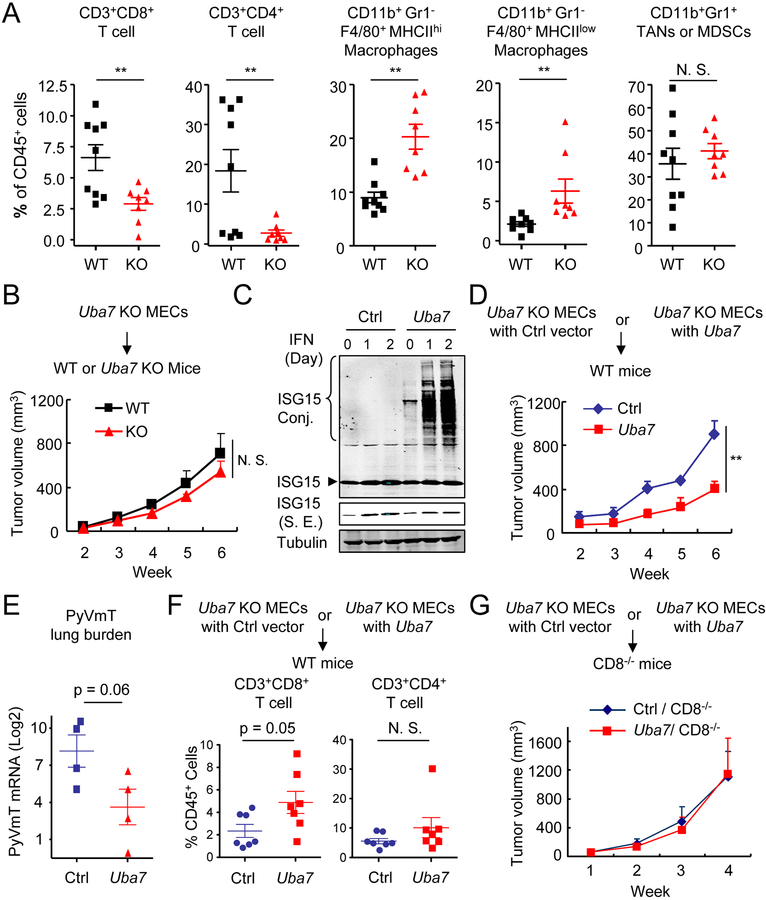
To advance this hypothesis, we asked whether the observation made in breast cancer patients could be mirrored in our MMTV-PyVmT breast cancer model. We firstly performed detailed analysis of T cell populations in tumors derived from these mice and observed that CD3+CD4+ and CD3+CD8+ T cells were all significantly reduced in tumors from Uba7 KO mice compared to tumors from WT mice (Figures 2A and S5). In contrast, tumors from KO mice showed an increase in the percentage of CD11b+Gr1−F4/80+ tumor-associated macrophages. No significant difference was observed in the percentage of intratumoral CD11b+Gr1+ cells, which could be tumor-associated neutrophils and/or myeloid-derived suppressor cells (Figure 2A). Considering the limited role of tumor-associated macrophages in regulating spontaneous PyVmT tumor growth, as established earlier (17,18), we chose to focus our efforts in understanding protein ISGylation-regulated T cell activity on the PyVmT tumor model. Staining with CD69 (a T cell activation maker, which is also expressed on anergic T cells) identified similar percentages of CD3+CD4+ and CD3+CD8+ T cells expressing such marker within tumors from both WT and Uba7 KO groups (Figure S6A). Further analysis revealed that there was no significant difference in the percentage of effector CD44hiCD62Llow T cells within the CD3+CD4+ or CD3+CD8+ T cell populations in tumors from WT and KO mice (Figure S6B). Additional analysis further suggested that intratumoral CD8+ T cells from Uba7 KO tumors were not compromised based either on cell proliferation or on the expression of the key cytotoxic effector molecule GZMB (Figures S7A and S7B). We interpreted these findings to indicate that UBA7 does not directly modulate T cell proliferation or function, but rather through increasing their infiltration into the tumor bed.
Figure 2. Protein ISGylation stimulates intratumoral infiltration of T lymphocytes.
A, Flow cytometric analysis of intratumoral leukocytes in the tumors from PyVmT/WT and PyVmT/Uba7 KO mice. Numbers expressed as percentages of total CD45+ leukocytes (n = 7–8). p, student’s t-test. TANs, tumor-associated neutrophils; MDSCs, myeloid-derived suppressor cells.
B, Tumor growth from PyVmT/Uba7 KO MECs in WT and Uba7 KO Mice (mean + SEM, n = 5). p, two-way ANOVA.
C, Restored Uba7 expression in PyVmT/Uba7 KO MECs. Cells were treated by type I IFN up to two days and cell lysate were immunoblotted by antibodies as indicated. S. E., short exposure.
D, Tumor growth from PyVmT/Uba7 KO + control vector (henceforth “Ctrl”) or PyVmT/Uba7 KO + Uba7 (henceforth “Uba7”) MECs in WT mice (mean + SEM, n = 4). A representative set from three independent experiments (n = 4–5) is shown. **, p<0.01, by two-way ANOVA.
E, PyVmT lung burden in mice from panel D (mean + SEM, n = 4). p, student’s t-test.
F, Flow cytometric analysis of CD4+ and CD8+ T cell in tumors from mice injected with Ctrl or Uba7 MECs after week 6. Numbers expressed as percentages of total CD45+ leukocytes. Pooled data are presented from two independent experiments (n = 7). p, student’s t-test.
G, Tumor growth from Ctrl or Uba7 MECs in CD8 deficient mice (mean + SEM, n = 6).
Since the expression of both UBA7 and ISG15 has been detected in tumor cells as well as in their stroma (Figures S8A and S8B), we next asked whether the suppressive tumor microenvironment was due to induced protein ISGylation in tumors versus stromal cells. We first established mouse mammary epithelial cells (MECs) from Uba7 KO tumors. When Uba7 KO MECs were injected into WT and Uba7 KO mice, these cells developed tumors of similar sizes (Figure 2B). This excludes the functional impairment of any tumor stroma cells due to lacking Uba7 expression in this tumor model. Conversely, restored expression of Uba7 expression rescued cellular protein ISGylation in MECs (Figure 2C), which led to smaller tumors and reduced lung metastasis when these MECs were injected into the mouse mammary fat pad of WT mice (Figures 2D–E). Notably, increased UBA7 expression did not affect cell growth when cultured in vitro (Figure S9). Meanwhile, knockdown of ISG15 in Uba7 KO MECs did not show further impact on tumor growth in vivo (Figures S10A and S10B), implying that intervening free ISG15 in Uba7 KO cells was insufficient to affect tumor growth. Furthermore, restored UBA7 expression greatly increased infiltration of CD3+CD8+ cytotoxic T lymphocytes in tumors derived from these cells (Figure 2F) and CD8 deficiency abrogated the tumor suppressive effect of Uba7-mediated ISGylation (Figure 2G). In combination, these results strongly implicate a tumor cell autonomous function of UBA7, suggesting that protein ISGylation may regulate T lymphocyte activities through a paracrine effect within the TME.
ISGylation Synergizes with TLR Signaling to Increase the Expression of CXCR3 Ligands
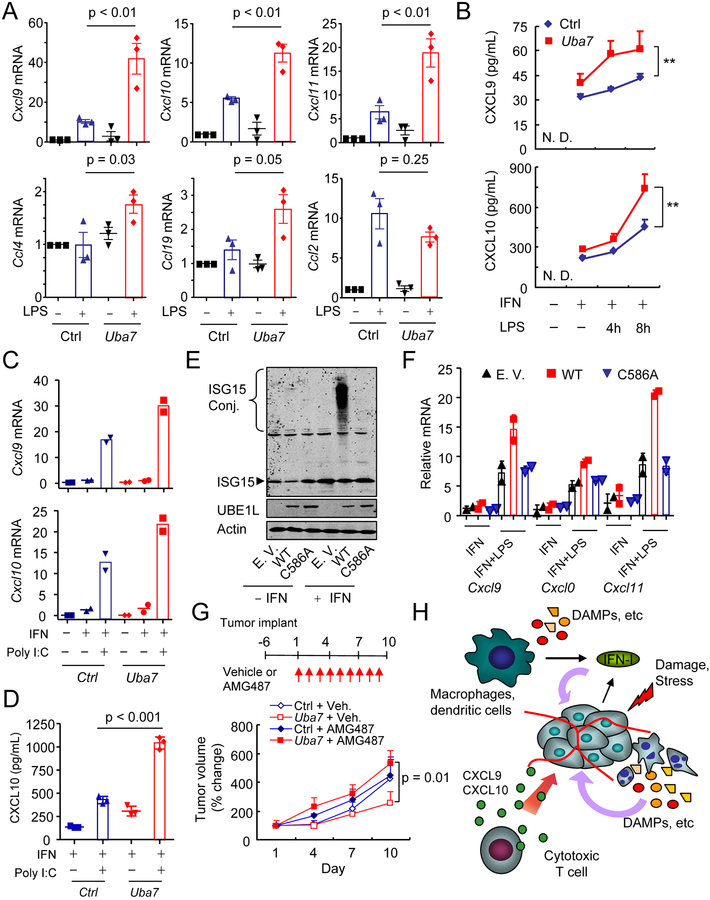
Having established the tumor cell autonomous effect, we prioritized our experimentation to understand the critical role of protein ISGylation in tumor cells. Considering the highest correlation between the expression of UBA7 and CXCR3 in breast cancer patients and the established role of the CXCL9/10/11-CXCR3 axis in the regulation of T lymphocyte trafficking and anti-tumor immunity (2,7,19), we postulated that UBA7-mediated protein ISGylation might directly modulate the CXCL9/10/11-CXCR3 signaling axis within the TME. Therefore, we next examined whether protein ISGylation regulates the production of CXCR3 ligands. Given the documented up-regulation of TLR signaling molecules (i.e. TLR3 and TLR4) in the MMTV-PyVmT breast cancer model (20) and their known functions in shaping anti-cancer immunity in human breast cancer (21,22), we firstly focused on TLR4 signaling and found TLR4 activation in combination with type I IFN priming greatly stimulated the expression of Cxcl9/10/11 (Figure S11). The observed synergy between IFN and TLR4 signaling prompted us to determine whether ISGylation directly contributed to the induction of CXCR3 ligands in our MEC model. Indeed, we found that UBA7-mediated ISGylation greatly enhanced the expression of Cxcl9/10/11 in response to TLR4 signaling in IFN-primed MECs (Figure 3A). We further confirmed ISGylation dependent increase in CXCL9/10 protein secretion from mouse MECs after TLR4 activation in IFN-primed cells (Figure 3B), although the CXCL11 protein was not detected under our conditions. We further showed that protein ISGylation also increased poly (I:C)-stimulated expression of Cxcl9/10 (Figures 3C and 3D), indicating that ISGylation synergized with both TLR3 and TLR4 signaling to increase the expression of CXCR3 ligands. In contrast, we detected little effect of ISGylation on TNFα, IL-1β or poly (dA:dT)-stimulated expression of CXCR3 ligands under investigated conditions (Figure S12A–C). Consistent with the importance of UBA7-catalyzed ISGylation in TLRs-stimulated expression of Cxcl9/10, we found that the expression of an enzyme-deficient form of UBA7 (C586A) failed to increase the expression of CXCR3 ligands (Figure 3E and 3F). Furthermore, CXCR3 blockade abrogated the tumor suppressive effect of ISGylation (Figure 3G). Together, these results demonstrated a vital role for cancer-autonomous protein ISGylation in facilitating cytotoxic T cell-mediated killing in breast cancer through the increased expression of CXCR3 ligands, as illustrated (Figure 3H).
Figure 3. ISGylation synergizes with TLRs signaling to increase expression of CXCR3 ligands.
A, RT-qPCR analyzing expression of multiple chemokines in IFN-primed Ctrl or Uba7 MECs after TLR4 activation (mean + SEM, n = 3). p, one-way ANOVA Post-hoc Tukey’s test.
B, Secreted CXCL9/10 in the supernatant from Ctrl or Uba7 MECs by ELISA (mean + SD, n = 3). **, p < 0.01, two-way ANOVA.
C, RT-qPCR analyzing expression of Cxcl9/10 in IFN-primed Ctrl or Uba7 MECs after poly I:C treatment (mean from biological duplicates).
D, Secreted CXCL10 in the supernatant from Ctrl or Uba7 MECs by ELISA (mean + SD, n = 3). p, one-way ANOVA Post-hoc Tukey’s test.
E, Restored expression of WT and UBA7 C586A in PyVmT/Uba7 KO MECs. Cells were treated by type I IFN (1000 U/ml) for 24h and cell lysate were immunoblotted for antibodies as indicated.
F, RT-qPCR analysis of expression of CXCR3 ligands from empty vector, WT UBA7, and UBA7 C586A expressing MECs after each perturbation (mean + SD from biological duplicates).
G, Tumor volume changes of orthotopic tumors by Ctrl or Uba7 MECs after treated with vehicle or AMG 487 (mean + SEM, n = 4–5). p, two-way ANOVA.
H, Model of protein ISGylation-regulated expression of CXCR3 ligands and their contribution to anti-tumor immunity. The presence of protein ISGylation facilitates IFN/TLRs-stimulated CXCL9/10 expression in the tumor microenvironment, therefore promotes CXCR3 signaling axis-guided intratumoral infiltration of cytotoxic T cell.
ISGylation Modulates the Function of Nuclear STATs in Chemokine Expression
With the established importance of CXCR3 signaling axis in protein ISGylation-regulated tumor suppression, we next investigated the molecular mechanisms underlying ISGylation-facilitated CXCR3 ligand expression in response to TLRs signaling in IFN-primed cells. Both TLR3 and TLR4 signaling are known to activate NF-κB, mitogen-activated protein kinase (MAPK), and TBK1/IKKε signaling to induce expression of many genes (23). In our experimental system, ISGylation induced by IFN priming did not seem to influence TLR4 activation-induced p38, JNK or IRF3 phosphorylation (Figure S13A). Although we did observe ISGylation impaired ERK phosphorylation and increased the abundance of IKBα (an inhibitor of NF-κB activation) (Figure S13A), treatment with an ERK inhibitor, U0126, did not increase Cxcl9/10/11 expression (Figure S13B). These data imply that protein ISGylation may not activate TLR4 signaling through any of the cytoplasmic TLR4 proximal signaling cascades.
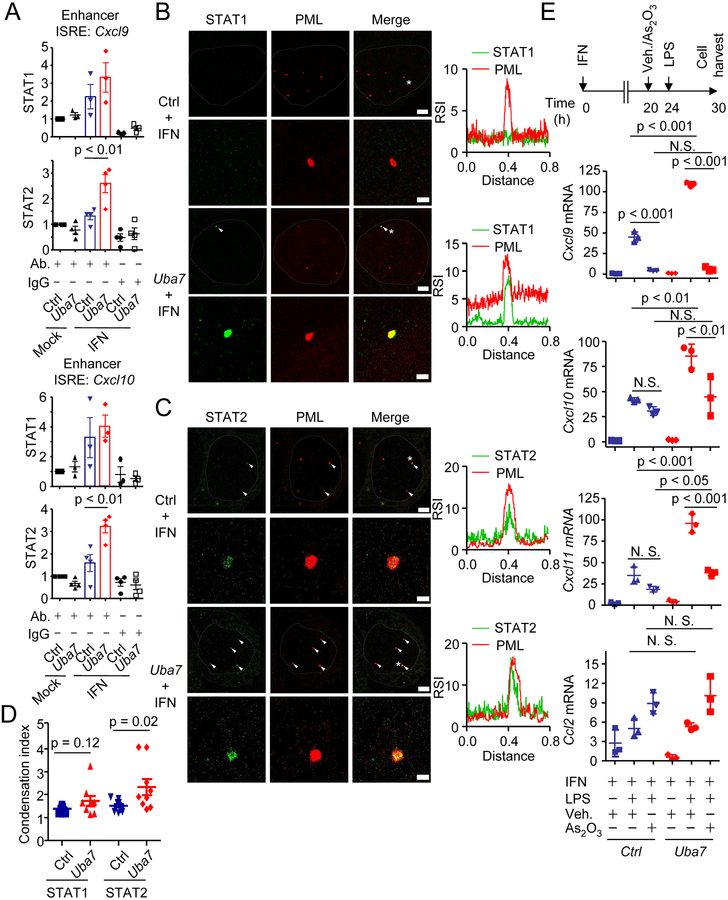
While it remains unclear how exactly TLRs activation licenses gene activation in IFN-primed cells, we decided to first focus on understanding how ISGylation contributes to the IFN-primed state, which is known to be critical for induction of CXCR3 ligands. In this regard, we noted that transcription factors STAT1 and STAT2 have been implicated in the regulation of CXCL9/10/11 expression. Although nuclear translocation is the primary mechanism for STAT activation in response to IFN signaling, we found that protein ISGylation had little influence on nuclear translocation of both STAT1 and STAT2 (Figure S14A). This directed us to investigate the possibility that ISGylation might regulate the function of STAT1 and STAT2 within the nucleus. Interestingly, both of these STATs are themselves ISGs and subject to protein ISGylation in response to IFN treatment (Figure S14B). Measuring the nuclear distribution of STAT1 by stochastic optical reconstruction microscopy (STORM), we observed the ISGylation-dependent redistribution of STAT1 to form clusters of various size and density in the nucleus (Figure S15A–D). We further observed increased interaction between nuclear STAT1 and STAT2 in the presence of ISGylation (Figure S15E). Importantly, ISGylation significantly increased STAT2 binding to interferon-stimulated response elements (ISREs) in Cxcl9/10 enhancers, and a similar trend was observed for STAT1 (Figures 4A and S15F). To determine the global impact of ISGylation on gene expression, we determined ISGylation-regulated gene expression by RNA-seq and found that the expression of a subset of IFN/TLR4 signaling-regulated genes was indeed enhanced by protein ISGylation (Figure S16A and S16B, Table S2), some of which were further confirmed by RT-qPCR (Figure S16C). Using specific pharmaceutical inhibitors, we further found that the expression of CXCR3 ligands required co-activation of multiple signaling pathways, including TBK1/IKKε and JAK-STAT pathways (Figure S17A and S17B). These data therefore attributed a subset of TLRs signaling-induced gene expression events, including Cxcl9 and Cxcl10, to ISGylation-induced clustering of STAT1 and STAT2 in the nucleus of IFN-primed cells.
Figure 4. ISGylation and PML cooperatively modulate the function of nuclear STATs and chemokine expression.
A, Quantitative ChIP-qPCR analysis of STAT1/STAT2 binding at enhancer ISREs of Cxcl9/10 in Ctrl or Uba7 MECs treated with or without IFN for 24h (mean + SEM, n = 3–4). p, One-way ANOVA Post-hoc Tukey’s test.
B & C, Colocalization analysis of PML bodies (red) and STAT1(B, green) and STAT2 (C, green) by expansion microscopy. PML bodies contain STAT1 or STAT2 were indicated by white arrows (Scale bar = 2 μm). Representative PML bodies (with asterisk) with higher magnification images and their plot profiling for the relative signal intensity of each protein are shown (Scale bar = 500 nm). Nucleus was outlined by dotted lines. RSI, relative signal intensity.
D, Comparison of STAT1/STAT2 signals around the PML bodies. The condensation index was defined as ratio of the mean intensity around PML bodies to the rest of nuclei (mean ± SD, n = 9–11). p, student’s t-test.
E, RT-qPCR analysis of chemokine expression upon arsenic treatment. Ctrl or Uba7 MECs were treated with IFN for 24h, and As2O3 or vehicle was added 4h prior to the LPS stimulation (mean ± SD, n = 3). p, One-way ANOVA Post-hoc Tukey’s test.
ISGylation Induces a Subset of STAT clusters to Associate with PML bodies
Given the substantial role of protein ISGylation in gene transcription, we further examined more detailed mechanisms. The observed clustering of both STAT1 and STAT2 prompted us to consider a role for ISGylation in mediating phase separation of the transcriptional machinery, a concept recently suggested to play critical roles in controlling compartmentalization and gene expression in the nucleus (24). Notably, PML is an ISG and PML nuclear bodies are membrane-less organelles, which can be induced by interferon signaling (25). We therefore hypothesized that protein ISGylation-mediated complex formation of STAT1 and STAT2 might also show increased association with IFN-induced PML bodies. Indeed, type I IFN dramatically induced the formation of PML bodies in MECs, although protein ISGylation did not affect the number of PML bodies (Figure S18A–C). Importantly, we found that protein ISGylation facilitated condensation of STAT1 and STAT2, especially STAT2, around IFN-induced PML bodies by expansion microscopy (Figure 4B–D). These observations suggest that ISGylation facilitates nuclear reprogramming.
To test the functional relevance of such IFN-induced nuclear compartmentalization, we took advantage of arsenic-mediated PML degradation as reported earlier (26), and observed that short-term treatment with arsenic trioxide depleted cellular PML protein without affecting the levels of STAT1, STAT2 or ISGylation (Figure S19). Importantly, however, such treatment diminished ISGylation-dependent expression of Cxcl9/10/11 (Figure 4E). However, Ccl2, whose expression is also regulated by IFN/TLR4 signaling, was not affected by either protein ISGylation or arsenic treatment (Figure 4E), indicating that the specificity of ISGylation and PML-controlled chemokine expression in our current biological context. These data thus provide evidence for STAT-dependent expression of CXCR3 ligands as a functional consequence of ISGylation-induced STAT clustering and redistribution in the nucleus. Importantly, these findings are corroborated by multiple clinical results, showing a significant association between the expression of both UBA7 and PML with CXCL9/10/11 expression and favorable overall survival (Figure S20A and S20B) in breast cancer patients. Together, these results highlighted the importance of the cooperation between ISGylation and PML in facilitating expression of CXCR3 ligands, whose expression are required for ISGylation-regulated tumor suppression.
Discussion
Type I IFNs activate expression of hundreds of ISGs and regulate tumorigenesis and response to cancer therapy through both tumor cell intrinsic and extrinsic mechanisms (5,6). However, it has remained unclear regarding the roles of many individual ISGs in cancer. Therefore, understanding the functional interplay between ISGs may provide novel therapeutic interventions in treating human cancers. Here, we identified the ISGylation activating enzyme UBA7 as a key ISG in regulating malignant progression. Interestingly, nearly all of the enzymes involved in protein ISGylation, including the E1, E2, E3 and the deconjugating enzyme, are ISGs themselves, suggesting their tight link to IFN-regulated cellular functions. Protein ISGylation thus affords a unique angle to look into IFN-induced gene networks that may synergistically function in the development of anti-tumor activities. Our current work uncovers that IFN-inducible protein ISGylation network has an immunostimulatory function by facilitating CXCR3 ligand expression and cytotoxic T cell infiltration in orchestrating a tumor suppressive microenvironment. It is worth noting that ISGylation-induced tumor microenvironment also decreased tumor-associated macrophages. Although TAMs are known to have a limited role in regulating spontaneous and allograft PyVmT tumor growth (17,18,27), they do play important roles in regulating metastatic potential (18) and response to cancer therapies in PyVmT tumors (17,27). Thus, future studies may be geared toward understanding the contribution of protein ISGylation to regulated TAMs during tumor metastasis and/or response to cancer therapies.
Several in vitro studies on human cancer cell lines have revealed both positive and negative roles of protein ISGylation in breast cancer (28,29). For example, ISG15 and protein ISGylation have been implicated in promoting invasive properties of malignant breast cancer cell lines through modulation of cytoskeletal architecture (29). On the other hand, ISGylation of ΔNp63α appears to ablate the ability of ΔNp63α to promote anchorage-independent cell growth and tumor formation (28). While these findings collectively suggest context-dependent effects of protein ISGylation in cancer, it is important to keep in mind that IFN responses are part of the adaptive and innate immune systems in the host, thus emphasizing the importance of studying the impact of protein ISGylation on cancer progression in immunocompetent hosts. Using immunocompetent hosts, our current work highlights the importance of protein ISGylation in orchestrating a tumor-suppressive microenvironment. This mechanism is well in line with our analysis of human cancer transcriptomes and associated clinical outcomes. While it is possible that ISGylation may enhance or suppress tumorigenesis in different contexts, just like IFN treatment, our data provide direct evidence to support the function of UBA7 as a key tumor suppressor in breast cancer. This is further corroborated by frequent chromosome 3p21.3 deletions where UBA7 resides in human cancer (30). Our findings suggest a mechanism underlying these genetic alterations in cancer.
We observed that the presence of protein ISGylation facilitates TLR¾ signaling-activated CXCR3 ligand expression in our current model. TLR4 signaling is activated by a variety of ligands, including DAMPs (22,31), and interestingly, certain ISGs (e.g. IFI35, NMI) are known to function as DAMPs (32). In addition, women with breast cancer have higher relative abundance of Bacillus, Enterobacteriaceae and Staphylococcus in their breast tissues (33) and TLR3 signaling-stimulated expression of CXCR3 ligands have also been well characterized to play critical roles in regulating tumor microenvironment and anti-tumor immunity (19,34). These prior observations are fully in line with the impact of loss-of-function TLR3 or TLR4 alleles in breast cancer patients who relapse more quickly and show reduced overall survival after therapeutic intervention (21,22), together indicating the importance of both TLRs in modulating anti-cancer immunity. Although we did not observe ISGylation-facilitated chemokine expression in the presence of polydAdT under investigated conditions, we cannot rule out the possibility that ISGylation may still contribute to signaling activation by other various endogenous stimuli, including some form of DNA released from damaged tissues, in the tumor microenvironment.
Our mechanistic studies have linked IFN priming to the dynamic regulation of PML bodies. PML is encoded by an ISG and IFN signaling is known to induce the formation of PML bodies in the nucleus (25). PML has been implicated in the regulation of type I IFN-induced ISGs in cell lines, albeit with unknown mechanisms (35). Using super-resolution microscopy, our current studies reveal that protein ISGylation enhances condensation of critical transcription factors, STAT1 and STAT2, and their association with PML bodies. We further observe that ISGylation-dependent chemokine expression requires the presence of PML nuclear bodies, together supporting a model in which PML nuclear bodies may contribute to the formation of a favorable nuclear environment for the expression of specific genes (36). It is entirely possible that microscopically detected PML bodies are indicative of IFN-induced condensation of both visible and invisible PML-containing complexes in the nucleus, which may underlie the IFN-induced and ISGylation-mediated gene expression program. Our findings join the recent discovery that sumoylation and the SUMO Interaction Motif (SIM) in PML bodies contributes to multivalent interactions within PML bodies (25,37), together suggesting that multiple post-transcriptional modifications are involved in modulating phase separation to account for the dynamic compositional control of PML bodies. It is important to point out that PML has been shown to function as a tumor suppressor in several cancer types, including breast cancer (38), but elevated PML expression has also been connected to the acquisition of aggressiveness and metastatic features in breast tumors (39). Additionally, the importance of PML in regulating the immune response has been reported in a mouse model of prostate cancer (40). These seemingly contradictory observations might reflect the winning versus losing battles of the host immune system to different tumors.
The most important concept conveyed in this work is a cooperative gene network of ISGs, which is shaped by UBA7-mediated ISGylation to form a favorable nuclear environment for STAT½-mediated chemokine expression. These cancer-cell autonomous effects provide mechanistic insights into the regulation of cytotoxic T cell infiltration into such tumor microenvironment when PRR-triggered type I IFN signaling is in synergy with activation of other inflammatory pathways, such as DMAPs-induced TLRs activation. Previous ChIP-seq studies have shown binding of AP-1, NF-κB, STAT½ and IRF3 on enhancers of CXCR3 ligands in TLRs-activated cells (41). Therefore, protein ISGylation regulated clustering of STATs may facilitate their assembly on IFN-stimulated response elements (ISREs) to form “enhanceosome” (42), thereby inducing gene expression according to our working model (Figure S21). Importantly, by dissecting the functions of some key ISGs in this network, we suggest a potential strategy to enhance the ISGylation-induced anti-cancer effect through deactivating its negative regulator USP18. Such enhanced ISGylation may also help divert tumor-prone activities of ISG15 by reducing free ISG15 in the TME. This and other potential modulations of the ISG network may therefore amplify the anti-tumor effects of IFN-based cancer therapies, while limiting other cancer-promoting events.
Materials and methods
Animals and tumor monitoring
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego (UCSD). All mice were housed and bred at the vivarium of the Moores Cancer Center at UCSD. Uba7 KO mice were backcrossed to FVB mice over 10 generations. CD8 KO mice in FVB background were kindly provided by Dr. Michael Karin (University of California, San Diego). USP18 C61A/C61A mice in C57BL/6 background were generated by Dr. Knobeloch Klaus-Peter (University of Freiburg). FVB mice expressing PyVmT under the control of the mouse MMTV-LTR were crossed with WT or Uba7 KO mice. Female mice heterozygous for the PyVmT transgene and homozygous null or homozygous wild-type for Uba7 were studied. C57BL/6 mice expressing PyVmT under the control of the mouse MMTV-LTR were crossed with WT or USP18 C61A/C61A mice. Female mice heterozygous for the PyVmT transgene and WT or homozygous for USP18 C61A/C61A were studied. Female mice were palpated twice weekly to monitor the development of mammary tumors. Tumor bearing mice were sacrificed for analysis at endpoint, which is 13 weeks (FVB) and 22 weeks (C57BL/6) of age after birth or whenever tumor burden reaches 20%. Approximate tumor volumes were calculated by the formula 4/3 × 3.14 × (long diameter/2) × (short diameter/2)2. For metastasis studies, mouse lungs were harvested at endpoint and the number of lung metastasis was determined by H&E staining of 10 serial lung sections per mouse. All studies were carried out following the NIH guidelines for the care and treatment of experimental laboratory rodents.
Isolation of mammary tumor cells for flow cytometric analysis and establishment of epithelial mammary cell lines
Single cell suspensions were established from spontaneous mammary tumors arising in FVB MMTV-PyVmT females as previously described (43). Tumors were minced and incubated at 37oC for 2 hours in 5 ml of Ham’s F12K medium containing 1 mg/ml collagenase (Roche), 2 mg/ml soybean trypsin inhibitor (Sigma) and 2% bovine serum albumin (BSA). After addition of fetal bovine serum (FCS)-containing medium, the suspension was passed through a 70 mm nylon filter (Fisher Scientific). Single cells were pelleted by centrifugation and resuspended in PBS/2% BSA for immediate flow cytometric analysis. Stained cells were analyzed by a FACS Canto cytometer (BD Biosciences). Antibodies for flow cytometric analysis are listed in the Table S3. For establishment of MEC cell lines, cells were cultured in DMEM: F12 (1:1) medium mix containing 5% FCS, 2.5 mg/ml amphotericin B, 10 mg/ml penicillin–streptomycin and MITO+ (BD Biosciences). Non-epithelial cells were removed by differential passaging and epithelial origin of established cell lines were confirmed by CD326 (Biolegend) staining after five passages (CD326+ > 98%). The cell lines have been tested as mycoplasma-free using mycosensor PCR assay kit (Agilent Technologies) with the latest test in 2018. The cell line authentication was not routinely performed. The cells were used within two month after thawing.
Orthotopic injections into the mammary fat pad
In vitro cultured MECs (1×106) re-suspended in 1 mg/ml matrigel (CORNING) solution were injected into the abdominal mammary fat pads of female mice. Growing tumors were monitored once a week with a caliper. Approximate tumor volumes were calculated by the formula 4/3 × 3.14 × (long diameter/2) × (short diameter/2)2.
Proteins, chemicals and antibodies
Universal type I IFN was purchased from PBL biomedical laboratories. Cells were generally treated by IFN at 1000 U/ml or as specified. Recombinant murine TNFα and IL-1β were purchased from Peprotech. LPS was purchased from List Biological Laboratories, Inc. MEK/ERK inhibitor U0126 and TBK1/IKKε inhibitor MRT67307 were purchased from Selleckchem. Polyinosinic-polycytidylic acid sodium salt [poly (I:C)], Arsenic(III) Oxide and JAK inhibitor I were purchased from Sigma-Aldrich. Poly (dA:dT)/LyoVec was purchased from Invivogen. Antibodies used in western blot analyses: Tubulin (Sigma-Aldrich), β-actin (Sigma-Aldrich), p-p38 (Cell Signaling Technology), p-JNK (Cell Signaling Technology) and IkBα (Biolegend) were purchased from the respective manufacturers. Rabbit anti-mouse ISG15 polyclonal antibodies have been described previously (44). Fc blocker (anti CD16/32) was purchased from eBiosciences. A full list of antibodies is shown in Table S3.
Plasmids
FLAG-mUBA7 and mUBA7 C586A cDNA was cloned into pMSCV-puro (Clontech). All the constructs were confirmed by DNA sequencing. shRNA plasmids (pLKO.1 vectors) for mIsg15 (TRCN0000077333 and TRCN TRCN0000077334) and control were obtained from The RNAi Consortium (TRC).
Cell culture, transfections and infection
293T cells have been purchased from ATCC and authenticated by this organization with certificates and been tested as mycoplasma-free using mycosensor PCR assay kit (Agilent Technologies) with the latest test in 2018. HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA) with 10% bovine calf serum (BCS) (HyClone, Logan, UT), 2 mM L-glutamine (Invitrogen) and Penicillin/streptomycin (100 U/ml, Invitrogen) and were used within two month after thawing. Cells were transfected using polyethylenimine (PEI) reagent as previously reported (45). For Uba7 expression in Uba7 KO MECs, 293T cells were transfected with pMSCV-mUBA7 and pIK6.1MCV ecopac using PEI reagent. Retroviruses from the culture medium of these cells were infected to Uba7 KO MECs and stable infected cells were selected by puromycin (1 μg/mL).
RNA isolation and RT-qPCR analysis
Total RNA from tumor tissue or MECs was isolated using Trizol reagent (ThermoFisher) according to the manufacturer’s instructions. For RT-qPCR analyses, equal amounts of RNA were reverse-transcribed by First Strand cDNA Synthesis Kit (MCLAB) and the resulting cDNA templates were subjected to RT-qPCR using SYBR Green detection system (Kapa Biosystems) on the CFX96 thermal cycler (BIO-RAD). Primer sequences are listed in Table S4.
ChIP-qPCR analysis
Chromatin immunoprecipitation (ChIP) was performed as described previously (46). A total of 1 μg anti-STAT2 (Cell signaling Technology) and 2 μg anti-STAT1 (Santa Cruz) antibodies were used per ChIP reaction. ChIP data were expressed as a percentage of input and presented as the relative fold enrichment to mock control. Primers used for qPCR of ISRE regions of CXCL9/10 are listed in Table S5.
Sandwich ELISA
For CXCL9/10/11 ELISA, MECs were seeded onto a 6-well plate, supernatants were collected at different time points as indicated, spun down to remove debris and analyzed according to manufacturer’s protocol (R&D Systems).
Sample preparation, image acquisition and data analysis for expansion microscopy
Cells were cultured on 18 × 18mm-1.5 microscope cover glass (Fisher Scientific) in 12-well plates. After IFN treatment, cells were fixed in 3.7% formaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 2 mg/mL BSA. Samples were incubated with primary antibodies against STAT1 (2 μg/ml), STAT2 (2 μg/ml) and PML (10 μg/ml) in blocking buffer at 4°C overnight. The procedure for sample gelation, digestion, and expansion has been described as previously reported (47–49). Briefly, the samples were incubated in 0.01% acrylic acid N-hydroxysuccinimide ester (Santa Cruz) in PBS for 2.5 hours. Amonomer solution (1x PBS, 2 M NaCl, 25% sodium acrylate (w/w), 4% acrylamide (w/w), 0.04% (w/w) N, N’-methylenebisacrylamide) was mixed with 0.2% (w/w) ammonium persulfate (APS) and 0.2% (w/w) tetramethylethylenediamine (TEMED) to promote polymerization. The monomer solution mixed with the APS and TEMED was then added to samples to a depth of 2 mm and incubated for 2 hours at 37°C to allow for the completed polymerization. The size of the gel was measured. The polymerized gel was then immersed in 4 ml of 0.2 mg/ml proteinase K (Roche) in digestion buffer (50 mM Tris pH8, 2 mM CaCl2, 3M NaCl, 0.5% Triton X-100, and 0.8 M guanidine HCl) and incubated at 37°C for 3 hours. Digested gels were next placed in excess volumes of PBS for 2 hour. The gels were then stained with Alexa488 or Alexa568 conjugated secondary antibodies at room temperature overnight, followed by washing in10 mM Tris buffer (pH 8.0), which achieved expansion ratio around 4 in each dimension. The expanded hydrogels were then placed onto poly-L-lysine coated cover glass for confocal imaging.
The confocal data were analyzed by ImageJ/Fiji. For each z-stack image, the nucleus region was first identified by Gaussian blurring the DAPI channel with 1 μm interval followed by thresholding and binarization. Then, PML bodies in each nucleus were identified by Gaussian blurring the PML channel signals with interval of 140 nm, followed by thresholding and binarization. STAT1/STAT2 signals within and outside of the PML body regions were compared, with the condensation index defined as ratio of the mean intensity within PML bodies to that outside of PML bodies in nuclei. A total of 9–11 cells in each sample conditions were analyzed.
Quantification and Statistical Analysis
Experiments were repeated at least twice. Results are expressed as mean + SD or SEM as indicated. Statistical significance was evaluated by a paired or unpaired t-test, or by one-way/two-away ANOVA as indicated in the figure legends, using p < 0.05 as indicative of statistical significance. Linear regression analyses in breast invasive carcinoma were performed by Rstudio using TCGA datasets. Data for ISG15, MX1 and UBA7-correlated genes were downloaded from cBioPotal (50,51). The gene expression data and survival information of 3554 patients were downloaded from GEO and analyzed as previously reported (52). Survival analysis was performed by drawing a Kaplan-Meier survival plot and computing a Cox proportional hazard regression as described previously (53). RNA sequencing data analyzed in this study are available from Gene Expression Ominibus (GSE112532).
Supplementary Material
Statement of significance.
We report a highly cooperative ISG network, in which UBA7-mediated ISGylation facilitates clustering of transcription factors and activates an anti-tumor gene expression program. These findings provide mechanistic insights into immune evasion in breast cancer associated with UBA7 loss, emphasizing the importance of a functional ISG–ISGylation network in tumor suppression.
Acknowledgments
We thank Dr. Sam Stoner for critical editing of this manuscript; Dr. Judith Varner and all the members of D. E. Z laboratory for discussion, suggestion and help. We gratefully acknowledge Drs. Ernest Borden and Deborah Lenschow for ISG15 antibodies, Dr. Lesley G. Ellies for C57BL/6 and FVB/N-Tg (MMTV-PyVmT) 634Mul/J mice, Dr. Michael Karin for CD8 KO mice. This work was supported by funding from National Institute of Health New Innovator Award to HC (1-DP2-EB020400) and funding from National Institutes of Health to DEZ (R01CA177305).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Patel SA, Minn AJ. Combination Cancer Therapy with Immune Checkpoint Blockade: Mechanisms and Strategies. Immunity 2018;48(3):417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nature Reviews Cancer 2018;18(3):139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stark GR, Darnell JE. The JAK-STAT Pathway at Twenty. Immunity 2012;36(4):503–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden EC. Interferons alpha and beta in cancer: therapeutic opportunities from new insights. Nature Reviews Drug Discovery 2019;18(3):219–34. [DOI] [PubMed] [Google Scholar]
- 5.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nature Reviews Cancer 2016;16(3):131–44. [DOI] [PubMed] [Google Scholar]
- 6.Zitvogel L, Galluzzi L, Kepp O, Smyth MJ, Kroemer G. Type I interferons in anticancer immunity. Nature Reviews Immunology 2015;15(7):405–14. [DOI] [PubMed] [Google Scholar]
- 7.Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and regulation in cancer immunotherapy. Cell Research 2017;27(1):96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benci JL, Xu BH, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor Interferon Signaling Regulates a Multigenic Resistance Program to Immune Checkpoint Blockade. Cell 2016;167(6):1540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minn AJ, Wherry EJ. Combination Cancer Therapies with Immune Checkpoint Blockade: Convergence on Interferon Signaling. Cell 2016;165(2):272–5. [DOI] [PubMed] [Google Scholar]
- 10.Yuan WM, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO Journal 2001;20(3):362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bektas N, Noetzel E, Veeck J, Press MF, Kristiansen G, Naami A, et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Research 2008;10(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang XQ, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, et al. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature 2015;517(7532):89–U229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Palma M, Mazzieri R, Politi LS, Pucci F, Zonari E, Sitia G, et al. Tumor-targeted interferon-alpha delivery by Tie2-expressing monocytes inhibits tumor growth and metastasis. Cancer Cell 2008;14(4):299–311. [DOI] [PubMed] [Google Scholar]
- 14.Escobar G, Moi D, Ranghetti A, Ozkal-Baydin P, Squadrito ML, Kajaste-Rudnitski A, et al. Genetic Engineering of Hematopoiesis for Targeted IFN-alpha Delivery Inhibits Breast Cancer Progression. Science Translational Medicine 2014;6(217):217ra3. [DOI] [PubMed] [Google Scholar]
- 15.Cai Y, Nogales-Cadenas R, Zhang QW, Lin JR, Zhang W, O’Brien K, et al. Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. Bmc Genomics 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketscher L, Hannss R, Morales DJ, Basters A, Guerra S, Goldmann T, et al. Selective inactivation of USP18 isopeptidase activity in vivo enhances ISG15 conjugation and viral resistance. Proceedings of the National Academy of Sciences of the United States of America 2015; 112(5):1577–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CMT, Pryer N, et al. Macrophage IL-10 Blocks CD8(+) T Cell-Dependent Responses to Chemotherapy by Suppressing IL-12 Expression in Intratumoral Dendritic Cells. Cancer Cell 2014;26(5):623–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. Journal of Experimental Medicine 2001;193(6):727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sistigu A, Yamazaki T, Vacchelli E, Chaba K, Enot DP, Adam J, et al. Cancer cell-autonomous contribution of type I interferon signaling to the efficacy of chemotherapy. Nature Medicine 2014;20(11):1301–9. [DOI] [PubMed] [Google Scholar]
- 20.Lee SH, Jia SD, Zhu YN, Utermark T, Signoretti S, Loda M, et al. Transgenic Expression of Polyomavirus Middle T Antigen in the Mouse Prostate Gives Rise to Carcinoma. Journal of Virology 2011;85(11):5581–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vacchelli E, Enot DP, Pietrocola F, Zitvogel L, Kroemer G. Impact of Pattern Recognition Receptors on the Prognosis of Breast Cancer Patients Undergoing Adjuvant Chemotherapy. Cancer Research 2016;76(11):3122–6. [DOI] [PubMed] [Google Scholar]
- 22.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nature Medicine 2007;13(9):1050–9. [DOI] [PubMed] [Google Scholar]
- 23.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Frontiers in Immunology 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell 2017;169(1):13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lallemand-Breitenbach V, de The H. PML nuclear bodies: from architecture to function. Current Opinion in Cell Biology 2018;52:154–61. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, et al. Arsenic Trioxide Controls the Fate of the PML-RAR alpha Oncoprotein by Directly Binding PML. Science 2010;328(5975):240–3. [DOI] [PubMed] [Google Scholar]
- 27.Shiao SL, Ruffell B, DeNardo DG, Faddegon BA, Park CC, Coussens LM. T(H)2-Polarized CD4(+) T Cells and Macrophages Limit Efficacy of Radiotherapy. Cancer Immunology Research 2015;3(5):518–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon YJ, Jo MG, Yoo HM, Hong SH, Park JM, Ka SH, et al. Chemosensitivity is controlled by p63 modification with ubiquitin-like protein ISG15. Journal of Clinical Investigation 2012;122(7):2622–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai SD, Reed RE, Burks J, Wood LM, Pullikuth AK, Haas AL, et al. ISG15 disrupts cytoskeletal architecture and promotes motility in human breast cancer cells. Experimental Biology and Medicine 2012;237(1):38–49. [DOI] [PubMed] [Google Scholar]
- 30.Sekido Y, Ahmadian M, Wistuba II, Latif F, Bader S, Wei MH, et al. Cloning of a breast cancer homozygous deletion junction narrows the region of search for a 3p21.3 tumor suppressor gene. Oncogene 1998;16(24):3151–7. [DOI] [PubMed] [Google Scholar]
- 31.Haricharan S, Brown P. TLR4 has a TP53-dependent dual role in regulating breast cancer cell growth. Proceedings of the National Academy of Sciences of the United States of America 2015;112(25):E3216–E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiahou ZK, Wang XL, Shen J, Zhu XX, Xu F, Hu R, et al. NMI and IFP35 serve as proinflammatory DAMPs during cellular infection and injury. Nature Communications 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Applied and Environmental Microbiology 2016;82(16):5039–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salaun B, Zitvogel L, Asselin-Paturel C, Morel Y, Chemin K, Dubois C, et al. TLR3 as a Biomarker for the Therapeutic Efficacy of Double-stranded RNA in Breast Cancer. Cancer Research 2011;71(5):1607–14. [DOI] [PubMed] [Google Scholar]
- 35.Kim YE, Ahn JH. Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus. Plos Pathogens 2015;11(3):e1004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boisvert FM, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. Journal of Cell Biology 2000;148(2):283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, et al. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016;166(3):651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell 2002;108(2):165–70. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Martin N, Piva M, Urosevic J, Aldaz P, Sutherland JD, Fernandez-Ruiz S, et al. Stratification and therapeutic potential of PML in metastatic breast cancer. Nature Communications 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bezzi M, Seitzer N, Ishikawa T, Reschke M, Chen M, Wang GC, et al. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nature Medicine 2018;24(2):165–75. [DOI] [PubMed] [Google Scholar]
- 41.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Molecular Cell 2013;51(3):310–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Au-Yeung N, Horvath CM. Transcriptional and chromatin regulation in interferon and innate antiviral gene expression. Cytokine & Growth Factor Reviews 2018;44:11–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burkart C, Arimoto KI, Tang TD, Cong XL, Xiao NM, Liu YC, et al. Usp18 deficient mammary epithelial cells create an antitumour environment driven by hypersensitivity to IFN-lambda and elevated secretion of Cxcl10. EMBO Molecular Medicine 2013;5(7):1035–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malakhova OA, Yan M, Malakhov MP, Yuan YZ, Ritchie KJ, Kim KI, et al. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes & Development 2003;17(4):455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Longo PA, Kavran JM, Kim MS, Leahy DJ. Transient Mammalian Cell Transfection with Polyethylenimine (PEI). Laboratory Methods in Enzymology: DNA 2013;529:227–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deliard S, Zhao JH, Xia QH, Grant SFA. Generation of High Quality Chromatin Immunoprecipitation DNA Template for High-throughput Sequencing (ChIP-seq). Jove-Journal of Visualized Experiments 2013(74). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chozinski TJ, Halpern AR, Okawa H, Kim HJ, Tremel GJ, Wong ROL, et al. Expansion microscopy with conventional antibodies and fluorescent proteins. Nature Methods 2016;13(6):485–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tillberg PW, Chen F, Piatkevich KD, Zhao YX, Yu CC, English BP, et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nature Biotechnology 2016;34(9):987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science 2015;347(6221):543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao JJ, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science Signaling 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerami E, Gao JJ, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2012;2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li QY, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment 2010;123(3):725–31. [DOI] [PubMed] [Google Scholar]
- 53.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-Related Cancer 2012;19(2):197–208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.