Abstract
Doxorubicin (DOX) is a potent anticancer agent that binds both DNA and cardiolipin (CL). To investigate DOX binding to CL versus DNA, aqueous soluble, CL-enriched nanoparticles, termed nanodisks (ND), were employed. Upon incubation with CL-ND, but not with phosphatidylcholine ND, DOX binding was detected. DOX binding to CL-ND was sensitive to buffer pH and ionic strength. To investigate if a DOX binding preference for DNA versus CL-ND exists, an agarose gel-based dye binding assay was developed. Under conditions wherein the commercial fluorescent dye, GelRed, detects a 636 bp DNA template following electrophoresis, DOX staining failed to visualize this DNA band. Incubation of the template DNA with DOX prior to electrophoresis resulted in a DOX concentration-dependent attenuation of GelRed staining intensity. When the template DNA was pre-incubated with equivalent amounts of free DOX or DOX-CL-ND, no differences in the extent of GelRed staining intensity attenuation were noted. When DOX was incubated with DNA alone, or a mixture of DNA and CL-ND, the extent of DOX-induced GelRed staining intensity attenuation was equivalent. Thus, DOX has a binding preference for DNA versus CL and, moreover, DOX-CL-ND offer a potential strategy to prevent DOX-induced cardiotoxicity while not affecting its affinity for DNA.
Keywords: Nanodisk, Phospholipid, GelRed, Fluorescence, Cardiotoxicity, Cancer
Graphical Abstract
Introduction
Doxorubicin (DOX) is an aqueous soluble anthracycline known for its bright red color and potent anti-tumor properties [1]. Anti-cancer properties of DOX manifest through intercalation of nuclear DNA, inhibiting topoisomerase II function and DNA replication [2]. While DOX is one of the most effective anti-cancer pharmaceuticals currently available, many patients suffer from a potentially lethal side effect referred to as ‘DOX-induced cardiotoxicity’ [3,4]. Evidence indicates that DOX-induced cardiotoxicity is unrelated to its ability to intercalate DNA but, rather, arises from a secondary DOX-induced phenomenon [5–8]. A prominent mechanism proposed to explain DOX-induced cardiotoxicity involves conversion of DOX’s anthraquinone moiety (Figure 1A) to a reactive semiquinone free radical, which induces oxidative stress via generation of reactive oxygen species [7]. Subsequent lipid peroxidation leads to membrane defects, alterations in molecular signaling pathways and, ultimately, cardiac cell death.
Figure 1. Effect of ND on DOX dialysis.
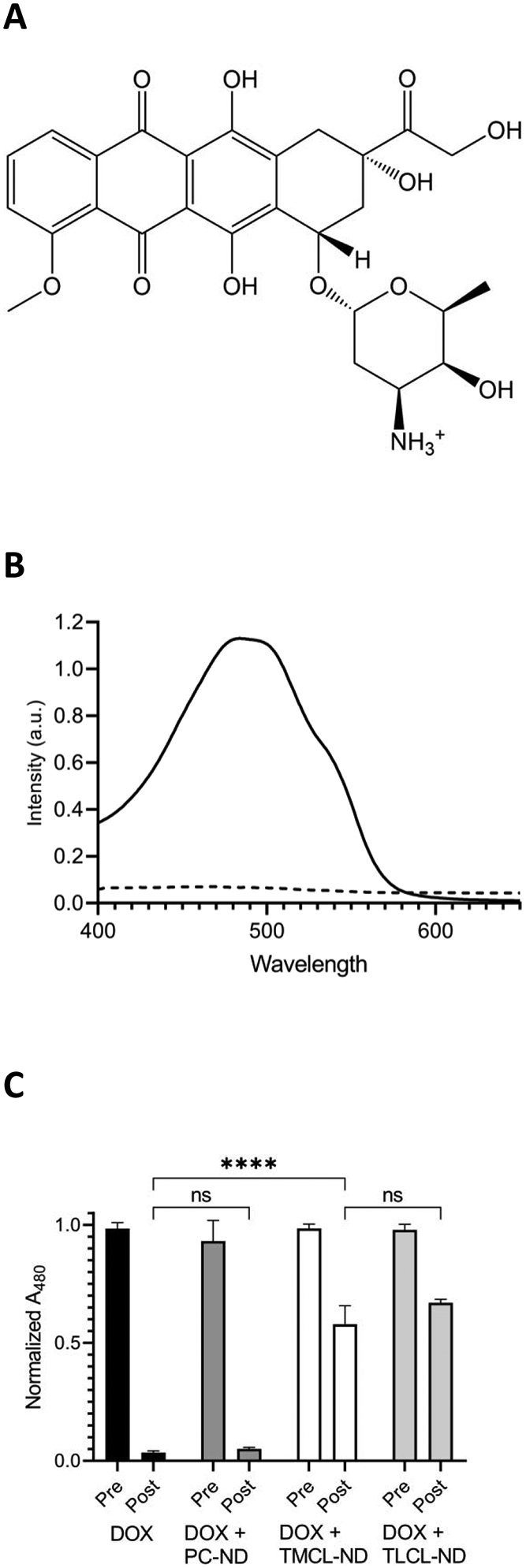
A) Structure of doxorubicin at physiological pH. B) Visible absorption spectra of 0.1 μmol free DOX (solid line) and 0.6 μmol CL-ND (dashed line) in 20 mM Tris HCl, pH 7.4, were collected on a Shimadzu UV-1800 spectrophotometer. C) Effect of dialysis on DOX sample absorbance. Samples containing free DOX, DOX plus PC-ND, DOX plus TMCL-ND and DOX + TLCL-ND were incubated for 30 min prior to 2 h dialysis against Tris HCl buffer. Sample absorbance at 480 nm was measured before and after dialysis. Data reported as normalized A480. Values reported are the mean ± standard error (n=3) ns, not significant; ****, p < .0001 versus free DOX control.
Cardiolipin (CL) is an anionic phospholipid that localizes exclusively to the inner mitochondrial membrane where it plays important roles in cristae structure and bioenergetics [9,10]. Despite the fact that DOX is water soluble, it displays a binding affinity for CL containing membranes [11,12]. Although the binding interaction between DOX and CL is unrelated to its DNA intercalation activity, it is plausible to consider that CL is involved in DOX-induced cardiotoxicity. With regard to the nature of the interaction between DOX and CL, it has been shown that the positively charged amino group on DOX’s carbohydrate constituent (Figure 1A) forms an electrostatic interaction with negatively charged phosphate groups on CL [13]. Others have proposed that, following an initial electrostatic attraction, DOX’s aromatic anthraquinone moiety intercalates between phospholipid fatty acyl chains in bilayer membranes [12, 14–16]. It is conceivable that the interaction between membrane localized DOX and CL leads to oxidative damage, initiating molecular events that lead to cardiotoxicity [17–19]. Toward the goal of developing strategies that minimize negative side effects of DOX, while retaining its beneficial anticancer effects, it is important to characterize the relative affinity of DOX for DNA versus CL.
To characterize this affinity, water soluble CL-enriched nanoparticles, termed CL nanodisks (CL-ND), were used. These self-assembled nanoscale complexes are comprised solely of cardiolipin and recombinant human apolipoprotein (apo) A-I [20]. The particles adopt a discoidal structure in which CL adopts a bilayer state while apoA-I molecules circumscribe the disk perimeter, creating a stable, water soluble complex [21]. In binding assays, DOX is shown to associate with CL-ND comprised of either tetramyristoyl (TM)- or tetralinoleoyl (TL)- CL. Subsequently, the relative affinity of DOX for DNA versus CL was investigated using a competitive agarose gel-based dye binding assay. Experiments employing this assay provide evidence that the affinity of DOX for DNA is greater than that for CL.
Materials and Methods
ND formulation.
Nanodisks (ND) were formulated as described previously [20]. Briefly, 25 mg dimyristoylphosphatidylcholine (PC), TMCL or TLCL (Avanti Polar Lipids) was dissolved in 1 mL CHCl3:CH3OH (3:1 v/v). Two hundred μL aliquots (5 mg) were transferred to glass tubes and dried under a stream of N2 gas. The samples were lyophilized overnight and stored at −20° C until use. For CL-ND formulation, 750 μl 20 mM Tris HCl pH 7.4 was added to 5 mg dried CL. The sample was vortexed, resulting in an opaque aqueous lipid dispersion. Subsequently, 2 mg recombinant human apolipoprotein (apo) A-I [22] in 20 mM Tris HCl was added. The mixture was brought to a final volume = 1.25 mL and bath sonicated at 48° C until the solution cleared (<10 min). PC-ND and TLCL-ND were formed in a similar manner, except that bath sonication was conducted at 25° C. TLCL-ND samples were sonicated under an N2 atmosphere. DOX was purchased from Cayman Chemical Company and solubilized in a) 20 mM Tris HCl buffer (1 mM solution for dialysis studies) or b) DMSO (42 mM stock solution for agarose gel studies). DOX in Tris HCl buffer was prepared immediately prior to use while DOX in DMSO was stored at −20 °C until use.
DOX binding studies.
To investigate DOX binding to different ND, 100 nmol DOX was incubated with PC-ND (1,090 nmol PC), TMCL-ND (622 nmol CL) or TLCL ND (622 nmol CL) at 22 °C for 30 min in Tris HCl, pH 7.4 (1.0 mL total volume). Following incubation, each sample was dialyzed against 1.66 L buffer. To examine the effect of pH on this binding interaction, samples were prepared as above, followed by dialysis against 1.66 L 25 mM B4Na2O7 (sodium tetraborate, anhydrous), pH 9.5, or 20 mM Tris HCl, pH 7.4. To test the effect of ionic strength on this binding interaction, samples were prepared as above, then dialyzed against 1.66 L 20 mM Tris HCl pH 7.4, in the presence or absence of 250 mM NaCl. At indicated time points, absorbance spectra from 650 nm to 400 nm were collected for all samples on a Shimadzu UV-1800 spectrophotometer.
DNA samples.
DOX binding experiments employed a 636 bp DNA gene amplification product generated from a plasmid encoding human glutaryl CoA dehydrogenase, using sequence specific forward and reverse primers. The amplification product was employed in gel-based DOX binding studies.
Competition binding studies.
One μg aliquots of the 636 bp gene amplification product were electrophoresed on 0.8% agarose gels (w/v). A 16 tooth comb was used for all experiments, and sample volumes were 8 μL per well. Samples were electrophoresed at 100 V in 1x TAE buffer for 70 min. Where indicated, the DNA template was pre-incubated with specified reagents. Following electrophoresis, gels were stained with 3x GelRed (Biotium) dye for 30 min and destained in deionized H2O for 2 h. Experiments were conducted a minimum of 3 times and gel data shown is representative of patterns observed between replicates.
Quantitative analysis.
Gel imaging was performed using a Typhoon FLA7000 (GE Life Sciences) or a Bio-Rad ChemiDoc MP (Bio-Rad). Typhoon images were obtained using a pixel size of 50 microns, power output of 450 V, blue laser (488 nm) and red filter (610/30). ChemiDoc images were obtained using the native GelRed protocol; transilluminator excitation 302 nm and red filter (630/30). Densitometric analysis was performed using tools native to Image Lab software (Bio-Rad, version 5.2.1). Band intensities were quantified on the basis of pixel volume and area (mm2) of individual bands.
Statistical Analysis.
Statistical analysis was performed by two-way ANOVA followed by Tukey multiple comparison test to compare samples from the same time point (Figures 1, 2). Statistical analysis was performed on agarose gel densitometry data by one-way ANOVA followed by Tukey test to compare control bands with unknown sample bands (Figures 4, 5, 6, 7). Statistical tests were performed using GraphPad Prism version 8.2.1 for Windows (GraphPad Software, San Diego, CA)
Figure 2. Effect of solution pH and ionic strength on DOX binding to CL-ND.
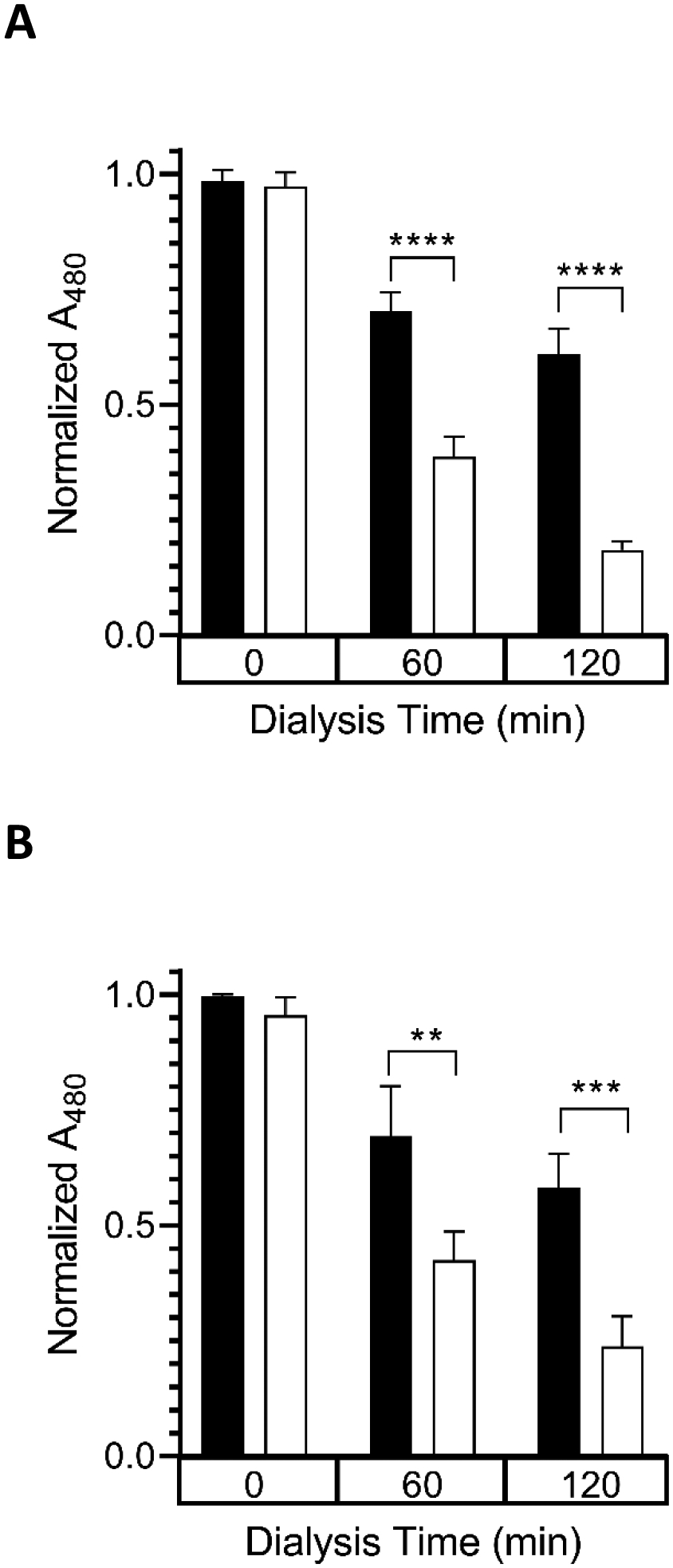
A) TMCL-ND were formulated and 0.6 μmol aliquots (as CL) were incubated with DOX (0.1 μmol DOX) in Tris HCl buffer (final volume = 1 mL). After 30 min incubation, the samples were dialyzed against 20 mM Tris, pH 7.4 (filled bars), or 25 mM sodium tetraborate, pH 9.5 (open bars). Dialysis was performed at 22° C for 2 h against 1.66 L buffer per sample. Sample absorbance was measured before dialysis, after 60 min dialysis and after 120 min dialysis. Data reported as normalized A480. Values reported are the mean ± standard error (n=3) ****, p < .0001 versus pH 7.4 samples; B) Aliquots of TMCL-ND (0.6 μmol CL) were incubated with DOX (0.1 μmol) in 20 mM Tris HCl, pH 7.4. After 30 min incubation, samples were dialyzed for 2 h at 22 °C against 20 mM Tris HCl, pH 7.4 (filled bars) or 20 mM Tris HCl, pH 7.4, 250 mM NaCl (open bars). Sample absorbance was measured before dialysis, after 60 min dialysis and 120 min dialysis. Data reported as normalized A480. Values reported are the mean ± standard error (n=3) **, p < 0.01 versus no NaCl control; ***, p < .001 versus no NaCl control.
Figure 4. Effect of CL-ND on DOX binding to DNA.
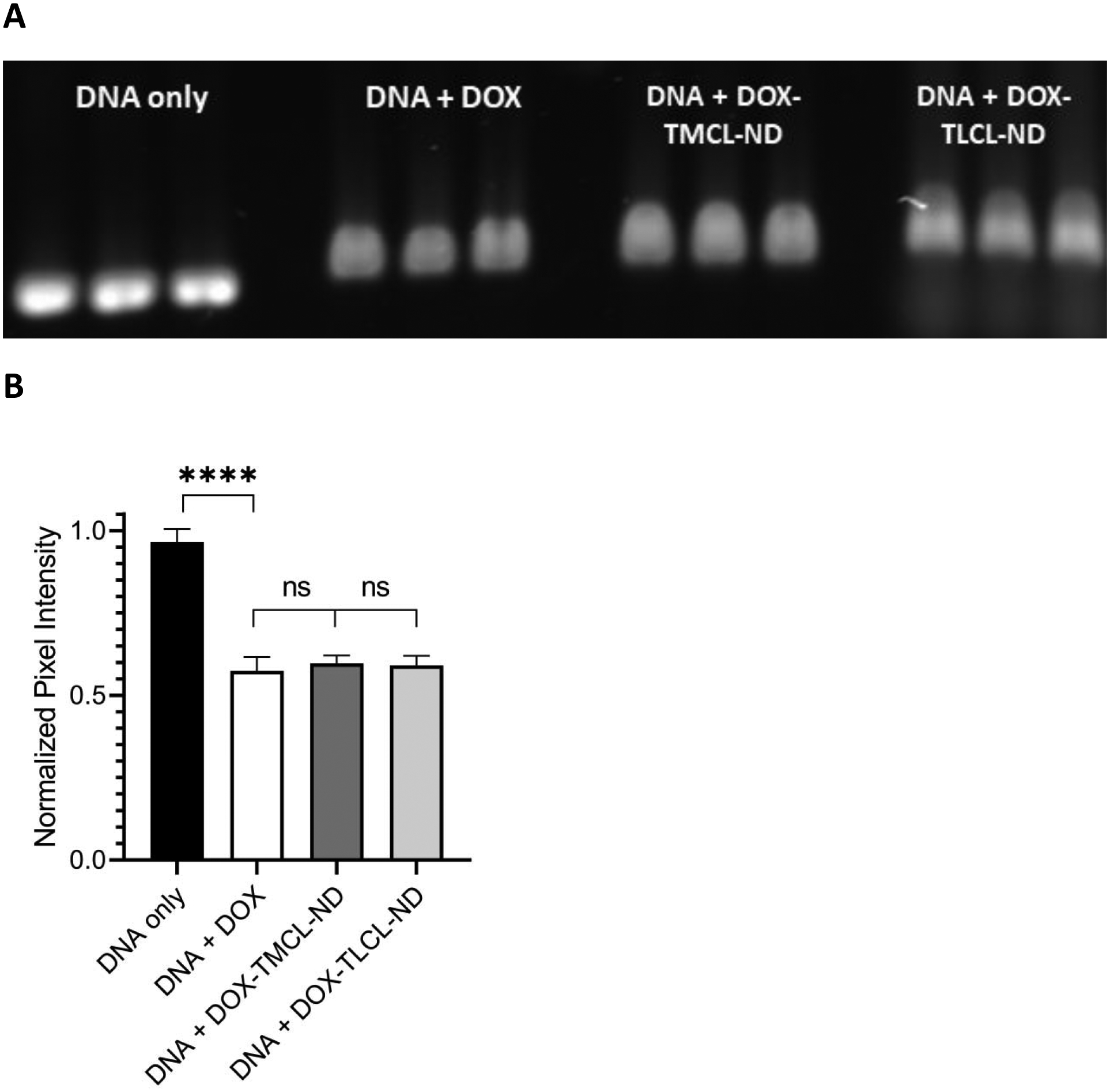
A) One μg aliquots of the 636 bp DNA template was incubated for 30 min in a) buffer alone; b) buffer containing 0.19 nmol free DOX; c) buffer containing 0.19 nmol DOX plus TMCL-ND and d) buffer containing 0.19 nmol free DOX plus TLCL-ND. Following incubation, the samples were applied to wells of a 0.8% agarose gel (w/v) and electrophoresed at 100 V for 70 min. Following electrophoresis, the gel was stained with GelRed for 30 min and destained for 2 h in deionized H2O. The gel was then imaged on a Bio-Rad ChemiDoc unit using the GelRed detection protocol; B) Densitometric analysis of gel bands from Panel A. Gel band pixel intensity was quantitated using ImageLab software analysis and defined as band pixel volume per band area (in mm2). The data were normalized to the “no DOX” control samples and presented as normalized pixel intensity. Values reported are the mean ± standard error (n=3) ns, not significant; ****, p < .0001 versus no DOX control.
Figure 5. Ability of CL-ND to extract DOX from DNA.
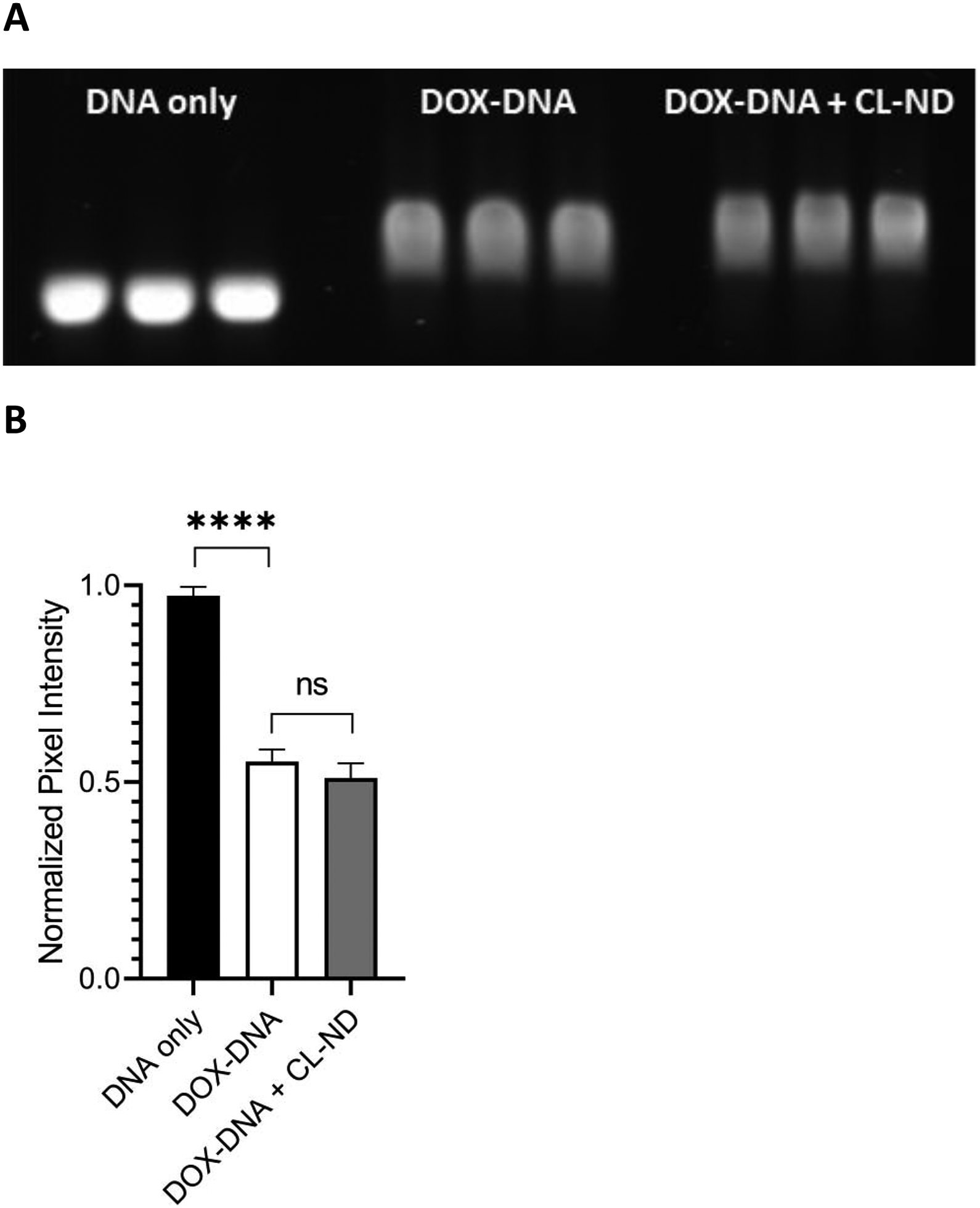
A) One μg aliquots of the 636 bp DNA template were incubated for 30 min in deionized H2O (n=3) or deionized H2O containing 0.19 nmol DOX (n=6). Following incubation, TMCL-ND (1.6 nmol CL) was added to 3 of the 6 DOX-treated DNA samples. After a further 30 min incubation, the samples were subjected to agarose gel electrophoresis and stained with GelRed. After destaining, the gel was imaged on a Bio-Rad ChemiDoc unit using the GelRed detection protocol. B) Densitometric analysis of gel bands from panel A. Gel band pixel intensity was quantitated using ImageLab software analysis and defined as band pixel volume per band area (in mm2). The data were normalized to the “no DOX” control samples and presented as normalized pixel intensity. Values reported are the mean ± standard error (n=3) ns, not significant; ****, p < .0001 versus no DOX control.
Figure 6. Effect of apoA-I on DOX binding to DNA.
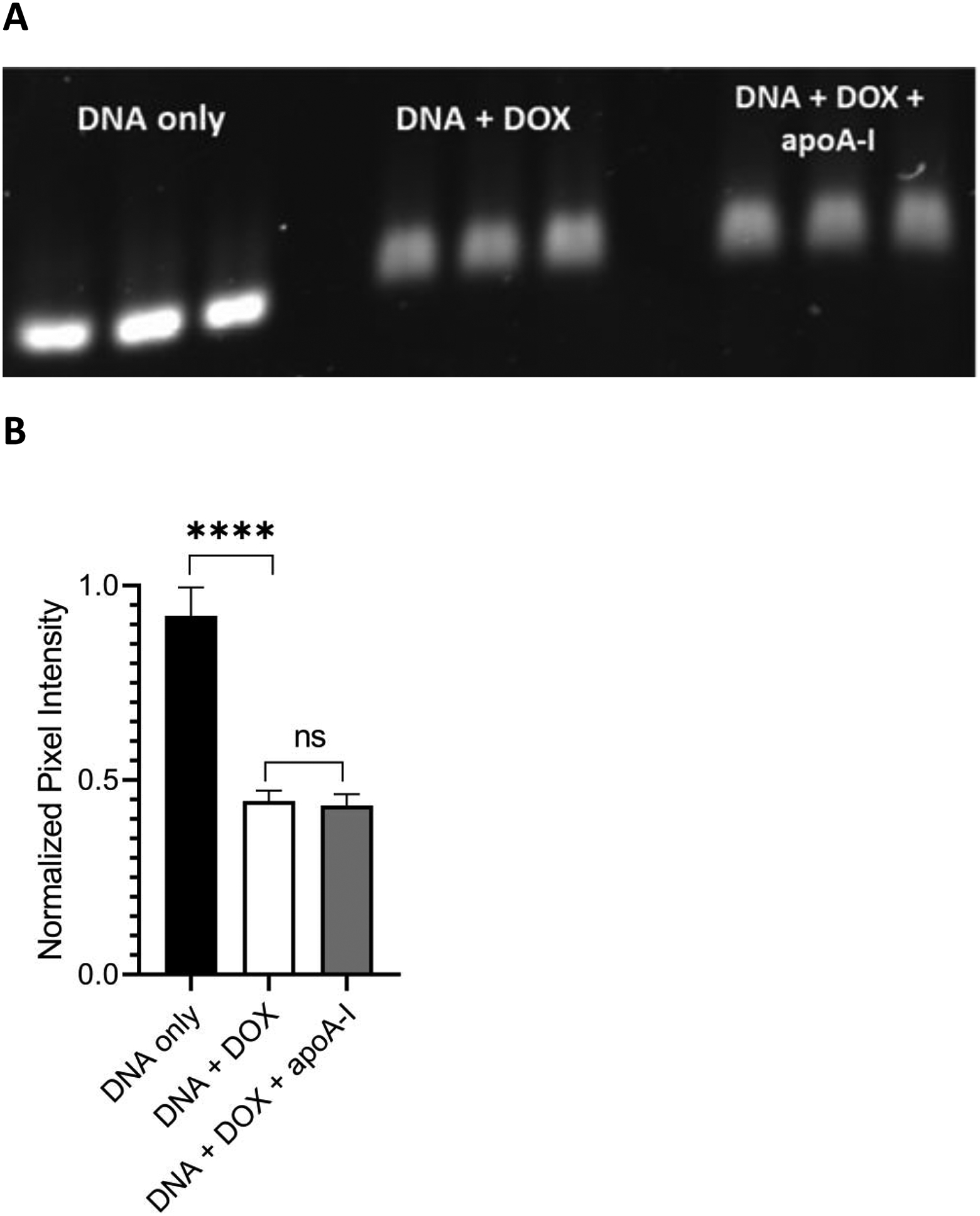
One μg aliquots of the 636 bp DNA template were incubated for 30 min in a) buffer alone, b) buffer containing 0.19 nmol free DOX (DNA + DOX) and c) buffer containing 0.19 nmol DOX plus 2.4 μg apoA-I (DNA + DOX + apoA-I). Following incubation, the samples were applied to wells of a 0.8% agarose gel (w/v) and electrophoresed at 100 V for 70 min (Panel A). The gel was stained with GelRed for 30 min, destained for 2 h in deionized H2O and imaged on a Bio-Rad ChemiDoc unit using the GelRed detection protocol; Panel B) Densitometric analysis of gel bands from Panel A. Gel band pixel intensity was quantitated using ImageLab software analysis and defined as band pixel volume per band area (in mm2). The data were normalized to the “DNA only” control samples and presented as normalized pixel intensity. Values reported are the mean ± standard error (n=3) ns, not significant; ****, p < .0001 versus no DOX control.
Figure 7. DOX competition binding assay for DNA versus CL.
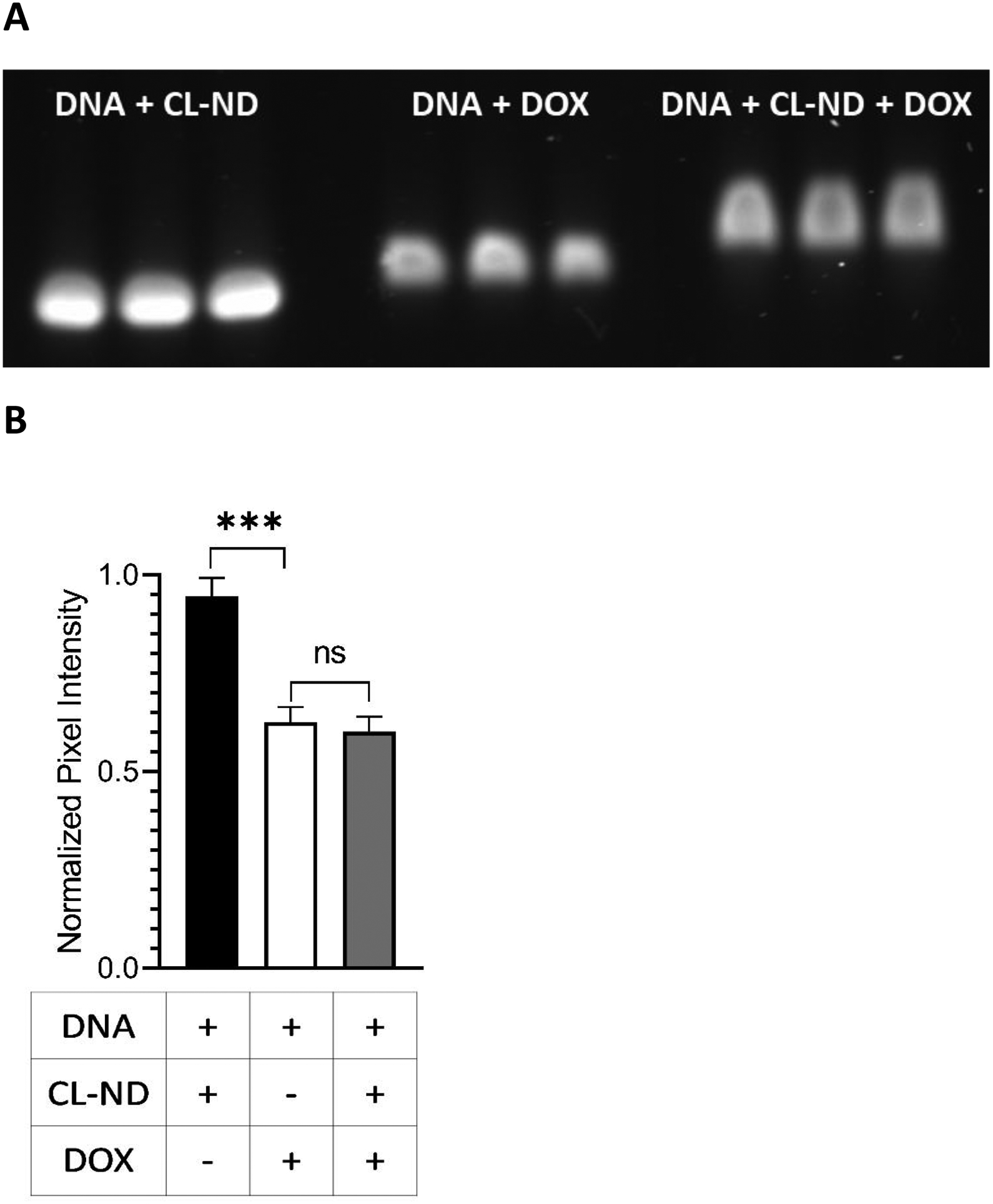
A) One μg aliquots of the 636 bp DNA template were incubated for 30 min in the absence (n=3) or presence (n=6) of TMCL-ND (1.6 nmol CL). DOX (0.19 nmol) was then added to the 3 DNA only samples and 3 of the DNA + CL-ND samples. Following a further 30 min incubation, the samples were subjected to agarose gel electrophoresis and stained with GelRed. After destaining, the gel was imaged on a Bio-Rad ChemiDoc unit using the GelRed detection protocol. B) Densitometric analysis of gel bands from Panel A. Gel band pixel intensity was quantitated using ImageLab software analysis and defined as band pixel volume per band area (in mm2). The data were normalized to the “no DOX” control lanes and presented as normalized pixel intensity. Values reported are the mean ± standard error (n=3) ns, not significant; ***, p < .0001 versus no DOX control.
Results
Cardiolipin ND.
CL-ND have a diameter in the range of 20 – 30 nm [23], corresponding to a particle molecular weight >200,000 Da. The CL component of CL-ND exists as a miniature bilayer membrane, similar to its organizational state in the inner mitochondrial membrane [10]. Surrounding the edge of this disk-shaped CL bilayer is apoA-I. Amphipathic a-helices of this scaffold protein contact otherwise exposed CL fatty acyl chains. This interaction serves to stabilize the CL bilayer while the polar face of apoA-I amphipathic α-helices project toward the aqueous environment.
DOX binding to CL-ND.
DOX has a characteristic absorbance spectrum with a main peak centered around 480 nm (Figure 1B). In contrast, CL-ND absorbance in this spectral region is negligible. Taking advantage of the unique spectral properties of DOX and large size difference versus CL-ND, a dialysis-based assay was used to investigate DOX interaction with CL. To determine the specificity of DOX binding to CL, indicated amounts of DOX were incubated in buffer alone (20 mM Tris HCl buffer, pH 7.4) and buffer containing PC-ND, TMCL-ND or TLCL-ND. After 30 min incubation at 22 °C, the samples were dialyzed against Tris HCl buffer for 2 h. DOX content in the retentate of each sample was measured spectrophotometrically (Figure 1C). Compared to pre-dialysis absorbance values, following 2 h dialysis the sample containing DOX in buffer only, as well as the DOX plus PC-ND sample, lost >90% of the original DOX content. Given its molecular weight (543 Da), it is evident that free DOX rapidly escapes the dialysis bag and moreover, PC-ND had no significant effect on the rate of DOX dialysis. On the other hand, upon incubation of DOX with either TMCL-ND or TLCL-ND, similar levels (~60–65%) of the added DOX were retained following dialysis.
Factors affecting DOX affinity for CL-ND.
To investigate the nature of the binding interaction between DOX and TMCL-ND, free DOX was incubated with TMCL-ND in 20 mM Tris HCl, pH 7.4. Following incubation, the samples were dialyzed against 20 mM Tris HCl buffer or 25 mM sodium tetraborate buffer, pH 9.5, and DOX content in the retentate determined spectrophotometrically as a function of time (Figure 2A). After 2 h dialysis, ~60% of the original absorbance intensity was retained in the DOX-CL-ND sample that was dialyzed against pH 7.4 buffer while the sample dialyzed against pH 9.5 buffer retained ~20%. To further investigate the binding interaction between DOX and TMCL-ND, free DOX, in 20 mM Tris buffer, was incubated with TMCL-ND to promote a binding interaction. The samples were then dialyzed against 20 mM Tris HCl buffer, in the presence or absence of 250 mM NaCl. DOX content in each retentate was then determined as a function of time (Figure 2B). Whereas ~60% of original absorbance intensity was retained in samples dialyzed against buffer in the absence of 250 mM NaCl, when DOX plus TMCL-ND was dialyzed against 20 mM Tris HCl buffer containing 250 mM NaCl, ~18% of the starting DOX was recovered in the retentate.
Effect of DOX on GelRed DNA staining intensity.
Having established that TMCL-ND constitute a useful platform for studies of DOX-CL interactions, a method was sought to determine whether DOX has a binding preference for DNA versus CL. To address this, an agarose gel-based assay was developed by taking advantage of the unique fluorescence properties of the commercial DNA binding dye, GelRed, versus those of DOX. GelRed is a nontoxic ethidium-based dye commonly used to visualize DNA following agarose gel electrophoresis [24]. When 1 μg of a 636 bp DNA template was electrophoresed on a 0.8% agarose gel and stained with GelRed, a strong positive band was observed (Figure 3A, left). However, pre-incubation of the DNA template with increasing amounts of DOX resulted in a DOX concentration-dependent attenuation of GelRed staining intensity (Figure 3A). Moreover, when pre-incubated with high concentrations of DOX, migration of the template DNA slowed and the band broadened. In control experiments (Figure 3B), when the template DNA was electrophoresed and stained directly with DOX, no band was visible under conditions used for optimal GelRed staining (excitation 302 nm) or when excited at 488 nm, the wavelength of maximum DOX fluorescence excitation [25]. On the other hand, when the DNA template was stained with GelRed, a band was visible under both conditions. These data confirm that DOX does not contribute to visualization of the DNA template band under GelRed imaging conditions. The apparent reduced electrophoretic mobility of the DNA template following pre-incubation with higher concentrations of DOX supports the interpretation that DOX intercalates base pairs in the DNA template and remains bound during electrophoresis [26]. The ability of DOX pre-incubation to attenuate GelRed-specific DNA staining intensity indicates that GelRed does not displace DOX from DNA. Moreover, DOX-dependent attenuation of GelRed staining intensity can be used as the basis for an assay to assess the relative binding affinity of DOX for DNA versus CL.
Figure 3. Effect of DOX preincubation on GelRed staining intensity of a DNA template.
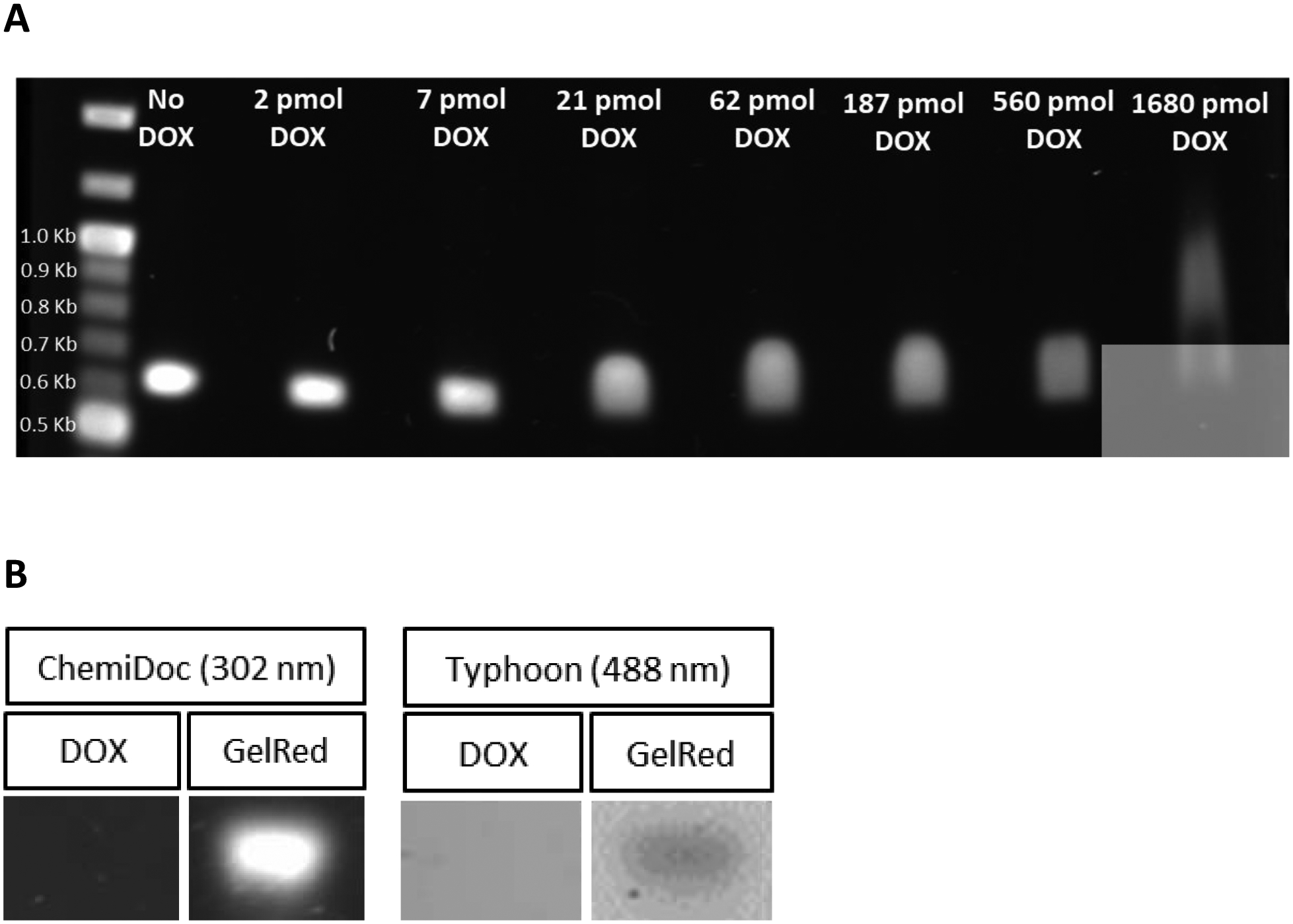
A) One μg aliquots of a 636 bp DNA template were incubated with indicated amounts of DOX for 30 min. Following incubation, samples were added to the wells of a 0.8% (w/v) agarose gel and electrophoresed at 100 V for 70 min. Following electrophoresis, the gel was stained with GelRed for 30 min and destained in deionized H2O for 2 h. The stained gel was imaged on a Bio-Rad ChemiDoc unit using the GelRed detection protocol. B) One μg aliquots of the 636 bp DNA template were applied to wells of a 0.8% (w/v) agarose gel and electrophoresed at 100 V for 70 min. Following electrophoresis, the gel was cut in half, and one half stained with GelRed while the other half was stained with DOX (50 μg/mL). Gels were then imaged using GelRed imaging conditions (302 nm excitation) on a Bio-Rad ChemiDoc and using DOX imaging conditions (488 nm excitation) on a GE Typhoon.
Effect of CL-ND on DOX binding to DNA.
To investigate the relative binding affinity of DOX for DNA versus CL, the gel-based dye binding assay described above was employed in conjunction with CL-ND comprised of either TMCL or TLCL. One μg of template DNA was incubated with free DOX or pre-formed DOX-CL-ND for 30 min. If prior association with CL prevents DOX binding to DNA, then it is expected that free DOX will result in a greater attenuation of GelRed staining intensity than that achieved by DOX-CL-ND. Additionally, by comparing ND formulated with TMCL versus TLCL in this assay, the effect of CL acyl chain saturation on DOX binding can be assessed. Following incubation, samples were subjected to agarose gel electrophoresis and stained with GelRed (Figure 4A). Densitometric analysis of the pixel intensity of the gel bands is depicted in Figure 4B. Compared to control DNA (no DOX incubation), the free DOX, DOX-TMCL-ND and DOX-TLCL-ND samples gave rise to a statistically significant attenuation of GelRed staining intensity. However, no differences were observed in the relative ability of free DOX, DOX-TMCL-ND or DOX-TLCL-ND to attenuate GelRed staining intensity of the template DNA band. Thus, it may be concluded that DOX displays preferential binding for DNA versus CL and prior association with CL does not affect DOX’s ability to bind DNA.
Effect of prior DNA binding on DOX affinity for CL-ND.
To further investigate the binding preference of DOX for DNA versus TMCL, the gel-based dye binding assay was adapted to assess whether pre-incubation of DOX with DNA is affected by subsequent incubation with TMCL-ND. Following incubation of free DOX with the template DNA, 1 μg aliquots (as DNA) were incubated for 30 min in Tris HCl buffer or Tris HCl buffer containing TMCL-ND. Following incubation, the samples were subjected to agarose gel electrophoresis and stained with GelRed (Figure 5A). Compared to a control DNA sample (no DOX), both the DNA-DOX alone and DNA-DOX plus TMCL-ND samples showed statistically significant attenuation in GelRed staining intensity. Densitometric analysis of the template DNA band pixel intensity revealed that incubation with TMCL-ND did not affect GelRed staining attenuation of the template DNA induced by pre-incubation with DOX (Figure 5B). These data indicate that, when bound to DNA, DOX is not available for interaction with CL-ND.
Effect of apoA-I on DOX binding to DNA.
To investigate whether lipid free apoA-I influences the interaction of DOX with DNA, the gel-based dye binding assay described above was employed. Samples of lipid free apoA-I were incubated with DOX for 30 min. One μg of template DNA was then incubated for an additional 30 min in buffer alone, in buffer containing free DOX or in buffer containing free DOX plus apoA-I. Following incubation, the samples were subjected to agarose gel electrophoresis and stained with GelRed (Figure 6A). Densitometric analysis of the pixel intensity of the gel bands is depicted in Figure 6B. Compared to the DNA only sample (no DOX incubation), both the free DOX and free DOX plus apoA-I samples gave rise to a statistically significant attenuation of GelRed staining intensity. At the same time, there was no statistically significant difference between the DNA plus DOX samples and the DNA plus DOX plus apoA-I samples in terms of their ability to attenuate GelRed staining intensity of the template DNA band. Thus, it may be concluded that lipid free apoA-I has no effect on the interaction of DOX with DNA.
DOX competition binding assay for DNA versus CL.
To further investigate the relative binding affinity of DOX for DNA versus CL, samples containing TMCL-ND and template DNA were incubated for 30 min. Following this, free DOX was added and the sample incubated a further 30 min. Control incubations, including DNA plus TMCL-ND (no DOX) and DNA plus DOX (no TMCL-ND) were run in parallel. Following incubation, the samples were subjected to agarose gel electrophoresis and stained with GelRed (Figure 7A). Band intensities were then analyzed by densitometry and the results depicted in Figure 7B. As expected, the DNA plus TMCL-ND (no DOX) control sample showed the strongest GelRed staining intensity, indicating that inclusion of TMCL-ND does not affect GelRed staining intensity. Compared to this sample, both DOX-containing samples (DNA alone and DNA plus TMCL-ND) gave rise to a statistically significant, DOX-dependent attenuation of GelRed staining intensity. At the same time, however, the extent of DOX-induced GelRed staining intensity attenuation observed in the sample containing DNA plus TMCL-ND was not statistically different from the sample containing DNA alone. One difference noted in this sample, however, was band broadening in incubations containing DNA and TMCL-ND, possibly due to the presence of excess TMCL-ND in the sample. Regardless, the data show that DOX displays preferential binding to DNA versus CL.
Discussion
CL-ND provide a versatile model membrane system to investigate the CL interactome. For example, recent studies provided insight into the effect of calcium binding on CL dependent membrane phase transitions [20]. This system is also amenable to studies of the interaction between cytochrome c and CL [27]. In the present study, the binding interaction between the fluorescent anthracycline, DOX, and the mitochondrial phospholipid, CL, were examined. DOX was shown to bind to CL-ND but not to PC-ND. This difference may be due to the fact that CL is an anionic phospholipid while PC is zwitterionic. The more exposed phosphate groups on CL may promote binding via electrostatic attraction with DOX’s single cationic amino group (see Figure 1A). Consistent with this interpretation, DOX binding to CL-ND was decreased at pH 9.5 (deprotonated, uncharged amino group) [28] versus pH 7.4 (protonated, positively charged amino group). The binding interaction between DOX and CL-ND was also reduced when solution ionic strength was increased from 0 mM to 250 mM NaCl (in 20 mM Tris HCl, pH 7.4). One possibility is that electrostatic attraction accounts for an initial step in DOX binding to CL containing membranes that is followed by a second step wherein the anthraquinone portion of the molecule integrates into the hydrophobic portion of the bilayer [14–16]. Despite these reports of a second step in DOX association with CL containing membranes, we observed no difference between TMCL- and TLCL-containing ND in terms of their relative ability to bind DOX or retain DOX upon incubation with DNA. If membrane insertion of the anthraquinone moiety occurred in the case of TLCL but not with TMCL, then a measurable difference in DOX binding behavior may be anticipated. Given that no effect of acyl chain saturation was observed, it may be that membrane insertion plays a less prominent role in DOX binding to membranes than previously reported or that some aspect of the ND particle structure, such as the presence of apoA-I as a scaffold protein, affects the ability of DOX to insert into the bilayer. This interpretation, however, does not align with the fact that numerous other hydrophobic molecules have been shown to intercalate between phospholipids in ND particles [21].
With regard to DOX-mediated membrane effects, it is conceivable that, when the drug binds to CL containing membranes, DOX may undergo conversion to a semiquinone radical that exerts pro-oxidant effects, leading to reactive oxygen species production, lipid peroxidation, membrane disruption and apoptosis. In fact, such a scenario may explain the well documented phenomenon of DOX-induced cardiotoxicity associated with DOX use as an anti-cancer agent [7]. Of interest to this concept is the fact that CL found in mitochondria from cardiac tissue is highly enriched in linoleic acid [29]. As such, DOX binding to endogenous CL containing membranes could elicit a direct effect on CL oxidation, promoting a transition from bilayer to non-bilayer phase, an event that is capable of triggering an apoptotic cascade [30].
Given that DOX-induced cardiotoxicity arises from a mechanism that is distinct from its DNA intercalation activity, we sought to employ CL-ND in studies designed to examine the intrinsic preference of DOX for DNA versus CL. To address this, a novel agarose gel-based competition binding assay was developed. Both DOX and GelRed function as DNA intercalating agents, such that upon incubation with a fixed amount of DNA, DOX reduces the intensity of GelRed staining intensity in a concentration-dependent manner (Figure 3A). Furthermore, due to differences in fluorescence emission properties and imaging conditions, it is possible to visualize GelRed stained DNA bands without any contribution from DOX, despite the fact that it is present (Figure 3B). The results presented show that pre-incubation of a 636 bp template DNA with DOX induces a concentration-dependent attenuation of GelRed staining intensity following electrophoresis. These data indicate that DOX interaction with DNA effectively prevents subsequent GelRed binding. Decreased GelRed binding under these conditions reduces GelRed-based signal intensity, which allows for the extent of DOX binding to be inferred.
When the ability of DNA to bind DOX, presented as free DOX or DOX-CL-ND, was investigated, no significant differences in GelRed signal intensity were observed. This indicates that DOX preference for DNA is greater than that for CL-ND and is consistent with results reported by Mustonen and Kinnunen [31] wherein the addition of DNA was shown to relieve DOX-induced quenching of liposomes that contain CL and a pyrene labeled phosphatidylcholine analog. When the ability of CL-ND to remove DOX pre-bound to DNA was investigated, no significant differences in GelRed signal intensity were observed, as compared to control incubations of DNA alone. These data indicate that, once DOX is complexed to DNA, its affinity for CL-ND is not high enough to induce dissociation. In addition, prior incubation of free DOX with lipid-free apoA-I had no effect on the ability of DOX to attenuate GelRed signal intensity of the DNA template. Lastly, upon comparison of DOX’s binding preference for DNA versus CL-ND in a direct competition binding assay, no significant difference in signal intensity was observed, as compared to control incubations of DNA without CL-ND. Overall, these data indicate DOX has a higher binding affinity for DNA than for CL-ND.
Given the prevalence of cardiotoxicity associated with DOX use in cancer chemotherapy, and evidence that interactions between DOX and CL may be responsible [32], the finding that DOX displays a strong binding preference for DNA versus CL-ND may be of key importance. For example, it is conceivable that administration of DOX as DOX-CL-ND may lead to a reduced incidence of cardiotoxicity because administered DOX will not be attracted to endogenous CL since it is already associated with this lipid. By the same token, given that DOX has a stronger affinity for DNA, its ability to intercalate DNA and, thereby, inhibit DNA replication, should be unaffected. While validation of this concept will require in vivo investigations, studies to date have revealed that CL-ND are well tolerated when injected into mice [33].
Highlights.
Doxorubicin(DOX) binds cardiolipin(CL) nanodisks(ND) but not phosphatidylcholine ND
Pre-incubation of DOX with DNA reduces GelRed staining intensity
DOX-mediated attenuation of GelRed-DNA staining formed the basis of an assay
Compared to free DOX, DOX-CL-ND showed similar GelRed staining attenuation of DNA
In competition binding experiments, DOX preferred binding to DNA versus CL-ND
Acknowledgements
The authors thank Christian Piscos, Irina Romenskaia and Sharon Young for contributing to the development of this project and helpful discussions. This work was supported by NIH grant R37 HL64159. CAF was supported by a Raymond H. Berner Graduate Scholarship and an American Heart Association Predoctoral Fellowship.
Abbreviations:
- CL
cardiolipin
- ND
nanodisks
- DOX
doxorubicin
- PC
phosphatidylcholine
- TM
tetramyristoyl
- TL
tetralinoleoyl
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no conflict of interest
References
- [1].Weiss RB, The anthracyclines: will we ever find a better doxorubicin? Semin. Oncol 19 (1992) 670–686. [PubMed] [Google Scholar]
- [2].Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, McLeod H, Klein TE, Altman RB, Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet. Genomics 21 (2011) 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chatterjee K, Zhang J, Honbo N, Karliner JS, Doxorubicin cardiomyopathy. Cardiology. 115 (2010) 155–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Al-Malky HS, Al Harthi SE, Osman AM, Major obstacles to doxorubicin therapy: Cardiotoxicity and drug resistance. J. Oncol. Pharm. Pract (2019) October 9 Epub ahead of print. doi: 10.1177/1078155219877931. [DOI] [PubMed] [Google Scholar]
- [5].Takemura G, Fujiwara H, Doxorubicin-induced cardiomyopathy from the cardiotoxic mechanisms to management. Prog. Cardiovasc. Dis 49 (2007) 330–352. [DOI] [PubMed] [Google Scholar]
- [6].Swain SM, Whaley FS, Ewer MS, Congestive heart failure in patients treated with doxorubicin. Cancer. 97 (2003) 2869–2879. [DOI] [PubMed] [Google Scholar]
- [7].Carvalho FS, Burgeiro A, Garcia R, Moreno AJ, Carvalho RA, Oliveira PJ, Doxorubicin-induced cardiotoxicity: from bioenergetics failure and cell death to cardiomyopathy. Med. Res. Rev 34 (2014) 106–135. [DOI] [PubMed] [Google Scholar]
- [8].Octavia Y, Tocchetti CG, Gabrielson KL, Janssens S, Crijns HJ, Moens AL, Doxorubicin-induced cardiomyopathy: from molecular mechanisms to therapeutic strategies. J. Mol. Cell Cardiol. 52 (2012) 1213–1225. [DOI] [PubMed] [Google Scholar]
- [9].Paradies G, Paradies V, Ruggiero FM, Petrosillo G, Role of cardiolipin in mitochondrial function and dynamics in health and disease: molecular and pharmacological aspects. Cells 8 (2019) 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ikon N, Ryan RO, Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 1859 (2017) 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].de Wolf FA, Binding of doxorubicin to cardiolipin as compared to other anionic phospholipids? An evaluation of electrostatic effects. Biosci. Rep 11 (1991) 275–284. [DOI] [PubMed] [Google Scholar]
- [12].Parker MA, King V, Howard KP, Nuclear magnetic resonance study of doxorubicin binding to cardiolipin containing magnetically oriented phospholipid bilayers. Biochem. Biophys. Acta 1514 (2001) 206–216. [DOI] [PubMed] [Google Scholar]
- [13].Burke TG, Sartorelli AC, Tritton TR, Selectivity of the anthracyclines for negatively charged model membranes: role of the amino group. Cancer Chemother. Pharmacol 21 (1988) 274–280. [DOI] [PubMed] [Google Scholar]
- [14].Henry N, Fantine EO, Bolard J, Garnier-Suillerot A, Interaction of Adriamycin with negatively charged model membranes: evidence of two types of binding sites. Biochemistry. 24 (1985) 7085–7092. [DOI] [PubMed] [Google Scholar]
- [15].Goormaghtigh E, Brasseur R, Huart P, Ruysschaert JM, Study of the Adriamycin-cardiolipin complex structure using attenuated total reflection infrared spectroscopy. Biochemistry. 26 (1987) 1789–1794. [DOI] [PubMed] [Google Scholar]
- [16].Goormaghtigh E, Huart P, Praet M, Brasseur R, Ruysschaert JM, Structure of the adriamycin-cardiolipin complex. Role in mitochondrial toxicity. Biophys. Chem 35 (1990) 247–257. [DOI] [PubMed] [Google Scholar]
- [17].Rahman A, Joher A, Neefe JR, Immunotoxicity of multiple dosing regimens of free doxorubicin and doxorubicin entrapped in cardiolipin liposomes. Br. J. Cancer 54 (1986) 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cheung KG, Cole LK, Xiang B, Chen K, Ma X, Myal Y, M Hatch G, Tong Q, Dolinsky VW, Sirtuin-3 (SIRT3) protein attenuates doxorubicin-induced oxidative stress and improves mitochondrial respiration in H9c2 cardiomyocytes. J. Biol. Chem 290 (2015) 10981–10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gorini S, De Angelis A, Berrino L, Malara N, Rosano G, Ferraro E, Chemotherapeutic drugs and mitochondrial dysfunction: focus on doxorubicin, trastuzumab, and sunitinib. Oxid Med. Cell Longev. 2018 (2018) 7582730, 10.1155/2018/7582730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fox CA, Ellison P, Ikon N, Ryan RO, Calcium-induced transformation of cardiolipin nanodisks. Biochim. Biophys. Acta Biomembr. 1861 (2019) 1030–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ryan RO, Nanobiotechnology applications of reconstituted high density lipoprotein. J. Nanobiotechnology 8 (2010) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ryan RO, Forte TM, Oda MN, Optimized bacterial expression of human apolipoprotein A-I. Protein Expr. Purif 27 (2003) 98–103. [DOI] [PubMed] [Google Scholar]
- [23].Ikon N, Su B, Hsu FF, Forte TM, Ryan RO, Exogenous cardiolipin localizes to mitochondria and prevents TAZ knockdown-induced apoptosis in myeloid progenitor cells. Biochem. Biophys. Res. Commun 464 (2015) 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang Q, Baum L, Fu WL, Simple and practical staining of DNA with GelRed in agarose gel electrophoresis. Clin. Lab 56 (2010) 149–152. [PubMed] [Google Scholar]
- [25].Mohan P, Rapoport N, Doxorubicin as a molecular nanotheranostic agent: effect of doxorubicin encapsulation in micelles or nanoemulsions on the ultrasound-mediated intracellular delivery and nuclear trafficking. Mol. Pharm 7 (2011) 1959–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sigmon J, Larcom LL, The effect of ethidium bromide on mobility of DNA fragments in agarose gel electrophoresis. Electrophoresis. 17 (1996) 1524–1527. [DOI] [PubMed] [Google Scholar]
- [27].Muenzner J, Pletneva EV, Structural transformations of cytochrome c upon interaction with cardiolipin. Chem. Phys. Lipids 179 (2014) 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang M, Fan K, Zhou M, Duan D, Zheng J, Yang D, Feng J, Yan X, H-ferritin-nanocaged doxorubicin nanoparticles specifically target and kill tumors with a single dose injection. Proc. Natl. Acad. Sci. USA 111 (2014) 14900–14905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Houtkooper RH, Vaz FM, Cardiolipin, the heart of mitochondrial metabolism. Cell Mol. Life Sci. 65 (2008) 2493–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Clementi ME, Giardina B, Di Stasio E, Mordente A, Misiti F. Doxorubicin-derived metabolites induce release of cytochrome C and inhibition of respiration on cardiac isolated mitochondria. Anticancer Res. 23 (2003) 2445–2450. [PubMed] [Google Scholar]
- [31].Mustonen P, Kinnunen PK, On the reversal by deoxyribonucleic acid of the binding of Adriamycin to cardiolipin-containing liposomes. J. Biol. Chem 268 (1993) 1074–1080. [PubMed] [Google Scholar]
- [32].Goormaghtigh E, Chatelain P, Caspers J, Ruysschaert JM, Evidence of a complex between adriamycin derivatives and cardiolipin: possible role in cardiotoxicity. Biochem. Pharmacol 29 (1980) 3003–3010. [DOI] [PubMed] [Google Scholar]
- [33].Ikon N, Hsu FF, Shearer J, Forte TM, Ryan RO, Evaluation of cardiolipin nanodisks as lipid replacement therapy for Barth syndrome. J. Biomed. Res 32 (2018) 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]


