Abstract
Glioblastoma (GBM) is a lethal brain tumor containing a subpopulation of glioma stem cells (GSCs). Pan-cancer analyses have revealed that stemness of cancer cells correlates positively with immunosuppressive pathways in many solid tumors, including GBM, prompting us to conduct a gain-of-function screen of epigenetic regulators that may influence GSC self-renewal and tumor immunity. The circadian regulator CLOCK emerged as a top hit in enhancing stem cell self-renewal, which was amplified in about 5% of human GBM cases. CLOCK and its heterodimeric partner BMAL1 enhanced GSC self-renewal and triggered pro-tumor immunity via transcriptional up-regulation of OLFML3, a novel chemokine recruiting immune-suppressive microglia into the tumor microenvironment. In GBM models, CLOCK or OLFML3 depletion reduced intratumoral microglia density and extended overall survival. We conclude that the CLOCK:BMAL1 complex contributes to key GBM hallmarks of GSC maintenance and immunosuppression and, together with its downstream target OLFML3, represents new therapeutic targets for this disease.
Keywords: Glioblastoma, CLOCK, Self-renewal, Microglia and OLFML3
INTRODUCTION
Glioblastoma (GBM) is the most aggressive and lethal form of adult brain cancer, wherein current standard-of-care offers minimal clinical benefit (1). Extensive genomic profiling has identified key alterations of distinct signaling pathways in GBM, including the RTK/RAS/PI3K/PTEN, RB/CDKN2A and P53/ARF/MDM2 pathways (2–5). Efforts to target these altered signaling pathways, e.g., EGFR or PI3K inhibition, have yielded minimal impact on GBM patient outcomes (6–9). While these genetic alterations impact on many intrinsic aspects of cancer cell biology, there is a growing recognition that these alterations also promote the expression of paracrine factors regulating the recruitment and activation of immune suppressive cells in the tumor microenvironment (TME) (10,11). In GBM, for example, we demonstrated that PTEN deletion/mutation can drive expression of lysyl oxidase (LOX), which promotes infiltration of immunosuppressive tumor-associated macrophages (TAMs) that in turn provide growth factor support for glioma cell survival (12). Such studies highlight the opportunities of identifying genetic alterations in glioma cells that establish symbiotic cancer-host interactions including immune suppression mechanisms in the TME.
In addition to the above genetic alterations, dysregulation of epigenetic programs is also known to impact tumor biology on many levels (13–15). In particular, various epigenetic regulators have been shown to play critical roles in the maintenance of glioma stem cells (GSCs) (such as N6-mA, EZH2 and DAXX) and in the regulation of tumor immunity (such as histone deacetylases) (16,17). These regulatory factors gain added significance as GSCs are critical to both tumor maintenance and therapeutic resistance in GBM (16). Moreover, pan-cancer computational analyses have demonstrated a positive correlation between stemness and immune signatures (18). Together, these insights prompted us to conduct a gain-of-function screen of known epigenetic regulators that may dually enhance GSC self-renewal and promote an immune suppressive TME. In this screen, the circadian regulator CLOCK (circadian locomotor output cycles kaput) emerged as the top hit.
The circadian rhythm serves as an important regulatory system maintaining homeostasis in normal cells and tissues (19,20) and has been shown to play a pivotal roles in cancer-relevant processes such as cell proliferation and survival, DNA repair, metabolism and inflammation (19–21). CLOCK and BMAL1 (brain and muscle ARNT-Like 1; also known as aryl hydrocarbon receptor nuclear translocator-like protein 1, ARNTL) are two key transcription factors of the circadian machinery, which constitute a heterodimeric complex (22). This complex can activate the expression of the PER and CRY genes, which ultimately forms a negative feedback loop to inhibit the activity of CLOCK:BMAL1 complex (22).
There is an increasing recognition that the impact of CLOCK and BMAL1 on cancer pathogenesis is highly context- and disease-dependent (21). For instance, CLOCK or BMAL1 provides tumor suppressor-like functions in prostate, breast, ovarian and pancreatic cancers, but exhibits tumor-promoting roles in colorectal cancer and acute myeloid leukemia (21,23). In GBM, CLOCK or BMAL1 is a tumor-promoting factor that regulates glioma cell proliferation and migration via regulation of the NF-κB pathway (24), and can support GSC function via regulation of anabolic metabolism (25). Here, we elucidate a novel function for CLOCK in supporting an immune suppressive TME via its upregulation of OLFML3, a novel and potent chemoattractant of immune suppressive microglia. Clinical-pathological correlations in human GBM point to CLOCK and OLFML3 as potential therapeutic targets for GBM.
RESULTS
CLOCK promotes GSC self-renewal and is amplified in human GBM
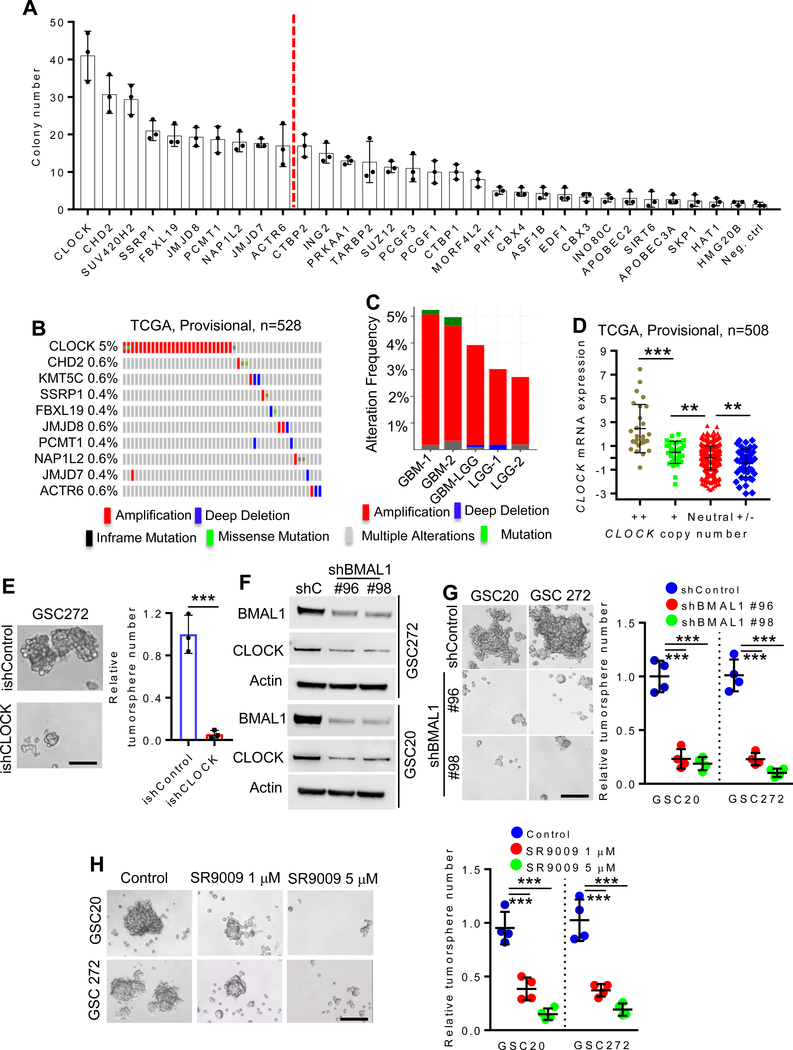
Employing previously characterized human neural stem cells (hNSC) (26), a gain-of-function screen revealed that 31 of 284 epigenetic regulators could enhance hNSC self-renewal activity (Fig. 1A). CLOCK exhibited the highest self-renewal activity which was comparable to the positive control myr-AKT, and CLOCK overexpression was confirmed by immunoblot analysis (Supplementary Fig. S1A). Examination of The Cancer Genome Atlas (TCGA) GBM datasets revealed that CLOCK, but not other epigenetic factors, is amplified in approximately 5% of GBM cases (Fig. 1B; Supplementary Fig. S1B) and 2.8% in low-grade glioma cases (Fig. 1C). Furthermore, increased gene copy number correlated positively with increased CLOCK mRNA levels (Fig. 1D). To further confirm the relevance of CLOCK in promoting GSC self-renewal and maintenance, we conducted shRNA-mediated depletion studies in human GSCs having relatively high CLOCK expression, such as GSC20, GSC167 and GSC272 (Supplementary Fig. S1C). Constitutive CLOCK depletion was associated with impaired self-renewal of GSC20 and GSC167 (Supplementary Fig. S1D,E) and an inducible shRNA knockdown system, termed ishCLOCK, reduced self-renewal in GSC272 and GSC20 (Fig. 1E; Supplementary Fig. S1F,G). Finally, we conducted shRNA-mediated CLOCK depletion in mouse QPP7 GSC having relatively high CLOCK expression (Supplementary Fig. S1C), and found that CLOCK depletion impaired QPP7 self-renewal, which can be rescued by re-expression of shRNA-resistant CLOCK cDNA (Supplementary Fig. S1H).
Figure 1. Clock is amplified in GBM and regulates GSC self-renewal.
(A) Soft agar colony formation of hNSCs overexpressing indicated epigenetic genes. n = 3 biological replicates.
(B) Genomic alterations of CLOCK and other epigenetic regulators (CHD2, KMT5C, SSRP1, FBXL19, JMJD8, PCMT1, NAP1L2, JMJD7 and ACTR6) in TCGA GBM database (Provisional dataset; n = 528).
(C) Genomic alteration frequency of CLOCK in TCGA GBM datasets, GBM-LGG merged dataset and LGG datasets.
(D) CLOCK copy number is significantly correlated with CLOCK mRNA expression in TCGA GBM patients (n = 508). ++, high level of amplification; +, gain; Neutral, no change; +/− homozygous deletion. **P <0.01 and ***P <0.001.
(E) Conditional depletion of CLOCK suppresses GBM tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC272 cells expressing ishCLOCK or ishControl. n = 3 biological replicates; ***P <0.001.
(F) Immunoblots for CLOCK and BMAL1 in cell lysates of GSC272 and GSC20 expressing shRNA control (shC) or BMAL1 shRNAs.
(G) BMAL1 depletion impairs GSC tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC20 and GSC 272 expressing two independent BMAL1 shRNAs or shControl. n = 4 biological replicates; ***P <0.001.
(H) SR9009 treatment impairs GSC tumorsphere formation. Representative images (left panel) and quantification (right panel) of tumorspheres in GSC20 and GSC272 treated with SR9009 at indicated concentrations. n = 4 biological replicates; ***P <0.001.
CLOCK heterodimerizes with BMAL1 to form a transcription factor complex that regulates core circadian clock genes (22). Within the heterodimer, depletion of one partner induces degradation of the other component (27). Indeed, we found that shRNA-mediated depletion of BMAL1 reduced CLOCK expression (Fig. 1F) and impaired self-renewal activity of GSC20 and GSC272 (Fig. 1G). SR9009 is an agonist of the nuclear receptors REV-ERBs which function as direct negative regulators of the CLOCK:BMAL1 complex (28). SR9009 treatment inhibited the self-renewal ability of GSC20 and GSC272 (Fig. 1H), reinforcing the role of CLOCK/BMAL1 in promotion of GSC self-renewal.
To determine the molecular basis of CLOCK’s support of GSC self-renewal, gene expression profiling and Gene Set Enrichment Analysis (GSEA) were compared in GSC272 with ishCLOCK versus ishControl. The major pathways affected were related to metabolism, including fatty acid (FA) metabolism and glycolysis (Supplementary Fig. S2A) which aligns well with previous work showing that FA and glucose metabolism play critical roles in the maintenance of GSC self-renewal (25). Specifically, CLOCK depletion resulted in reduced expression of key glycolysis and TCA enzymes such as PGM1, HK2, LDHA, ACO2, SUCLA2, OGDH and CS as well as FA enzymes such as ACACA, HSD17B, RPP14, ACAT1, HADH (with PGM1 and ACACA showing the most dramatic reduction) (Supplementary Fig. S2B,C). Treatment with PGM1 inhibitor (lithium) or ACACA inhibitor (CP-640186) significantly impaired GSC272 self-renewal, and CLOCK-induced upregulation of self-renewal in GSC17 was blocked by inhibition of PGM1 or ACACA (Supplementary Fig. S2D,E). These findings, are consistent with previous reports establishing CLOCK as a major regulator of metabolic pathways shown to be critical in supporting GSC self-renewal (25).
CLOCK promotes microglia infiltration in GBM
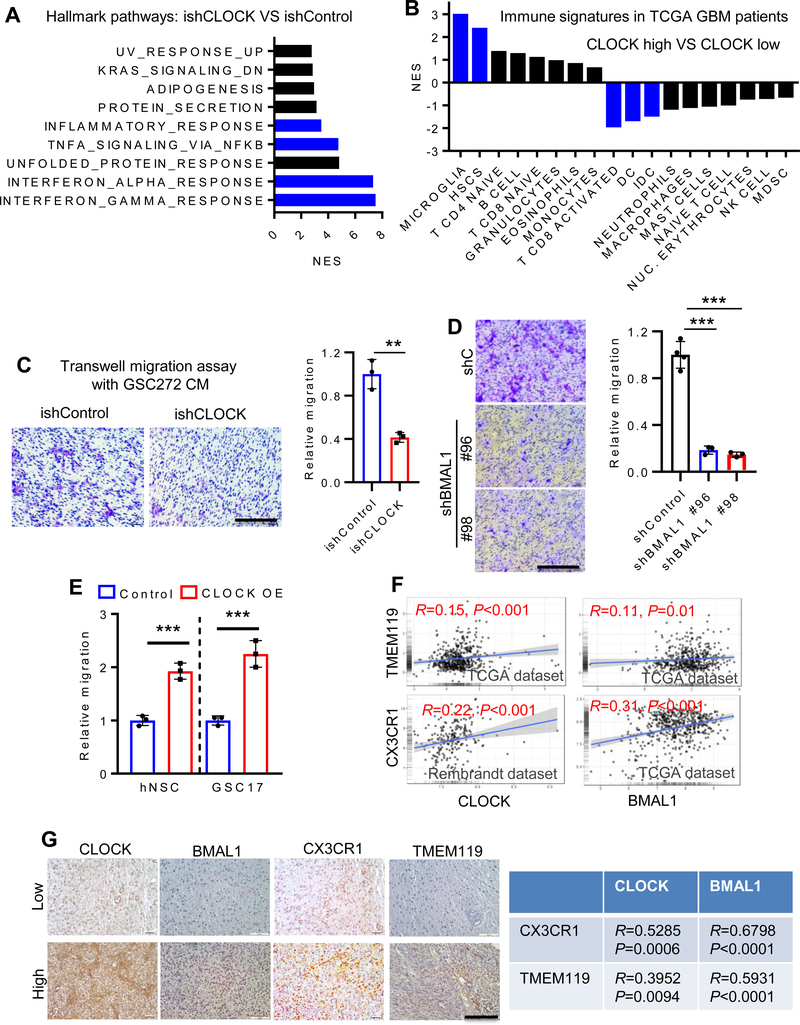
In addition to therapeutic resistance, high stemness of cancer cells has been shown to correlate positively with immunosuppressive pathways in 21 types of solid tumors, including GBM (18). Indeed, in CLOCK-depleted GSCs, GSEA revealed prominent representation of immune suppressive signatures, including interferon γ/α response, TNFα/NF-κB signaling and inflammatory response (Fig. 2A). These immune signatures prompted in silico immune cell auditing of TCGA GBM datasets using validated gene set signatures for 18 types of immune cells (10,29–31). Analysis of immune cell signatures showed that high CLOCK expression correlated positively with increased microglia and, to a lesser extent, hematopoietic stem cells (HSC), and with decreased CD8 activated T cells and dendritic cells (DC); other immune cell types were not significantly changed (Fig. 2B; Supplementary Fig. S3A). Correspondingly, using transwell migration assays, conditioned media (CM) from CLOCK shRNA knockdown GSC272, GSC20, U87 or QPP7 cells exhibited reduced microglia migration relative to CM from shRNA control cells (Fig. 2C; Supplementary Fig. S3B–D). Moreover, the impaired microglia migration in CLOCK shRNA knockdown QPP7 cells can be rescued by re-expression of shRNA-resistant CLOCK cDNA (Supplementary Fig. S3D). Similarly, CM from shBMAL1 GSC20 cells inhibited microglia migration compared to CM from shControl cells (Fig. 2D). Conversely, CM from hNSC and GSC17 with enforced CLOCK expression increased microglia migration relative to controls (Fig. 2E; Supplementary Fig. S3E,F). Finally, in human GBM tissue microarrays (TMAs), CLOCK and BMAL1 signals showed a strong positive correlation with expression of microglia markers, TMEM119 and CX3CR1 (Fig. 2F,G). Together, these findings point to a potential link between high CLOCK expression and infiltration of immune suppressive microglia into the GBM TME.
Figure 2. CLOCK promotes microglia infiltration in GBM.
(A) Transcriptomic profiling in GSC272 cells following CLOCK depletion shows top ten enriched hallmark pathways by GSEA. Blue bars indicate the signatures relate to immune response.
(B) GSEA analysis shows the normalized enrichment score of various types of immune cells in CLOCK-high and CLOCK-low patients in TCGA GBM Provisional dataset (n = 528). Microglia are the most enriched immune cells in CLOCK-high patients. The lower quartile was set to the cut-off value. Blue bars indicate FDR<0.25. (C) Conditioned media from CLOCK-depleted GSC272 cells shows reduced ability to attract human HMC3 microglia in a transwell assay. Representative images (left panel) and quantification (right panel). Scale bar, 200 μm; n = 3 biological replicates; **P <0.01.
(D) Conditioned media from BMAL1-depleted GSC20 cells shows reduced ability to attract HMC3 microglia in a transwell assay. Representative images (left panel) and quantification (right panel). Scale bar, 200 μm; n = 3–4 biological replicates; ***P <0.001.
(E) Conditioned media from CLOCK-overexpressed hNSCs and GSC17 promotes HMC3 microglia migration in a transwell assay. n = 3 biological replicates; ***P <0.001.
(F) CLOCK and BMAL1 expression is positively correlated with microglia markers (TMEM119 and CX3CR1) in TCGA and Rembrandt GBM databases.
(G) CLOCK and BMAL1 expression is positively correlated with CX3CR1 and TMEM119 expression in human GBM TMA samples. Left panel, representative images showing low and high expression levels of CLOCK, BMAL1, CX3CR1 and TMEM119 in human GBM TMA (n = 35). Scale bar, 100 μm; Right panel, quantification data showing strong positive correlations between CLOCK/BMAL1 and CX3CR1/TMEM119in human GBM TMA. R and P values are shown.
CLOCK-regulated OLFML3 promotes microglia migration
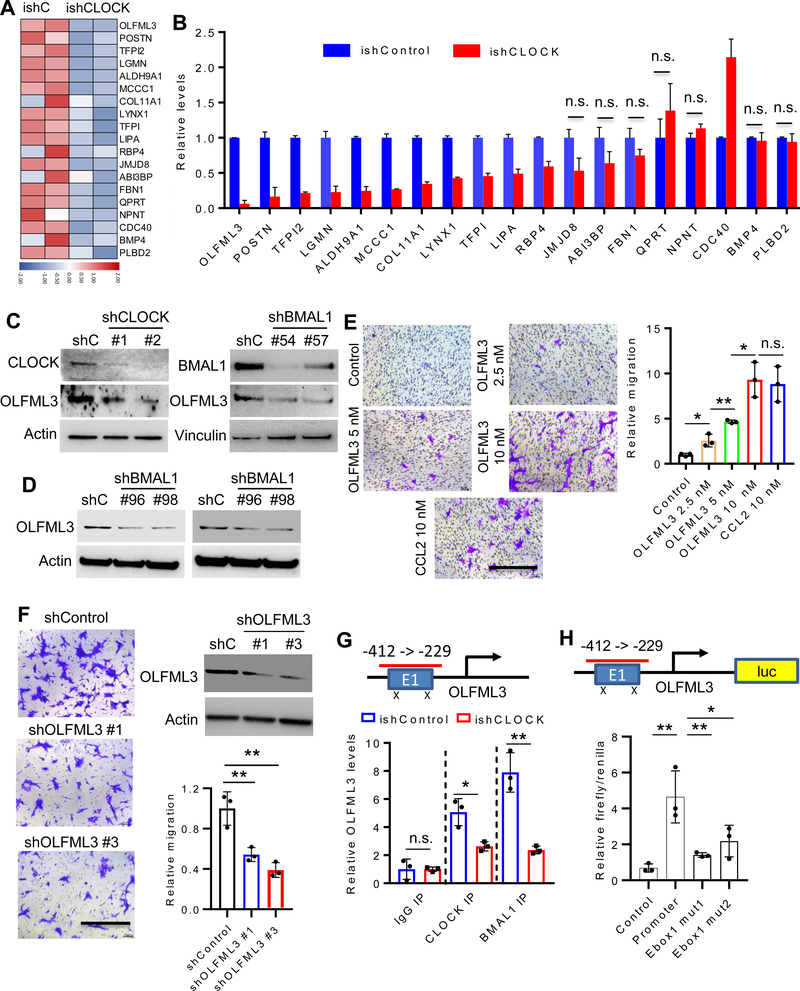
To identify putative CLOCK-regulated secreted factors governing microglia recruitment, our microarray profiling data were intersected with a secreted protein database (32). Using a ≥4.0-fold change in expression, coupled with qRT-PCR validation, 11 genes tracked positively with CLOCK expression including OLFML3, POSTN, TFPI2, LGMN, ALDH9A1, MCCC1, COL11A1, LYNX1, TFPI, LIPA and RBP4. OLFML3 showed the most dramatic decrease in ishCLOCK GSC272 cells (Fig. 3A,B). Gene Ontology Enrichment Analysis (GOEA) on the sub-ontology of Biological Process in TCGA GBM patients showed that OLFML3, LGMN, and LIPA, but not other factors, correlated with leukocyte migration and chemotaxis (Supplementary Fig. S4). Among these three genes, only OLFML3 was reduced by CLOCK depletion in GSC20 (Supplementary Fig. S5A). Moreover, TCGA GBM bioinformatics analysis demonstrated that the expression of OLFML3, LGMN and LIPA correlated positively with microglia markers (CX3CR1 and TMEM119), with OLFML3 showing the most significant correlation prompting further in-depth analysis (Supplementary Fig. S5B). Further studies using immunoblotting demonstrated that shRNA-mediated depletion of CLOCK or BMAL1 reduced OLFML3 expression in several GSC models, including mouse QPP7 (Fig. 3C), and human GSC20 and GSC272 (Fig. 3D). CLOCK depletion-induced decrease of OLFML3 expression in QPP7 cells was rescued by re-expression of shRNA-resistant CLOCK cDNA (Supplementary Fig. 5C).
Figure 3. CLOCK-regulated OLFML3 promotes microglia migration.
(A) Heat map representation of the microarray data of ishControl and ishCLOCK GSC272 cells shows the most downregulated genes (exhibiting a ≥4.0-fold change) encoding secreted proteins following CLOCK depletion. Red and blue indicate higher and low expression, respectively.
(B) qRT-qPCR validation of downregulated genes as in (A). n.s. indicates not significant (P >0.05).
(C) Immunoblots for CLOCK and OLFML3 in cell lysates of QPP7 GSC expressing shRNA control (shC), CLOCK shRNAs or BMAIL1 shRNAs.
(D) Immunoblots for OLFML3 in cell lysates of GSC272 (left panel) and GSC20 (right panel) expressing shRNA control (shC) or BMAL1 shRNAs.
(E) Recombinant OLFML3 protein at indicated concentrations promotes HMC3 microglia migration in transwell assay, and is comparable to positive control CCL2 (10 nM). Representative images (left panel) and quantification (right panel). Scale bar, 200 μm; n = 3 biological replicates; *P <0.05 and **P <0.01; n.s. indicates not significant (P >0.05).
(F) OLFML3-depleted GSC272 conditioned media impairs HMC3 microglia migration in transwell assay. Representative images (left panel), shRNA knockdown efficiency and quantification (right panel). Scale bar, 200 μm; n = 3 biological replicates; **P <0.001.
(G) ChIP-PCR shows that CLOCK and BMAL1 bind to OLFML3 promoter and that this binding was diminished following CLOCK depletion. n = 3 biological replicates; *P <0.05 and **P <0.001; n.s. indicates not significant (P >0.05).
(H) Luciferase reporter assay shows that mutations in OLFML3 promoter E-box sites reduce the transcriptional activity of CLOCK for OLFML3. n = 3 biological replicates; *P <0.05 and **P <0.001.
Using transwell migration assays, recombinant OLFML3-supplemented media dramatically increased microglia migration in a dose dependent manner, which was comparable to the activity of the prototypical microglia chemokine CCL2 (aka, MCP-1) (Fig. 3E). Conversely, CM from shRNA-mediated depletion of OLFML3 in GSC272 or U87 cells showed reduced microglia migration (Fig. 3F; Supplementary Fig. S5D,E). To assess whether CLOCK and BMAL1 directly regulate OLFML3 expression, ChIP-PCR assays were performed, showing that CLOCK and BMAL1 bound to the OLFML3 promoter and that this binding was reduced in CLOCK-depleted GSC272 cells (Fig. 3G). Moreover, luciferase reporter assays showed that CLOCK-induced transcriptional activity was abolished by E-box mutations in the OLFML3 promoter region (Fig. 3H). We conclude that OLFML3 is a novel CLOCK-regulated chemokine with potent microglia recruitment activity.
CLOCK depletion inhibits GSC self-renewal and intratumoral microglia infiltration and extends survival
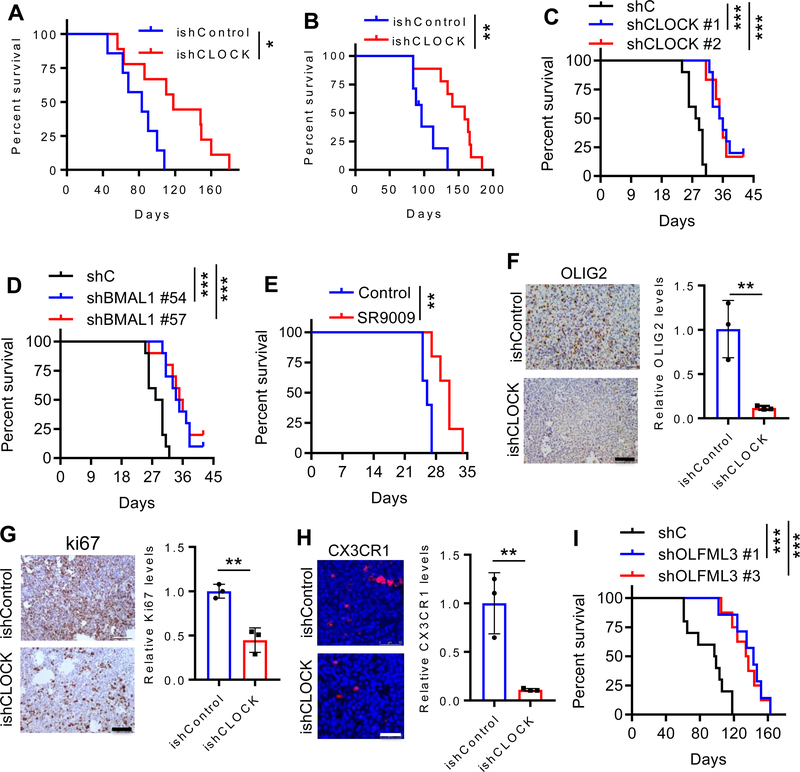
To further investigate the role of CLOCK in GBM tumor biology, we utilized the ishCLOCK system to inducibly deplete CLOCK in GSC272 and GSC20 tumors implanted into SCID mice, revealing that CLOCK depletion significantly extended survival (Fig. 4A,B; Supplementary Fig. S6A). Using a murine model CT2A which was isolated from a carcinogen-induced glioma and possesses a GSC-like phenotype (33), depletion of CLOCK or BMAL1 resulted in a significant extension of survival in C57BL/6 mice (Fig. 4C,D). Similarly, pharmacological inhibition of the CLOCK:BMAL1 complex extended the survival of C57BL/6 mice implanted with CT2A cells (Fig. 4E). On the histological level, GSC stem cell markers OLIG2 and nestin, and proliferation marker Ki67 were dramatically reduced, whereas apoptosis was increased upon CLOCK depletion (Fig. 4F,G; Supplementary Fig. S6B,C). In addition, infiltrating microglia were profoundly reduced (10-fold) in the CLOCK-depleted tumors (Fig. 4H; Supplementary Fig. S6D). The microglia phenotype which can be immune-stimulatory (M1) or immunosuppressive (M2) (34), is strongly biased towards the M2 phenotype in both mouse and human GBM tumors (Supplementary Fig. S7A,B). M2 microglia were significantly reduced in CLOCK-depleted tumors (Supplementary Fig. S7C) and, conversely, the M2 signature correlated positively with high expression levels of CLOCK and BMAL1 in TCGA GBM patients (Supplementary Fig. S7D,E). Since OLFML3 plays a prominent role in microglia migration, we also explored the impact of shRNA-mediated depletion of OLFML3 on GBM growth, and found that decreased OLFML3 in the GSC272 model significantly extended survival (Fig. 4I). Together, these in vivo results confirm the role of CLOCK in promoting GBM tumor maintenance which correlates with CLOCK-induced enhancement of stemness, proliferation and survival, as well as increased recruitment of microglia into the GBM TME.
Figure 4. CLOCK depletion extends survival, and inhibits GSC self-renewal and microglia infiltration.
(A) Survival curves of SCID mice implanted with ishCLOCK and ishControl GSC272 cells (5×105 cells). Dox food was provided at day 30 post-orthotopic injection to induce CLOCK knockdown in vivo. (n = 7 and 9 mice for ishControl and ishCLOCK groups, respectively). *P <0.05.
(B) Survival curves of SCID mice implanted with ishCLOCK and ishControl GSC20 cells (5×105 cells). Dox food was provided at day 30 post-orthotopic injection to induce CLOCK knockdown in vivo. (n = 7 and 9 mice for ishControl and ishCLOCK groups, respectively). **P <0.01.
(C) Survival curves of C57BL/6 mice implanted with CT2A cells (2×104 cells) expressing shRNA control (shC) or CLOCK shRNAs. (n = 10, 10 and 6 mice for shC, shCLOCK #1 and shCLOCK #2 groups, respectively). ***P <0.001.
(D) Survival curves of C57BL/6 mice implanted with CT2A cells (2×104 cells) expressing shRNA control (shC) or BMAL1 shRNAs. (n = 10 mice per group). ***P <0.001.
(E) Survival curves of C57BL/6 mice implanted with CT2A cells (2×104 cells). Mice were treated with SR9009 (100 mg/kg, i.p., daily) for 10 days beginning at day 7 post-orthotopic injection. (n = 5 mice per group). **P <0.01.
(F) IHC (left panel) and quantification (right panel) of OLIG2 in mouse tumors from ishCLOCK and ishControl GSC272 models. Scale bar, 100 μm; n = 3 biological replicates; **P <0.01.
(G) IHC (left panel) and quantification (right panel) of Ki67 in mouse tumors from ishCLOCK and ishControl GSC272 models. Scale bar, 100 μm; n = 3 biological replicates; **P <0.01.
(H) Immunofluorescence (left panel) and quantification (right panel) of microglia marker CX3CR1 in mouse tumors from ishCLOCK and ishControl GSC272 models. Scale bar, 100 μm; n = 3 biological replicates; **P <0.01.
(I) Kaplan-Meier survival curves of SCID mice implanted with GSC272 cells (5×105 cells) expressing shRNA control (shC) or OLFML3 shRNAs (n=10, 7 and 8 mice for shC, shOLFML3 #1 and shOLFML3 #3 groups, respectively). ***P <0.001.
DISCUSSION
In this study, we uncovered the role and underlying mechanisms of the core circadian regulators CLOCK and BMAL1 in GBM tumor maintenance via its regulation of GSC self-renewal and immunity. We identified OLFML3 as a novel and potent CLOCK-regulated microglia chemoattractant in GBM and demonstrated that OLFML3 depletion can increase survival. The key role of the CLOCK:BMAL1 complex in GBM tumor biology, particularly its regulation of specific metabolic and immunity genes such as OLFML3, illuminates potential therapeutic targets governing key cancer hallmarks of stemness and immune suppression.
Circadian rhythm regulators have been extensively studied in model organisms (35), and have been linked to the development of cancers, including breast, lung, and colorectal cancers (36,37). For example, depletion of CLOCK or BMAL1 has been shown to impair leukemia stem cell proliferation and enhance myeloid differentiation in acute myeloid leukemia (23), as well as suppress glioma cell proliferation and migration (24). Moreover, pharmacological activation of circadian clock components REV-ERBs, which repress transcription of CLOCK and BMAL1, have been shown to impair the growth of multiple cancer types including GBM (28). Specifically, activation of REV-ERBs is selectively lethal to cancer cells by affecting oncogenic drivers (such as HRAS, BRAF, PIK3CA and others), inducing apoptosis and inhibiting autophagy (28). In this current study, we extend the actions of CLOCK in GBM as a promoter of GSC self-renewal, suppressor of anti-tumor immunity and, consistent with recent reports, regulator of fatty acid metabolism and glycolysis (25).
A hallmark feature of the GBM TME is an abundance of infiltrating immune cells (38) wherein microglia are known to contribute to an immunosuppressive microenvironment and support GBM progression (39). Here, our findings of CLOCK-regulated microglia recruitment are consistent with previous observations that the immune system can be regulated by circadian components (40) and that dysregulation of the intrinsic circadian clock can alter inflammatory responses (41,42). In addition, our work also aligns with previous tumor biology findings showing that CLOCK can influence T cell infiltration in melanoma (43) and that BMAL1 deficiency in endothelial cells impairs the migration of leukocytes in mice (44). Moreover, our mechanistic work reinforces this intimate link by demonstrating the capacity of CLOCK to specifically and directly regulate the chemokine OLFML3, which in turn recruits microglia into the GBM TME.
OLFML3 belongs to the family of olfactomedin domain-containing proteins, which have important roles in tumorigenesis and embryonic patterning (45). Previous work has shown that OLFML3 is a proangiogenic factor in the TME where it promotes endothelial cell migration and sprouting through activation of the canonical SMAD1/5/8 signaling pathway (45). Along similar lines, it would be useful to determine potential druggable molecular pathways in microglia that are activated by OLFML3, thus expanding therapeutic targets for GBM. Intriguingly, microglia are known to express OLFML3 (46), suggesting that following a CLOCK-directed program of microglia recruitment, microglia themselves could further increase the recruitment of additional microglia through their own secretion of OLFML3 in a feed-forward manner.
TAMs play an important role in GBM tumor biology, prompting assessment of the therapeutic benefit of targeting TAMs in GBM. To date, however, CSF1R inhibitor BLZ945 treatment of mouse GBM models have failed to deplete TAMs and elicited transient anti-tumor responses (47,48). Correspondingly, a phase II clinical trial with CSF1R inhibitor PLX3397 has shown minimal activity in recurrent GBM patients (49). The basis for these meager responses is not clear, although it is worth noting that 2 of 37 GBM patients who experienced extended progression-free survival (49) are of the mesenchymal subtype which typically harbors PTEN deficiency. Along these lines, our recent studies demonstrated that inhibition of macrophage recruitment by LOX inhibitor specifically impairs PTEN-deficient GBM progression, establishing a synthetic lethal interaction between PTEN deficient LOX-expressing glioma cells and SPP1-expressing TAMs which support glioma cell survival (12). Thus, with respect to CSF1R inhibitors, it is tempting to speculate that PTEN deficient GBM patients may be particularly susceptible to such agents targeting TAMs. Along similar lines, our discovery here of the CLOCK-OLFML3-microglia axis and the correlative studies in human GBM TMAs showing high CLOCK and abundant microglia encourages the design of clinical trials targeting OLFML3 in high CLOCK GBM patients. We believe that targeting CLOCK:BMAL1 downstream targets as opposed to CLOCK:BMAL1 directly, provides a superior therapeutic strategy given the likelihood of disturbed sleep cycles by targeting circadian regulators. Finally, microglia are well known to be immunosuppressive cells in the GBM TME and may therefore dampened immune checkpoint blockade activity (50); thus, it is tempting to speculate that combined inhibition of OLFML3 and immune checkpoint blockade may also prove beneficial for GBM patients.
METHODS
Cell Culture
HMC3 microglia cells were cultured in Eagle’s Minimum Essential Medium (EMEM). CT2A, U87 and 293T cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM). All cell lines were cultured in indicated medium containing 10% FBS (Sigma) and 1:100 antibiotic-antimycotic (Gibco), and were purchased from the American Type Culture Collection (ATCC). p53DN-hNSCs were generated by our laboratory as described recently (26). Patient-derived GSCs were provided by Dr. Erik P. Sulman and Dr. Frederick F. Lang from the Brain Tumor Center (MD Anderson Cancer Center, Houston, TX). Mouse 005 and QPP7 GSCs were provided by Dr. Samuel D. Rabkin and Dr. Jian Hu. All GSCs and NSCs were cultured in NSC proliferation media (Millipore Corporation, Billerica, MA) containing 20 ng/ml EGF and 20 ng/ml bFGF. These GSCs and NSC have been validated through fingerprinting by the MD Anderson Cell Line Core Facility. All cells were confirmed to be mycoplasma-free, and maintained at 37°C and 5% CO2. Conditioned media were collected from treated or untreated cells as indicated after culturing for 24 h in FBS-free culture medium.
Tumorsphere Formation Assay
Soft agar colony formation assay and tumorsphere formation were performed as previously described (51).
Epigenetic Screen
The open reading frame (ORF) lentiviral vectors in the Precision LentiORF collection were obtained from the Functional Genomics Facility at MD Anderson Cancer Center. In 96-well plates, we packaged 284 ORF lentiviruses (encoding known epigenetic factors) individually and infected with p53DN-hNSCs. Stable sublines were generated by blasticidin selection, and then subjected to soft agar colony formation assay.
Plasmids, Viral Transfections and Cloning
shRNAs targeting human and mouse CLOCK, BMAL1 and OLFML3 in the pLKO.1 vector (Sigma) were used in the current study. Lentiviral particles (8 μg) were generated by transfecting 293T cells with the packaging vectors psPAX2 (4 μg) and pMD2.G (2 μg). Lentiviral particles were collected 48 and 72 hr after transfection of 293T cells, filtered through a 0.45 μm filter (Corning), and then used to treat cells in culture. After 48 hr, cells were selected by Puromycin (2 μg/mL). The following human shRNA sequences [CLOCK: #74: TRCN0000018974 and #75: TRCN0000018975; BMAL1: #96: TRCN0000019096 and #98: TRCN0000019098; and OLFML3: #1: TRCN0000186745 and #3: TRCN0000203502] and mouse shRNA sequences [Clock: #1:TRCN0000095686 and #2 TRCN0000306474; Bmal1: #54: TRCN0000095054 and #57: TRCN0000095057] were selected for further use following the validation. Doxycycline-inducible plasmids were generated by cloning the desired shRNA sequences (shCLOCK #75) into a pLKO.1 vector through the Gateway Cloning System (Thermo Fisher Scientific). Following transfection, cells were treated with doxycycline (Dox, 2 μg/ml) for 48 hr to knockdown CLOCK. For rescue experiments, CLOCK shRNA knockdown QPP7 cells were transfected with human CLOCK construct that is resistant to CLOCK shRNAs (shCLOCK #1 and #2).
Immunoblotting
Immunoblotting was performed following standard protocol (12). Antibodies were purchased from the indicated companies, including β-actin (Sigma, #A3854), Vinculin (EMD Millipore, #05–386), CLOCK (CST, #5157S), BMAL1 (CST, #14020S) and OLFML3 (Invitrogen, #PA5–31581).
Immunohistochemistry and immunofluorescence
Immunohistochemistry was performed as standard protocol. In brief, a pressure cooker (95°C for 30 min followed by 120°C for 10 s) was used for antigen retrieval using antigen unmasking solution (Vector Laboratories). Antibodies specific to CLOCK (CST, #5157S), BMAL1 (CST, #14020S), CX3CR1 (Invitrogen, #702321), TMEM119 (BioLegend, #853302), CD206 (R&D, #AF2535) cleaved caspase 3 (CST, #9661S), OLIG2 (Millipore, #AB15328), Nestin (Millipore, #MAB5326), Ki67 (Thermo Scientific, #RM-9106-S1) were used in this study. The human and mouse tumor tissue sections were reviewed and scored by TMARKER software (52). Slides were scanned using Pannoramic 250 Flash III (3DHISTECH Ltd) and images were captured through Pannoramic Viewer software (3DHISTECH Ltd). The studies related to human specimens were approved by the MD Anderson Institutional Review Board under protocol #PA14–0420. Immunofluorescence was performed as previously described (12), and antibodies specific to CX3CR1 (Invitrogen, #702321) were used. Images were captured using a fluorescence microscope (Leica DMi8).
Migration assay
Human microglia HMC3 cells (5 × 105) were suspended in serum-free culture medium and seeded into 24-well Transwell inserts (8.0 μm). Medium with indicated factors or conditioned media was added to the remaining receiver wells. After 24 h, the migrated microglia were fixed and stained with crystal violet (0.05%, Sigma), and then counted as cells per field of view under microscope.
ChIP-PCR and Luciferase Reporter Assay
ChIP-PCR was performed using the standard protocol. Briefly, GSC272 cells were cross-linked using 1% PFA (10 min) and then reactions were quenched using glycine (5 min) at room temperature. Cells were lysed with ChIP lysis buffer for 30 min on ice. Chromatin fragmentation was performed using a Diagenode BioruptorPico sonicator (45 cycles, each with 30 sec on and 30 sec off). Solubilized chromatin was then incubated with a mixture of antibody [CLOCK (Abcam, #ab3517) or BAML1 (CST, #14020S)] and Dynabeads (Life Technologies) overnight. Immune complexes were then washed with RIPA buffer three times, once with RIPA-500, and once with LiCl wash buffer. Elution and reverse-crosslinking were performed in direct elution buffer containing proteinase K (20 mg/ml) at 65°C overnight. Eluted DNA was purified using AMPure beads (Beckman-Coulter), and then was used to perform qPCR. The OLFML3 primer was designed according to the E-box of human OLFML3 gene (−412 to −229 bp) (Forward: TGACCACTTGGGCCATTGTT; Reverse: CAGCAAACGCCATTCCTGTT). To perform the luciferase reporter assay, the promoter region of human OLFML3 (−412 to −229 bp to ATG) was amplified by PCR and inserted into the BglII/HindIII sites of the pGL3 vector to generate the corresponding reporter constructs with or without point mutations in human OLFML3 E-box sites. The luciferase reporter assay was conducted by transfecting the reporter constructs, CLOCK expression vector and renilla luciferase vector into 293T cells. Cells were harvested after 24 hr of transfection, and luciferase activities were measured.
Quantitative Real-Time PCR (qRT-PCR)
Cells were pelleted, and RNA was isolated with the RNeasy Mini Kit (Qiagen). RNA was reverse-transcribed into cDNA by following the ABM cDNA Synthesis Kit. qRT-PCR was performed using SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a 7500 Fast Real-Time PCR Machine (Applied Biosystems). qRT-PCR primers are listed in Supplementary Table S1. The expression of each gene was normalized to that of GAPDH.
Microarray Analysis
RNA was isolated as described above, with slight modifications. ishControl and ishCLOCK GSC272 cells (n = 2 biological replicates) were first lysed with Buffer RLT, then purified with TRIzol Reagent (Life Technologies) and chloroform. The remaining steps of the RNeasy Mini Kit were then followed. Microarray experiments were conducted by the MD Anderson Sequencing and Microarray Core Facility using the Clariom D Assay (Thermo Fisher Scientific). Microarray experiments were performed in duplicate. The raw data were processed and analyzed by GenePattern using Transcriptome Analysis Console. Genes that were differentially expressed between ishControl and ishCLOCK GSC272 were subjected to GSEA.
Mice and intracranial xenograft tumor models
Female ICR SCID mice (3–4 weeks age) were purchased from Taconic Biosciences. Mice were grouped by 5 animals in large plastic cages and were maintained under pathogen-free conditions. All animal experiments were performed with the approval of MD Anderson Cancer Center’s Institutional Animal Care and Use Committee (IACUC). The intracranial xenograft tumor model in SCID mice was established as we described recently (26). The mice were bolted and intracranially implanted with cells at MD Anderson’s Brain Tumor Center Animal Core. Mice with neurological deficits or moribund appearance were sacrificed, and the tumor tissues were harvested for histological analysis. Following transcranial perfusion with 4% paraformaldehyde (PFA), brains were removed and fixed in formalin, and were processed for paraffin embedded blocks.
Human Samples
Tissue microarrays containing 35 human GBM samples and 5 normal brain tissues were purchased from US Biomax (Cat#GL806f).
Computational analysis of human GBM data
For analysis of human GBM data, we downloaded the gene expression and copy number data of TCGA datasets or other available datasets from GlioVis: http://gliovis.bioinfo.cnio.es/ or cBioPortal: https://www.cbioportal.org/. The expression and correlation of interesting genes in GBM, and Gene ontology enrichment analysis were analyzed using GlioVis.
Statistical analysis
All statistical analyses were performed with Student’s t-Test and represented as mean ± SD unless noted otherwise. The analysis of GBM TCGA database and TAM IHC staining for the correlation between genes or proteins was performed using Pearson’s Correlation test (GraphPad Prism 7). The analysis of the survival data from GBM TCGA database was performed using Log-rank (Mantel-Cox) test (GraphPad Prism 7). The P values were designated as: *P <0.05; **P <0.01 and ***P <0.001; n.s. non-significant (P >0.05).
Data and Software Availability
The newly generated microarray data have been submitted to the GEO repository and the accession number is GSE140409.
Supplementary Material
SIGNIFICANCE.
Circadian regulator CLOCK drives GSC self-renewal and metabolism, and promotes microglia infiltration through direct regulation of a novel microglia attracting chemokine, OLFML3. Targeting CLOCK and/or OLFML3 may represent novel therapeutic targets for GBM.
ACKNOWLEDGMENTS
The authors thank Dr. Michael D. Peoples for providing shRNAs, Dr. Erik P. Sulman for providing human derived GSCs and Drs. Samuel D. Rabkin and Jian Hu for providing mouse GSCs.
Financial support: This work was supported by the Cancer Research Institute Irvington Postdoctoral Fellowship (P.C.), The Harold C. and Mary L. Daily Endowment Fellowship (P.C.), the Caroline Ross Endowed Fellowship (P.C.), Emerson Collective Award (Y.A.W.), NIH R01 CA231349 (Y.A.W.), Clayton & Modesta William Cancer Research Fund (R.A.D.), NIH P01 CA117969 (R.A.D.), NIH R01 CA084628 (R.A.D.) and the Burkhart III Distinguished University Chair in Cancer Research Endowment (R.A.D.).
Footnotes
Conflict of interest: RAD is a co-founder, advisor and director of Tvardi Therapeutics focused on the development of STAT3 inhibitors. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Khosla D Concurrent therapy to enhance radiotherapeutic outcomes in glioblastoma. Annals of translational medicine 2016;4(3):54 doi 10.3978/j.issn.2305-5839.2016.01.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, et al. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harbor symposia on quantitative biology 2008;73:427–37 doi 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 3.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes & development 2012;26(8):756–84 doi 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455(7216):1061–8 doi 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155(2):462–77 doi 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352(10):987–96 doi 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 7.McNamara MG, Lwin Z, Jiang H, Chung C, Millar BA, Sahgal A, et al. Conditional probability of survival and post-progression survival in patients with glioblastoma in the temozolomide treatment era. J Neurooncol 2014;117(1):153–60 doi 10.1007/s11060-014-1368-7. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Wu C, Chen N, Gu H, Yen A, Cao L, et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 2016;7(22):33440–50 doi 10.18632/oncotarget.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westphal M, Maire CL, Lamszus K. EGFR as a Target for Glioblastoma Treatment: An Unfulfilled Promise. Cns Drugs 2017;31(9):723–35 doi 10.1007/s40263-017-0456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, et al. Targeting YAP-Dependent MDSC Infiltration Impairs Tumor Progression. Cancer discovery 2016;6(1):80–95 doi 10.1158/2159-8290.CD-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer cell 2019;35(4):559–72 e7 doi 10.1016/j.ccell.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen P, Zhao D, Li J, Liang X, Li J, Chang A, et al. Symbiotic Macrophage-Glioma Cell Interactions Reveal Synthetic Lethality in PTEN-Null Glioma. Cancer cell 2019;35(6):868–84 e6 doi 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romani M, Pistillo MP, Banelli B. Epigenetic Targeting of Glioblastoma. Front Oncol 2018;8:448 doi 10.3389/fonc.2018.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol 2009;19(3):188–97 doi 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Kondo Y, Katsushima K, Ohka F, Natsume A, Shinjo K. Epigenetic dysregulation in glioma. Cancer Sci 2014;105(4):363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes & development 2019;33(11–12):591–609 doi 10.1101/gad.324301.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yelton CJ, Ray SK. Histone deacetylase enzymes and selective histone deacetylase inhibitors for antitumor effects and enhancement of antitumor immunity in glioblastoma. Neuroimmunol Neuroinflamm 2018;5:46 doi 10.20517/2347-8659.2018.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miranda A, Hamilton PT, Zhang AW, Pattnaik S, Becht E, Mezheyeuski A, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proceedings of the National Academy of Sciences of the United States of America 2019;116(18):9020–9 doi 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulli G, Lam MTY, Panda S. Interplay between Circadian Clock and Cancer: New Frontiers for Cancer Treatment. Trends in cancer 2019;5(8):475–94 doi 10.1016/j.trecan.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masri S, Sassone-Corsi P. The emerging link between cancer, metabolism, and circadian rhythms. Nature medicine 2018;24(12):1795–803 doi 10.1038/s41591-018-0271-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shafi AA, Knudsen KE. Cancer and the Circadian Clock. Cancer research 2019;79(15):3806–14 doi 10.1158/0008-5472.CAN-19-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buhr ED, Takahashi JS. Molecular components of the Mammalian circadian clock. Handb Exp Pharmacol 2013(217):3–27 doi 10.1007/978-3-642-25950-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, et al. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell 2016;165(2):303–16 doi 10.1016/j.cell.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li A, Lin X, Tan X, Yin B, Han W, Zhao J, et al. Circadian gene Clock contributes to cell proliferation and migration of glioma and is directly regulated by tumor-suppressive miR-124. FEBS letters 2013;587(15):2455–60 doi 10.1016/j.febslet.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 25.Dong Z, Zhang G, Qu M, Gimple RC, Wu Q, Qiu Z, et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer discovery 2019;9(11):1556–73 doi 10.1158/2159-8290.CD-19-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu B, Wang Q, Wang YA, Hua S, Sauve CG, Ong D, et al. Epigenetic Activation of WNT5A Drives Glioblastoma Stem Cell Differentiation and Invasive Growth. Cell 2016;167(5):1281–95 e18 doi 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, et al. Clock-Talk: Interactions between Central and Peripheral Circadian Oscillators in Mammals. Cold Spring Harbor symposia on quantitative biology 2015;80:223–32 doi 10.1101/sqb.2015.80.027490. [DOI] [PubMed] [Google Scholar]
- 28.Sulli G, Rommel A, Wang XJ, Kolar MJ, Puca F, Saghatelian A, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 2018;553(7688):351–+ doi 10.1038/nature25170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engler JR, Robinson AE, Smirnov I, Hodgson JG, Berger MS, Gupta N, et al. Increased microglia/macrophage gene expression in a subset of adult and pediatric astrocytomas. PloS one 2012;7(8):e43339 doi 10.1371/journal.pone.0043339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39(4):782–95 doi 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 31.Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, et al. Macrophage Ontogeny Underlies Differences in Tumor-Specific Education in Brain Malignancies. Cell reports 2016;17(9):2445–59 doi 10.1016/j.celrep.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Zhang Y, Yin Y, Gao G, Li S, Jiang Y, et al. SPD--a web-based secreted protein database. Nucleic acids research 2005;33(Database issue):D169–73 doi 10.1093/nar/gki093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saha D, Martuza RL, Rabkin SD. Macrophage Polarization Contributes to Glioblastoma Eradication by Combination Immunovirotherapy and Immune Checkpoint Blockade. Cancer cell 2017;32(2):253–67 e5 doi 10.1016/j.ccell.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nature neuroscience 2016;19(1):20–7 doi 10.1038/nn.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronauer RE, Gunzelmann G, Van Dongen HPA, Doyle FJ, Klerman EB. Uncovering physiologic mechanisms of circadian rhythms and sleep/wake regulation through mathematical modeling. J Biol Rhythm 2007;22(3):233–45 doi 10.1177/0748730407301237. [DOI] [PubMed] [Google Scholar]
- 36.Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. Night-shift work and risk of colorectal cancer in the nurses’ health study. Journal of the National Cancer Institute 2003;95(11):825–8. [DOI] [PubMed] [Google Scholar]
- 37.Fu L, Kettner NM. The circadian clock in cancer development and therapy. Prog Mol Biol Transl Sci 2013;119:221–82 doi 10.1016/B978-0-12-396971-2.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nature medicine 2013;19(11):1423–37 doi 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matias D, Predes D, Niemeyer Filho P, Lopes MC, Abreu JG, Lima FRS, et al. Microglia-glioblastoma interactions: New role for Wnt signaling. Biochim Biophys Acta Rev Cancer 2017;1868(1):333–40 doi 10.1016/j.bbcan.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Scheiermann C, Kunisaki Y, Frenette PS. Circadian control of the immune system. Nat Rev Immunol 2013;13(3):190–8 doi 10.1038/nri3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fonken LK, Kitt MM, Gaudet AD, Barrientos RM, Watkins LR, Maier SF. Diminished circadian rhythms in hippocampal microglia may contribute to age-related neuroinflammatory sensitization. Neurobiol Aging 2016;47:102–12 doi 10.1016/j.neurobiolaging.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fonken LK, Frank MG, Kitt MM, Barrientos RM, Watkins LR, Maier SF. Microglia inflammatory responses are controlled by an intrinsic circadian clock. Brain Behav Immun 2015;45:171–9 doi 10.1016/j.bbi.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Assis LVM, Kinker GS, Moraes MN, Markus RP, Fernandes PA, Castrucci AML. Expression of the Circadian Clock Gene BMAL1 Positively Correlates With Antitumor Immunity and Patient Survival in Metastatic Melanoma. Front Oncol 2018;8:185 doi 10.3389/fonc.2018.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He WY, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, et al. Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues. Immunity 2018;49(6):1175–+ doi 10.1016/j.immuni.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miljkovic-Licina M, Hammel P, Garrido-Urbani S, Lee BP, Meguenani M, Chaabane C, et al. Targeting olfactomedin-like 3 inhibits tumor growth by impairing angiogenesis and pericyte coverage. Molecular cancer therapeutics 2012;11(12):2588–99 doi 10.1158/1535-7163.MCT-12-0245. [DOI] [PubMed] [Google Scholar]
- 46.Neidert N, von Ehr A, Zoller T, Spittau B. Microglia-specific expression of Olfml3 is Directly regulated by Transforming growth Factor beta 1-induced smad2 signaling. Front Immunol 2018;9:1728 doi 10.3389/fimmu.2018.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 2016;352(6288):aad3018 doi 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine 2013;19(10):1264–72 doi 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butowski N, Colman H, De Groot JF, Omuro AM, Nayak L, Wen PY, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-oncology 2016;18(4):557–64 doi 10.1093/neuonc/nov245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.See AP, Parker JJ, Waziri A. The role of regulatory T cells and microglia in glioblastoma-associated immunosuppression. J Neurooncol 2015;123(3):405–12 doi 10.1007/s11060-015-1849-3. [DOI] [PubMed] [Google Scholar]
- 51.Ong DST, Hu B, Ho YW, Sauve CG, Bristow CA, Wang Q, et al. PAF promotes stemness and radioresistance of glioma stem cells. Proceedings of the National Academy of Sciences of the United States of America 2017;114(43):E9086–E95 doi 10.1073/pnas.1708122114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schuffler PJ, Fuchs TJ, Ong CS, Wild PJ, Rupp NJ, Buhmann JM. TMARKER: A free software toolkit for histopathological cell counting and staining estimation. J Pathol Inform 2013;4(Suppl):S2 doi 10.4103/2153-3539.109804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The newly generated microarray data have been submitted to the GEO repository and the accession number is GSE140409.