Abstract
Gut dysbiosis, defined as a maladaptive gut microbial imbalance, has been demonstrated in patients with end-stage liver disease, defined as a contributor to disease progression, and associated clinically with severity of disease and liver-related morbidity and mortality. Despite this well-recognized phenomena in patients with end-stage liver disease, the impact of gut dysbiosis and its rate of recovery following liver transplantation (LT) remains incompletely understood. The mechanisms by which alterations in the gut microbiota impact allograft metabolism and immunity, both directly and indirectly, are multifactorial and reflect the complexity of the gut-liver axis. Importantly, while research has largely focused on quantitative and qualitative changes in gut microbial composition, changes in microbial functionality (in the presence or absence of compositional changes) are of critical importance. Therefore, to translate functional microbiomics into clinical practice, one must understand not only the compositional but also the functional changes associated with gut dysbiosis and its resolution post-LT. In this review, we will summarize critical advances in functional microbiomics in LT recipients as they apply to immune-mediated allograft injury, posttransplant complications, and disease recurrence, while highlighting potential areas for microbial-based therapeutics in LT recipients.
INTRODUCTION
The human intestinal microbiome, composed of 1013 to 1014 individual microbes (or microbiota) and their genomes, is a central symbiotic participant in modulating host metabolic and immune homeostasis along the gut-liver axis.1–3 There is a bidirectional cross talk between the gut and the liver; intestinal dysbiosis (imbalance in intestinal microbiota) and increased bacterial translocation across the intestinal barrier are ubiquitous in patients with end-stage liver disease (ESLD) and have been associated with disease severity, complications including hepatic encephalopathy and hepatocellular carcinoma (HCC), and mortality.4–8 These maladaptive changes in the microbiome persist following liver transplantation (LT) though the impact of the severity and rate of recovery of dysbiosis on patient outcomes remains incompletely understood.
With the advent of advanced techniques to facilitate culture-independent characterization of microbiota and their products, such as large-scale metagenomics and metabolomics, our understanding of the degree and nature of gut dysbiosis in the clinical setting is advancing at a rapid pace. These tools are complemented by animal models, including germ-free and gnotobiotic models, which use the same techniques and allow further mechanistic investigation. As our technical expertise evolves, so too does our understanding of the functional consequences of dysbiosis in the LT recipient, highlighting potential avenues for microbial-based interventions to improve clinical outcomes. In this review, we will summarize critical advances in functional microbiomics in LT recipients as they apply to immune-mediated allograft injury, posttransplant complications, and disease recurrence, while highlighting potential areas for microbial-based therapeutics in LT recipients.
FORM VERSUS FUNCTION IN GUT DYSBIOSIS
Given its complexity, characterizing the composition of gut microbiota to correlate with clinical events can be challenging. Diversity measurements are used to characterize the complexity of microbial communities and provide an index for comparison and correlation. Alpha-diversity indices, such as the Shannon diversity index (SDI), are qualitative markers of the richness (number of bacterial species) and evenness (relative abundance of bacterial species) of gut microbiota in an individual ecosystem (eg, gut microbiota in a LT recipient).9 Higher SDI values indicate a larger number of bacterial taxa with a relatively even distribution of abundance, whereas low SDI values indicate a smaller number of bacterial taxa where a limited number are particularly dominant.10 Alpha-diversity indices such as SDI are used to characterize clinical subsets of patients (eg, patients with cirrhosis) and correlate them to a clinical event or outcome (eg, in-hospital mortality). In general, and specifically in liver disease, low-diversity dysbiosis (characterized by a low SDI) is associated with inflammation and disease progression.11 For instance, low microbial alpha diversity is associated with hepatic inflammation in nonobese nonalcoholic fatty liver disease (NAFLD) patients,12 with cirrhosis and subsequent severity of clinical decompensation,8,13 and with 30-day mortality in patients with cirrhosis.5
Beta-diversity indices, such as weighted and unweighted UniFrac, quantify compositional differences of microbial communities between independent samples (as opposed to within an individual ecosystem).14 Beta-diversity indices have been critical in advancing our understanding of the microbiome in the context of specific clinical conditions including liver disease. For instance, UniFrac principal component analysis has been essential to demonstrate differences in gut microbiota between healthy controls and patients with cirrhosis as well as longitudinal changes in individual patients pre- and post-LT.13,15 However, they represent a qualitative description of microbial composition rather than an assessment of the functional changes in the microbiome that will be required for mechanistic understanding and targeted therapy.
Metagenomics and (meta)metabolomics studies (describing changes in the functional potential of gut microbiota and microbial products, respectively) are now a focus to better understand functional consequences of dysbiosis.16,17 Advances in this area have led to a better understanding of the functional consequences of dysbiosis and led to recognition of clinically relevant changes that may have not otherwise been appreciated by more general compositional data approaches. Further integration of omics data with functional immune assays has also correlated immunophenotypes to specific bacterial taxa, further honing our ability to generate hypotheses on important immunomodulatory microbes for experimental follow-up and potential therapeutic targets in LT recipients.18 Ultimately, the impact of dysbiosis on allograft biology is due to both compositional and functional changes in the microbiome with both serving as potential targets for microbial-based therapies.
MEDIATORS OF GUT-LIVER CROSS TALK
Gut microbiota modulate systemic metabolic and immune functions via multiple mechanisms that may drive the impact of dysbiosis on subsequent allograft function.19 In addition to contributions from impaired mucosal immunity and portal hypertension, gut dysbiosis can result in a breakdown of the mucosal barrier, facilitating increased translocation of microbes and microbial products into the portal circulation.20,21 Commensal bacteria are essential for maintenance of the mucosal barrier, both directly via pathogen-associated molecular patterns (PAMPs) binding to pattern recognition receptors (PRRs; eg, Toll-like receptors) facilitating mucosal cell-mediated immunity22 and indirectly via production of short-chain fatty acids (SCFA), notably butyrate, which are essential for enterocyte health.23 Additional barrier functions are altered in dysbiosis and associated with ESLD, notably reduced mucin production coinciding with loss of autochthonous Akkermansia muciniphila.24
Once in the portal circulation, PAMPs (eg, lipopolysaccharide [LPS]) activate innate immune cells, including Kupffer cells, liver sinusoidal endothelial cells, and stellate cells in hepatic sinusoids, via PRR binding (eg, Toll-like receptor-4) and induce inflammation and fibrogenesis.25–28 Liver-related gut dysbiosis increases delivery of pro-inflammatory PAMPs to the liver and induces an inflammatory cytokine milieu, which in turn can shape adaptive immune function, for instance polarizing CD4+ T cells to a Th1 phenotype.29,30
Microbial metabolites also play a critical role in the gut-liver axis.31 Dietary choline is converted into trimethylamine (TMA) by gut bacteria and subsequently converted to TMA-N-oxide (TMAO) after entering the portal circulation.32 While originally identified as an atherogenic microbial metabolite,33 TMAO has now been associated with hepatic injury via increased triglyceride production and promotion of hepatic steatosis and elevated levels were associated with shorter transplant-free survival in patients with primary sclerosing cholangitis (PSC).34,35 Furthermore, bacterial conversion to TMA reduces conversion to phosphatidylcholine, a metabolite that when deficient contributes to chronic liver disease in both nonalcoholic steatohepatitis (NASH) and alcoholic steatohepatitis models.36,37
In addition to their importance for maintaining intestinal barrier function, SCFA (butyrate, propionate, and acetate) are potent mediators of both hepatic immunity and metabolism. This occurs both by SCFA binding to G-protein coupled receptors and by transcriptional regulation via inhibition of histone deacetylases.38 Both butyrate and propionate have been shown to reduce nuclear factor κB gene expression via histone deacetylase inhibition, ameliorating macrophage activation in ischemic models of tissue injury.39–41 SCFA have also been shown to facilitate neutrophil recruitment, both directly via induction of neutrophil chemotaxis and indirectly via chemokine induction (eg, interleukin-8) in endothelial cells.38,41
SCFA can also mediate systemic adaptive immune responses. Butyrate is known to impair maturation of dendritic cells, reduce costimulatory molecules, and impair cytokine production, collectively reducing subsequent T-cell activation and promoting induction of regulatory T cells (TREG).38 Reduced butyrate can in turn lead to decreased TREG/Th17 ratio, which has been associated with liver allograft rejection.42,43 Saturated long-chain fatty acids are also produced via gut microbiota, notably Lactobacillus, and are essential to maintenance of intestinal barrier function. Long-chain fatty acids supplementation and Lactobacillus probiotic supplementation have both been shown to reduce mucosal and hepatic inflammation in a mouse model of alcohol-induced liver injury.44
Gut microbiota metabolize ethanol into acetaldehyde and acetate via aldehyde dehydrogenase. Bacterial taxa rich in aldehyde dehydrogenase activity include Enterobacteriaceae, and abundance of this bacterial family can lead to accumulation of acetaldehyde, resulting in further breakdown of the mucosal barrier, increased inflammation, and adaptive immune activation.45,46
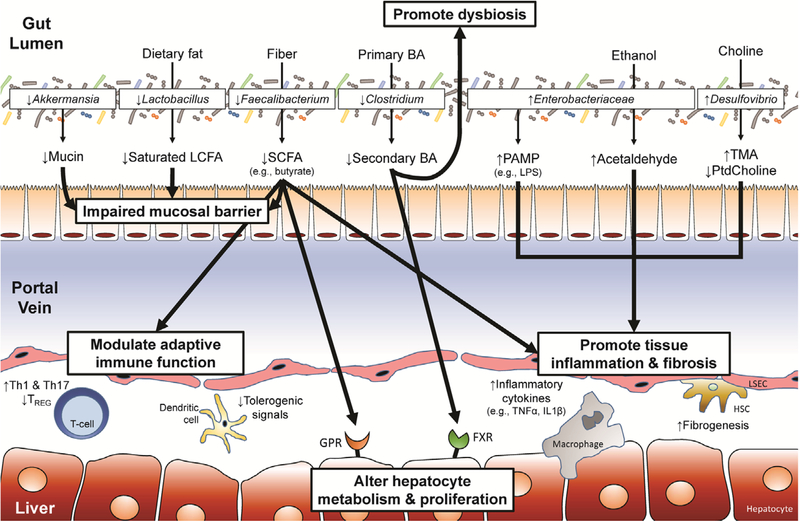
Enterohepatic circulation of bile acids (BAs) also plays a critical role in the gut-liver axis. Gut microbiota convert primary BAs into secondary BAs via deconjugation, dehydrogenation, and dehydroxylation.47 The balance of primary versus secondary BAs both in the intestinal lumen and portal circulation regulate metabolic functions, notably glucose and lipid metabolism, via binding to farsenoid X receptor (FXR) and Takeda G-protein coupled receptor-5 (TGR5).48 Furthermore, both receptors are expressed ubiquitously on circulating monocytes/macrophages, and therefore dysbiosis-induced alterations in the circulating BA pool can be associated with tissue inflammation.49 FXR-binding by BAs also regulates hepatic conversion of cholesterol to primary BAs via hepatic cholesterol 7α-monooxygenase.50 In dysbiosis, this production is decreased resulting in a decreased BA pool, further promoting expansion of gram-negative organisms, increasing the LPS burden, and promoting inflammation.51,52 Collectively, enterohepatic circulation of BAs plays an essential homeostatic role in hepatic immunity and metabolism with gut dysbiosis disturbing this balance in pathologic conditions. Mediators of gut-liver cross talk are summarized in Figure 1.
FIGURE 1.
Mechanisms of altered hepatic immunity and metabolism secondary to gut dysbiosis. Gut dysbiosis leads to an imbalance in bacterial microbes that results in alterations in microbial metabolites delivered to the liver via the portal vein that ultimately lead to changes in hepatic immunity and metabolism. Depicted are examples of specific bacteria that have been shown to be altered in gut dysbiosis related to end-stage liver disease or postliver transplant complications in human and mouse models. Corresponding alterations in microbial metabolites are also noted. Collectively, these changes further promote dysbiosis (via reduced secondary bile acid [BA] conversion), impair mucosal barrier function (via reduction in mucin, long-chain fatty acid [LCFA], and short-chain fatty acid [SCFA]), promote tissue inflammation (via increased pathogen-associated molecular pattern [PAMP], acetaldehyde, and trimethylamine (TMA)/trimethylamine-N-oxide (TMAO) with reduced SCFA and phosphatidylcholine [PtdCholine]), impair adaptive immune tolerance (via reduced SCFA and increase in inflammatory cytokines/chemokines), and alter hepatic metabolism and cellular profileration (via G-protein coupled receptor [GPR] and farsenoid X receptor [FXR] signaling by SCFA and BAs, respectively). These functional changes due to gut dysbiosis highlight the mechanisms by which an altered microbiome can potentially impact allograft function. HSC, hepatic stellate cell; IL, interleukin; LSEC, liver sinusoidal endothelial cell; LPS, lipopolysaccharide; TNF, tumor necrosis factor; TREG, regulatory T cells.
ALTERATIONS IN MICROBIOME POST-LT
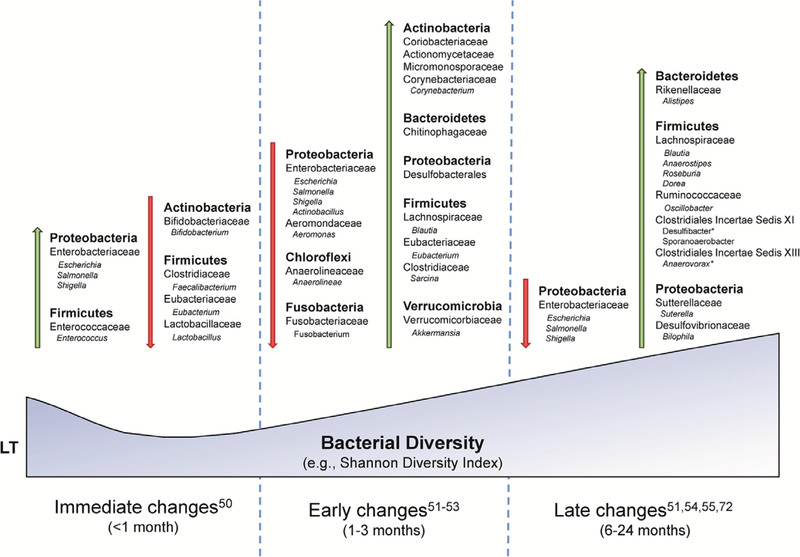
LT recipients have multiple factors (eg, abdominal surgery, antibiotics, hospitalizations, and immunosuppression) that can further compound the baseline gut dysbiosis present in ESLD, underscoring the critical importance of understanding the impact of gut dysbiosis post-LT. Several longitudinal studies have now assessed gut dysbiosis in patients post-LT.15,53–57 An initial quantitative polymerase chain reaction (qPCR)-based study of 111 LT recipients with hepatitis B virus (HBV)-related liver disease established persistence of dysbiosis in LT recipients.53 Both HBV cirrhotics and LT recipients (with HBV-related liver disease) showed quantitative changes in specific genera consistent with dysbiosis, though importantly, in LT recipients, there was further reduction in Faecalibacterium, a bacterial genus shown to have potent anti-inflammatory properties via nuclear factor κB inhibition and induction of TREG.58 This coincided with an increase in pathogenic genera (Enterococcus and Enterobacteriaceae family), suggesting LT resulted in initial worsening of dysbiosis with subsequent partial recovery seen in patients 12 to 24 months posttransplant.53
Two subsequent studies assessed perioperative (pre-transplant and within 3 mo of posttransplant) changes in gut microbial composition comparing pre-LT versus post-LT fecal samples in LT recipients using 16S ribosomal RNA gene sequencing. In a small pilot study by Sun et al15 that assessed perioperative changes in 8 LT recipients at 3 months posttransplant, there were no changes in microbial alpha diversity (richness) noted; however, there were differences in the relative abundance of specific bacterial taxa, notably increases in taxonomic groups that contain many putatively beneficial bacteria, such as Clostridiales cluster XIVa, Blautia, Akkermansia, and Eubacteriaceae family (includes Eubacterium), with a reduction in taxonomic groups that contain many pathogenic organisms, notably Enterobacteriaceae family (includes Escherichia). This finding underscores the potential limitations on diversity indices as markers of dysbiosis. Kato et al55 studied 38 LT recipients and demonstrated for the first time, an initial decrease in SDI early post-LT (<7 d) with subsequent improvement in diversity nearing pretransplant levels by week 8 post-LT. While this study did not report genus-specific changes post-LT, it correlated for the first time low SDI to both acute cellular rejection (ACR) and blood stream infections.
In a study of 45 patients by Bajaj et al,56 similar increases in SDI were observed at 6 months post-LT with reduction in the potentially pathogenic family Enterobacteriaceae and increases in potentially beneficial families Ruminococcaceae and Lachnospiraceae (Clostridiales cluster XIVa). Interestingly, in this study, there were no differences in SDI in post-LT patients compared with controls; however, there was a greater relative abundance of beneficial bacteria including Lachnospiraceae (Clostridiales cluster XIVa), Ruminococcaceae, and Eubacteriaceae in the healthy controls, indicating incomplete recovery of the microbiome post-LT. A follow-up study in a subset of 40 patients assessed functional consequences of these changes via metabolomics analysis.57 Improvement in SDI post-LT was associated with reduction in serum endotoxin levels with taxonomic changes similar to those previously reported. Further metabolomic consequences of these compositional microbiota changes included increased microbial conversion of primary to secondary BAs and nontoxic iso- and oxo-BA post-LT, collectively generating an antimicrobial gut milieu (secondary BA) and decreased BA toxicity profile (iso- and oxo-BA). Interestingly, improvement in diversity resulted in increased conversion of TMA to its atherogenic metabolite, TMAO, though the relevance of this functional consequence remains unclear. The compositional changes in microbiota post-LT are summarized in Figure 2.
FIGURE 2.
Changes in gut microbiome in liver transplant (LT) recipients. All changes in gut microbiome are relative to pre-LT cirrhotic patients at the designated time point post-LT. All bacteria are identified by phyla and further classified by family and genus (with the exception of desulfobacterales).
IMPACT OF DYSBIOSIS ON IMMUNE-MEDIATED ALLOGRAFT INJURY
Animal models and human studies support a role for the gut microbiome as a modulator of both innate and adaptive liver allograft immunity, presenting a potential role for microbial-based therapies to reduce the risk of immune-mediated allograft injury.59,60 Ischemia-reperfusion injury (IRI) is universal following LT with initial ischemic injury leading to parenchymal metabolic disturbance and cell death. This results in the release of danger-associated molecular patterns that signal via PRR to activate innate immune cells (eg, Kupffer cells). Subsequent reperfusion potentiates this proinflammatory innate immune response that, if persistent, can impact the adaptive immune response.61,62 The severity of IRI predicts early allograft dysfunction and long-term graft survival and is of particular relevance to study the impact of the gut microbiome on early innate immune activation.63,64 Administration of probiotics, specifically Bifidobacterium and Lactobacillus, has been shown to ameliorate IRI in a mouse model via reduction in plasma endotoxin levels and restoration of intestinal barrier function.65 Furthermore, in a rat model of LT, liver ischemic preconditioning (short periods of ischemia-reperfusion to condition tissue for a more prolonged IRI) restores gut microbial composition and ameliorates IRI, specifically increasing Lactobacillus, Bifidobacterium, and Clostridiales, with decreased Proteobacteria compared with untreated controls.66 Gut bacteria–derived SCFA are potent immunomodulators and can inhibit macrophage activation, a critical mediator of IRI.39,61 Intravenous administration of butyrate alleviates IRI in mouse models and administration of acetate-producing Bifidobacterium probiotic strains alleviates kidney IRI in a mouse model, supporting a role for targeted administration of SCFA-converting bacteria.39,40
BA signaling via FXR has also been shown to ameliorate IRI, and therefore targeting early restoration of autochthonous bacteria responsible for BA conversion to secondary BA, which include FXR agonists deoxycholic acid and lithocholic acid, is another potential therapeutic approach.57,67 While no studies are yet reported in humans, these findings support a role for microbial-based interventions to reduce IRI and subsequent early allograft dysfunction.
The microbiome can also shape adaptive immunity, and changes in the gut microbiota have been associated with ACR. Initial clinical trials investigating the impact of gut decontamination and administration of pre/probiotics showed no impact on liver allograft rejection.68 However, subsequent animal models of LT have established a role for an altered microbiome in ACR, with initial observations demonstrating an altered microbiome as early as 1 week post-LT in rats with ACR.69 In a more discrete analysis with serial fecal sampling in the first week following LT in rats, Ren et al70 demonstrated that there was a dramatic shift in gut microbiome composition between rats that developed ACR and those that did not at post-LT day 3 (coinciding with early ACR) and day 7 (coinciding with severe ACR). The SDI was decreased in all rats post-LT compared with controls with a trend toward lower diversity at day 7 in rats with ACR. Quantification of dominant bacterial species by qPCR in rats with ACR at day 7 revealed reduction in beneficial bacteria Faecalibacterium prausnitzii and Lactobacillus coinciding with an increase in Clostridium bolteae, an opportunistic pathogen. These findings support dysbiosis as a potential precursor to clinically apparent ACR post-LT.
Low-diversity dysbiosis has also been associated with ACR in humans post-LT. In a single longitudinal study, lower SDI was seen in 8 patients who developed ACR compared with those who did not develop ACR.55 Specific bacterial families altered in patients who developed ACR included increases in Bacteroides, Enterobacteriaceae, Streptococcaceae, and Bifidobacteriaceae, with decreases in Enterococcaceae, Lactobacillaceae, Clostridiaceae, Ruminococcaceae (which includes Faecalibacterium), and Peptostreptococcaceae. Importantly, 5 patients had received antibiotics 7 days before the diagnosis, making generalization of these findings difficult. The impact of prebiotics on ACR has been assessed in a meta-analysis of 3 randomized controlled trials, all of which incorporated Lactobacillus into their probiotic formulation, though differences in specific species, dose, timing, and additional probiotics were present (Table 1).71–74 While there was a difference in ACR (20% in probiotic group vs 28% in control group), this was not statistically significant. Importantly, observational clinical trials are ongoing, and while no further data are yet available correlating rate of recovery of dysbiosis to ACR, initial reports from a longitudinal study from Columbia show that persistence of dysbiosis posttransplant is associated with 1-year mortality post-LT.77
TABLE 1.
Summary of probiotic clinical trials in liver transplant recipients
| Probiotic | Dose, CFU | Duration | ACR (risk ratio) | Infection (risk ratio) | LOS (mean difference), days | |
|---|---|---|---|---|---|---|
| Rayes et al72 | Lactobacillus plantarum 299 | 1 × 109 | Day 1 to day 14 | 0.69 (0.37:1.29) | 0.38 (0.13:1.05) | −1.00 (−2.26 to 0.26) |
| Rayes et al73 |
Pediacoccus pentosaceus 5–33:3 Leuconostoc mesenteroides 77:1 Lactobacillus paracasei species paracasei F19 Lactobacillus plantarum 2362 |
10 × 109 | Day 0 to day 14 | 0.86 (0.32:2.28) | 0.06 (0.01:0.44) | −1.40 (−2.09 to −0.71) |
| Eguchi et al75 |
Lactobacillus casei strain Shirota Bifidobacterium breve strain Yakult |
20 mg 15 mg |
Day −2 to day 14 | 0.17 (0.02:1.29) | ||
| Zhang et al74 |
Lactobacillus acidophilus (LA-14) Lactobacillus plantarum (LP-115) Bifidobacterium lactis (BL-04) Lactobacillus casei (LC-11) Lactobacillus rhamnosus (LR-32) Lactobacillus brevis (LBr-35) |
27 × 109 | Day 1 to day 7+ | 0.67 (0.12:3.65) | 0.29 (0.09:0.96) | −2.00 (−3.44 to −0.56) |
| Grat et al76 |
Lactococcus lactis PB411 Lactobacillus casei PB121 Lactobacillus acidophilus PB111 Bifidobacterium bifidum PB211 |
3 × 109 | Day of listing to day 0 (16.7% <2 weeks) | 0.18 (0.02:1.37) | ||
| Cumulative data | 0.73 (0.44:1.22) | 0.20 (0.11:0.38) | −1.41 (−1.97 to −0.86) |
The findings of interventional probiotic clinical trials in liver transplant recipients are summarized. All patients received enteral nutrition with fiber supplementation in addition to the listed probiotics with the exception of Eguchi et al75 (enteral nutrition alone) and Grat et al76 (no additional nutritional supplementation).
ACR, acute cellular rejection; CFU, colony-forming units; LOS, length of stay.
IMPACT ON INFECTIOUS POSTTRANSPLANT COMPLICATIONS
Modulation of the gut microbiota has been shown to impact the risk of infection post-LT; however, specific qualitative and functional predictors of infection remain elusive. Single-center studies have consistently shown reduction in post-LT infectious complications with administration of probiotics ± prebiotics.78 In a systematic review and meta-analysis examining the pooled outcomes of 4 controlled studies that are assessing the impact of probiotics, findings revealed a reduction in infection rate (7% in probiotics group vs 35% in controls) and decreased length of hospitalization and intensive care unit stay (mean reduction 1.41 d for both in probiotic group).71 All 4 studies incorporated Lactobacillus into their probiotic formulations and 2 incorporated Bifidobacterium, both genera reduced in post-LT dysbiosis (Table 1).53,57,72–75 A subsequent single-center study showed similar reduction in posttransplant infectious complications with Lactobacillus lactus, Lactobacillus casei, Lactobacillus acidophilus, and Bifidobacterium bifidum probiotic administration (4.8% in probiotic group vs 34.8% in controls; P = 0.02).76 Furthermore, in a small cohort of human LT recipients, low-diversity dysbiosis defined by low SDI was associated with blood stream infections when compared with time-matched controls, supporting the importance of dysbiosis as a predictor of infection and as a target for intervention.55 Additional studies are needed to identify specific microbial-based targets for intervention as well as to define mechanisms by which probiotic use (with or without prebiotics) abrogates infection risk.
IMPACT OF DYSBIOSIS ON REGENERATION FOLLOWING PARTIAL GRAFT TRANSPLANTS
Innate immune activation is known to be a critical regulator of hepatic regeneration, and therefore targeting the microbiota to promote hepatic regeneration following transplantation with partial liver grafts (including living donor LT) is promising. Early mouse studies demonstrated that LPS is a critical trigger of hepatic regeneration following partial hepatectomy via increased deoxyribonucleic acid synthesis.79 Subsequent studies involving bowel decontamination with antibiotics have revealed differing effects on hepatic regeneration, likely owing to different spectrums of activity.80,81 Commensal bacteria that are ampicillin-sensitive, specifically Eubacteria, Lactobacillus, and Clostridium families, appear to be necessary for promotion of hepatic regeneration.81 Interestingly, loss of these commensal bacteria led to a loss of Kupffer cell tolerance and an increase in activation in natural killer T (NKT) cells, the latter resulting in impaired hepatic regeneration. Furthermore, partial hepatectomy itself induces dynamic changes in gut microbiota characterized by decreased Firmicutes families (Clostridiales, Lachnospiraceae, and Ruminococcaceae) with increased Bacteroidetes families (Bacteroidetes S24–7 and Rikenellaceae), mirroring changes seen in decompensated cirrhosis.5,82
BAs play a critical role in hepatic regeneration and their homeostasis is regulated in large part by microbial modification in the gut. BA signaling via FXR is necessary for normal hepatic regeneration in mice.83 In a cross-sectional study of cirrhotics, dysbiosis is associated with reduced bacterial conversion of primary to secondary BAs (via bacterial 7α-dehydroxylase) with an overall reduction in circulating BA pool.84 Importantly, loss of autochthonous Firmicutes families (Ruminococcaceae and Lachnospiraceae) correlated with reduced conversion of primary to secondary BAs. Priming mice with retinoic acid reverses these maladaptive changes, increasing Ruminococcaceae and Lachnospiraceae, and leads to increased diversity of BAs with increases in enterohepatic circulation that promotes regeneration.85
Human studies investigating the impact of dysbiois and microbial-based interventions in the context of hepatic regeneration are limited, though one small single-center study did show improvement in liver function following right hepatectomy in patients receiving synbiotics (probiotics plus prebiotics; Pediacoccus pentosaceus 5–33:3, Leuconostoc mesenteroides 77:1, Lactobacillus paracasei species paracasei F19, and Lactobacillus plantarum 2362 plus beta-glucan, inulin, pectin, and resistant starch) with uncomplicated postoperative courses.86 Additional cross-sectional and therapeutic studies are needed to better understand the impact of microbial-based therapies on hepatic regeneration for both partial liver graft recipients as well as living liver donors.
IMPACT OF DYSBIOSIS ON DISEASE RECURRENCE
Nonalcoholic Fatty Liver Disease
Approaches aimed to facilitate recovery of dysbiosis and subsequent maintenance of eubiosis may help reduce the risk of recurrent disease post-LT. Gut dysbiosis is central to the pathogenesis of NAFLD and severity of dysbiosis correlates with both disease onset and progression.16,87–91 Therefore, the gut microbiome is a potential target for prevention of recurrent NAFLD post-LT. Low-diversity dysbiosis characterized by an increase in Escherichia and Enterococcus and decrease in Lactobacillus and Bifidobacterium is present in animal models of NASH progression, suggesting probiotic interventions beneficial to allograft outcomes may also address dysbiosis related to NASH recurrence.92,93 A small study in humans with NASH assessing the impact of probiotics that included Lactobacillus and Bifidobacterium showed reduction in aspartate aminotransferase and intrahepatic triglyceride content, however, with no change in abundance of Lactobacillus or Bifidobacterium in the gut.94,95 Additional observational studies have demonstrated conflicting results about relative abundance of specific genera in adult patients with NASH, likely owing in large part to dietary and disease severity differences, and underscore the complexity of microbial-based interventions for recurrent NASH post-LT.87 There is a growing body of evidence for metabolic consequences of dysbiosis, both in the context of NAFLD and independent of liver disease, suggesting that microbial-based therapies may not only prevent the risk of de novo or recurrent NAFLD but also reduce the risk of metabolic complications associated with immunosuppressant side effects.36
Hepatocellular Carcinoma
The gut microbiome has been shown to impact hepatocarcinogenesis in disease models; therefore, microbial-based therapies may be useful to reduce the risk of recurrent HCC post-LT. Gut bacteria–derived LPS has been demonstrated to promote inflammation and accelerate hepatocarcinogenesis in mouse models via its action on both parenchymal cells and nonparenchymal cells, supporting a central role for the gut-liver axis in the development of HCC.96,97 In a small cohort of 15 patients with cirrhosis and HCC, culture-based analysis revealed an increase in Escherichia coli compared with patients with cirrhosis but without HCC, supporting this hypothesis.98 Subsequent culture-independent analysis in 20 patients with NAFLD-related HCC revealed relative decreases of Bifidobacterium and Blautia compared with NAFLD-related cirrhosis without HCC coinciding with decreased circulating inflammatory markers and peripheral mononuclear cell activation.7 Interestingly, Bifidobacterium has been shown to modulate antitumor immunity and administration of probiotic formulation containing Bifodobacterium strains ameliorated tumor size and progression in a rat model of HCC.99,100 Taken together, these data support a possible role for Bifidobacterium-based therapy to reduce the risk of recurrent HCC posttransplant.
Additional microbial-based mechanisms of HCC have been recognized. Using qPCR, Faecalibacterium prausnitzii was noted to be reduced in a cohort of cirrhotic patients with HCC compared to those without, suggesting a potential additional role for SCFA as a risk modifier via maintenance of intestinal barrier function and systemic anti-inflammatory effects.53 Modulation of gut microbiota may also impact antitumor surveillance as evidenced by a mouse model that demonstrated that antitumor NKT cell recruitment is dependent on microbial conversion of primary to secondary BAs.101 While more studies are needed, there is growing evidence that microbial-based therapies can impact recurrence of HCC in LT recipients.
Cholestatic Liver Disease
Low-diversity dysbiosis has been associated with cholestatic liver diseases and microbial-based therapies may hold promise to reduce the risk of recurrence of PSC and primary biliary cholangitis (PBC) post-LT. Mouse models of PSC have established the importance of dysbiosis in disease progression, in part through alterations in microbial-mediated BA metabolism.102,103 Alterations in BA composition and gut microbial composition have also been noted in patients with both PSC and PBC.104–108 PSC has been consistently associated with low-diversity dysbiosis with enrichment in Veillonella as well as Enterococcus.105,106,109,110 Similarly, in a single study of PBC patients, PBC was associated with reduced microbial diversity, increases in 8 genera including Veillonella, and decreases in 4 genera including Faecelibacterium, when compared with healthy controls. These changes were partially reversed with administration of ursodeoxycholic acid, a secondary BA.108 Multiple antibiotic and probiotic trials have demonstrated biochemical improvement in patients with PBC and PSC,111 though ongoing studies are needed for targeted interventions that may be applicable to prevention of recurrent disease post-LT. Interestingly, a small single-center study described alterations in the gut microbiota in post-LT patients with nonanastomotic strictures compared to those without, notably increased Enterococcaceae, Streptococcaceae, and Enterobacteriaceae with decreased Bacteroidaceae, suggesting a potential role for microbial-based interventions to prevent post-LT biliary complications independent of cholestatic liver disease.112
OPERATIONALIZING FUNCTIONAL MICROBIOMICS IN LT
Moving forward, several issues warrant attention to ensure sustained progress in translating our evolving wealth of knowledge on the microbial changes described into clinically meaningful interventions. Diversity indices, while helpful as coarse measures to associate dysbiosis with an outcome or specific patient population, do not incorporate taxa-specific changes that mechanistically may be essential to causation of a specific clinical end point. For instance, Enterococcus species have been associated with severity of PSC and, given their abundance in bile, may be of particular importance for biliary complications. Alternatively, loss of butyrate-producing species, such as Faecalibacterium, may be specifically relevant to immune-mediated allograft injury given their association with regulatory T-cell induction. Therefore, future studies in LT recipients will need to be designed and powered to focus on specific posttransplant complications.
In addition, ongoing microbiota-based studies in LT recipients will require integration of microbiota, metabolomics, functional data, and confirmatory experiments in in vitro and in vivo model systems to move beyond clinical associations and establish causative roles and associated mechanisms. This will require multicenter collaboration for comprehensive clinical and biological phenotyping to elucidate the multiscalar interactions at play across the gut-liver axis. The FLORINASH (the role of intestinal microflora in nonalcoholic fatty liver disease) study, a multicenter European clinical registry and biorepository, serves as model for this approach, integrating clinical; fecal 16S ribosomal RNA and metagenomics microbiome assessment; liver transcriptome; serum metabolomics, proteomics, and lipidomics; and urinary metabolomics data. The power of this resource is evident in the recent identification of the novel role of microbial-driven branched-chain amino acid metabolism in hepatic steatosis.113 Discoveries such as this can then provide specific targets for intervention, via either induction or inhibition of specific microbial pathways. For instance, the latter approach has recently been used to inhibit gut microbial TMA production from choline to reduce atherogenic TMAO production and atherosclerotic disease burden in a mouse model.114 While limited, single-center prospective clinical trials investigating the impact of microbiota on LT outcomes are ongoing, there is a critical need for comprehensive, multicenter studies to refine specific microbial targets for intervention.
Importantly, the magnitude of impact of changes in microbiota on post-LT clinical outcomes must also be considered. For instance, is the relative abundance of E coli posttransplant a more powerful prognostic tool for HCC recurrence than the presence of viable tumor in the explant? Can microbiome composition data be considered together with well known predictors of HCC recurrence post–OLT, such as Milan/University of California, San Francisco criteria and microvascular invasion, to improve prognostics? These prognostic inferences will require more robust prospective studies and incorporation of currently validated clinical predictors to determine the true impact of dysbiosis on clinical outcomes in LT recipients. Collectively, using these principles will eliminate the heterogeneity observed in nontargeted pre- and probiotic interventional studies to date and provide novel, targeted, and impactful microbial-based interventions moving forward.
FUTURE DIRECTIONS
Microbiome-based research has to date focused on bacteria; however, the microbiome includes fungi and viruses, both of which may have additional metabolic and immunologic consequences on the liver allograft. For instance, chronic alcohol ingestion results in increased intestinal fungi, notably Candida species, coinciding with increased portal 1,3-β-d-glucan that leads to activation of hepatic macrophages. This finding correlated with severity of alcoholic liver disease in a small cohort, suggesting this may be a critical mediator and potential target to ameliorate hepatic inflammation.115
The interplay between immunosuppression agents and gut microbiome is another evolving area of interest. Tacrolimus has been associated with dysbiosis, decreasing overall gut microbiota diversity associated with loss of butyrate-producing species. Furthermore, when compared with everolimus and mycophenolate mofetil, patients on tacrolimus and mycophenolate mofetil had a distinct bacterial metagenome, suggesting functional differences in gut microbiota based on immunousppression regimen were present independent of taxonomic changes.116,117 Bacterial species may also impact drug absorption and metabolism and, conversely, can ameliorate drug-specific side effects as has been shown with Lactobacillus and tacrolimus-induced hypertension.116,118
Given the abundance of donor passenger lymphocytes and liver-resident innate immune cells transferred from the donor to recipient in LT, the donor microbiome may also be of clinical significance. Interestingly, there is evidence that the donor microbiome may impact alloimmunity in a cohort of patients following bone marrow transplant with increased donor microbial diversity associated with lower rates of acute graft-versus-host disease in recipients, supporting a possible role for donor-based interventions for the liver allograft.119
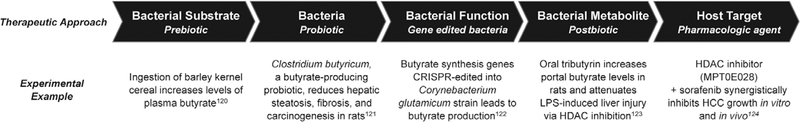
Ultimately, understanding the evolution of the functionality of the microbiome both pre- and post-LT will be critical to translate our current knowledge into therapeutic targets. While clinical investigation has thus far focused largely on administration of pre- and probiotics, more sophisticated and focused approaches, including bacterial gene editing to modify specific metabolic pathways are being developed.120 Combined with ongoing usage of gnotobiotic animal models to delineate specific functional consequences of individual bacterial taxa, more targeted, microbial-based interventions are likely to evolve (Figure 3).121
FIGURE 3.
Potential approaches to microbial-based therapy in the liver allograft. While clinical trials to date have focused on administration of pre- and probiotics, our physiologic understanding of the functional impact of an altered gut microbiome as well as our ability to therapeutically manipulate the gut microbiome and microbial metabolites is rapidly evolving. Administration of pre- and probiotics is shifting to target taxa-specific microbial pathways; functional modification of bacteria via gene editing or pathway inhibition allows for modulation of specific metabolites; and targeted post-biotics and synthetic analogs present opportunities for novel therapeutics informed by microbial pathways. Depicted are experimental examples of targeting microbiota-mediated butyrate production at multiple levels along the microbial pathway.122–126 This construct can be applied to other microbial pathways for therapeutic intervention specific to the effects of individual microbiota and microbial metabolites. HCC, hepatocellular carcinoma; HDAC, histone deacetylase; LPS, lipopolysaccharide.
CONCLUSIONS
The gut microbiome is a central participant in regulating hepatic metabolism and immunity and our understanding of its impact on post-LT physiology is evolving at an increasingly rapid pace. As we continue to obtain data from longitudinal studies of post-LT patients, therapeutic targets for microbial-based interventions will continue to evolve to improve the allograft function and to reduce the risk of post-LT complications. Our hope is that microbial-based therapies will be used in concert with our current standard of care to improve clinical outcomes in post-LT patients.
Acknowledgments
M.K. was supported by NIH/NCATS Colorado CTSA (grant UL1 TR002535). Contents are the authors’ sole responsibility and do not necessarily represent official NIH views. E.C.V. was supported by NIH (grant K23 DK101827). C.A.L. was supported by NIH (grants R01 DK104047, RO1 DK108366, and RO1 HL138639).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tripathi A, Debelius J, Brenner DA, et al. Publisher correction: the gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj JS, Betrapally NS, Hylemon PB, et al. Gut microbiota alterations can predict hospitalizations in cirrhosis independent of diabetes mellitus. Sci Rep. 2015;5:18559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponziani FR, Bhoori S, Castelli C, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology. 2019;69:107–120. [DOI] [PubMed] [Google Scholar]
- 8.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- 9.Kim BR, Shin J, Guevarra R, et al. Deciphering diversity indices for a better understanding of microbial communities. J Microbiol Biotechnol. 2017;27:2089–2093. [DOI] [PubMed] [Google Scholar]
- 10.Bromberg JS, Fricke WF, Brinkman CC, et al. Microbiota-implications for immunity and transplantation. Nat Rev Nephrol. 2015;11:342–353. [DOI] [PubMed] [Google Scholar]
- 11.Kriss M, Hazleton K, Nusbacher NM, et al. Low diversity gut microbiota dysbiosis: drivers, functional implications, and recovery. Curr Opin Microbiol. 2018;44:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Jiang X, Cao M, et al. Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sci Rep. 2016;6:32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Yang F, Lu H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. [DOI] [PubMed] [Google Scholar]
- 14.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun LY, Yang YS, Qu W, et al. Gut microbiota of liver transplantation recipients. Sci Rep. 2017;7:3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loomba R, Seguritan V, Li W, et al. Gut microbiome-based metagenomic signature for non-invasive detection of advanced fibrosis in human nonalcoholic fatty liver disease. Cell Metab. 2017;25:1054–1062.e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei X, Jiang S, Chen Y, et al. Cirrhosis related functionality characteristic of the fecal microbiota as revealed by a metaproteomic approach. BMC Gastroenterol. 2016;16:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geva-Zatorsky N, Sefik E, Kua L, et al. Mining the human gut microbiota for immunomodulatory organisms. Cell. 2017;168:928–943.e911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tripathi A, Debelius J, Brenner DA, et al. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellot P, Frances R, Such J. Pathological bacterial translocation in cirrhosis: pathophysiology, diagnosis and clinical implications. Liver Int. 2013;33:31–39. [DOI] [PubMed] [Google Scholar]
- 21.Tuomisto S, Pessi T, Collin P, et al. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. [DOI] [PubMed] [Google Scholar]
- 24.Grander C, Adolph TE, Wieser V, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891–901. [DOI] [PubMed] [Google Scholar]
- 25.Paik YH, Schwabe RF, Bataller R, et al. Toll-like receptor 4 mediates inflammatory signaling by bacterial lipopolysaccharide in human hepatic stellate cells. Hepatology. 2003;37:1043–1055. [DOI] [PubMed] [Google Scholar]
- 26.Schnabl B, Brandl K, Fink M, et al. A TLR4/MD2 fusion protein inhibits LPS-induced pro-inflammatory signaling in hepatic stellate cells. Biochem Biophys Res Commun. 2008;375:210–214. [DOI] [PubMed] [Google Scholar]
- 27.Wu J, Meng Z, Jiang M, et al. Toll-like receptor-induced innate immune responses in non-parenchymal liver cells are cell type-specific. Immunology. 2010;129:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Q, Zou L, Jagavelu K, et al. Intestinal decontamination inhibits TLR4 dependent fibronectin-mediated cross-talk between stellate cells and endothelial cells in liver fibrosis in mice. J Hepatol. 2012;56:893–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe M, Tokita D, Raimondi G, et al. Endotoxin modulates the capacity of CpG-activated liver myeloid DC to direct Th1-type responses. Eur J Immunol. 2006;36:2483–2493. [DOI] [PubMed] [Google Scholar]
- 30.De Creus A, Abe M, Lau AH, et al. Low TLR4 expression by liver dendritic cells correlates with reduced capacity to activate allogeneic T cells in response to endotoxin. J Immunol. 2005;174:2037–2045. [DOI] [PubMed] [Google Scholar]
- 31.Blacher E, Levy M, Tatirovsky E, et al. Microbiome-modulated metabolites at the interface of host immunity. J Immunol. 2017;198:572–580. [DOI] [PubMed] [Google Scholar]
- 32.Velasquez MT, Ramezani A, Manal A, et al. Trimethylamine N-oxide: the good, the bad and the unknown. Toxins (Basel). 2016;8(11):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang WH, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circ Res. 2017;120:1183–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen YM, Liu Y, Zhou RF, et al. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep. 2016;6:19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kummen M, Vesterhus M, Troseid M, et al. Elevated trimethylamine-N-oxide (TMAO) is associated with poor prognosis in primary sclerosing cholangitis patients with normal liver function. United European Gastroenterol J. 2017;5:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung C, Rivera L, Furness JB, et al. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–425. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Zou W, Cui F, et al. Protective effect of phosphatidylcholine on restoration of ethanol-injured hepatocytes related with caveolin-1. J Membr Biol. 2014;247:73–80. [DOI] [PubMed] [Google Scholar]
- 38.Correa-Oliveira R, Fachi JL, Vieira A, et al. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol. 2016;5:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiao YL, Qian JM, Wang FR, et al. Butyrate protects liver against ischemia reperfusion injury by inhibiting nuclear factor kappa B activation in Kupffer cells. J Surg Res. 2014;187:653–659. [DOI] [PubMed] [Google Scholar]
- 40.Andrade-Oliveira V, Amano MT, Correa-Costa M, et al. Gut bacteria products prevent AKI induced by ischemia-reperfusion. J Am Soc Nephrol. 2015;26:1877–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vinolo MA, Ferguson GJ, Kulkarni S, et al. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011;6:e21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Asarat M, Apostolopoulos V, Vasiljevic T, et al. Short-chain fatty acids regulate cytokines and Th17/Treg cells in human peripheral blood mononuclear cells in vitro. Immunol Invest. 2016;45:205–222. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Zhang M, Liu ZW, et al. The ratio of circulating regulatory T cells (Tregs)/Th17 cells is associated with acute allograft rejection in liver transplantation. PLoS One. 2014;9:e112135. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Chen P, Torralba M, Tan J, et al. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148:203–214. e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: role of oxidative metabolism. World J Gastroenterol. 2014;20:17756–17772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nosova T, Jousimies-Somer H, Kaihovaara P, et al. Characteristics of alcohol dehydrogenases of certain aerobic bacteria representing human colonic flora. Alcohol Clin Exp Res. 1997;21:489–494. [PubMed] [Google Scholar]
- 47.Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baars A, Oosting A, Knol J, et al. The gut microbiota as a therapeutic target in IBD and metabolic disease: a role for the bile acid receptors FXR and TGR5. Microorganisms. 2015;3:641–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calmus Y, Poupon R. Shaping macrophages function and innate immunity by bile acids: mechanisms and implication in cholestatic liver diseases. Clin Res Hepatol Gastroenterol. 2014;38:550–556. [DOI] [PubMed] [Google Scholar]
- 50.Xu G, Li H, Pan LX, et al. FXR-mediated down-regulation of CYP7A1 dominates LXRalpha in long-term cholesterol-fed NZW rabbits. J Lipid Res. 2003;44:1956–1962. [DOI] [PubMed] [Google Scholar]
- 51.Sayin SI, Wahlstrom A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. [DOI] [PubMed] [Google Scholar]
- 52.Ramirez-Perez O, Cruz-Ramon V, Chinchilla-Lopez P, et al. The role of the gut microbiota in bile acid metabolism. Ann Hepatol. 2017;16:s15–s20. [DOI] [PubMed] [Google Scholar]
- 53.Wu ZW, Ling ZX, Lu HF, et al. Changes of gut bacteria and immune parameters in liver transplant recipients. Hepatobiliary Pancreat Dis Int. 2012;11:40–50. [DOI] [PubMed] [Google Scholar]
- 54.Lu H, He J, Wu Z, et al. Assessment of microbiome variation during the perioperative period in liver transplant patients: a retrospective analysis. Microb Ecol. 2013;65:781–791. [DOI] [PubMed] [Google Scholar]
- 55.Kato K, Nagao M, Miyamoto K, et al. Longitudinal analysis of the intestinal microbiota in liver transplantation. Transplant Direct. 2017;3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bajaj JS, Fagan A, Sikaroodi M, et al. Liver transplant modulates gut microbial dysbiosis and cognitive function in cirrhosis. Liver Transpl. 2017;23:907–914. [DOI] [PubMed] [Google Scholar]
- 57.Bajaj JS, Kakiyama G, Cox IJ, et al. Alterations in gut microbial function following liver transplant. Liver Transpl. 2018;24:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miquel S, Martin R, Rossi O, et al. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–261. [DOI] [PubMed] [Google Scholar]
- 59.Doycheva I, Leise MD, Watt KD. The intestinal microbiome and the liver transplant recipient: what we know and what we need to know. Transplantation. 2016;100:61–68. [DOI] [PubMed] [Google Scholar]
- 60.Tabibian JH, Kenderian SS. The microbiome and immune regulation after transplantation. Transplantation. 2017;101:56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai Y, Petrowsky H, Hong JC, et al. Ischaemia-reperfusion injury in liver transplantation—from bench to bedside. Nat Rev Gastroenterol Hepatol. 2013;10:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Land WG. The role of postischemic reperfusion injury and other nonantigen-dependent inflammatory pathways in transplantation. Transplantation. 2005;79:505–514. [DOI] [PubMed] [Google Scholar]
- 63.Ali JM, Davies SE, Brais RJ, et al. Analysis of ischemia/reperfusion injury in time-zero biopsies predicts liver allograft outcomes. Liver Transpl. 2015;21:487–499. [DOI] [PubMed] [Google Scholar]
- 64.Lu L, Zhou H, Ni M, et al. Innate immune regulations and liver ischemia-reperfusion injury. Transplantation. 2016;100:2601–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xing HC, Li LJ, Xu KJ, et al. Protective role of supplement with foreign Bifidobacterium and Lactobacillus in experimental hepatic ischemia-reperfusion injury. J Gastroenterol Hepatol. 2006;21:647–656. [DOI] [PubMed] [Google Scholar]
- 66.Ren Z, Cui G, Lu H, et al. Liver ischemic preconditioning (IPC) improves intestinal microbiota following liver transplantation in rats through 16s rDNA-based analysis of microbial structure shift. PLoS One. 2013;8:e75950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ceulemans LJ, Verbeke L, Decuypere JP, et al. Farnesoid X receptor activation attenuates intestinal ischemia reperfusion injury in rats. PLoS One. 2017;12:e0169331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gurusamy KS, Nagendran M, Davidson BR. Methods of preventing bacterial sepsis and wound complications after liver transplantation. Cochrane Database Syst Rev. 2014:CD006660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie Y, Luo Z, Li Z, et al. Structural shifts of fecal microbial communities in rats with acute rejection after liver transplantation. Microb Ecol. 2012;64:546–554. [DOI] [PubMed] [Google Scholar]
- 70.Ren Z, Jiang J, Lu H, et al. Intestinal microbial variation may predict early acute rejection after liver transplantation in rats. Transplantation. 2014;98:844–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawas T, Al Halabi S, Hernaez R, et al. Patients receiving prebiotics and probiotics before liver transplantation develop fewer infections than controls: a systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2015;13:1567–1574.e1563; quiz e1143–1564. [DOI] [PubMed] [Google Scholar]
- 72.Rayes N, Seehofer D, Hansen S, et al. Early enteral supply of lactobacillus and fiber versus selective bowel decontamination: a controlled trial in liver transplant recipients. Transplantation. 2002;74:123–127. [DOI] [PubMed] [Google Scholar]
- 73.Rayes N, Seehofer D, Theruvath T, et al. Supply of pre- and probiotics reduces bacterial infection rates after liver transplantation—a randomized, double-blind trial. Am J Transplant. 2005;5:125–130. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Chen J, Wu J, et al. Probiotic use in preventing postoperative infection in liver transplant patients. Hepatobiliary Surg Nutr. 2013;2:142–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eguchi S, Takatsuki M, Hidaka M, et al. Perioperative synbiotic treatment to prevent infectious complications in patients after elective living donor liver transplantation: a prospective randomized study. Am J Surg. 2011;201:498–502. [DOI] [PubMed] [Google Scholar]
- 76.Grat M, Wronka KM, Lewandowski Z, et al. Effects of continuous use of probiotics before liver transplantation: a randomized, double-blind, placebo-controlled trial. Clin Nutr. 2017;36:1530–1539. [DOI] [PubMed] [Google Scholar]
- 77.Verna E, Annavajhala M, Nenad M, et al. Intestinal microbiome diversity is associated with liver disease etiology and predicts post-liver transplant mortality [abstract]. Available at https://atcmeetingabstracts.com/abstract/intestinal-microbiome-diversity-is-associated-with-liver-disease-etiology-and-predicts-post-liver-transplant-mortality/. Accessed June 14, 2018.
- 78.Jorgenson MR, Descourouez JL, Siodlak M, et al. Efficacy and safety of probiotics and synbiotics in liver transplantation. Pharmacotherapy. 2018;38:758–768. [DOI] [PubMed] [Google Scholar]
- 79.Cornell RP, Liljequist BL, Bartizal KF. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology. 1990;11:916–922. [DOI] [PubMed] [Google Scholar]
- 80.MacIntosh EL, Gauthier T, Harding GK, Minuk GY. Selective bowel decontamination does not alter hepatic regeneration in rats. Gastroenterology. 1992;102(4 Pt 1):1403–1405. [PubMed] [Google Scholar]
- 81.Wu X, Sun R, Chen Y, et al. Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology. 2015;62:253–264. [DOI] [PubMed] [Google Scholar]
- 82.Liu HX, Rocha CS, Dandekar S, et al. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J Hepatol. 2016;64:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang W, Ma K, Zhang J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. [DOI] [PubMed] [Google Scholar]
- 84.Bajaj JS, Heuman DM, Sanyal AJ, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu HX, Hu Y, Wan YJ. Microbiota and bile acid profiles in retinoic acid-primed mice that exhibit accelerated liver regeneration. Oncotarget. 2016;7:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rayes N, Pilarski T, Stockmann M, et al. Effect of pre- and probiotics on liver regeneration after resection: a randomised, double-blind pilot study. Benef Microbes. 2012;3:237–244. [DOI] [PubMed] [Google Scholar]
- 87.Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33:128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Da Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep. 2018;8:1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. [DOI] [PubMed] [Google Scholar]
- 90.Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. [DOI] [PubMed] [Google Scholar]
- 91.Stanislawski MA, Lozupone CA, Wagner BD, et al. Gut microbiota in adolescents and the association with fatty liver: the EPOCH study. Pediatr Res. 2018;84:219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie G, Wang X, Liu P, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget. 2016;7:19355–19366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xue L, He J, Gao N, et al. Probiotics may delay the progression of nonalcoholic fatty liver disease by restoring the gut microbiota structure and improving intestinal endotoxemia. Sci Rep. 2017;7:45176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong VW, Tse CH, Lam TT, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis—a longitudinal study. PLoS One. 2013;8:e62885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wong VW, Won GL, Chim AM, et al. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann Hepatol. 2013;12:256–262. [PubMed] [Google Scholar]
- 96.Dapito DH, Mencin A, Gwak GY, et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu LX, Yan HX, Liu Q, et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52:1322–1333. [DOI] [PubMed] [Google Scholar]
- 98.Grat M, Wronka KM, Krasnodebski M, et al. Profile of gut microbiota associated with the presence of hepatocellular cancer in patients with liver cirrhosis. Transplant Proc. 2016;48:1687–1691. [DOI] [PubMed] [Google Scholar]
- 99.Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang HL, Yu LX, Yang W, et al. Profound impact of gut homeostasis on chemically-induced pro-tumorigenic inflammation and hepatocarcinogenesis in rats. J Hepatol. 2012;57:803–812. [DOI] [PubMed] [Google Scholar]
- 101.Ma C, Han M, Heinrich B, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schrumpf E, Kummen M, Valestrand L, et al. The gut microbiota contributes to a mouse model of spontaneous bile duct inflammation. J Hepatol. 2017;66:382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tabibian JH, O’Hara SP, Trussoni CE, et al. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology. 2016;63:185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bell LN, Wulff J, Comerford M, et al. Serum metabolic signatures of primary biliary cirrhosis and primary sclerosing cholangitis. Liver Int. 2015;35:263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kummen M, Holm K, Anmarkrud JA, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611–619. [DOI] [PubMed] [Google Scholar]
- 106.Sabino J, Vieira-Silva S, Machiels K, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Trottier J, Bialek A, Caron P, et al. Metabolomic profiling of 17 bile acids in serum from patients with primary biliary cirrhosis and primary sclerosing cholangitis: a pilot study. Dig Liver Dis. 2012;44:303–310. [DOI] [PubMed] [Google Scholar]
- 108.Tang R, Wei Y, Li Y, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534–541. [DOI] [PubMed] [Google Scholar]
- 109.Iwasawa K, Suda W, Tsunoda T, et al. Characterisation of the faecal microbiota in Japanese patients with paediatric-onset primary sclerosing cholangitis. Gut. 2017;66:1344–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruhlemann MC, Heinsen FA, Zenouzi R, et al. Faecal microbiota profiles as diagnostic biomarkers in primary sclerosing cholangitis. Gut. 2017;66:753–754. [DOI] [PubMed] [Google Scholar]
- 111.Li Y, Tang R, Leung PSC, et al. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun Rev. 2017;16:885–896. [DOI] [PubMed] [Google Scholar]
- 112.Zhang J, Ren FG, Liu P, et al. Characteristics of fecal microbial communities in patients with non-anastomotic biliary strictures after liver transplantation. World J Gastroenterol. 2017;23:8217–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hoyles L, Fernandez-Real JM, Federici M, et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat Med. 2018;24:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Z, Roberts AB, Buffa JA, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163:1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang AM, Inamine T, Hochrath K, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Invest. 2017;127:2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Toral M, Romero M, Rodriguez-Nogales A, et al. Lactobacillus fermentum improves tacrolimus-induced hypertension by restoring vascular redox state and improving eNOS coupling. Mol Nutr Food Res. 2018;62:e1800033. [DOI] [PubMed] [Google Scholar]
- 117.Zaza G, Dalla Gassa A, Felis G, et al. Impact of maintenance immunosuppressive therapy on the fecal microbiome of renal transplant recipients: comparison between an everolimus- and a standard tacrolimus-based regimen. PLoS One. 2017;12:e0178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee JR, Muthukumar T, Dadhania D, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS One. 2015;10:e0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu C, Frank DN, Horch M, et al. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant. 2017;52:1643–1650. [DOI] [PubMed] [Google Scholar]
- 120.Shen TC, Albenberg L, Bittinger K, et al. Engineering the gut microbiota to treat hyperammonemia. J Clin Invest. 2015;125: 2841–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shaffer M, Armstrong AJS, Phelan VV, et al. Microbiome and metabolome data integration provides insight into health and disease. Transl Res. 2017;189:51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nilsson AC, Ostman EM, Knudsen KE, et al. A cereal-based evening meal rich in indigestible carbohydrates increases plasma butyrate the next morning. J Nutr. 2010;140:1932–1936. [DOI] [PubMed] [Google Scholar]
- 123.Endo H, Niioka M, Kobayashi N, et al. Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS One. 2013;8:e63388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yoon J, Woo HM. CRISPR interference-mediated metabolic engineering of Corynebacterium glutamicum for homo-butyrate production. Biotechnol Bioeng. 2018;115:2067–2074. [DOI] [PubMed] [Google Scholar]
- 125.Miyoshi M, Sakaki H, Usami M, et al. Oral administration of tributyrin increases concentration of butyrate in the portal vein and prevents lipopolysaccharide-induced liver injury in rats. Clin Nutr. 2011;30:252–258. [DOI] [PubMed] [Google Scholar]
- 126.Chen CH, Chen MC, Wang JC, et al. Synergistic interaction between the HDAC inhibitor, MPT0E028, and sorafenib in liver cancer cells in vitro and in vivo. Clin Cancer Res. 2014;20:1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]