Abstract
Bacterial endophytes are well known inhabitants of living plant system and perform important assignments in maintaining plant growth and health. Currently, limited reports are available on the endophytes of pearl millet (Pennisetum glaucum) reflecting antagonistic and plant growth promoting (PGP) attributes. Therefore, the major objectives of current investigation were to identify antagonistic strains of endophytic Bacillus from pearl millet and further illustrate their PGP capabilities. In this study, 19 endophytic Bacillus strains (EPP5, EPP21, EPP30, EPP32, EPP35, EPP42, EPP49, EPP55, EPP62, EPP65, EPP70, EPP71, EPP74, EPP78, EPP83, EPP86, EPP93, EPP100, and EPP102) displaying antagonistic activity towards Rhizoctonia solani (RS), Sclerotium rolfsii (SR), and Fusarium solani (FS) were isolated from different sections (root, leaf, stem, and root) of pearl millet. Phenotypic (shape, colony, gram staining reaction, endospore formation, and motility) and biochemical features (catalase, oxidase, citrate, gelatinase, urease, Voges Proskauer’s, methyl red, indole, and nitrate reduction), along with the similarly comparison of 16S rRNA gene sequence with type strains identified eight antagonistic endophyhtes as B. amyloliquefaciens (EPP35, EPP 42, EPP62, and EPP 102), Bacillus subtilis subsp. subtilis (EPP65), and Bacillus cereus (EPP5, EPP71, and EPP74). The production of indole acetic acid and siderophores was varied among the isolated endophytes. Besides displaying enzymatic activities, these isolates varied in solubilizing capabilities of phosphate, potassium, and zinc. The presence of three antimicrobial peptide genes (ituD, bmyC, and srfA) also confirmed their antifungal nature. Further, single treatment of three promising strains (EPP5, EPP62, and EPP65) offered protection ranging from 35.68 to 45.74% under greenhouse conditions. However, microbial consortium (EPP5+ EPP62 + EPP65) provided the highest protection (71.96%) against root rot and wilt infection with significant increase in plant biomass. Overall, the current study indicated that pearl millet plant harbors various species of endophytic Bacillus that possess excellent biocontrol and growth promotion activities.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00172-5) contains supplementary material, which is available to authorized users.
Keywords: Antagonism, Bacillomycin, Iturin, Root rot, Siderophores, Stress, Surfactin, Zinc solubilization
Introduction
Pearl millet (Pennisetum glaucum) is one of the most imperative and extensively cultivated millet crops throughout the world. In India, it is one of the prime millet crops grown widely under rainfed conditions and consumed as a major dietary source for millions of peoples living in the resource deprived regions. Moreover, pearl millet plant has inherent capability to grow luxuriantly on unfertile and low nutrient soils [1, 2]. Besides this, pearl millet host has a remarkable ability to tolerate the negative impacts of climate change because of the unique and diverse type of endophytic microbiota present inside the host system [3]. Generally, internal parts of the plant provide homogeneous and sound environment to microbes in contrast to rhizosphere and phylloplane, where microbes struggle for their food and undergo drastic temperature shift and ultra violet (UV) revelation [4]. Several studies reported mammoth diversity of endophytic bacteria associated with plant system and identified their prospective role in plant growth promotion through multiple mechanisms, including rapid synthesis of growth hormones, siderophores, hydrolytic enzymes, secondary metabolites, and activation of induced plant resistance (IPR) [5–9]. Emerging research evidences clearly pointed out the significance of microbiota dwelling inside the pearl millet host in offering stress tolerance as well as assistance in enhancing the survival and fitness attributes under unfavorable conditions [3, 8]. So far, a wide range of bacterial species from Bacillus, Agrobacterium, Burkholderia, Azospirillum, Enterobacter, Rhizobium, Pseudomonas, and Azotobacter genera have been documented as an excellent growth-promoting endophytes in different types of crops [8–15]. Unfortunately, limited reports are available on the plant growth promoting and antifungal activity in endophytic strains of Bacillus species from pearl millet. Thus, a better understanding of plant growth promoting and antagonistic potential of bacterial endophytes from pearl millet will be useful for establishing an environment friendly and sustainable crop production system.
Endophytic Bacillus is recognized as one of the most promising bacterium in agriculture sector due to its unique features of endospore formation, multifarious plant growth promoting attributes and biocontrol activities towards different kinds of fungal and bacterial pathogens [9, 16, 17]. Several studies highlighted the significant correlation of antimicrobial peptides (AMPs) released by Bacillus with the biological management of several crop diseases and in their indirect role in plant growth enhancement [18–22]. Generally, AMPs encompass cyclic lipopeptides such as iturin, fengycin, bacillomycin, and surfactin. Koumoutsi et al. [23] mentioned that B. amyloliquefaciens FZB42 secretes fengycin and bacillomycin D for displaying antagonistic activity against Fusarium oxysporum. Later, it has been observed that the production of mixtures of bacillomycin, fengycin, and iturin A by B. subtilis effectively control Podosphaera fusca in cucurbits [24]. Similar results of effective suppression of Fusarium wilt of cucumber and Phytophthora blight of pepper due to iturin secretion by B. subtilis ME488 have been observed [25]. Mora et al. [22] demonstrated the prevalence of surfactin, fengycin, bacillomycin, subtilin, and iturin in plant-associated antagonistic populations of Bacillus. Later, Mora et al. [26] revealed that the antimicrobial capabilities of plant-associated Bacillus strains towards crop pathogens is linked with the presence of AMPs genes and to the production of the corresponding cyclic lipopeptides, and it is mainly associated to the species B. subtilis and B. amyloliquefaciens. On parallel lines, Cao et al. [20] reported strong association of iturin and fengycin with antagonistic activity towards R. solanacearum and F. oxysporum. These findings clearly highlighted that an understanding of the role of AMPs in the ecological fitness of pearl millet requires an appreciation of the distribution of AMP genes in bacterial endophytes. So far, limited information is available about the presence of AMP biosynthetic genes in endophytic strains of Bacillus species from pearl millet.
The endophytic Bacillus strains are emerging players as a stimulator for plant growth and as biological control agent for sustainable plant health management. Zhao et al. [27] illustrated promising application of endophytic strains of Paenibacillus and Bacillus species from Lonicera japonica in escalating wheat growth and biological control of plant pathogens. Similarly, Zhang et al. [28] also highlighted the antagonistic capabilities of endophytic strains of B. pumilus and B. subtilis in protecting grapevine seedlings from downy mildew infection. Later, Haidar et al. [29] explored population structure and diversity of bacteria from internal parts of Corchorus olitorius plant and established endophytic B. subtilis as a promising bioinoculant based on its antifungal and PGP attributes. Thus, owing to the relevance of endophytic bacilli in crop growth promotion and disease control, it is decided to execute the study on the identification and characterization of endophytic strains of Bacillus species with antifungal and plant growth promoting activity. In addition, attempts have been made to study the additive plant growth promoting effects of inoculation of these antagonistic endophytic strains in pearl millet plant.
Materials and methods
Sampling and isolation of endophytic Bacillus
Field surveys of four distinct sites (Mau, Mirzapur, Varanasi, and Gazipur) representing Indo-Ganagetic plain (IGP) province of India (Table 1) were conducted to collect healthy and stress free samples of pearl millet from 40–50 day old crop. Each plant sample comprised of three plants. Different plant sections (root, stem, and leaf) were collected in sterile plastic bags to avoid contamination. The protocol of complete surface sterilization of plant materials mentioned by Coombs and Franco [30] with few minor modifications was used for the isolation of endophytic bacteria from pearl millet. Briefly, exterior portions of the collected stem, root, and leaf samples were washed with sterile tap water followed by double-distilled sterile water. Soaking of the sterile water-treated plant samples was performed in ethanol (70%) for 60 s and later dipped in sodium hypochlorite (3%) for 3 min followed by ethanol (70%) treatment for 30 s. The final washing of the treated plant samples was made three times with autoclaved double-distilled sterile water. Air drying of the processed plant samples was done on the sterilized Whatman filter paper under laminar air flow cabinet. The completely sterilized plant tissue (1 g) from each representative sample was macerated and grinded in sterile mortar pestle using sterile phosphate buffer saline (PBS) solution (Hi-Media Laboratories Pvt Ltd., India). Tissue extracts were collected in the tubes, and heat treatment was performed in shaking water bath according to procedure mentioned by Sharma et al. [31]. Heat-treated tissue extracts were then serially diluted (10−2 to 10−9), and an equal volume (100 μl) of aliquot from each dilution was plated onto Luria-Bertani (LB) agar, nutrient agar (NA), nutrient broth yeast extract (NBYE), and Tryptic Soya Agar (TSA). The inoculated Petri plates were incubated for 48 h at 30 ± 2 °C. The sterilization efficiency was confirmed and validated by inoculating the last rinse of washing water (100 μl) on Petri plates containing TSA medium and later inspected for the appearance of colonies. Colonies with distinct morphotypes were identified and each strain was preserved in 50% (v:v) glycerol at − 80 °C [32].
Table 1.
Sampling site description and in vitro evaluation of antifungal properties of endophytic Bacillus strains on inhibition of fungal plant pathogens
| Strain(s) | Plant tissue used for isolation/geographical information coordinates | Mycelial growth inhibition (%) | ||
|---|---|---|---|---|
| Rhizoctonia solani (RS) | Sclerotium rolfsii (SR) | Fusarium solani (FS) | ||
| EPP5 | Stems/25.8696° N, 83.4386° E | *63.21 ± 1.02a | 58.11 ± 2.11a | 58.65 ± 2.14a |
| EPP21 | Stems/25.2876° N, 82.9239° E | 46.43 ± 1.29f | 48.32 ± 2.18d | 49.76 ± 2.76c |
| EPP30 | Stem/25.8696° N, 83.4386° E | 48.56 ± 1.62e | 45.65 ± 1.95e | 45.98 ± 1.98d |
| EPP32 | Root/25.8696° N, 83.4386° E | 43.29 ± 1.95g | 42.65 ± 2.22f | 38.65 ± 1.65f |
| EPP35 | Root/25.8696° N, 83.4386° E | 55.21 ± 2.12c | 51.26 ± 1.14c | 50.32 ± 1.41c |
| EPP42 | Root/25.5835° N, 83.4870° E | 54.12 ± 2.46c | 50.41 ± 3.19c | 51.95 ± 1.46c |
| EPP49 | Root/25.5835° N, 83.4870° E | 26.32 ± 1.11l | 44.28 ± 1.06ef | 50.95 ± 1.42c |
| EPP55 | Root/25.2876° N, 82.9239° E | 35.62 ± 1.65i | 44.26 ± 3.26ef | 38.46 ± 1.65f |
| EPP62 | Root / 25.1137° N, 82.5444° E | 58.42 ± 2.13b | 50.21 ± 2.85c | 55.38 ± 2.95b |
| EPP65 | Root / 25.1137° N, 82.5444° E | 50.18 ± 2.19d | 53.23 ± 1.42b | 50.17 ± 2.22c |
| EPP70 | Stem/25.1137° N, 82.5444° E | 39.43 ± 2.31h | 35.32 ± 2.12h | 38.32 ± 1.76f |
| EPP71 | Stem/25.5835° N, 83.4870° E | 51.24 ± 2.10d | 52.23 ± 1.95cd | 50.11 ± 2.91c |
| EPP74 | Stem/25.5835° N, 83.4870° E | 54.18 ± 2.13c | 51.32 ± 2.28cd | 50.13 ± 2.32c |
| EPP78 | Leaf/25.5835° N, 83.4870° E | 36.44 ± 2.11i | 29.74 ± 1.98i | 31.65 ± 1.33g |
| EPP83 | Leaf / 25.5835° N, 83.4870° E | 27.13 ± 2.32k | 23.32 ± 1.65j | 25.85 ± 1.08h |
| EPP86 | Leaf/25.2876° N, 82.9239° E | 32.98 ± 2.97j | 39.85 ± 2.98g | 35.43 ± 1.95f |
| EPP93 | Leaf/25.2876° N, 82.9239° E | 41.23 ± 3.11h | 46.37 ± 1.46e | 43.13 ± 2.08e |
| EPP100 | Leaf/25.1137° N, 82.5444° E | 29.85 ± 2.79k | 49.65 ± 2.65d | 45.13 ± 2.11d |
| EPP102 | Leaf/25.1137° N, 82.5444° E | 51.41 ± 1.94d | 51.73 ± 1.98cd | 52.11 ± 2.98c |
*Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference
In vitro screening for antagonistic activity
Antagonistic capabilities of bacterial strains of endophytic origin were confirmed by monitoring mycelium growth inhibition of three highly aggressive and virulent crop pathogens viz., Fusarium solani (FS), Rhizoctonia solani (RS), and Sclerotium rolfsii (SR) in dual-culture plate assay. A small piece of fungal mass (5 mm in diameter) from active culture (FS/RS/SR) was placed onto the center of Petri plate containing PDA:NA (1:1) media. Each bacterial strain (~ 6 × 108 cfu ml−1) was streaked in a straight line along the outer edge of Petri plate and incubated at 30 ± 2 °C for 5 days. Fungus (FS/RS/SR) inoculated Petri plate without endophytic strain was maintained as a control. The inhibition diameter (mm) and the percentage of mycelial growth inhibition by each strain of endophytic bacteria were determined by using the following formula proposed by Sharma et al. [33].
where, C = radial growth (mm) of fungus in control, and T = Radial growth of the fungus (mm) in the presence of endophytic strain in dual culture plate
PCR amplification of AMP gene (s), sequencing, and phylogenetic analysis
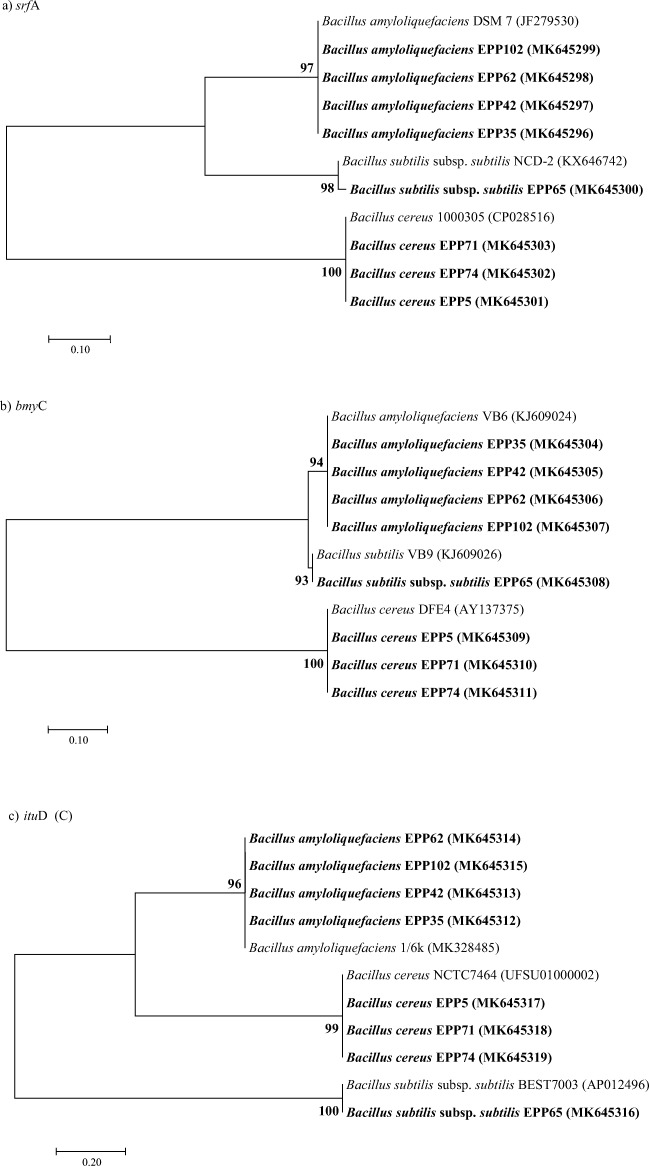
Eight Bacillus strains (EPP5, EPP35, EPP42, EPP62, EPP65, EPP71, EPP74, and EPP102) displaying strong antifungal activities (Table 1) were further screened for the presence of various AMP genes viz., surfactin (srfA), iturin (ituD), and bacillomycin (bmyC). The PCR amplification of AMP genes was performed according to the information mentioned in Table S1. The generated PCR products were purified by using Nucleo-pore sure extract PCR clean-up kit (Genetix Biotech Asia Pvt. Ltd) and sequenced by using BigDye terminator chemistry version 3.1 with an automated capillary sequencer (Applied Biosystems). National Center for Biotechnology Information (NCBI) GenBank was used to recognize the best similarly match for obtained gene sequence data and submitted in NCBI GenBank under the following gene sequence accession numbers: MK645296-MK645303 for surfactin (srfA), MK645304- MK645311 for iturin (ituD), and MK645312-MK645319 for bacillomycin (bmyC). Nucleotide sequences encoding for AMP genes in phylogenetically closely related bacterial species were retrieved from NCBI GenBank for performing phylogenetic analysis. Sequence reads were aligned with Clustal W [34], and resulted alignment profiles were used to construct best fit phylogenetic tree using the neighbor-joining method [35] with Kimura’s two-parameter model [36] executed in MEGA 7 software [37]. Bootstrap analysis was performed to assess confidence levels for the branches with 1000 replicates [38] (Fig. 1).
Fig. 1.
Neighbor-joining phylogenetic tree based on the srfA (a), bmyC (b), and ituD (c) gene sequences of shortlisted antagonistic bacterial endophytes from pearl millet host and those of closely related strain sequences. The significance of each branch is indicated by a bootstrap value (%) estimated for 1000 subsets; only values above 50% are shown. Scale (bar) indicated substitutions per site. The accession numbers of the strains are presented in parentheses. Bold accessions indicate the strains identified in the present study
Identification of bacterial endophytes
Phenotypic identification and characterization
For phenotypic identification, colony morphology (gram-staining reaction, shape, colony, endospore formation, and motility, etc.) of each endophytic bacterial strain was recorded. Gram and endospore staining was performed according to Prescott et al. [39]. Motility of each bacterial strain was examined by cultivating bacterial strain in semi-solid motility test medium [40].
Biochemical characterization
Biochemical characterization (catalase, oxidase, citrate, gelatinase, urease, Voges-Proskauer’s, methyl red, indole, and nitrate reduction) was executed by employing Bacillus identification test kit (Himedia, India), following the manufacturer’s instructions. Qualitative determination of ammonia and hydrogen cyanide (HCN) production by each endophytic bacterial strain was performed according to the outline mentioned by Cappuccino and Sherman [41] and Bakker and Schippers [42], respectively.
Molecular identification
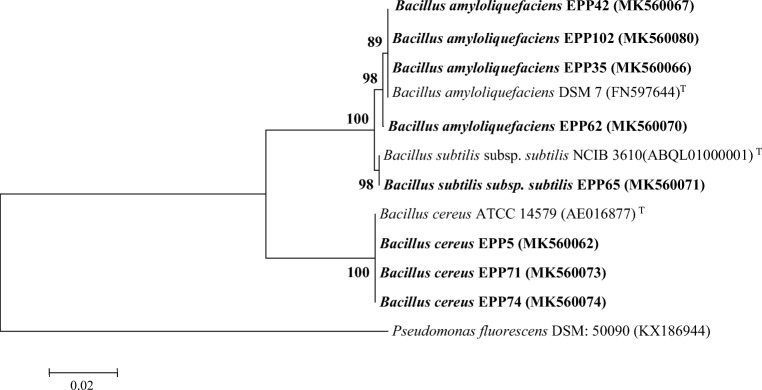
For molecular identification of endophytic bacteria, total genomic DNA of each pure strain was isolated as per the procedure adopted by Solanki et al. [43]. The 16S rRNA genomic region of the each bacterial strain was amplified by using primer pair (27F and 1525R) mentioned in Table S1. Polymerase chain reaction (PCR) was carried out in a thermal cycler (G-STORM, UK) and thermal profiles mentioned in Table S1 were employed for amplicon generation. The generated PCR product (~ 1500 bp) was gel purified by Nucleo-pore sure extract PCR clean-up kit (Genetix Biotech Asia Pvt. Ltd) and later sequenced by using BigDye terminator chemistry version 3.1 with an automated capillary sequencer (Applied Biosystems). NCBI database was used to recognize the best similarly match type strain for obtained 16S rRNA gene sequences and submitted in NCBI GenBank database under following accession numbers: MK560062, MK560066, MK560067, MK560070, MK560071, MK560073, MK560074, and MK560080 for EPP5, EPP35, EPP42, EPP62, PP65, EPP71, EPP74, and EPP102 strains, respectively. Phylogenetic analysis and tree construction was performed as per the methodology mentioned in earlier section.
Determination of plant growth promoting (PGP) features
The chromeazurol (CAS) agar assay described by Schwyn and Neiland [44] was used to test the ability of endophytic bacterial strains for siderophore production. Briefly, test strains were spot inoculated on CAS agar plates and incubated at 30 ± 2 °C for 5 days. Development of yellow-orange halo around the growth was identified as positive for siderophore production. Similarly, the production of IAA in selected endophytic bacterial strains was determined according to Bric et al. [45]. Endophytic strains grown for 3 days in nutrient broth at 30 ± 2 °C were centrifuged at 3000 rpm for 30 min. The supernatant (2 ml) was mixed with two drops of orthophosphoric acid and 4 ml of the Salkowski reagent and incubated at room temperature for 20 min, after which the absorbance was determined at 530 nm using pure IAA (Sigma- Aldrich, USA) as a standard. For assessing the zinc (Zn), potassium (K), and phosphorous (P) solubilization potential; each endophytic bacterial strain was spot inoculated in agar Petri plates comprising LGI-P semi-solid medium [46] amended with 0.1 % zinc oxide (ZnO), modified solid Aleksandrov medium [47] supplemented with potassium feldspar powder, and tricalcium phosphate [Ca3(PO4)2] as insoluble phosphate source [48], respectively. All inoculated Petri plates were incubated at 30 ± 2 °C for 3–7 days. The mineral solubilization potential of each endophytic bacterial strain was confirmed by transparent halo zone formation around the bacterial colony. The diameter of halo zone and colony growth was recorded in mm. The ability of endophytic bacterial strains for the production of extracellular hydrolytic enzymes (amylase, cellulose, protease, and lipase) were studied by growing each strain of endophytic bacteria on NA medium supplemented with a specific ingredient substrate [49–52]. Different substrate sources such as soluble starch (1%), carboxy methyl cellulose (CMC; 1%), casein (1%), and tributerin were utilized for the detection of amylase, cellulase, protease, and lipase activities, respectively. Each inoculated Petri plate was independently treated with iodine reagent, congo red, and Coomassie Brilliant Blue stains and incubated at 30 ± 2 °C for 5 days. A positive enzymatic activity was reflected by transparent halo zone formation around the bacterial colony. The diameter of halo zone and colony growth was recorded in mm. All the assays were executed in three independent replicates and means were determined.
Greenhouse experiment
The greenhouse experiments were carried out in sterile sandy loam soil [organic C (0.60%); EC = 6.5 dS m−1; NPK (160: 70: 44, Kg ha−1); pH = 7.2 and sodium absorption ratio (SAR) of 75]. Depending on the antifungal activity and detection of antimicrobial peptide genes (AMP) and PGP attributes, three endophytic Bacillus strains were chosen for pot study. FS was cultivated on sterilized pearl millet seeds. Later, pathogen inoculum was added in sterile soil and incubated for 2 days at 25 ± 2 °C inside the polythene bag to maintain the moisture level. The sterilized soil was inoculated with virulent FS conidia (3 × 106 conidia gram−1) into the sterilized soil at a rate of 5% (w:w) and filled in pots (45 × 60 cm). Before sowing, seeds of pearl millet (cv. NDFB-2) were rinsed with sterile tap water and later dipped in sodium hypochlorite (1%) for 5 min to ensure complete surface sterilization. For seed treatment, suspension of overnight grown active culture (108 cfu ml−1) of each shortlisted strain of endophytic bacteria in PBS solution was prepared. Surface sterilized pearl millet (cv. NDFB-2) seeds were soaked in bacterium culture for 12 h. The treatments were (T1) EPP5 + FS, (T2) EPP62 + FS, (T3) EPP65 + FS, (T4) EPP5 + EPP62 + FS, (T5) EPP5 + EPP65 + FS, (T6) EPP62 + EPP65 + FS, (T7) EPP5 + EPP 62 + EPP65 + FS, (T8) Healthy control (sterilized soil without FS only), and (T9) disease control (FS alone). Three replications were maintained for each treatment, and each replication composed of three pots in a completely randomized design (CRD) under greenhouse condition. Plant growth characteristics like length and weight (both fresh and dry) of the leaf and root were monitored after 30 days of seed sowing. Disease symptoms were recorded and disease severity was scored on a 0–4 scale after 30 days, where 0, 1, 2, 3, and 4 indicate no infection (completely healthy root tissue), 0–25% infection (minute pin pointed dark-brown lesions on root tissue), 25–50 % (bigger size, superficial dark-brown lesions on root tissue), 50–75 % infection (necrotic root), and 75–100% infection (tissue death), respectively [53]. The data was further employed to calculate the percent disease index (DI) using the following formula:
Statistical analysis
All the laboratory and greenhouse experiments were executed in three replicates and followed complete randomized design (CRD). The level of significance and effects of each treatment were measured by performing one-way ANOVA. Post hoc comparative analysis of mean values was conducted by Duncan’s multiple range test (DMRT) at 5% probability level.
Results
Isolation and screening of endophytic bacterial strains for antifungal activity
The results of in vitro antagonistic screening of endophytic strains isolated from the leaves, stems, and roots of pearl millet host (Table 1) indicated that nineteen endophytic bacterial strains reflected positive inhibitory effects on fungal mycelium growth towards all the three phytopathogenic fungi (FS/RS/SR). Eight strains (EPP5, EPP35, EPP42, EPP62, EPP65, EPP71, EPP74 and EPP102) displayed antagonistic effect against the growth of three tested pathogens (> 50.0% mycelium inhibition), where endophyte EPP5 was recorded as the highest inhibitor for all the three fungi (Table 1).
Identification and characterization of bacterial endophytes
For phenotypic identification, colony morphology (gram staining reaction, shape, colony, endospore formation, and motility etc.) of each endophytic bacterial strain was recorded and listed in Table S2. Morphological characters indicated that all the isolated endophytic bacterial strains belonged to genus Bacillus. Further, molecular phylogeny of all the eight strains of endophytic bacteria was performed, and a combined dendrogram based on 16S rRNA gene sequences of endophytic strains and their best match type strains (T) from NCBI database was generated (Fig 2). All screened strains of endophytic bacteria from pearl millet host showed 99–100% resemblance with Bacillus genus. The 16S rRNA gene sequences of endophytic bacterial strains were submitted in NCBI GenBank with following accession numbers: EPP5 (MK560062), EPP35 (MK560066), EPP42 (MK560067), EPP62 (MK560070), EPP65 (MK560071), EPP71 (MK560073), EPP74 (MK560074), and EPP102 (MK560080). The antagonistic endophytic strains were classified as: B. amyloliquefaciens (EPP35, EPP42, EPP62, and EPP 102), Bacillus subtilis subsp. subtilis (EPP65), and Bacillus cereus (EPP5, EPP71, and EPP74).
Fig 2.
Neighbor-joining phylogenetic tree based on the 16S rRNA gene sequence of shortlisted antagonistic bacterial endophytes from pearl millet host and those of closely related type strain (T) sequences. The significance of each branch is indicated by a bootstrap value (%) estimated for 1000 subsets; only values above 50% are shown. All positions containing gaps and missing data were eliminated. Scale (bar) indicated 0.02 substitutions per site. The accession numbers of the strains are presented in parentheses. Bold accessions indicated the strains identified in the present study. Pseudomonas fluorescens DSM: 50090 was used as an outgroup
Detection and phylogenetic analysis of AMP genes
The PCR amplification of AMP genes from shortlisted antagonistic endophytic Bacillus strains was shown in Fig S1. Surfactin (srfA), bacillomycin (bmyC), and iturin (ituD) gene amplification showed one specific band of 201 bp, 395 bp, and 647 bp, respectively, in all the eight shortlisted endophytic strains. Phylogenetic tree based on srfA (Fig. 2a), bmyC (Fig. 2b), and ituD (Fig 2c) and genes reflected 93–100% homology among each Bacillus strain and further categorized into three clades according to their species. All AMPs gene sequences from different strains were submitted in NCBI with accession number MK645296-MK645319.
Functional characterization of endophytes for plant growth promotion attributes
The statistical analysis and evaluation of examination of results (Table 2) demonstrated variable range of IAA production (49.04–152.32 μg ml−1) by endophytic Bacillus strains. Strain EPP5 was identified as maximum IAA producer (152.32 μg ml−1). Strain EPP71 did not show IAA production. Among eight shortlisted strains, only seven strains (EPP5, EPP42, EPP62, EPP65, EPP71, EPP74, and EPP102) indicated siderophore production, which is reflected by variable size transparent halo zone (10.62–25.65 mm) formation around the bacterial colony. Strain EPP5 was observed as highest siderophore producer and resulted in large and transparent halo zone (25.65 mm) formation. No transparent halo zone formation was observed in case of EPP35.
Table 2.
The production of the phytohormone (IAA), siderophores, and solubilization of phosphorous (P), potassium (K), and zinc (Zn) by shortlisted endophytic Bacillus strains
| Strain | IAA (μg ml−1) | Diameter of clear zone (mm) | |||
|---|---|---|---|---|---|
| Siderophore | P solubilization | K solubilization | Zn solubilization | ||
| EPP5 | *152.32 ± 1.02a | 25.65 ± 2.52a | 21.21 ± 1.21a | 19.45 ± 1.84a | 25.13 ± 1.12a |
| EPP35 | 69.52 ± 2.16d | - | - | 14.34 ± 2.43b | 11.17 ± 1.21d |
| EPP42 | 114.11 ± 1.08b | 10.62 ± 2.41d | 11.25 ± 1.65d | 11.08 ± 1.41c | - |
| EPP62 | 77.35 ± 0.98c | 14.64 ± 1.40b | 10.41 ± 0.95d | 11.95 ± 2.13c | 20.12 ± 1.13b |
| EPP65 | 42.04 ± 1.22f | 11.21 ± 1.10cd | 13.21 ± 1.21c | 16.85 ± 2.98b | 10.95 ± 2.10d |
| EPP71 | - | 13.21 ± 1.18c | 16.38 ± 1.08b | 9.86 ± 0.95e | 14.95 ± 2.76c |
| EPP74 | 115.18 ± 1.14 b | 15.62 ± 0.98b | 13.02 ± 1.18 c | - | - |
| EPP102 | 49.04 ± 1.45e | 12.31 ± 1.95cd | - | 10.95 ± 0.98d | - |
*Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference
Endophytic bacterial strains were assessed for their Zn, K, and P solubilization potential under controlled laboratory conditions and statistically analyzed results were mentioned in Table 2. Apart from four strains (EPP5, EPP62, EPP65, and EPP71) of endophytic bacteria, none of the other strain was able to demonstrate transparent halo zone against all the three tested nutrient substrates. Further, it was noticed that all the tested strains of endophytic bacteria differed in their capabilities to solubilize Zn, K, and P substrates. Strain EPP35 and EPP102 did not illustrate any type of transparent halo zone for P nutrient. Only six strains (EPP5, EPP42, EPP62, EPP65, EPP71, and EPP74) were categorized as the phosphate solubilizers, displaying prominent transparent halo zone (13.02–21.21 mm). Interestingly, seven endophytic bacterial strains (EPP5, EPP35, EPP42, EPP62, EPP65, EPP71, and EPP102) were reported as promising K solubilizer (Table 2). Strain EPP5 displayed strong K solubilizing potential and produced highest halo zone (19.45 mm) in contrast to EPP74 strain reflecting no transparent and prominent halo zone formation (Table 2). Similarly, in case of Zn solubilization, only five endophytic bacterial strains (EPP5, EPP35 EPP62, EPP65, and EPP71) showed prominent and variable size transparent halo zones around their bacterial colony. Statistically, two strains (EPP65 and EPP35) showed least Zn solubilization (10.95–11.17 mm zone) in comparison to strain EPP5, which showed maximum halo zone diameter (25.13 mm) around bacterial colony.
It was remarkable that shortlisted bacterial endophytes produced several extracellular enzymes; however, different endophytic strains displayed variable enzymatic activities (Table 3). Bacterial strain EPP74 showed the maximum protease (21.70 mm zone) and lipase (32.07 mm zone) activities compared to other strains (Table 3). Strains EPP5, EPP71, and EPP65 were recorded as the highest producer for amylase (23.12 mm zone), chitinase (31.43 mm zone), and pectinase (25.74 mm zone) enzymes, respectively.
Table 3.
The in vitro hydrolytic enzyme activities displayed by shortlisted endophytic Bacillus strains
| Strain | Clear zone formation (diameter in mm) | ||||
|---|---|---|---|---|---|
| Protease | Amylase | Lipase | Chitinase | Pectinase | |
| EPP5 | *11.20 ± 0.71c | 23.12 ± 1.02a | 22.47 ± 1.71b | 31.43 ± 0.85a | 21.04 ± 1.01cd |
| EPP35 | 10.08 ± 0.91d | 13.42 ± 1.52 d | 19.08 ± 1.27c | 23.12 ± 1.42b | 24.04 ± 2.05b |
| EPP42 | 11.14 ± 2.10cd | 13.62 ± 2.21d | 19.10 ± 1.04c | 13.42 ± 1.18e | 18.42 ± 2.20e |
| EPP62 | 12.42 ± 0.95bc | 18.02 ± 1.07c | 13.71 ± 1.43e | 10.12 ± 1.09g | 13.62 ± 2.28f |
| EPP65 | 14.21 ± 2.01b | 20.32 ± 1.72b | 17.07 ± 1.76d | 14.30 ± 1.43e | 25.74 ± 1.19ab |
| EPP71 | 12.41 ± 0.74bc | 23.62 ± 2.08a | 12.27 ± 1.08e | 11.40 ± 1.07f | 23.12 ± 1.12b |
| EPP74 | 21.70 ± 0.32a | 12.22 ± 1.31d | 32.07 ± 1.85a | 21.46 ± 0.76c | 12.10 ± 1.82f |
| EPP102 | 11.14 ± 2.08c | 13.22 ± 1.09d | 18.54 ± 1.23d | 18.73 ± 0.95d | 20.85 ± 2.21d |
*Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT (p = 0.05). Values with different letter indications represent a statistically significant difference
Effect of endophytic Bacillus strains on biocontrol and growth promotion of pearl millet
In order to study the effect of endophytes Bacillus strains on seedling growth under pathogen presence, three endophytes (EPP5, EPP62, and EPP65) were selected based on their potential antagonistic activity (Table 1), presence AMPs genes (Fig 1), and PGP attributes (Tables 2 and 3). The results of glass house study (Table 4) revealed significant inhibition of FS, in endophytic Bacillus-treated seeds when compared with T8 and T9 control treatment. Treatment T4 (EPP5 + EPP62), T5 (EPP5 + EPP65), T6 (EPP62 + EPP65), and T7 (EPP5 + EPP62 + EPP65) reflected synergistic effect on the plant growth promotion and disease suppression. Treatment T7 (EPP5 + EPP62 + EPP65 + FS) reflected higher value of means in terms of plant fresh weight (6.594 g), dry weight (2.854 g), shoot length (18.95 cm), and root length (9.85 cm). FS treatment (T9) had the lowest mean for root (1.45 cm) and shoot length (1.10 cm) followed by T8 (control without endophyte + FS). In case of root length, FS treatment (T6) was found significantly different in comparison to other treatments. Among individual application of endophytic strains, T1 (EPP5 + FS) accounts for the highest mean for shoot length (16.52 cm), root length (8.52 cm), fresh weight (5.65 g), and dry weight (2.654 g) followed by T2 (EPP62 + FS) and T3 (EPP65 + FS), respectively (Table 4). The seedlings from seeds treated with the bacterium displayed significant increment in percent disease control (35–72%) compared to FS control (T9). Maximum disease index was observed in T9 (76.51%) followed by T2 (49.21 %) and T3 (48.98 %) treatments (Table 4), whereas maximum disease reduction over control was reported in T7 (71.96 %) followed by T6 (56.11), T5 (53.67 %), and T4 (49.90 %).
Table 4.
The in vivo inhibitory and plant growth-promoting activity of endophytic Bacillus strains on pearl millet plants under glasshouse conditions
| Treatment (s) | Shoot length (cm) | Root length (cm) | Fresh weight (g) | Dry weight (g) | PDI (%) | PDC (%) |
|---|---|---|---|---|---|---|
| T1: EPP5 + FS | *16.52 ± 0.98c | 8.52 ± 0.46bc | 5.651 ± 0.95abc | 2.654 ± 0.54ab | 41.51 ± 1.51c | 45.74 |
| T2: EPP62 + FS | 14.95 ± 1.18e | 7.46 ± 1.11c | 5.214 ± 0.66bcd | 2.252 ± 0.58bc | 49.21 ± 3.31b | 35.68 |
| T3: EPP65 + FS | 15.20 ± 1.02d | 7.97 ± 1.10b | 4.821 ± 0.46d | 1.952 ± 0.49d | 48.98 ± 3.11b | 35.98 |
| T4: EPP5 + EPP62 + FS | 16.98 ± 0.98c | 8.75 ± 0.97b | 5.947 ± 0.43abc | 2.784 ± 0.28ab | 38.33 ± 3.01c | 49.90 |
| T5: EPP5 + EPP65 + FS | 16.77 ± 0.92c | 8.65 ± 0. 74b | 5.827 ± 0.31ab | 2.624 ± 0.38ab | 35.45 ± 2.91c | 53.67 |
| T6: EPP62 + EPP65 + FS | 17.52 ± 0.28bc | 8.94 ± 0.66ab | 6.121 ± 0.94abc | 2.722 ± 0.62ab | 33.58 ± 1.91d | 56.11 |
| T7: EPP5 + EPP62 + EPP65 + FS | 18.95 ± 1.20a | 9.85 ± 0.97a | 6.594 ± 1.05a | 2.854 ± 0.31a | 21.45 ± 2.91e | 71.96 |
| T8: Control (sterilized soil without FS) | 12.41 ± 0.79f | 6.13 ± 0.85d | 5.028 ± 1.27cd | 2.101 ± 0.44cd | - | - |
| T9: FS only | 1.10 ± 0.52g | 1.45 ± 0.74e | 1.114 ± 1.11e | 0.85 ± 0.05e | 76.51 ± 1.91a | - |
*Data were analyzed for significance with analysis of variance (ANOVA) followed by DMRT test (p = 0.05). Values with different letter indications represent a statistically significant difference; PDI, per cent disease index; PDC, percent disease over control
Discussion
Bacillus is recognized as one of the most dominating and prospective genus as cultivable endophytes in myriads of crops including cereals and vegetable crops [9, 16, 17, 54–58]. Due to their exceptional feature of endospore formation, they can withstand harsh and hostile environment, and this attribute is clearly evident in the endophytic strains of Bacillus recovered from pearl millet, which has inherent capability to grow luxuriantly on unfertile and low nutrient soils. In the present study, eight endophytic bacterial strains displaying excellent antagonistic activity from pearl millet host were assessed on the basis of dual culture assay and presence of AMP genes and further evaluated for their prospective role as a plant growth stimulator. The present investigation also confirms the existence of phenotypic and metabolic assorted population of antagonistic endophytic Bacillus inside pearl millet host. Morphological, biochemical, and molecular identification of endophytic bacterial strains based on 16S rRNA sequence analysis and their inferences clearly demonstrated that pearl millet host contained different strains of B. amyloliquefaciens, B. subtilis subsp. subtilis, and B. cereus. Similarly, analogous reports of endophytic and ubiquitous presence of Bacillus strains in various plant species have been documented [5, 16, 17, 59].
In the current study, majority of the endophytic strains of Bacillus species dwelling inside pearl millet host revealed their antifungal potentialities towards three agriculturally important plant pathogens (RS, SR, and FS). These fungal pathogens are preferred as test organisms for antagonistic confrontation assay due to their wide host range, prolific growth, and ability to cause major economic loss in various crops [60–62]. Further, in India, wilt and root rot caused by FS, is a major disease of pearl millet that significantly reduces production, and causes severe yield losses under congenial environmental conditions [63]. So, microbial management of FS by utilizing endophytic Bacillus strains may offer a potential and viable solution to replace health hazardous and environment-polluting chemicals. Endophytic B. amyloliquefaciens, B. subtilis, and B. cereus strains with antifungal activity towards diverse types of crop pathogens have been well documented [64–67]. The present investigation revealed that endophytic B. amyloliquefaciens (EPP35, EPP 42, EPP62, and EPP 102), B. subtilis subsp. subtilis (EPP65), and B. cereus (EPP5, EPP71, and EPP74) strains are highly promising and proficient antagonists showing ≥ 50% fungal mycelium growth inhibition against all the tested phytopathogenic fungi (RS, SR, and FS). One of the most significant characters of these endophytic bacterial strains reflecting strong antagonistic behavior may be explained by prevalence of structurally different antibiotics (surfactin, bacillomycin, and iturin) with manifold mode of action [14, 21, 26, 68–70]. PCR based screening of AMP gene(s) in the present study clearly reflected the presence srfA, bmyC, and ituD genes in all the endophytic Bacillus strains. These results are in line with other researchers that correlated presence of multiple antifungal peptide genes with antagonistic capabilities of Bacillus strains [20, 22, 25, 26, 43, 71]. Thus, these AMP genes and their secretions by antagonists act as one of the vital components for effective pathogen inhibition. For instance, itu gene has been reported to enhance the permeability of fungal cell membranes by pore formation and reducing the loss of essential macromolecular compounds. Similarly, srf genes(s) facilitated bacterium to produce biofilm synthesis [72]. Several reports indicated that bmy gene(s) cause severe damage to the plasma membranes and alterations to both cell wall and cell membrane of fungal spores and hypha, resulting in cell death [69, 73]. In general, a large number of antifungal peptide-producing microbes of fungal and bacterial nature have been identified as effective biocontrol agents (BCAs) against myriads of fungal crop pathogens [19, 20, 23, 43, 74, 75]. Analogous to earlier reported research findings, the present study also confirmed the role of antifungal peptide-mediated mechanism in the effective inhibition of RS, FS, and SR. Besides, PCR based amplification of AMP biosynthetic genes (srfA, bmyC, and ituD) in all the endophytic strains has established the occurrence of these gene(s) at the molecular level, and reflected homology with earlier reported antibiotic genes in Bacillus strains [43, 76]. Phylogenetic tree based on these three distinct AMP genes also bestow a clear cut connotation of the homology. With these supportive evidences of the previous research findings [43, 77], it seems that AMPs could be responsible for the noteworthy inhibition of fungal plant pathogens in current investigation. However, the exact mechanism of antagonistic capabilities generated by endophytic bacterial strains need to be explored in detail, in light of present clues that hydrolytic enzyme (protease, amylase, lipase, and cellulose) synthesis, siderophore release, and presence of lipopeptide genes (surfA, bmyC, and ituD) in the identified antagonistic strains of endophytic bacteria reveal their competitiveness which can be clubbed with other crop protection and PGP features for improving the plant growth.
Generally, plant growth is drastically disturbed by poor availability of soil nutrients [78]. The inadequate solubilization of mineral nutrients like P, K, Zn, and N resulted in nutrient poor agricultural soil [79, 80]. Four endophytic antagonistic Bacillus strains (EPP5, EPP62, EPP65, and EPP71) identified in the present study were able to transform insoluble form of nutrients (P, Zn, and K) to soluble form as evident from their capabilities to form clear halo zone. These observations are in conformity with prior reports that bacterial strains differ significantly in their level of mineral nutrient (Zn, K, and P) solubilization [80, 81]. Dias et al. [82] also noticed the prominent phosphate solubilizing trait in endophytic strains of Bacillus subtilis and B. megaterium from strawberry host. Further, they also concluded that a transparent and big halo zone formation clearly confirmed their strong solubilization potential of insoluble P, Zn, and K compounds due to pH change. Similarly, in the present study, P, K, and Zn solubilizing strains such as B. amyloliquefaciens EPP62, B. subtilis subsp. subtilis EPP65, B. cereus EPP5, and B. cereus EPP71 have been reported as antagonistic endophytes in pearl millet host. These results are in conformity with previous published research findings of Ramesh et al. [83], Yuan et al. [84], and Nair and Padmavathy [85] that highlighted the K and Zn solubilizing potential of endophytic Bacillus strains. There might be a possibility that seed soaking with endophytic strain of Bacillus can result in rapid introduction, penetration, and colonization of internal sections of root radicals, as observed by Algam et al. [86]. In the present study, it has been observed that strain B. cereus EPP5 remarkably improves plant biomass by escalating nutrient uptake efficiency, releasing plant growth hormones, and hampering fungal mycelium growth by synthesizing siderophores and extracellular fungal cell wall degrading enzymes as reflected by different in vitro experiments mentioned in the current study. Parallel reports regarding the siderophore producing capability of endophytic strains of Bacillus species have been presented by Ginting et al. [87]. Recently, Ribeiro et al. [13] also obtained encouraging results of inoculation of endophytic strains of Bacillus species on plant biomass augmentation and nutrient (N, P, K) availability in the stem. Similarly, in the present study, a significant increment in plant biomass as a consequence of B. cereus EPP5 inoculation has been observed. Thus, the major processes responsible for growth promotion of pearl millet by endophytic strains of Bacillus attributed to the rapid activation of various biochemical and molecular processes during the period of plant- Bacillus interactions, which as a consequences lead to the synthesis of hormones (IAA and siderophores), rapid nutrient solubilization and mobilization (Zn, K, and P), excessive production of extracellular hydrolytic enzymes, and secretion of AMP genes, etc.
Wilt and root rot caused by FS is an atrocious soil-borne fungal disease of pearl millet in India with limited disease management success [63]. The present investigation clearly evidenced the effectiveness of EPP5, EPP62, and EPP65 strains in promoting growth and suppressing FS infection in pearl millet. Positive and significant results of antagonistic and plant growth promoting traits were achieved with these strains when applied singly or in consortium mode during in vivo bio-control experiments. Seeds treated with antagonistic endophytic strains (B. cereus EPP5, B. amyloliquefaciens EPP62, and B. subtilis subsp. subtilis EPP65) effectively inhibit the growth of the FS as well as enhanced the plant biomass including shoot and root length. These research findings are in conformity with previous reports, where significant growth improvement in wheat, rice, pearl millet, sorghum, maize, tomato, and oilseed crops have been documented by inoculating seeds with endophytic bacteria [8, 88–90]. An important observation was that the combined application of B. cereus EPP5 +B. amyloliquefaciens EPP62 + B. subtilis subsp. subtilis EPP65 strains has additive effect relative to single strain treatments on FS suppression. Similarly, Raupach and Kloepper [91] also documented that, consortium of antagonistic strains (B. pumilus + B. subtilis + Curtobacterium flaccumfaciens) improved growth promotion and reduced disease incidence in cucumber in comparison to individual strain application.
In conclusion, the present research identified and characterized endophytic strains of Bacillus species displaying PGP attributes along with strong and wide spectrum antagonistic traits. Further, research findings advocate the potential application of these antagonistic endophytic Bacillus strains in consortium mode (B. cereus EPP5 + B. amyloliquefaciens EPP62 + B. subtilis subsp. subtilis EPP65) in transforming current pearl millet production system as well as health management strategies. Although, the research findings of current study are at beginning stage for developing field effective biocontrol measures, and further detailed field experimentation is required to support the current findings.
Electronic supplementary material
(DOC 763 kb)
Acknowledgments
The authors thanks to Director, ICAR-NBAIM, Maunath Bhanjan (Uttar Pradesh) for providing necessary support for conducting the research work and acknowledge AMITY Institute of Biotechnology, Amity University, Lucknow, India.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Yadav OP, Rai KN. Genetic improvement of pearl millet in India. Agric Res. 2013;2:275–292. [Google Scholar]
- 2.Serba DD, Yadav RS. Genomic tools in pearl millet breeding for drought tolerance: status and prospects. Front Plant Sci. 2016;7:1724. doi: 10.3389/fpls.2016.01724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manjunatha BS, Paul S, Aggarwal C, Rathi M. Effect of osmotic stress on growth and plant growth promoting activities of osmotolerant endophytic bacteria from pearl millet. Environ Ecol. 2018;34:1223–1228. [Google Scholar]
- 4.Abraham A, Philip S, Jacob CK, Jayachandran K. Novel bacterial endophytes from Hevea brasiliensis as biocontrol agent against Phytophthora leaf fall disease. BioControl. 2013;58:675–684. [Google Scholar]
- 5.Santoyo G, Hagelsieb GM, Orozco-Mosqueda MC, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Qin S, Feng WW, Wang TT, Ding P, Xing K, Jiang JH. Plant growth promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil. 2017;416:117–132. [Google Scholar]
- 7.Wang T-T, Ding P, Pan C, Xing K, Bai J-L, Wan W, Jiang J-H, Qin S. Complete genome sequence of endophyte Bacillus flexus KLBMP 4941 reveals its plant growth promotion mechanism and genetic basis for salt tolerance. J Biotechnol. 2017;260:38–41. doi: 10.1016/j.jbiotec.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 8.Kushwaha P, Kashyap PL, Kuppusamy P, Srivastava AK, Tiwari RK (2019) Functional characterization of endophytic bacilli from pearl millet (Pennisetum glaucum) and their possible role in multiple stress tolerance. Plant Biosyst. 10.1080/11263504.2019.1651773
- 9.Gond SK, Bergen MS, Torres MS, White JF., Jr Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol Res. 2015;172:79–87. doi: 10.1016/j.micres.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 10.Slama HB, Cherif-Silini H, Bouket AC, Qader M, Silini A, Yahiaoui B, Alenezi FN, Luptakova L, Triki MA, Vallat A, Oszako O, Rateb Mostafa E, Belbahri L. Screening for Fusarium antagonistic bacteria from contrasting niches designated the endophyte Bacillus halotolerans as plant warden against Fusarium. Front Microbiol. 2019;9:3236. doi: 10.3389/fmicb.2018.03236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borah A, Das R, Mazumdar R, Thakur D. Culturable endophytic bacteria of Camellia species endowed with plant growth promoting characteristics. J Appl Microbiol. 2019;127(3):825–844. doi: 10.1111/jam.14356. [DOI] [PubMed] [Google Scholar]
- 12.Cheffi Manel, Bouket Ali Chenari, Alenezi Faizah N., Luptakova Lenka, Belka Marta, Vallat Armelle, Rateb Mostafa E., Tounsi Slim, Triki Mohamed Ali, Belbahri Lassaad. Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms. 2019;7(9):314. doi: 10.3390/microorganisms7090314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribeiro VP, Marriel IE, deSousa SM, Lana UGP, Mattos BB, deOliveira CA, Gomes EA. Endophytic Bacillus strains enhance pearl millet growth and nutrient uptake under low-P. Braz J Microbiol. 2018;49(1):40–46. doi: 10.1016/j.bjm.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandel SL, Joubert PM, Doty SL. Bacterial endophyte colonization and distribution within plants. Microorganisms. 2017;5:77. doi: 10.3390/microorganisms5040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi G, Zhang XF, Zhao X. Endophytic Bacillus subtilis WH2 containing Pinellia ternate agglutinin showed insecticidal activity against white backed plant hopper Sogatella furcifera. BioControl. 2013;58:233–246. [Google Scholar]
- 16.Lopes R, Tsui S, Gonçalves PJRO, de Queiroz MV. A look into a multifunctional toolbox: endophytic Bacillus species provide broad and underexploited benefits for plants. World J Microbiol Biotechnol. 2018;34(7):94. doi: 10.1007/s11274-018-2479-7. [DOI] [PubMed] [Google Scholar]
- 17.Hassan SE. Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J Adv Res. 2017;8:687–695. doi: 10.1016/j.jare.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol. 2019;10:302. doi: 10.3389/fmicb.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamali H, Sharma A, Prity Kushwaha P, Kashyap PL, Roohi, Srivastava AK. Exploitation of multifarious abiotic stresses, antagonistic activity and plant growth promoting attributes of Bacillus amyloliquefaciens AH53 for sustainable agriculture production. Int J Curr Microbiol App Sci. 2018;7(10):751–763. [Google Scholar]
- 20.Cao Y, Pi H, Chandrangsu P, Li Y, Wang Y, Zhou H, Xiong H, Helmann JD, Cai Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep. 2018;8:4360. doi: 10.1038/s41598-018-22782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solanki MK, Singh RK, Srivastava S, Kumar S, Kashyap PL, Srivastava AK. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctonia solani in tomato. J Basic Microbiol. 2015;55(1):82–90. doi: 10.1002/jobm.201300528. [DOI] [PubMed] [Google Scholar]
- 22.Mora I, Cabrefiga J, Montesinos E. Antimicrobial peptide genes in Bacillus strains from plant environments. Int Microbiol. 2011;14:213–223. doi: 10.2436/20.1501.01.151. [DOI] [PubMed] [Google Scholar]
- 23.Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186:1084–1096. doi: 10.1128/JB.186.4.1084-1096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romero D, de Vicente A, Rakotoal RH, Dufour SE, Veening JW, Arrebola E, Cazorla FM, Kuipers OP, Paquot M, Pérez-García A. The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant-Microbe Interact. 2007;20:430–440. doi: 10.1094/MPMI-20-4-0430. [DOI] [PubMed] [Google Scholar]
- 25.Chung S, Kong H, Buyer JS, Lakshman DK, Lydon J, Kim SD, Roberts DP. Isolation and partial characterization of Bacillus subtilis ME488 for suppression of soilborne pathogens of cucumber and pepper. Appl Microbiol Biotechnol. 2008;80(1):115–123. doi: 10.1007/s00253-008-1520-4. [DOI] [PubMed] [Google Scholar]
- 26.Mora I, Cabrefiga J, Montesinos E. Cyclic Lipopeptide biosynthetic genes and products, and inhibitory activity of plant-associated Bacillus against phytopathogenic bacteria. PLoS One. 2015;10(5):e0127738. doi: 10.1371/journal.pone.0127738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Xu Y, Lai X-H, Shan C, Deng Z, Ji Y. Screening and characterization of endophytic Bacillus and Paenibacillus strains from medicinal plant Lonicera japonica for use as potential plant growth promoters. Braz J Microbiol. 2015;46(4):977–989. doi: 10.1590/S1517-838246420140024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Yingying Z, Yan L, Xuechi F. Screening and characterization of endophytic Bacillus for biocontrol of grapevine downy mildew. Crop Prot. 2017;96:173–179. [Google Scholar]
- 29.Haidar B, Ferdous M, Fatema B, Ferdous AS, Islam MR, Khan H. Population diversity of bacterial endophytes from jute (Corchorus olitorius) and evaluation of their potential role as bioinoculants. Microbiol Res. 2018;208:43–53. doi: 10.1016/j.micres.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Coombs JT, Franco CM. isolation and identification of Actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69:5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma A, Singh P, Kumar S, Kashyap PL, Srivastava AK, Chakdar H, Singh RN, Kaushik R, Saxena AK, Sharma AK. Deciphering diversity of salt-tolerant bacilli from saline soils of eastern Indo-gangetic plains of India. Geomicrobiology. 2015;32:170–180. [Google Scholar]
- 32.Ramnani P, Gupta R. Optimization of medium composition for keratinase production on feather by Bacillus licheniformis RG1 using statistical methods involving response surface methodology. Biotechnol Appl Biochem. 2004;40:191–196. doi: 10.1042/BA20030228. [DOI] [PubMed] [Google Scholar]
- 33.Sharma A, Kashyap PL, Srivastava AK, Bansal YK, Kaushik R. Isolation and characterization of halotolerant bacilli from chickpea (Cicer arietinum L.) rhizosphere for plant growth promotion and biocontrol traits. Eur J Plant Pathol. 2019;153(3):787–800. [Google Scholar]
- 34.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 36.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 37.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 39.Prescott LM, Harley JP, Klein DA. Microbiology. 3. London: WCB Publishers; 1996. Antimicrobial chemotherapy; pp. 657–671. [Google Scholar]
- 40.Soutourina OA, Semenova EA, Parfenova VV, Danchin A, Bertin P. Control of bacterial motility by environmental factors in polarly flagellated and peritrichous bacteria isolated from lake Baikal. Appl Environ Microbiol. 2001;67(9):3852. doi: 10.1128/AEM.67.9.3852-3859.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cappuccino JG, Sherman N. Biochemical activities of microorganisms. In: Harvey AR, Champe PC, editors. Microbiology. California: The Benjamin/Cummings Publishing Co.; 1992. pp. 49–197. [Google Scholar]
- 42.Bakker AW, Schippers B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth reduction. Soil Biol Biochem. 1987;19:452–458. [Google Scholar]
- 43.Solanki MK, Singh RK, Srivastava S, Kumar S, Kashyap PL, Srivastava AK, Arora DK. Isolation and characterization of siderophore producing antagonistic rhizobacteria against Rhizoctonia solani. J Basic Microbiol. 2014;54(6):585–597. doi: 10.1002/jobm.201200564. [DOI] [PubMed] [Google Scholar]
- 44.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 45.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic Acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;7(2):535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavalcante VA, Dobereiner A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. J Plant Soil. 1988;108:23–31. [Google Scholar]
- 47.Hu XF, Chen J, Guo JF. Two phosphate and potassium solubilizing bacterial isolated from Tiannu Mountain, Zhijiang, China. World J Microbiol Biotechnol. 2006;22:983–990. [Google Scholar]
- 48.Pikovskaya RL. Mobilzation of phosphorus in soil. In: Connection with vital activity of some microbial species. Microbiologiya. 1948;17:362–370. [Google Scholar]
- 49.Claus H, Filip Z. Behaviour of phenoloxidases in the presence of clays and other soil-related adsorbents. Appl Microbiol Biotechnol. 1988;28(4):506–511. [Google Scholar]
- 50.Teather RM, Wood PJ. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vermelho AB, Meirelles MNL, Lopes A, Petinate SDG, Chaia AA, Branquinha MH. Detection of extracellular proteases from microorganisms on agar plates. Mem Inst Oswaldo Cruz. 1996;91(6):755–760. doi: 10.1590/s0074-02761996000600020. [DOI] [PubMed] [Google Scholar]
- 52.Lusty CJ, Doudoroff M. Poly-β-Hydroxybutyrate depolymerases of Pseudomonas Lemoignei. Proc Natl Acad Sci U S A. 1966;56:960–965. doi: 10.1073/pnas.56.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amira MB, Lopez D, Mohamed AT, Khouaja A, Chaar H, Fumanal B, Gousset-Dupont A, Bonhomme L, Label P, Goupil P, Ribeiro S, Pujade-Renaud V, Julien JL, Auguin D, Venisse JS. Beneficial effect of Trichoderma harzianum strain Ths97 in biocontrolling Fusarium solani causal agent of root rot disease in olive trees. Biol Control. 2017;110:70–78. [Google Scholar]
- 54.Chandrashekhara, Niranjanraj S, Deepak SA, Amruthesh KN, Shetty NP, Shetty HS. Endophytic bacteria from different plant origin enhance growth and induce downy mildew resistance in pearl millet. Asian J Plant Pathol. 2007;1:1–11. [Google Scholar]
- 55.Romero FM, Marina M, Pieckenstain FL. The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiol Lett. 2014;351:187–194. doi: 10.1111/1574-6968.12377. [DOI] [PubMed] [Google Scholar]
- 56.Bai Y, Zhou X, Smith DL. Enhanced soybean plant growth resulting from coinoculation of Bacillus strains with Bradyrhizobium japonicum. Crop Sci. 2003;43:1774–1781. [Google Scholar]
- 57.Lin L, Qiao YS, Ju ZY, Ma CW, Liu YH, Zhou YJ, Dong HS. Isolation and characterization of endophytic Bacillus subtilis Jaas ed1 antagonist of eggplant Verticillium wilt. Biosci Biotechnol Biochem. 2009;73(7):1489–1493. doi: 10.1271/bbb.80812. [DOI] [PubMed] [Google Scholar]
- 58.Okunishi S, Sako K, Mano H, Imamura, Morisaki H. Bacterial flora of endophytes in the maturing seeds of cultivated rice (Oryza sativa) Microbes Environ. 2005;20:168–177. [Google Scholar]
- 59.Panchal H, Ingle S. Isolation and characterization of endophytes from the root of the medicinal plant Chlorophytum borivilianum (Safed musli) J Adv Dev Res. 2011;2:205–209. [Google Scholar]
- 60.Goswami SK, Singh V, Kashyap PL. Population genetic structure of Rhizoctonia solani AG1IA from rice field in North India. Phytoparasitica. 2017;45(3):299–316. [Google Scholar]
- 61.Elisabeth ZP, Paco S, Vibeke L, Philippe S, Irenee S, Adama N. Importance of seed-borne fungi of sorghum and pearl millet in burkina faso and their control using plant extracts. Pak J Biol Sci. 2008;11:321–331. doi: 10.3923/pjbs.2008.321.331. [DOI] [PubMed] [Google Scholar]
- 62.Pleban S, Ingel F, Chet I. Control of Rhizoctonia solani and Sclerotium rolfsii in the greenhouse using endophytic Bacillus spp. Eur J Plant Pathol. 1995;101(6):665–672. [Google Scholar]
- 63.Prasad G, Kumar V, Dwivedi SK. Antifungal activity of some selected medicinal plants against Fusarium solani causing wilt and rot in Pearl millet. Asian J Bio Sci. 2018;13(1):21–27. [Google Scholar]
- 64.Vinayarani G, Prakash HS. Growth promoting rhizospheric and endophytic bacteria from Curcuma longa L. as biocontrol agents against rhizome rot and leaf blight diseases. Plant Pathol J. 2018;34(3):218–235. doi: 10.5423/PPJ.OA.11.2017.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao T, Foulston L, Chai Y, Wang Q, Losick R. Alternative modes of biofilm formation by plant-associated Bacillus cereus. Microbiologyopen. 2015;4(3):452–464. doi: 10.1002/mbo3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu HJ, Chen YL, Wang YF, Tang YY, Chen SL, Yan SZ. Endophytic Bacillus cereus effectively controls Meloidogyne incognita on tomato plants through rapid rhizosphere occupation and repellent action. Plant Dis. 2017;101(3):448–455. doi: 10.1094/PDIS-06-16-0871-RE. [DOI] [PubMed] [Google Scholar]
- 67.Ahmad Z, Wu J, Chen L, Dong W. Isolated Bacillus subtilis strain 330-2 and its antagonistic genes identified by the removing PCR. Sci Rep. 2017;7:1777. doi: 10.1038/s41598-017-01940-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ek-Ramos MJ, Gomez-Flores R, Orozco-Flores AA, Rodríguez-Padilla C, González-Ochoa GP. Bioactive products from plant-endophytic gram-positive bacteria. Front Microbiol. 2019;10:463. doi: 10.3389/fmicb.2019.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shafi J, Tian H, Ji M. Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotechnol Equip. 2017;31:446–459. [Google Scholar]
- 70.Singh RK, Kumar DP, Singh P, Solanki MK, Srivastava S, Kashyap PL, Kumar S, Srivastava AK, Singhal PK, Arora DK. Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. J Plant Growth Regul. 2014;73(1):91–101. [Google Scholar]
- 71.Hazarika DJ, Goswami G, Gautom T, Parveen A, Das P, Barooah M, Boro RC. Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 2019;19:71. doi: 10.1186/s12866-019-1440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim PI, Ryu J, Kim YH, Chi YT. Production of biosurfactant lipopeptides iturin A, fengycin and surfactin A from Bacillus subtilis CMB32 for control of Colletotrichum gloeosporioides. J Microbiol Biotechnol. 2010;20:138–145. [PubMed] [Google Scholar]
- 73.Toral L, Rodríguez M, Béjar V, Sampedro I. Antifungal activity of lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea. Front Microbiol. 2018;9:1315. doi: 10.3389/fmicb.2018.01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raaijmakers JM, de Bruijn I, Nybroe O, Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiol Rev. 2010;34:1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 75.Arguelles-Arias A, Ongena M, Halimi B, Lara Y. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb Cell Factories. 2009;8:63–74. doi: 10.1186/1475-2859-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vinodkumar S, Nakkeeran S, Renukadevi P, Malathi VG. Biocontrol potentials of antimicrobial peptide producing Bacillus species: multifaceted antagonists for the management of stem rot of carnation caused by Sclerotinia sclerotiorum. Front Microbiol. 2017;8:446. doi: 10.3389/fmicb.2017.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jasim B, Sreelakshmi S, Mathew J, Radhakrishnan EK (2016) Identification of endophytic Bacillus mojavensis with highly specialized broad spectrum antibacterial activity. 3 Biotech 6:187 [DOI] [PMC free article] [PubMed]
- 78.Jasrotia P, Kashyap PL, Bhardwaj AK, Kumar S, Singh GP. Scope and applications of nanotechnology for wheat production: a review of recent advances. Wheat Barley Res. 2018;10(1):1–14. [Google Scholar]
- 79.Vaid SK, Kumar B, Sharma A, Shukla AK, Srivastava PC. Effect of zinc solubilizing bacteria on growth promotion and zinc nutrition of rice. J Soil Sci Plant Nutr. 2014;14(4):889–910. [Google Scholar]
- 80.Shree N, Kashyap PL, Chakdar H, Srivastava AK, Sharma AK. Isolation and characterization of potassium solubilizing bacteria from forest soils of Meghalaya. Indian J Environ Sci. 2015;19:43–48. [Google Scholar]
- 81.Dinesh R, Srinivasan V, Hamza S, Sarathambal C, Anke Gowda SJ, Ganeshamurthy AN, Gupta SB, Aparna Nair V, Subila KP, Lijina A, Divya VC. Isolation and characterization of potential Zn solubilizing bacteria from soil and its effects on soil Zn release rates, soil available Zn and plant Zn content. Geoderma. 2018;321:173–186. [Google Scholar]
- 82.Dias ACF, Costa FEC, Andreote FD, Lacava PT, Teixeira MA, Assumpção LC, Araújo WL, Azevedo JL, Melo IS. Isolation of micropropagated strawberry endophytic bacteria and assessment of their potential for plant growth promotion. World J Microbiol Biotechnol. 2009;25:189–195. [Google Scholar]
- 83.Ramesh A, Sharma SK, Sharma MP, Yadav N, Joshi OP. Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in vertisols of central India. Appl Soil Ecol. 2014;73:87–96. [Google Scholar]
- 84.Yuan ZS, Liu F, Zhang GF. Characteristics and biodiversity of endophytic phosphorus- and potassium-solubilizing bacteria in Moso Bamboo (Phyllostachys edulis) Acta Biol Hung. 2015;66(4):449–459. doi: 10.1556/018.66.2015.4.9. [DOI] [PubMed] [Google Scholar]
- 85.Nair D, Padmavathy S (2014) Impact of endophytic microorganisms on plants environment and humans. Sci World J:250693 [DOI] [PMC free article] [PubMed]
- 86.Algam SA, Guan-lin X, Coosemans J. Delivery Methods for introducing endophytic Bacillus into tomato and their effect on growth promotion and suppression of tomato wilt. Plant Pathol J. 2005;4:69–74. [Google Scholar]
- 87.Ginting RCB, Sukarno N, Widyastuti U, Darusman LK, Kanaya S. Diversity of endophytic fungi from red ginger (Zingiber officinale Rosc.) plant and their inhibitory effect to Fusarium oxysporum plant pathogenic fungi. Hayati J Biosci. 2013;20:127–137. [Google Scholar]
- 88.Prabhukarthikeyan R, Saravanakumar D, Raguchander T. Combination of endophytic Bacillus and Beauveria for the management of Fusarium wilt and fruit borer in tomato. Pest Manag Sci. 2013;70(11):1742–1750. doi: 10.1002/ps.3719. [DOI] [PubMed] [Google Scholar]
- 89.Nejad P, Johnson PA. Endophytic bacteria induce growth promotion and wilt disease suppression in oilseed rape and tomato. Biol Control. 2000;18:208–215. [Google Scholar]
- 90.Gutiérrez-Zamora ML, Martínez-Romero E. Natural endophytic association between Rhizobium etli and maize (Zea mays L.) J Biotechnol. 2001;91:117–126. doi: 10.1016/s0168-1656(01)00332-7. [DOI] [PubMed] [Google Scholar]
- 91.Raupach GS, Kloepper JW. Mixtures of plant growth promoting rhizobacteria enhance biological control of multiple cucumber pathogens. Phytopathology. 1998;88:1158–1164. doi: 10.1094/PHYTO.1998.88.11.1158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 763 kb)