Abstract
Characterization of methicillin-resistant Staphylococcus (MRS) is a continuous challenge at diagnostic laboratories. The phenotypic methods present heterogeneous results and the occurrence of variants of mecA gene turned this goal even more challenging to achieve. The present study provided an accurate and highly discriminatory screening tool for MRS, improving its detection.
Keywords: mecA genes, mec variants, Methicillin-resistant Staphylococcus, Antimicrobial resistance
Methicillin-resistant Staphylococcus (MRS) spp. are important human pathogens that are also a concern in veterinary medicine and animal agriculture. The Staphylococcus species are present in a wide range of animal species, including dogs, cats, rabbits, horses, cattle, pigs, poultry, and exotic species, both in healthy carriers and as a cause of infection [1–3]. Besides the broad host range distribution and pathogenicity, its significant antimicrobial resistance levels are of great concern [4]. The high antimicrobial resistance level to beta-lactams favors treatment failures and its persistence in the environment. Bacterial resistance mechanisms to this antimicrobial class include a low-affinity penicillin-binding protein 2a (PBP2a) determined by the expression of mecA gene [5].
The phenotypic methicillin-resistant expression does not depend only on the mecA gene. This expression is under a more complex control and is only beginning to be better understood since it is expressed in a peculiar and heterogeneous way [6]. Because of this phenotypic heterogeneity, detection of the mecA gene is considered the gold standard method for confirmation of methicillin-resistant isolates by Clinical and Laboratory Standards Institute [7, 8]. However, for samples of animal origin, this proposition is not reliable, since variants of the mec gene impair this detection [9].
In the UK, methicillin-resistant S. aureus (MRSA) strains carrying a novel mecA gene, firstly named mecALGA251, and mecC have been identified in cattle. These strains were only 70% identical at the nucleotide level to the classical mecA gene and so escaped detection by routine PCR assays [10]. MRSA carrying mecC gene have been reported from 13 European countries and have been isolated from 14 different host species [11]. Recently, the occurrence of mecC gene in coagulase-negative staphylococci (CoNS) isolated from various wild and domestic animals was reported [12].
The discovery of this homolog of mecA gene emphasizes the possibility of the circulation of variants of this gene in the animal production environment and the consequent acquisition by other species of Staphylococcus leading to the emergence of new methicillin-resistant strains [13]. Even though there are several reports of the occurrence of mecC gene in Staphylococcus species from human and animals, the puzzling question is that they are all restricted to European countries.
Our previous studies [14, 15] reported several phenotypic methicillin-resistant Staphylococcus spp. isolates, but none tested positive for the mecA gene. Otherwise, we detected a mecA gene variant from bovine samples [9]. This variant contained mutations in the annealing region that does not allow detection of the gene with the already described primers. These results justified the absence of a correlation of beta-lactam resistance in phenotypic and genotypic methods observed in these isolates. Another controversial aspect that should be considered when analyzing methicillin resistance is the phenotypic marker used to predict this resistance. Depending on the Staphylococcus species involved, a specific marker, oxacillin or cefoxitin, and a specific method must be used to determine resistance to the beta-lactam class [7, 8]. The present study aimed to develop and to validate a universal PCR primer set to ensure adequate detection of the mec genes (classical and variant), to investigate the presence of mecC gene, and also to compare phenotypic assays to screen MRS.
A total of 563 Staphylococcus spp. were isolated from different origins, such as livestock (pigs (84), sheep (11), cattle (19), and lamb (1)), processed meat (ham (4), turkey breast (4), pork (23), beef (31), and chicken (11)), companion animals (horse (1) and cat (1)), human (hospital patients (99) and nasal (26)) from the USA collected in 2011 until 2015, and 248 isolates from Brazil, 173 from bovine intramammary infection, and 75 from milking environment, collected in 2014–2015. All isolates were previously identified at the species level by either phenotypic assays and PCR of the genes (16S rRNA for Staphylococcus spp.); nuc for Staphylococcus species) and confirmed by MALDI-TOF. The prevalent species analyzed was S. aureus (67.1%), followed by CoNS (24.9%), and other coagulase-positive Staphylococcus (8.0%).
All S. aureus were screened for detection of phenotypic methicillin resistance in oxacillin salt agar, cefoxitin disk diffusion, and microdilution broth method. For S. lugdunensis, cefoxitin disk diffusion and microdilution broth method were used. Methicillin-resistant CoNS were detected by cefoxitin disk diffusion, except for S. pseudintermedius and S. schleiferi, for which the oxacillin disk was used, according to standardized protocols [7, 8].
A pair of universal primers was designed for the detection of classical and variant mecA. Sequences of mec gene of Staphylococcus spp. from different origins (KF058904, KF058903, KF058901, KF058902, KF058900, KF058905, KF058906, KF058907) were aligned using the ClustalW sequence alignment tool. The resultant alignment, based on the highly conserved regions of the mec gene, was used to design the universal primers (Fig. 1). Universal primer was validated by in silico analysis using the software tool Primer-Blast [16].
Fig. 1.
Alignment of the mec genes sequences (KF058900, KF058901, KF058902 from variant mecA, and KF058903, KF058904, KF058905, KF058906, KF058907 from classical mecA). Gray areas represent the region of forward and reverse primers annealing
The DNA extraction was performed as previously described [17]. All isolates were submitted to PCR assays to detect mec genes using the primers listed in Table 1. Staphylococcus aureus ATCC® 43300™ (methicillin resistant and mecA positive), Staphylococcus aureus ATCC® 29213™ (methicillin susceptible), and Staphylococcus aureus BAA 2312 (methicillin resistant and mecC positive) were used as reference strains.
Table 1.
Sequence of primers used for mec gene amplification in this study
| Primers | Sequence (5′–3′) | PCR product | Program/references |
|---|---|---|---|
| Universal (classical and variant) |
ACG TTA CAA GAT ATG AAG ACA TTA ATA GCC ATC ATC |
574 bp | 94 °C 5 min (94 °C 1 min, 55 °C 1 min and 72 °C 1 min) × 30 cycles and 72 °C 10 min (developed in this study). |
| variant mecA |
CAG GCA TGC AGA AAA ATC AA TTG AGT CGA ACC AGG TGA TG |
809 bp | 95 °C 5 min (94 °C 1 min, 55 °C 1 min, 72 °C 1 min) × 30 cycles and 72 °C 10 min [9]. |
| Classical mecA |
AAA ATC GAT GGT AAA GGT TGG C AGT TCT GCA GTA CCG GAT TTG C |
533 bp | (94 °C 30 s, 55 °C 30 s, 72 °C 1 min) × 40 cycles and 72 °C 5 min [18]. |
| mecC | GAA AAA AAG GCT TAG AAC GCC TC | 718 bp | 94 °C 15 min (94 °C 30 s, 50 °C 1 min, 72 °C 1 min) × 35 cycles and 10 min at 72 °C [19] |
| CCT GAA TC[W] GCT AAT AAT ATT TC |
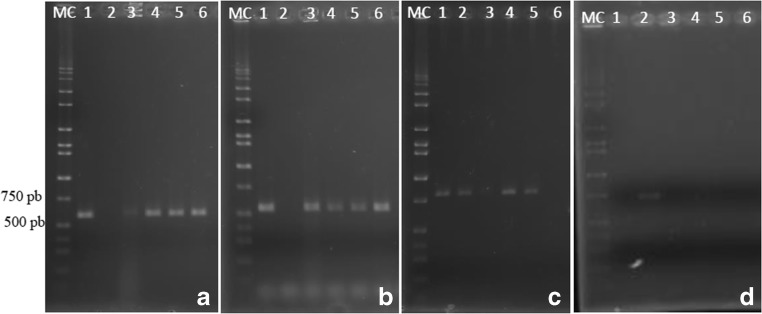
Among the 563 Staphylococcus spp. isolates tested, 220 (39.1%) were confirmed as MRS by amplification using classical, variant, and universal primers. None of the isolates tested positive for the mecC gene. Figure 2 shows the amplification pattern using the primers described to mec gene detection.
Fig. 2.
Amplification pattern of mec gene. (A) Universal (574 bp), (1) positive control, (2) blank, (3–6) positive strains. (B) Classical mecA (533 bp), (1) positive control, (2) blank, (3–6) positive strains. (C) Variant mecA (809 bp), (1) positive control, (2–5) positive strains, (6) blank. (D)mecC (718 bp), (1) blank, (2) positive control, (3–5) negative strains. MC, ladder (Hi-Lo DNA marker—10,000 bp)
The classical mecA gene was detected in 91.4% (201/220), being 177 S. aureus from human clinical (95), meat pork (6), and nasal pig (76), and 24 CoNS from meat beef (1), deli meat ham (1), deli meat turkey (3), deli meat pork (15), horse nasal (1), milker’s hands (1), milk line (1), and human nose (1). The variant mecA was detected in 6.4% of isolates (14/220), being 2 S. aureus isolates (meat pork and a bovine nasal sample), and 12 CoNS isolates from meat beef (1), milk (9), milker’s hands (1), and meat chicken (1). These results reinforce that the variant mecA is widespread in the animal environment. A strain of S. xylosus isolated from a porcine nasal swab carried both mec genes (classical and variant). Further investigation is required to understand the regulation and expression of these genes in these isolates.
The developed universal primer set successfully amplified mec genes in 205 isolates (93.2%), even four isolates did not amplify any classical or variant mecA using conventional primers. These isolates were identified as one S. saprophyticus (from pork meat) and three S. sciuri (milk (2) and milker’s hands (1)). It presented sensitivity, specificity, positive predictive, and negative predictive values of 93.1%, 98.8%, 98%, and 95.8% when using the classical and variant mec gene detection as the gold standard (Table 2). Also, it presented a higher discriminatory power once four isolates just amplified mec genes using this primer set. The occurrence of deletions, insertions, or point mutations could alter the mec gene sequence impairing its detection [9, 20], so the development of this new tool has provided a fast and reliable result, reducing costs and optimizing the diagnostic at routine laboratories.
Table 2.
Percent of sensitivity, specificity, positive predictive value, and negative predictive value observed
| Test result (universal primer) | mec gene (classical and variant) | Total | Predictive value | |
|---|---|---|---|---|
| + | - | |||
| + | 201 | 4 | 205 | PPV = 98% |
| - | 15 | 343 | 358 | NPV = 95.8% |
| Total | 216 | 347 | ||
| S = 93.1% | Sp = 98.8% | |||
S, sensitivity; Sp, specificity; PPV, positive predictive value; NPV, negative predictive value; +, positive; -, negative
The phenotypic prediction of mec-positive strains was performed according to standardized protocols [7, 8] for the different Staphylococcus species (see Table 3). Staphylococcus aureus positive for either classical or variant mecA presented the highest pheno-genotypical correlation using cefoxitin disk diffusion (98.9% (175/177)). Our results match those of several previous reports that found that cefoxitin is superior to oxacillin in detecting low-level heterogeneous methicillin-resistant Staphylococcus populations and could be an alternative to PCR assays in diagnostic routine [21–23].
Table 3.
Results of mec gene detection and phenotypic methods for detection of methicillin resistance in staphylococci examined in this study
| Genotypics analyses/no. of isolates | Phenotypic resistance results by species | |
|---|---|---|
| mec gene | Universal primer/species | |
| classical mecA (201) | (+) 193 (170 S. aureus; 23 CoNS) |
S. aureus (n = 177) Cefoxitin disk diffusion 97.7% (173/177) Cefoxitin broth microdilution 88.7% (157/177) Oxacillin salt agar with 6 mg/mL 72.3% (128/177) |
| (-) 8 (7 S. aureus; 1 CoNS) |
CoNS (n = 24) Cefoxitin disk diffusion 87.5% (21/24) |
|
| variant mecA (14) | (+) 7 (1 S. aureus; 6 CoNS) |
S. aureus (n = 2) Cefoxitin disk diffusion 100% (2/2) Cefoxitin broth microdilution 50% (1/2) Oxacillin salt agar with 6 mg/mL—no resistance was detected |
| (-) 7 (1 S. aureus; 6 CoNS) |
CoNS (n = 12) Cefoxitin disk diffusion 16.7% (2/12) |
|
| N.A | (+) 4* (CoNS = 4) |
CoNS (n = 4) Cefoxitin disk diffusion 50% (2/4) |
| Classical and variant mecA (1) | (+) 1 (CoNS = 1) |
CoNS (n = 1) Cefoxitin disk diffusion 100% (1/1) |
N.A, not amplified; (+), number of isolates that amplified using universal primer; (-), number of isolates that not amplified using universal primer. *Isolates that only amplified using universal primer
The phenotypic resistance assays presented significant different results for the CoNS isolates. The cefoxitin disk diffusion test yielded 87.5% resistance to the classical mecA-positive isolates, while just 8.3% resistance for the variant mecA positive. Most susceptible variant mecA CoNS was isolated from milk line samples and identified as S. sciuri, except for an S. epidermidis strain, isolated from chicken meat. Previous studies related hetero-resistance in the phenotype of methicillin-resistant isolates [3, 6, 24].
The twelve variant mecA isolates were submitted to complementary tests to understand if the observed heterogeneity was related with a peculiar resistance expression codified by this variant. The cefoxitin broth microdilution yielded 50% (6/12) resistance, the oxacillin disk diffusion yielded 25% (3/12), and the oxacillin salt agar with 6 μg/mL test yielded 9.1% (1/12). Additional studies on the use of cefoxitin or oxacillin to improve the accuracy in detection of methicillin resistance mediated by variant mecA will be necessary.
The cefoxitin disk diffusion yielded a 50% level of resistance for the four CoNS isolates that just amplified the mec gene by universal primer. The S. xylosus strain that amplified simultaneously classical and variant mecA was phenotypically resistant to cefoxitin.
The development of tools to improve MRS diagnosis is crucial for its accurate and rapid identification. The universal primer set developed in this study is an excellent option when screening for mec-positive isolates. Further, the development of methods using appropriate phenotypic markers according to Staphylococcus species is necessary for accurate detection of methicillin resistance.
Acknowledgments
We thank Dr. C. M. Logue and Dr. L.K. Nolan for supporting this study at Veterinary Microbiology Research Institute, Iowa State University.
Funding information
Foundation for Research Support of the State of Rio de Janeiro, scholarship protocol number E26/200.101/2016, and National Council for Scientific and Technological Development supported this work.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weese JS. Methicillin resistance Staphylococcus aureus in animals. ILAR J. 2010;51:233–244. doi: 10.1093/ilar.51.3.233. [DOI] [PubMed] [Google Scholar]
- 2.Calazans-Silva AC, Medeiros PTC, Melo DA, Carvalho BO, Coelho IS, Coelho SMO, Souza MMS. Short communication: genetic analysis of mecA gene and detection of homologue pbpD in Stahylococcus sciuri group. Braz J Microbiol. 2014;45:651–655. doi: 10.1590/S1517-83822014000200038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melo DA, Motta CC, Rojas ACCM, Soares BS, Coelho IS, Coelho SMO, Souza MMS. Characterization of coagulase-negative staphylococci and pheno-genotypic beta lactam resistance evaluation in samples from bovine intramammary infection. Arq Bras Med Vet Zootec. 2018;70:368–374. doi: 10.1590/1678-4162-9209. [DOI] [Google Scholar]
- 4.Pantosti A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol. 2012;3:127. doi: 10.3389/fmicb.2012.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fishovitz J, Hermoso JA, Chang M, Mobashery S. Penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. IUBMB Life. 2014;66:572–577. doi: 10.1002/iub.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aedo S, Tomasz A. The role of stringent stress response – in the antibiotic resistant phenotype of MRSA. Antimicrob Agents Chemother. 2016;60:2311–2317. doi: 10.1128/AAC.02697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing. 28. Wayne: CLSI; 2018. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. 5. Wayne: CLSI; 2018. [Google Scholar]
- 9.Melo DA, Coelho IS, Motta CC, Rojas ACCM, Dubenczuk FC, Coelho SMO, Souza MMS. Impairments of mecA gene detection in bovine Staphylococcus spp. Braz J Microbiol. 2014;45:1075–1082. doi: 10.1590/S1517-83822014000300041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhil J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paterson GK, Morgan FJE, Harrison EM, Cartwright EJP, TöRöK ME, Zadoks RN, Parkhill J, Peacock SJ, Holmes MA. The emergence of mecC methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2014;22:42–47. doi: 10.1016/j.tim.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loncaric I, Kübber-Heiss A, Posautz A, Ruppitsch W, Lepuschitz S, Schauer B, Feßler AT, Krametter-Frötscher R, Harrison EM, Holmes MA, Künzel F, Szostak MP, Hauschild T, Desvars-Larrive A, Misic D, Rosengarten R, Walzer C, Slickers P, Monecke S, Ehricht R, Schwarz S, Spergser J. Characterization of mecC gene-carrying coagulase-negative Staphylococcus spp. isolated from various animals. Vet Microbiol. 2019;230:138–144. doi: 10.1016/j.vetmic.2019.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald JR. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 2010;20:192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Soares LC, Pereira IA, Pribul BR, Oliva MS, Coelho SMO, Souza MMS. Antimicrobial resistance and detection of mecA and blaZ genes in coagulase-negative Staphylococcus isolated from bovine mastitis. Pesqui Vet Bras. 2012;32:692–696. doi: 10.1590/S0100-736X2012000800002. [DOI] [Google Scholar]
- 15.Mendonça ECL, Marques VF, Melo DA, Alencar TA, Coelho IS, Coelho SMO, Souza MMS. Caracterização fenogenotípica da resistência antimicrobiana em Staphylococcus spp. isolados de mastite bovina. Pesqui Vet Bras. 2012;31:859–864. doi: 10.1590/S0100-736X2012000900008. [DOI] [Google Scholar]
- 16.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velasco V, Buyukcangaz E, Sherwood JS, Stepan RM, Koslofsky RJ, Logue CM. Characterization of Staphylococcus aureus from humans and a Comparison with isolates of animal origin, in North Dakota, United States. PLoS One. 2015;10(10):e0140497. doi: 10.1371/journal.pone.0140497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami KW, Minamide K, Wada W, Nakamura E, Teraoka H, Watanbe S. Identification of methicillin resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/JCM.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steeger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Shov R, Larsen AR. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecALGA251. Clin Microbiol Infect. 2012;18:395–400. doi: 10.1111/j.1469-0691.2011.03715.x. [DOI] [PubMed] [Google Scholar]
- 20.Sakoulas G, Gold HS, Venkataraman L, Degirolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol. 2001;39:3946–3951. doi: 10.1128/JCM.39.11.3946-3951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain A, Agarwal A, Verma RK. Cefoxitin disc diffusion test for detection of methicillin-resistant staphylococci. J Med Microbiol. 2008;57:957–961. doi: 10.1099/jmm.0.47152-0. [DOI] [PubMed] [Google Scholar]
- 22.Broekman NM, Van TT, Monson TA, Marshall SA, Warshauer DM. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus. J Clin Microbiol. 2009;47:217–219. doi: 10.1128/JCM.01506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews AA, Thomas M, Appalaraju B, Jayalakshmi J. Evaluation and comparison of tests to detect methicillin resistant S. aureus. Indian J Pathol Microbiol. 2010;53:79–82. doi: 10.4103/0377-4929.59189. [DOI] [PubMed] [Google Scholar]
- 24.Kim C, Mwangi M, Chung M, Milheiriço C, de Lencastre H, Tomasz A. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One. 2013;8:1–10. doi: 10.1371/annotation/c8b2e360-b78a-4c2f-a1a3-c53325f18211. [DOI] [PMC free article] [PubMed] [Google Scholar]