Abstract
Preterm birth remains the main contributor to early childhood mortality. The vaginal environment, including microbiota composition, might contribute to the risk of preterm delivery. Alterations in the vaginal microbial community structure might represent a risk factor for preterm birth. Here, we aimed to (a) investigate the association between preterm birth and the vaginal microbial community and (b) identify microbial biomarkers for risk of preterm birth. Microbial DNA was isolated from vaginal swabs in a cohort of 69 women enrolled at hospital admission for their delivery. Microbiota was analyzed by high-throughput 16S rRNA sequencing. While no differences in microbial diversity measures appeared associated with the spontaneous preterm and full-term outcomes, the microbial composition was distinct for these groups. Differential abundance analysis showed Lactobacillus species to be associated with full-term birth whereas an unknown Prevotella species was more abundant in the spontaneous preterm group. Although we studied a very miscegenated population from Brazil, our findings were similar to evidence pointed by other studies in different countries. The role of Lactobacillus species as a protector in the vaginal microbiome is demonstrated to be also a protector of spontaneous preterm outcome whereas the presence of pathogenic species, such as Prevotella spp., is endorsed as a factor of risk for spontaneous preterm delivery.
Keywords: Next-generation sequencing, Vaginal microbiome, Lactobacillus, Brazilian microbiome
Introduction
According to the World Health Organization, every birth before 37 weeks of pregnancy is considered preterm. Each year, about 15 million babies are born prematurely in the world [1]. Prematurity is the leading cause of mortality before 4 weeks of life and the second until 5 years of age [1, 2]. Preterm birth also leads to disorders related to brain development, the deficit of attention, hyperactivity [3–5], autism [6], and respiratory problems [7]. In the USA, the preterm delivery rate is around 9.6% [8] while in Europe and other developed countries it is between 5 and 9% [9]. In developing countries, especially South Asia and sub-Saharan Africa, the preterm birth rates are above 15% [1].
The epidemiological and clinical natures of preterm birth are not yet fully understood [2, 10, 11]. Nevertheless, preterm delivery is associated with type 2 diabetes [12], weight gain, chronic postpartum hypertension [13], air pollution [14], psychological and social conditions, physical exertion during pregnancy [15], diet, hygiene, and access to health care [2, 11]. Several studies attempted to map the endemicity of this disease and their results indicate a higher incidence in black women, in women under low socioeconomic levels, in smokers, in pregnancies of twins, and/or more advanced age [1, 2, 11, 16–20].
Different microbes also have been correlated with preterm delivery [21, 22], but microbial community-level studies represent a suitable and fast alternative to better understand the relationship between the microbial community and the preterm birth. As women from different ethnic backgrounds have different vaginal microbial communities [23–25], local attempts to detect and associate microbial communities with preterm birth are required [26]. Such regional attempts might sum up with other worldwide initiatives to elaborate a prediction risk assessment plan based on the vaginal microbial community. Within this work we aimed to (a) investigate the association between preterm birth and the vaginal microbial community and (b) identify microbial biomarkers for risk of preterm birth.
Material and methods
Experimental design
This study was carried out with samples collected from women attending the Hospital de Clínicas de Porto Alegre (HCPA). Experimentation used a convenience sampling strategy. Expecting mothers were enrolled at hospital admission for their delivery between May 2014 and March 2016. All women provided written informed consent to allow their samples to be used in the study. The ethics committee of HCPA approved the study protocol. Exclusion criteria were: (1) HIV or congenital infections,( 2) drug user or alcoholic, (3) urinary tract infections or (4) antibiotic usage in the third trimester of gestation, (5) urogenital infection in the last 3 months, and (6) gestational diabetes.
Sixty-nine pregnant women were analyzed in this study. Twenty-three of them had spontaneous preterm labor (before 33 weeks of gestation), whereas 29 had spontaneous term labor. Another 17 women had non-spontaneous labor but, due to medical reasons affecting pregnancy, had cesarean delivery before 33 gestational weeks. Those subjects called hereinafter “non-spontaneous preterm” group, were used as a second control because they present a microbial community that might not be associated with spontaneous preterm delivery but have a better match in terms of gestational age with the spontaneous preterm group. All pregnant women sampled on this work had vaginal swab (Sterile Specimen Collection Swabs to collect specimens from soft tissue surfaces-Labor swab®) collected up to 4 hours before labor begins, as described by Roesch and colleagues [27]. Collected swab samples were stored at − 80 °C until DNA extraction. The characteristics from the mothers enrolled in this experiment include maternal age, previous pregnancies, gestational age, incidence of chorioamnionitis, preeclampsia, infection by Group B Streptococcus, intrapartum penicillin administration, and delivery mode.
Microbial DNA extraction, 16S rRNA amplification and sequencing, and data processing
Microbial DNA was extracted from frozen swab samples as previously described by Roesch et al. [27]. All DNA samples were kept at − 20 °C until use in PCR reactions. Vaginal microbiota was determined by amplification of the V4 region of the 16S rRNA gene and downstream sequencing on the Ion PGM Platform (Thermo Fisher Scientific, Waltham, MA, USA) with the bacterial/archaeal primers 515F and 806R [28]. All samples were PCR-amplified using barcoded primers linked with the Ion adapter “A” sequence (5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′) and Ion adapter “P1” sequence (5′-CCTCTCTATGGGCAGTCGGTGAT-3′) to obtain a sequence of primer composed by A-barcode-806R and P1-515F adapter and primers. PCRs were carried out in 25 μL reactions contained 2 U of Platinum® Taq DNA High Fidelity Polymerase (Invitrogen, Carlsbad, CA, USA), 4 μL 10X High Fidelity PCR Buffer, 2 mM MgSO4, 0.2 mM dNTP’s, 0.1 μM of both the 806R barcoded primer and the 515F primer, 25 μg of Ultrapure BSA (Invitrogen, Carlsbad, CA, United States) and approximately 50 ng of DNA template. PCR conditions used were: 95 °C for 5 min; 30 cycles of 94 °C per 45 s denaturation, 56 °C per 45 s annealing, and 72 °C per 1 min extension; followed by 72 °C per 10 min for final extension. Fragments presenting around 400 base pairs from resulting PCR products were purified with the Agencourt® AMPure® XP Reagent (Beckman Coulter, Brea, CA, USA), and final concentration of the PCR products was quantified by using the Qubit Fluorometer kit (Invitrogen, Carlsbad, CA, United States) following manufacturer’s instructions. Finally, reactions were combined in equimolar concentrations to create a mixture composed of amplicon fragments of each sample. This composite sample was used for library preparation with Ion OneTouch™ 2 System with the Ion PGM™ Template OT2 400 Kit Template (Thermo Fisher Scientific, Waltham, MA, USA). The sequencing was performed using Ion PGM™ Sequencing 400 on Ion PGM™ System using Ion 318™ Chip v2 with a maximum of 40 samples per microchip.
Raw reads were analyzed according to the pipeline proposed by the Brazilian Microbiome Project [29]. A table of operational taxonomic units (OTUs) was constructed by using the UPARSE pipeline [30], with a minimum similarity cutoff value of 97% for clustering and a maximum expected error of 0.5%. Taxonomic classifications were made on QIIME 1.9.0 [31], based on UCLUST method against the SILVA ribosomal RNA gene database version 128[32] with a confidence interval of 95%. Sampling effort was measured by the Good’s coverage [33].
Data analyses
Maternal variables were analyzed into the R environment [34]. Numeric variables were summarized as average ± SEM and compared using the Kruskal-Wallis followed by a post hoc Dunn test. Categorical variables were compared using chi square post hoc test.
The 16S rRNA database was analyzed through the phyloseq [35] and the Microbiome [36] packages after removing singletons and rarefying the dataset to the minimum library size. Possible confounding variables were tested by permutational multivariate analysis of variance (PERMANOVA) into the vegan package [37].
Initial insights about general microbial structure were provided by analyses of relative abundance (measured by Kruskall-Wallis post hoc Dunn test) of the most frequent genera and alpha diversity tests.
The differential abundance analysis, applied to find microbial biomarkers of preterm birth, was performed by using the DESeq2 [38] with the raw (non-rarefied) dataset. Briefly, after removing the samples from mothers treated with intrapartum penicillin from the dataset, the OTU table was conglomerated at species level. Taxa not seen more than 3 times in at least 20% of the samples were removed and the number of sequences per OTU was transformed by calculating the geometric mean. Two different contrasts were applied: (a) term birth versus preterm birth and (b) false-preterm birth versus preterm birth. The FDR method was used to control for false discovery rate. Additional correlation analysis between OTUs was tested by using SparCC approach [39].
Results
Maternal variables used for comparison between groups
The characteristics of the three groups are shown in Table 1. Maternal age ranged from 23 to 30 years but was not significantly different between women with subsequent term or preterm labor. Nonetheless, women significantly older composed the non-spontaneous preterm group. The number of previous pregnancies was also similar in all, spontaneous term labor and spontaneous preterm labor groups as well as in non-spontaneous preterm group. Moreover, as expected, gestational age was significantly different between term and preterm groups, but not different between spontaneous preterm and non-spontaneous preterm groups. Women who had preterm labor presented a higher incidence of chorioamnionitis than the term group. This condition did not differ between term and non-spontaneous preterm groups. Only the preterm and the non-spontaneous preterm groups presented cases of infection by Group B Streptococcus (GBS). Although the GBS infection rate was higher in the non-spontaneous preterm group, the incidence of GBS infection was not statistically different between spontaneous preterm and non-spontaneous preterm groups. Most cases of non-spontaneous preterm labor presented preeclampsia, whereas there were no cases in the term group and only three cases in the spontaneous preterm labor group. The intrapartum penicillin was used in 15 preterm cases. The term and non-spontaneous preterm groups did not receive prophylactic antibiotics. Intravenous penicillin was administered approximately 4 h before labor in many preterm samples due to a positive test for GBS or due a suspicion of infection in absence of a test. Finally, the three groups differed in terms of vaginal or cesarean delivery. The preterm condition was the major driver of cesarean, especially in the non-spontaneous preterm labor group.
Table 1.
Maternal variables used for comparison between groups
| Variables | Spontaneous term labor (n = 29) | Spontaneous preterm labor (n = 23) | Non-spontaneous preterm labor (n = 17) |
|---|---|---|---|
| Maternal age (years) | 25.03 (± 1.13)a | 23.60 (± 1.28)a | 30.24 (± 1.75)b |
| Previous pregnancies | 2.00 (± 0.17)a | 1.87 (± 0.34)a | 2.12 (± 0.28)a |
| Gestational age (weeks) | 39.60 (± 0.20)a | 30.70 (± 0.39)b | 29.42 (± 0.59)b |
| Chorioamnionitis | 0a | 8b | 2b |
| Preeclampsia | 0a | 3a | 14b |
| GBS* infection | 0a | 5b | 4b |
| Intrapartum penicillin | 0a | 15b | 0a |
| Delivery mode (cesarean) | 1a | 10b | 17c |
*GBS, Group B Streptococcus. Numeric variables were summarized as average ± SEM and compared using the Kruskal-Wallis followed by a post hoc Dunn test. Categorical variables were compared using chi square post hoc test. Data followed by the same letter in the line represent groups without significant statistical difference (p > 0.05) whereas data followed by different letters in the line represent statistically different groups at the significance level of 95% (p ≤ 0.05)
Controlling for confounding variables
Permutational analysis of variance was applied to test the effect of confounding variables on the microbiota analyses (Table 2). As significant reduction in taxonomic diversity of vaginal microbial community was already observed as pregnancy advances [40], we first attempted to verify the influence of gestational age on the vaginal microbiota. A pairwise analysis among the spontaneous term labor, spontaneous preterm labor, and non-spontaneous preterm labor groups revealed undetectable microbial community differences among those groups in our dataset (Table 2). In all comparisons, R2 was smaller than 1% and the p value was greater than 0.05. Fifteen out of 23 women from the spontaneous preterm labor group received prophylactic antibiotics during labor, whereas no women from the term group received antibiotics. The R2 for antibiotic usage was 0.034 and the p value was 0.043 indicating that about 3.4% of the variation in the microbial community between groups was explained by the prophylactic use of antibiotics during labor. Intrapartum antibiotics were administrated only in cases with preterm labor. For this reason, this factor could not be used in a multi-factor design with interactions. All OTUs under intrapartum antibiotics influence were removed from the dataset prior to diversity and differential abundance analysis.
Table 2.
Permutational analysis of variance (PERMANOVA) of the Bray-Curtis dissimilarities for bacterial OTU community structure used for detection of possible confounding variables associated with preterm labor
| Confounding variables | R2 | p value |
|---|---|---|
| Mother’s age | 0.029 | 0.482 |
| Preview pregnancies | 0.014 | 0.748 |
| Gestational age | 0.017 | 0.540 |
| Corioamniotitis | 0.011 | 0.895 |
| Preeclampsia | 0.030 | 0.106 |
| GBS infection | 0.013 | 0.734 |
| Intrapartum penicillin | 0.034 | 0.043 |
| Delivery mode (cesarean/vaginal) | 0.014 | 0.711 |
Significant value is set in italics. p values are based on 999 permutations
Different microbial community structure but similar vaginal microbial diversity within treatments
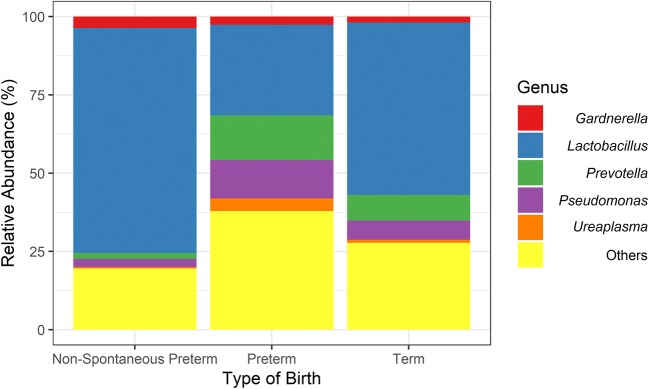
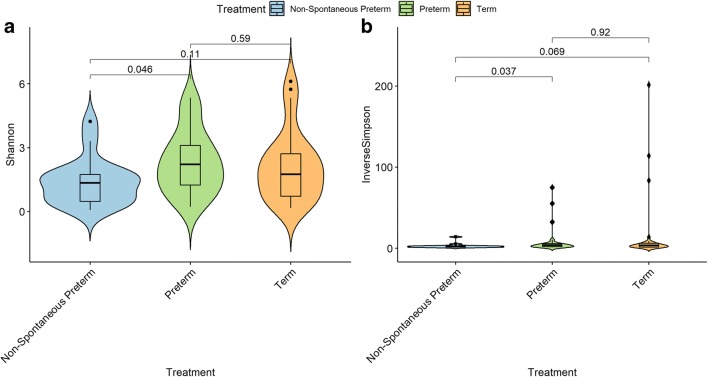
The mean of organisms’ abundance was particularly similar between term and non-spontaneous preterm groups as indicated by the Kruskal-Wallis post hoc Dunn test. On the other hand, preterm group presented a low mean of OTUs closest related to the genus Lactobacillus among its samples jointly with a tendency to a high mean of OTUs with best hit to Prevotella and Pseudomonas when compared with terms and non-spontaneous preterm (Fig. 1). Overall, the alpha diversity was low among all samples. The non-parametric Wilcox test indicated no differences in microbial diversity among the three groups tested using either Shannon or inverse of Simpson diversity indexes (Fig. 2).
Fig. 1.
Percentage of the five most abundant microbial genera found in the three tested groups. The genus Lactobacillus appeared with a low mean in preterm group when compared with term (Kruskal-Wallis post hoc Dunn test, p = 0.003) and with marginally low mean compared with non-spontaneous preterm (p = 0.080). Although Prevotella tended to be in high abundance in preterm samples, the test only found such difference when comparing preterm with non-spontaneous preterm group (p = 0.016). Pseudomonas, Ureaplasma, and Gardnerella did not present significant difference neither sample-to-sample or among samples (p > 0.05)
Fig. 2.
Alpha diversity measurements of microbial communities in the spontaneous preterm labor and control groups. a Shannon diversity index. b Inverse of Simpson index. Boxes span the first to third quartiles; the horizontal line inside the boxes represents the median. Whiskers extending vertically from the boxes indicate variability outside the upper and lower quartiles, and the single circles indicate outliers. No significant difference was found among the three groups (p > 0.05) according to the non-parametric Wilcoxon test
Defining the main biomarkers associated with term and preterm delivery
To outline the main microbes associated with the term and preterm labor we performed a differential abundance analysis. Pairwise comparisons among spontaneous term and spontaneous preterm groups revealed the abundance of OTUs closest related to two species of Lactobacillus associated with the term birth (Table 3). They were Lactobacillus iners and Lactobacillus jensenii. An unknown species related to the Prevotella genus was more abundant in the spontaneous preterm group. Similar tendency was also observed when comparing non-spontaneous preterm labor and spontaneous preterm labor groups. An OTU with the best hit to Lactobacillus jensenii was associated with subjects with non-spontaneous preterm labor while two taxa related to the Prevotella genus were associated with the spontaneous preterm labor. In an attempt to verify whether Prevotella was associated with Lactobacillus, we perform a correlation analysis at the genus level by using the SparCC approach [39]. No significant strong correlation (correlation coefficient = − 0.17 and p = 0.09) was found involving either Lactobacillus or Prevotella.
Table 3.
Differential abundance analysis depicting vaginal microbial biomarkers associated with term or preterm labor
| Base mean | Log2-fold change | lfcSE | Stat | p value | p adj | Closest microbial relative | Increased in |
|---|---|---|---|---|---|---|---|
| Spontaneous term versus spontaneous preterm | |||||||
| 22769 | 3.48 | 1.37 | 2.54 | 0.011 | 0.016 | Lactobacillus iners | Spontaneous term labor |
| 229 | 7.84 | 2.32 | 3.39 | 0.001 | 0.030 | Lactobacillus jensenii | Spontaneous term labor |
| 106 | 4.25 | 1.30 | − 3.27 | 0.001 | 0.031 | Prevotella sp. | Spontaneous preterm labor |
| Spontaneous preterm versus non-spontaneous preterm | |||||||
| 229 | 7.40 | 2.46 | 3.00 | 0.003 | 0.045 | Lactobacillus jensenii | Non-spontaneous preterm labor |
| 218 | 2.90 | 1.48 | − 1.96 | 0.050 | 0.353 | Prevotella bivia | Spontaneous preterm labor |
| 106 | 5.03 | 1.39 | − 3.61 | 0.000 | 0.026 | Prevotella sp. | Spontaneous preterm labor |
Base mean, the average of the normalized counts taken over all samples; log2-fold change, log2 fold change between the groups; lfcSE, standard error of the log2-fold change; Stat, Wald statistic; p value; Wald test p value; p adj, FDR-adjusted p value
The results indicated that the absence of high numbers of OTUs classified as Lactobacillus, particularly as Lactobacillus iners and Lactobacillus jensenii, might be the main difference between the vaginal microbial community of pregnant women following term or spontaneous preterm labor.
Discussion
In this work, we attempted to detect biomarkers for preterm labor on the vaginal microbiota of pregnant women. Several studies have described the vaginal microbiota of pregnant women; however, most of them were based in the USA, Canada, Europe, or Mexico [23–25, 41–43] and came to very incongruent results. A North American NGS-based study performed by Romero and colleagues, for example, concluded that there was no difference between abundance and structure of the vaginal microbiome, independent of the type of birth [23]. On the other hand, the efforts by Hyman and collaborators, whom worked with chain-termination sequencing, conclude that mothers whom deliver prematurely present a high diverse vaginal microbiota [42]. In addition, a Canadian study by Freitas et al. not only correlates high diversity on vaginal microbiota to preterm delivery but also the presence of Mollicutes [43].
Here, we used next-generation sequencing to analyze a Brazilian cohort composed of 69 pregnant women. The unique feature of this work is the high miscegenation rates of the Brazilian population. The aforementioned studies suggest that women from different ethnic backgrounds have different vaginal microbial communities. Therefore, investigations using cohorts with different ethnic backgrounds are important to better understand the etiology of preterm labor and its relationship with microbes.
Vaginal microorganisms possess a known key role in states of health and disease acting as both generators/stimulators and protectors from diseases [24, 44, 45]. Interactions between the microbiota and human diseases occur in a two-way process. Bacteria can cause diseases as much as states of diseases can cause changes in the normal microbiota. An example is the increase in bacterial pathogen abundance in cases of depression. Gut microbes can produce identical hormones and neurotransmitters produced by humans. In turn, the bacterial receptors for these hormones influence microbial growth [46].
In this context, we presented multiple lines that lead to the presence of different vaginal microbial communities associated with the full-term and spontaneous preterm labor. The first evidence was provided by overall abundance analysis (Fig. 1). Preterm group represented differences when compared with other ones, mainly related to the decrease of general abundance of Lactobacillus. The second and most strong evidence was obtained by a differential abundance analysis (Table 3). Lower numbers of OTUs with best hit to species from the genus Lactobacillus were associated with the spontaneous preterm labor while vaginal bacterial communities rich in these microbial species (e.g., spontaneous term and non-spontaneous preterm groups) were associated with the full-term outcome. Non-spontaneous preterm labor presented similar microbial communities composition to those subjects with spontaneous term labor. Indeed, the healthy vaginal microbiota in the Brazilian pregnant woman has low microbial diversity and is dominated by Lactobacillus species [27]. Besides, Lactobacillus species are very often correlated to states of health in the vaginal environment [24, 47–50]. Bacteria from this genus present a fermentative metabolism with lactate and usually acetate, ethanol, CO2, formate, or succinate as products [51]. These compounds acts lowering the vaginal pH to levels around 4,5 and creating an inhospitable environment for most of pathogenic species [49, 52, 53].
On the other hand, we were able to detect the presence of OTUs closest related to the genus Prevotella in association with the spontaneous preterm labor. In fact, many microorganisms, just like Prevotella species, can produce pro-inflammatory substances that can also lead to a preterm birth [54]. Studies point to adaptation of specific Prevotella strains at different niches. The report by Gupta and colleagues, for example, showed 83% of the Prevotella genome may contribute to singletons and flexible sequences and this condition performs a key role in the adaptation to many body sites [55]. Indeed, several works indicate Prevotella strains related dysbiosis in states of disease in highly different body parts, i.e., asthma and bacterial vaginosis [56, 57]. Prevotella is still correlated with inflammatory processes by the activation of Toll-like receptor 2, which leads to production of T helper type 17 cells (Th17) and increase of interleukin 8 (IL-8), interleukin 6 (IL-6), and chemokine (C-C motif) ligand 20 (CCL20) [58]. In addition, the intrauterine infection, which may have originated in the vaginal cavity, might account for 25–40% of preterm births [59]. The most commonly associated bacteria are bacteria from the class Mollicutes (Ureaplasma species, Mycoplasma genitalium, and M. hominis, for example) [60, 61], but many other microbial species have been identified in cases of bacterial vaginosis, including Prevotella [62–64]. Those microbes might invade the uterus by migrating from the passage through the cervix from the vagina and infect the amniotic fluid [59]. The metabolism of some of these bacteria may also produce urease, an enzyme that catalyzes the hydrolysis of urea into carbon dioxide and ammonia. Its activity increases the vaginal pH, a stress environmental condition for the mother and the fetus that may influence in a spontaneous premature outcome [65, 66].
Callahan et al. [67] recently presented similar results. The authors studied two cohorts from different locations of the USA. A lower abundance of L. crispatus was significantly associated with the preterm birth in both cohorts. In line with our results, the cohort from Birmingham, AL, presented decreased abundance of L. jensenii associated with the preterm birth. But contrary to our results, no significant association was detected for L. iners. Moreover, among the women with lower levels of Lactobacillus, a higher abundance of Gardnerella and Ureaplasma was associated with the increased risk of preterm labor. According to Baldwin et al. [68], Lactobacillus spp. were markedly decreased when compared with vaginal swabs collected from uncomplicated pregnancy subjects with a matched gestational time. As observed in our dataset, the authors also observed deficiency of Lactobacillus and persistence of known pathogenic species, such as Prevotella sp., as a risk factor for preterm birth.
In short, reports from the aforementioned studies as well as from this one converge to a pattern of bacteria either pathogenic or related to stress conditions as increased in preterm cases. Considering this fact and the niche adaptation performed by Prevotella spp. [55], we are able to suggest Prevotella as a microbial biomarker for preterm labor in the vaginal microbiota.
Conclusion
The relationship between the vaginal microbes and the spontaneous preterm labor was already described in racially distinct cohorts. In spite of this, to the best of our knowledge, this is the first study to describe and correlate the vaginal microbiota with the spontaneous preterm labor in a Brazilian cohort. This is especially important because: (i) preterm-microbiota associations are population dependent [67] and (ii) the Brazilian population presents high rates of miscegenation. As so, this population cannot be classified using standard stratifications of Caucasian/white and black/African American. Our results add to the ecological theory of the protective effect of Lactobacillus and the occurrence of other pathogenic taxa (e.g., Prevotella) as a possible risk factor for preterm labor.
Funding information
This work was supported by the Bill & Melinda Gates Foundation (OPP1107597), CNPQ, and DECIT/Ministério da Saúde do Brasil. This study was also financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES) (Finance Code 001). L.F.W. Roesch and AS de Freitas received research fellowships from the CNPq.
Compliance with ethical standards
The ethics committee of HCPA approved the study protocol.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Althabe F, Howson CP, Kinney M, Lawn J (2012) World Health Organization. Born too soon: The global action report on preterm birth. http://www.who.int/pmnch/media/news/2012/201204_borntoosoon-report.pdf. Accessed March 24, 2017
- 2.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371(9606):75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Larroque B, Ancel P-Y, Marret S, Marchand L, André M, Arnaud C, Pierrat V, Rozé JC, Messer J, Thiriez G, Burguet A, Picaud JC, Bréart G, Kaminski M. Neurodevelopmental disabilities and special care of 5-year-old children born before 33 weeks of gestation (the EPIPAGE study): a longitudinal cohort study. Lancet. 2008;371(9615):813–820. doi: 10.1016/S0140-6736(08)60380-3. [DOI] [PubMed] [Google Scholar]
- 4.Ment LR, Vohr BR. Preterm birth and the developing brain. Lancet Neurol. 2008;7(5):378–379. doi: 10.1016/S1474-4422(08)70073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rommel A-S, James S-N, McLoughlin G, Brandeis D, Banaschewski T, Asherson P, Kuntsi J. Association of preterm birth with attention-deficit/hyperactivity disorder–like and wider-ranging neurophysiological impairments of attention and inhibition. J Am Acad Child Adolesc Psychiatry. 2017;56(1):40–50. doi: 10.1016/j.jaac.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson S, Hollis C, Kochhar P, Hennessy E, Wolke D, Marlow N. Autism spectrum disorders in extremely preterm children. The Journal of Pediatrics. 2010;156(4):525–531.e2. doi: 10.1016/j.jpeds.2009.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Rosen CL, Larkin EK, Kirchner HL, Emancipator JL, Bivins SF, Surovec SA, Martin RJ, Redline S. Prevalence and risk factors for sleep-disordered breathing in 8- to 11-year-old children: association with race and prematurity. J Pediatr. 2003;142(4):383–389. doi: 10.1067/mpd.2003.28. [DOI] [PubMed] [Google Scholar]
- 8.Martin JA, Hamilton BE, Osterman MJK, Driscoll AK, Mathews TJ. Births: final data for 2015. Natl Vital Stat Rep. 2017;66(1):1. [PubMed] [Google Scholar]
- 9.Zeitlin J, Szamotulska K, Drewniak N, Mohangoo AD, Chalmers J, Sakkeus L, Irgens L, Gatt M, Gissler M, Blondel B, The Euro-Peristat Preterm Study Group Preterm birth time trends in Europe: a study of 19 countries. BJOG Int J Obstet Gynaecol. 2013;120(11):1356–1365. doi: 10.1111/1471-0528.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howson CP, Kinney MV, McDougall L, Lawn JE. Born too soon: preterm birth matters. Reprod Health. 2013;10(1):S1. doi: 10.1186/1742-4755-10-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slattery MM, Morrison JJ. Preterm delivery. Lancet. 2002;360(9344):1489–1497. doi: 10.1016/S0140-6736(02)11476-0. [DOI] [PubMed] [Google Scholar]
- 12.Kajantie E, Osmond C, Barker DJP, Eriksson JG. Preterm birth--a risk factor for type 2 diabetes?: the Helsinki birth cohort study. Diabetes Care. 2010;33(12):2623–2625. doi: 10.2337/dc10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanz LJ, Stuart JJ, Williams PL, Rimm EB, Missmer SA, Rexrode KM, Mukamal KJ, Rich-Edwards JW. Preterm delivery and maternal cardiovascular disease in young and middle-aged adult women clinical perspective. Circulation. 2017;135(6):578–589. doi: 10.1161/CIRCULATIONAHA.116.025954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huynh M, Woodruff TJ, Parker JD, Schoendorf KC. Relationships between air pollution and preterm birth in California. Paediatr Perinat Epidemiol. 2006;20(6):454–461. doi: 10.1111/j.1365-3016.2006.00759.x. [DOI] [PubMed] [Google Scholar]
- 15.Steer P. The epidemiology of preterm labour. BJOG Int J Obstet Gynaecol. 2005;112(s1):1–3. doi: 10.1111/j.1471-0528.2005.00575.x. [DOI] [PubMed] [Google Scholar]
- 16.Ahern J, Pickett KE, Selvin S, Abrams B. Preterm birth among African American and white women: a multilevel analysis of socioeconomic characteristics and cigarette smoking. J Epidemiol Community Health. 2003;57(8):606–611. doi: 10.1136/jech.57.8.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ananth CV, Misra DP, Demissie K, Smulian JC. Rates of preterm delivery among black women and white women in the United States over two decades: an age-period-cohort analysis. Am J Epidemiol. 2001;154(7):657–665. doi: 10.1093/aje/154.7.657. [DOI] [PubMed] [Google Scholar]
- 18.Kindinger LM, Bennett PR, Lee YS, et al. (2017) The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome 5(1). 10.1186/s40168-016-0223-9 [DOI] [PMC free article] [PubMed]
- 19.Kramer MS, Goulet L, Lydon J, Seguin L, McNamara H, Dassa C, Platt RW, Fong Chen M, Gauthier H, Genest J, Kahn S, Libman M, Rozen R, Masse A, Miner L, Asselin G, Benjamin A, Klein J, Koren G. Socio-economic disparities in preterm birth: causal pathways and mechanisms. Paediatr Perinat Epidemiol. 2001;15(s2):104–123. doi: 10.1046/j.1365-3016.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 20.Smith LK, Draper ES, Manktelow BN, Dorling JS, Field DJ. Socioeconomic inequalities in very preterm birth rates. Arch Dis Child Fetal Neonatal Ed. 2007;92(1):F11–F14. doi: 10.1136/adc.2005.090308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son K-A, Kim M, Kim YM, Kim SH, Choi SJ, Oh SY, Roh CR, Kim JH. Prevalence of vaginal microorganisms among pregnant women according to trimester and association with preterm birth. Obstet Gynecol Sci. 2018;61(1):38–47. doi: 10.5468/ogs.2018.61.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson DB, Hanlon A, Hassan S, Britto J, Geifman-Holtzman O, Haggerty C, Fredricks DN. Preterm labor and bacterial vaginosis-associated bacteria among urban women. J Perinat Med. 2009;37(2):130–134. doi: 10.1515/JPM.2009.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2(1):18. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernández-Rodríguez C, Romero-González R, Albani-Campanario M, Figueroa-Damián R, Meraz-Cruz N, Hernández-Guerrero C. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated Bacteria. Infect Dis Obstet Gynecol. 2011;2011:1–9. doi: 10.1155/2011/851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:8988. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pylro VS, Morais DK, Roesch LFW. Microbiome studies need local leaders. Nature. 2015;528:39. doi: 10.1038/528039e. [DOI] [PubMed] [Google Scholar]
- 27.Roesch LFW, Silveira RC, Corso AL, Dobbler PT, Mai V, Rojas BS, Laureano ÁM, Procianoy RS. Diversity and composition of vaginal microbiota of pregnant women at risk for transmitting group B Streptococcus treated with intrapartum penicillin. PLoS One. 2017;12(2):e0169916. doi: 10.1371/journal.pone.0169916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal. 2012;2012(6):1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pylro VS, Roesch LFW, Morais DK, Clark IM, Hirsch PR, Tótola MR. Data analysis for 16S microbial profiling from different benchtop sequencing platforms. J Microbiol Methods. 2014;107:30–37. doi: 10.1016/j.mimet.2014.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40(3/4):237. doi: 10.2307/2333344. [DOI] [Google Scholar]
- 34.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. http://www.R-project.org
- 35.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lahti L, Shetty S, Blake T, Salojarvi J (2017) Microbiome r package. Tools Microbiome Anal R
- 37.Oksanen J, Blanchet FG, Kindt R, et al. (2015) Vegan: community ecology package. R Package Vegan, Vers. 2.2–1. World Agroforestry Centre Nairobi, Kenya
- 38.Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15(12). 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed]
- 39.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. von Mering C, ed. PLoS Comput Biol. 2012;8(9):e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aagaard K, Riehle K, Ma J, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. Ratner AJ, ed. PLoS ONE. 2012;7(6):e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2(1):4. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, Vo KC, Caughey AB, Hilton JF, Davis RW, Giudice LC. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21(1):32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.the VOGUE Research Group, Freitas AC, Bocking A, Hill JE, Money DM (2018) Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth. Microbiome 6(1). 10.1186/s40168-018-0502-8 [DOI] [PMC free article] [PubMed]
- 44.Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci. 2015;112(35):11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galland L. The gut microbiome and the brain. J Med Food. 2014;17(12):1261–1272. doi: 10.1089/jmf.2014.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Science Translational Medicine. 2012;4(132):132ra52–132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravel J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S. K., McCulle S. L., Karlebach S., Gorle R., Russell J., Tacket C. O., Brotman R. M., Davis C. C., Ault K., Peralta L., Forney L. J. Vaginal microbiome of reproductive-age women. Proceedings of the National Academy of Sciences. 2010;108(Supplement_1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma B, Forney LJ, Ravel J. Vaginal microbiome: rethinking health and disease. Annu Rev Microbiol. 2012;66(1):371–389. doi: 10.1146/annurev-micro-092611-150157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamont R, Sobel J, Akins R, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R. The vaginal microbiome: new information about genital tract flora using molecular based techniques: vaginal microbiome using molecular tools. BJOG Int J Obstet Gynaecol. 2011;118(5):533–549. doi: 10.1111/j.1471-0528.2010.02840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammes WP, Hertel C (2015) Lactobacillus. In: Whitman WB, Rainey F, Kämpfer P, et al., eds. Bergey’s manual of systematics of archaea and bacteria. Chichester, UK: John Wiley & Sons, Ltd:1–76. 10.1002/9781118960608.gbm00604
- 52.Boskey ER, Telsch KM, Whaley KJ, Moench TR, Cone RA. Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect Immun. 1999;67(10):5170. doi: 10.1128/IAI.67.10.5170-5175.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Linhares IM, Summers PR, Larsen B, Giraldo PC, Witkin SS. Contemporary perspectives on vaginal pH and lactobacilli. American Journal of Obstetrics and Gynecology. 2011;204(2):120.e1–120.e5. doi: 10.1016/j.ajog.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Yang S, Reid G, Challis JRG, Kim SO, Gloor GB, Bocking AD (2015) Is there a role for probiotics in the prevention of preterm birth? Front Immunol 6. 10.3389/fimmu.2015.00062 [DOI] [PMC free article] [PubMed]
- 55.Gupta VK, Chaudhari NM, Iskepalli S, Dutta C. Divergences in gene repertoire among the reference Prevotella genomes derived from distinct body sites of human. BMC Genomics. 2015;16(1):153–153. doi: 10.1186/s12864-015-1350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, Davies J, Ervine A, Poulter L, Pachter L, Moffatt MF, Cookson WOC. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578–e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zozaya-Hinchliffe M, Lillis R, Martin DH, Ferris MJ. Quantitative PCR assessments of bacterial species in women with and without bacterial vaginosis. J Clin Microbiol. 2010;48(5):1812–1819. doi: 10.1128/JCM.00851-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342(20):1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 60.Kataoka S, Yamada T, Chou K, Nishida R, Morikawa M, Minami M, Yamada H, Sakuragi N, Minakami H. Association between preterm birth and vaginal colonization by mycoplasmas in early pregnancy. J Clin Microbiol. 2006;44(1):51–55. doi: 10.1128/JCM.44.1.51-55.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Viscardi RM. Ureaplasma species: role in diseases of prematurity. Clin Perinatol. 2010;37(2):393–409. doi: 10.1016/j.clp.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hillier SL, Nugent RP, Eschenbach DA, Krohn MA, Gibbs RS, Martin DH, Cotch MF, Edelman R, Pastorek JG, Rao AV, McNellis D, Regan JA, Carey JC, Klebanoff MA. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The vaginal infections and prematurity study group. N Engl J Med. 1995;333(26):1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 63.Tebes CC, Lynch C, Sinnott J. The effect of treating bacterial vaginosis on preterm labor. Infect Dis Obstet Gynecol. 2003;11(2):123–129. doi: 10.1080/10647440300025509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Onderdonk AB, Delaney ML, Fichorova RN. The human microbiome during bacterial vaginosis. Clin Microbiol Rev. 2016;29(2):223–238. doi: 10.1128/CMR.00075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphries C (2017) Detecting diversity. Nature Publishing Group Macmillan Building, 4 Crinan St, London N1 9xw, England
- 66.Kenny GE, Cartwright FD. Effect of urea concentration on growth of Ureaplasma urealyticum (T strain mycoplasma) J Bacteriol. 1977;132(1):144–150. doi: 10.1128/JB.132.1.144-150.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Callahan BJ, DiGiulio DB, Goltsman DSA, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci U S A. 2017;114(37):9966–9971. doi: 10.1073/pnas.1705899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baldwin EA, Walther-Antonio M, MacLean AM, et al. Persistent microbial dysbiosis in preterm premature rupture of membranes from onset until delivery. PeerJ. 2015;3:e1398. doi: 10.7717/peerj.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]