Abstract
Lactococcus lactis subsp. lactis bv. diacetylactis strains are often used as starter cultures by the dairy industry due to their production of acetoin and diacetyl, important substances that add buttery flavor notes in dairy products. Twenty-three L. lactis subsp. lactis isolates were obtained from dairy products (milk and cheese) and dairy farms (silage), identified at a biovar level, fingerprinted by rep-PCR and characterized for some technological features. Fifteen isolates presented molecular and phenotypical (diacetyl and citrate) characteristics coherent with L. lactis subsp. lactis bv. diacetylactis and rep-PCR allowed the identification of 12 distinct profiles (minimum similarity of 90%). Based on technological features, only two isolates were not able to coagulate skim milk and 10 were able to produce proteases. All isolates were able to acidify skim milk: two isolates, in special, presented high acidifying ability due to their ability in reducing more than two pH units after 24 h. All isolates were also able to grow at different NaCl concentrations (0 to 10%, w/v), and isolates obtained from peanut and grass silages presented the highest NaCl tolerance (10%, w/v). These results indicate that the L. lactis subsp. lactis bv. diacetylactis isolates presented interesting technological features for potential application in fermented foods production. Despite presenting promising technological features, the isolates must be assessed according to their safety before being considered as starter cultures.
Keywords: Lactic acid bacteria, Diacetyl, Starter culture, Technological potential
Introduction
Lactococcus lactis is a lactic acid bacteria (LAB) of particular interest in the dairy industry due to its technological potential as starter culture, being used to produce fermented milks and ripened cheeses [1, 2]. Different L. lactis subspecies are described and two, lactis and cremoris, have special interest for being usually associated with fermentation processes and, therefore, being the target of many studies aiming the isolation and characterization of promising and novel starter cultures [2–6].
Besides their fermentation abilities, some L. lactis strains are able to ferment citrate and produce diacetyl and acetoin, desirable flavor compounds in specific ripened cheeses: these strains are referred as L. lactis subsp. lactis bv. diacetylactis [3, 7]. These strains can convert citrate to aroma compounds (C4) and carbon dioxide, substances that improve the organoleptic characteristics of fermented foods [8]. Moreover, diacetyl is an essential component of many dairy products, since it determines a creamy and buttery aroma when present at low concentrations, and it is responsible for typical characteristics of specific cheeses, such as Camembert, Cheddar, and Emmental [2, 9]. Based on these features, L. lactis subsp. lactis bv. diacetylactis is often mixed with other LAB during cheese production, at an usual ratio of 20% of the whole starter culture population [1, 10, 11]. Besides conferring these organoleptic features, diacetyl is also considered to be an antimicrobial compound that enhances product safety [12, 13].
Laroute et al. [6] explain that aroma-producing strains with potential use in the dairy industry must be screened to determine their citrate-depleting potential. This characteristic can be assessed by studying the strains’ growth in a Kempler and McKay (KMK) medium, followed by a study of their acetoin and diacetyl production using the Voges-Proskauer reaction. Besides this phenotypical approach, PCR can also be used to detect genes related to citrate pathway and to characterize the mosaic structure of the histidine biosynthesis operon, what is specific for each Lactococcus species and biovar [14–16].
This study aimed to present a comprehensive characterization of L. lactis subsp. lactis isolates obtained from dairy products (milk and cheese) and dairy farms (silage) by using phenotypical and molecular methods to identify bv. diacetylactis strains and to characterize some of their technological features (lactofermentation, proteolysis, acidifying ability, and NaCl resistance), aiming the selection of promising and potential starter cultures that could be used by the dairy industry.
Material and methods
Lactococcus lactis subsp. lactis isolates
Twenty-three L. lactis subsp. lactis isolates were obtained from the bacteria culture collection at InovaLeite (Laboratory of Milk and Dairy Products, Universidade Federal de Viçosa) and included in this study. The isolates were obtained from different samples and ecosystems, all related to dairy production: raw milk, raw milk cheeses (Amazonas, Brazil and Marajó Island, Brazil) and dairy farm silages (peanut and grass). All isolates were previously identified as L. lactis by sequencing of the 16S rRNA, and further subspecies-specific PCR assays which targeted L. lactis subsp. lactis [17, 18].
Identification of L. lactis subsp. lactis bv. diacetylactis
The selected strains were cultured in de Man, Rogosa, and Sharpe (MRS) broth (Oxoid Ltd., Basingstoke, England) at 30 °C for 18 h. Then, 1 mL aliquots of the cultures were centrifuged at 14,000×g for 2 min at room temperature, and the cell pellets were subjected to DNA extraction using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). A PCR assay was conducted to identify the specific region of the histidine biosynthesis operon for bv. diacetylactis isolates, using primers Lhis5F (5'-CTTCGTTATGATTTTACA-3′) and Lhis6R (5'-AATATCAACAATTCCATG-3′), as described by Beimfohr et al. [15]. PCR conditions were (1) 2 min at 93 °C, (2) 30 cycles of 30 s at 94 °C; 90 s at 46 °C and 2 min at 72 °C, and (3) final extension of 5 min at 72 °C. The obtained PCR products were analyzed on 1.5% (w/v) agarose gels, stained using GelRed (Biotium Inc., Hayward, CA, USA) and visualized using a transilluminator LPIX (Loccus Biotecnologia, São Paulo, SP, Brazil), in order to detect a 934-bp PCR product, as typical for bv. diacetylactis [15].
Isolates were also characterized by their ability to ferment citrate and produce diacetyl. Aliquots of the obtained cultures were streaked onto Kempler and McKay agar and incubated at 30 °C for 48 h; isolates that presented blue colonies were considered as citrate-fermenting [16, 19]. Diacetyl production was assessed by transferring 1 mL aliquots of the cultures to sterile skim milk (10% w/v, Nestlé, São Paulo, SP, Brazil), followed by incubation at 30 °C for 24 h; then, 1 mL aliquots of the obtained cultures in milk were added to 0.5 mL of α-naphthol (1% w/v) and KOH (16% w/v) and incubated at 30 °C for 10 min: diacetyl production was indicated by the formation of a red ring at the top of the tubes [20].
L. lactis subsp. lactis bv. diacetylactis ATCC 13675 and L. lactis subsp. cremoris ATCC 19257 were used as positive and negative controls, respectively [15]. Isolates that presented the specific 934 bp band after PCR amplification, diacetyl production, and citrate fermentation were identified as L. lactis subsp. lactis bv. diacetylactis, and then subjected to rep-PCR fingerprinting and characterized for some technological features, as described below.
Rep-PCR fingerprinting
Rep-PCR was performed according to Dal Bello et al. [21] using a single primer (GTG)5 (5′-GTGGTGGTGGTGGTG-3′). PCR reactions contained 12.5 μL of Go Taq Green Master Mix 2x (Promega), 50 pMol of the primer, 2 μL of DNA (50 ng/μL), and ultra-pure PCR water (Promega) to a final volume of 25 μL. PCR conditions were (1) 5 min at 95 °C, (2) 30 cycles of 30 s at 95 °C; 30 s at 40 °C and 8 min at 65 °C, and (3) final extension of 16 min at 65 °C. PCR products were analyzed in 2% (w/v) agarose gels for 6 h at a constant voltage of 75 V, in 0.5 × Tris/Borate/EDTA buffer (TBE). Gels were stained using GelRed (Biotium) and recorded using a transilluminator LPIX (Loccus). The fingerprints were analyzed using BioNumerics 6.6.11 (Applied Maths, Kortrijk, Belgium). The similarities among profiles were calculated using the Pearson correlation, and a dendrogram was constructed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA).
Technological features of L. lactis subsp. lactis bv. diacetylactis
Isolates identified as L. lactis subsp. lactis bv. diacetylactis were subjected to phenotypic assays to characterize some of their technological features. Before each assay, the isolates were cultured in MRS (Oxoid) at 30 °C for 18 h and centrifuged at 14,000×g for 2 min (4 °C). After discarding the supernatant, the cell pellets were suspended in NaCl 0.85% (w/v) until a turbidity similar to McFarland tube 1, corresponding to approximately 3 × 108 CFU/mL. The obtained cultures were used in the assays described below, which were conducted in three independent repetitions.
Lactofermentation patterns were assessed by inoculating 0.1 mL aliquots of the isolate cultures into 10 mL of skim milk (10% w/v, Nestlé), followed by incubation at 30 °C for 24 h. Based on the formed clot characteristics, the lactofermentation patterns were described using an empirical analysis and classified as: uniform, uniform with presence of serum, uniform and fragile (appearance), broken with presence of serum, and absence of clot.
A screening assay was conducted to identify the extracellular proteolytic activity of the isolates, as described by Franciosi et al. [20]. Two μL aliquots of bacterial cultures were spotted onto the surface of a Plate Count Agar (PCA, HiMedia, Mumbai, MH, India) enhanced with skim milk (10% w/v, Nestlé), and incubated at 30 °C for 4 days; presumed proteolytic activity was indicated by a clear zone around the colonies. Pseudomonas fluorescens 07A [22] was considered as positive control.
Isolates that presented presumed proteolytic activity in the screening assay were subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) to confirm this feature. Isolate cultures in exponential growth phase were inoculated (1%, v/v) in UHT skim milk, incubated at 30 °C for 24 h, and treated for SDS-PAGE as described by Adams et al. [23], with some modifications: the cultures were acidified to pH 4.6 with HCl (3 M) and centrifuged at 10,000×g for 15 min at 4 °C; then, the pellets were resuspended to the original volumes in 0.5 mM Tris-HCl buffer, pH 9.0 and diluted 1:20 (v/v) in Milli-Q water: 20 μL aliquots were mixed in 5 μL of sample buffer 5× (1% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 20% (v/v) glycerol, 0.2 M Tris–HCl pH 6.8, 0.05% (w/v) bromophenol blue and heated at 90 °C for 5 min. The treated cultures were then subjected to a SDS-PAGE with 12% polyacrylamide [24] in a Mini-Protean® Tetra System (BioRad, California, CA, USA). Proteins were stained with 0.01% Coomassie Brilliant Blue R-250 solution. As in the screening assay, P. fluorescens 07A was considered as the positive control [22], and non-inoculated UHT milk as negative control: clearer α- and β-casein related bands were indicative of proteolytic activity, when compared to negative control [22].
The acidifying ability of L. lactis subsp. lactis bv. diacetylactis isolates was assessed by adding culture aliquots to skim milk (10% w/v, Nestlé), followed by incubation at 30 °C for 24 h. pH values were measured at the time of inoculation (T0), then after 6 (T6), 12 (T12), and 24 h (T24) of incubation using a digital pHmeter (Hanna Instruments, São Paulo, SP, Brazil). Based on the cultures’ abilities to acidify milk pH after 6, 12, and 24 h, the tested isolates were categorized into 3 main groups: (I) high acidifying ability—more than 2 pH units decrease; (II) medium acidifying ability—pH decrease of 1.5 to 2.0 pH units; and (III) low acidifying ability—less than 1.5 pH units decrease [25].
Finally, resistance to NaCl was assessed as described by Dal Bello et al. [5]. Two μL aliquots of the selected isolates were transferred to 188 μL of MRS broth (Oxoid) prepared with different concentrations of NaCl (0, 2, 4, 6, 8, and 10%, w/v), previously distributed into 96-well microtiter plates. In each prepared microtiter plate, 2 blank wells with only 190 μL of MRS broth (Oxoid) were prepared for each NaCl concentration. The microplates were then incubated in MultiskanTM GO Microplate Spectrophotometer (Thermo Fisher Scientific, Madison, WI, USA) at 30 °C, agitated for 24 h, and measured for optical density (OD) of cultures every 30 min (λ = 650 nm). Mean values of OD readings were plotted in graphs in order to demonstrate the growth curves of L. lactis subsp. lactis bv. diacetylactis isolates at different NaCl concentrations.
Results and discussion
Among the 23 selected L. lactis subsp. lactis isolates, 17 presented 934 bp PCR products amplification (Table 1), typical for bv. diacetylactis according to Beimfohr et al. [15]. Based on the adopted phenotypical assays, 17 isolates were shown to be able to ferment citrate and 20 were able to produce diacetyl (Table 1). BUF1 showed a typical PCR result for bv. diacetylactis but it was not able to ferment citrate or produce diacetyl, whereas SBR3 was only unable to produce diacetyl. Conversely, LVTC8MRS and Q1C4 did not present positive PCR results, although they were able to ferment citrate and produce diacetyl. Q13C4 was able to produce diacetyl even though it did not present a typical PCR result for bv. diacetylactis (Table 1). Despite only phenotypical assays being usually considered for bv. diacetylactis identification [4, 26, 27] in the present study, we associated this approach with a bv-specific PCR assay for this purpose. In this sense, we were able to identify isolates that present both the genetical and the phenotypical features that characterize L. lactis subsp. lactis bv diacetylactis: based on these criteria, 15 isolates were identified as bv. diacetylactis (Table 1). According to Siezen et al. [28], strains that can utilize citrate and stimulate the production of acetoin and diacetyl present an interesting phenotypic trait for potential use in the dairy industry. Diacetyl production is usually described as a common feature of L. lactis subsp. lactis [5, 20, 29, 30] being strain-dependent and particularly associated with bv. diacetylactis [7, 11]; the acetoin/diacetyl pathway is essential for the production of compounds that result in the buttery aroma in some cheeses [31]. Therefore, the ability to produce diacetyl must be also considered as an important criterion for identification of isolates as bv. diacetylactis. However, these phenotypical characteristics must be associated to a molecular approach, as a PCR assay, targeting a specific mosaic structure of the histidine biosynthesis operon that is typical for L. lactis subsp. lactis bv. diacetylactis [15].
Table 1.
Molecular and phenotypic characterization of L. lactis subsp. lactis isolates targeting the identification of L. lactis subsp. lactis bv. diacetylactis
| Isolate | Sample origin | Identification protocol1 | Identification | ||
|---|---|---|---|---|---|
| PCR2 | citrate | diacetyl | |||
| LVA2.1 | Cow milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| LVA2.2 | Cow milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| LVA2VACA | Cow milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| LCA1 | Goat milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| LCA2 | Goat milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| LCA4 | Goat milk | − | − | − | − |
| LCA5 | Goat milk | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| BUF1 | Buffalo milk | + | − | − | − |
| LVTC8MRS | Cream milk (cow) | − | + | + | − |
| Q1C2 | Artisanal cheese (Amazon) | − | − | + | − |
| Q1C4 | Artisanal cheese (Amazon) | − | + | + | − |
| Q1C5 | Artisanal cheese (Amazon) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| Q1C7 | Artisanal cheese (Amazon) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| Q1C10 | Artisanal cheese (Amazon) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| Q4C8 | Artisanal cheese (Amazon) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| Q5C6 | Artisanal cheese (Amazon) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| Q6C2 | Artisanal cheese (Amazon) | − | − | − | − |
| Q13C4 | Artisanal cheese (Marajó) | − | − | + | − |
| Q15C3 | Artisanal cheese (Marajó) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| SAM12 | Peanut silage (dairy farm) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| SBR1 | Grass silage (dairy farm) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
| SBR3 | Grass silage (dairy farm) | + | − | + | - |
| SBR4 | Grass silage (dairy farm) | + | + | + | L. lactis subsp. lactis bv. diacetylactis |
1Identification protocol described in the Material and methods section; 2PCR product of 934 bp was indicative as typical of L. lactis subsp. lactis bv. diacetylactis, as described by Beimfohr et al. [15]; Results: +, positive; −, negative
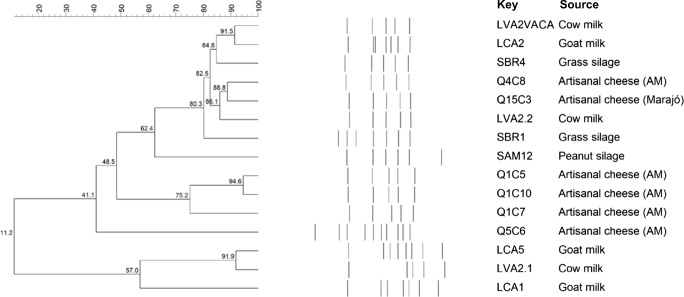
Based on rep-PCR profiles, isolates identified as L. lactis subsp. lactis bv. diacetylactis were selected and grouped considering a 90% of similarity or above (Fig, 1). The 15 isolates were grouped in 12 profiles, and the groups formed showed low homology to each other, indicating a high diversity among the strains of L. lactis subsp. lactis bv. diacetylactis. There were no profiles with more than two similar isolates, and the maximum homology was found between isolates Q1C5 and Q1C10 that shared 94.6% of homology: both were isolated from artisanal cheeses from the Amazon region. LVA2.1 and LCA5 were isolated from cow and goat milk, respectively, presented a 91.9% similarity. LVA2VACA (cow milk) and LCA2 (goat milk) isolates shared 91.5% of similarity. Q5C6 isolate, obtained from artisanal cheese from the Amazon region, showed low similarity (41.1%) in relation to the other milk strains. Q15C3 was obtained from an artisanal cheese from Marajó Island and presented 88.8% of homology with Q4C8, isolated also from an artisanal cheese (Amazon); this result can indicate a potential similarity among artisanal cheeses isolated from different regions. It was expected that isolates obtained from dairy products, like milk, cheese, and cream, would present a higher genetic similarity when compared to isolates obtained from non-dairy products, such as silage; in such conditions, bacterial strains are subjected to gene losses, mutations, and acquisitions that allow them to adapt to these new habitats [6, 28]. Regarding the isolates obtained from silage samples, SBR1 and SBR4 were grouped together and shared 80.3% of similarity, while SMA12 presented the lowest similarity to the other silage-isolates (62.4%, Fig. 1). The particular genetic profiles of SAM12 and Q5C6 (Fig. 1) can indicate their potential as novel strains to be explored as starter cultures in the dairy industry, leading to further assays to assess their technological potential.
Fig. 1.
Schematic representation of the obtained Rep-PCR profiles of 15 Lactococcus lactis subsp. lactis bv. diacetylactis obtained from different dairy related samples (source). The similarity analysis was performed using the Pearson correlation and the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) method (tolerance 5%)
The technological features of the isolates identified as L. lactis subsp. lactis bv. diacetylactis are presented in Table 2 (lactofermentation, proteolysis, and acidifying ability) and in Fig. 2 (growth at different NaCl concentrations). From all evaluated isolates, only 2 did not present coagulation abilities, a feature evidenced by the lack of clot formation in skim milk (Table 2). Milk coagulation is the first stage of cheese production, determined by the destabilization of casein micelles and leading to curd formation; this stage can occur due to the activity of coagulant enzymes and/or due to the activity of bacterial strains that will release lactic acid. Lactic acid reduces pH and increases the milk coagulation through serum expulsion, and also inhibits the growth of microorganisms [32]. The obtained results indicate the potential use of the characterized isolates as coagulant agents during cheese production.
Table 2.
Technological properties of isolates identified as L. lactis subsp. lactis bv. diacetylactis
| Isolate | Lactofermentation | Proteolysis | pH | Acidifying potential group2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Milk-agar | SDS-PAGE1 | T0 | T6 | T12 | T24 | T6-T0 | T12-T0 | T24-T0 | ||
| LVA2.1 | Uniform | + | + | 6.70 | 6.40 | 6.03 | 6.01 | III | III | III |
| LVA2.2 | Uniform | + | + | 6.70 | 6.19 | 5.96 | 5.78 | III | III | III |
| LVA2VACA | Broken with serum | + | − | 6.70 | 5.98 | 5.53 | 4.96 | III | III | II |
| LCA1 | Uniform | + | − | 6.70 | 6.26 | 6.06 | 5.17 | III | III | II |
| LCA2 | Uniform | + | − | 6.70 | 6.37 | 5.80 | 5.66 | III | III | III |
| LCA5 | Uniform | + | + | 6.70 | 5.99 | 5.64 | 5.38 | III | III | III |
| Q1C5 | Uniform with serum | + | + | 6.70 | 5.88 | 5.45 | 4.54 | III | III | I |
| Q1C7 | Uniform | + | + | 6.70 | 6.28 | 6.17 | 6.13 | III | III | III |
| Q1C10 | Uniform | + | + | 6.70 | 6.25 | 6.07 | 5.79 | III | III | III |
| Q4C8 | Uniform with serum | + | + | 6.70 | 5.91 | 5.37 | 4.66 | III | III | I |
| Q5C6 | Uniform with serum | + | + | 6.70 | 6.23 | 5.87 | 5.45 | III | III | III |
| Q15C3 | Broken with serum | + | + | 6.70 | 6.13 | 5.61 | 4.91 | III | III | II |
| SAM12 | Absence of clot | − | NC | 6.70 | 6.33 | 6.22 | 5.91 | III | III | III |
| SBR1 | Absence of clot | − | NC | 6.70 | 6.33 | 6.22 | 5.91 | III | III | III |
| SBR4 | Uniform and fragile | + | + | 6.70 | 6.33 | 6.12 | 6.04 | III | III | III |
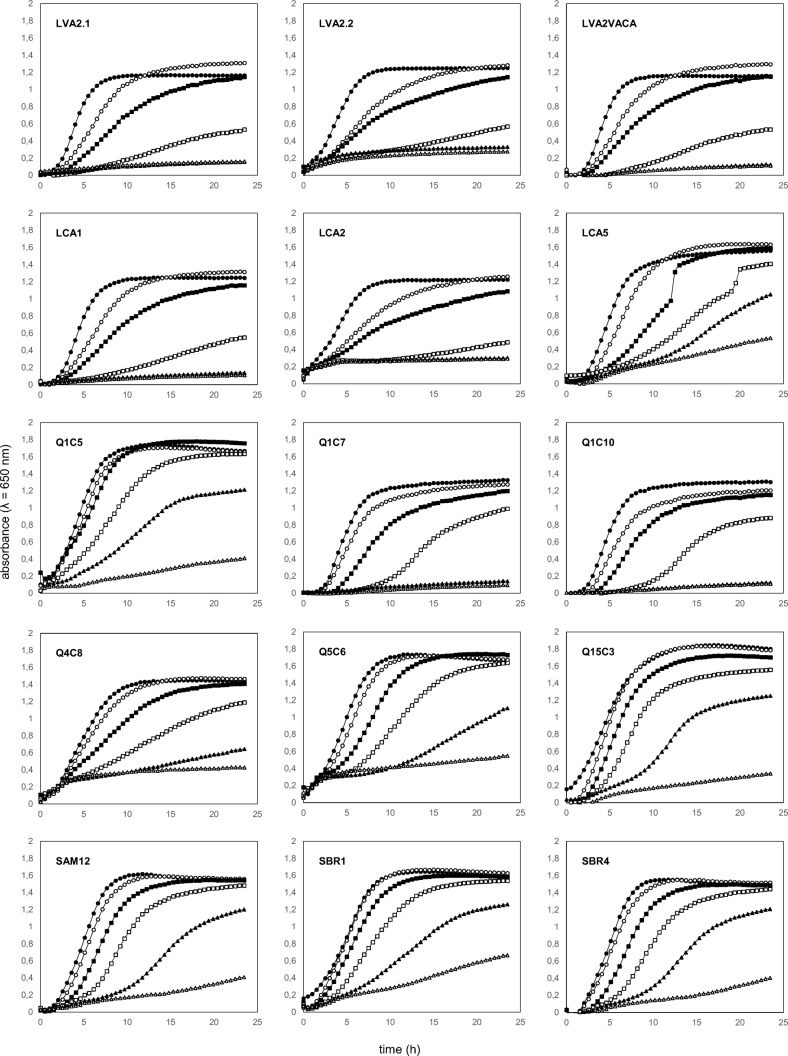
Fig. 2.
Growth curves of L. lactis subsp. lactis bv. diacetylactis isolates at different concentrations of NaCl. Growth in: black circle, 0% NaCl; white circle, 2% NaCl; black square, 4% NaCl; white square, 6% NaCl; black triangle, 8% NaCl; upper-case delta, 10% NaCl
Extracellular proteolysis is considered as the most important biochemical event in cheese production because it leads to the development substances that are either important for flavor or act as aroma precursors [33]. In this study, 13 isolates presented clear zones around their colonies when grown in milk-agar, indicating their proteolytic potential; the ability to hydrolase casein was confirmed for 10 isolates as observed by SDS-PAGE (Table 2). Isolates from silage samples did not present proteolytic activity, except for the SBR4 strain (Table 2); this can be considered an unexpected result, as this activity would not be necessarily required in this original niche (Table 2). These results were similar to those recorded for several L. lactis subsp. lactis isolates obtained from dairy-related samples [5, 20, 30]. Nomura et al. [4] described the proteolytic activity in L. lactis subsp. lactis strains obtained from fermented vegetables and cheese, though no proteolytic activity was observed with the bv. diacetylactis strains. Herreros et al. [34] analyzed L. lactis subsp. lactis and L. lactis subsp. lactis bv. diacetylactis isolated from Armada cheese (a Spanish goat’s milk cheese), and low proteolytic activity was recorded. Extracellular proteolytic activity is an essential property for starter cultures: LAB proteolytic system allows their optimal growth in milk due to the consumption of caseins and peptides, leading to the development of texture, aroma, and flavor in cheese during fermentation and ripening [35, 36]. Nevertheless, it is of great importance to assure a well-balanced breakdown of caseins by the strains that can be considered as starter cultures in a dairy product, once excessive proteolysis may develop some undesirable attributes in cheese, as low viscosity and high bitterness [37, 38].
Despite presenting different profiles at 6, 12, and 24 h, all L. lactis subsp. lactis bv. diacetylactis isolates were considered as able to acidify skim milk at 30 °C (Table 2). A rapid drop in pH is crucial during cheese production because it contributes to changes in cheese texture and helps to control the development of undesirable microorganisms; two out of the 15 isolates lowered milk pH by more than 1.25 pH units, but only after 24 h (Q1C5 and Q4C8, Table 2). Studies have shown that most strains of L. lactis are initially slow in acid production [5, 20, 39], as observed for the high acidifying isolates described in this study (Table 2). Herreros et al. [34] presented a similar acidification capacity of L. lactis subsp. lactis and bv. diacetylactis strains, that showed a pH drop to 4.2 after 24 h of incubation. L. lactis subsp. lactis bv. diacetylactis strains are globally used as starter cultures that present the citrate and acetoin/diacetyl pathways regulated and expressed at low pH [40]. Isolates obtained from dairy-related samples were characterized as capable to reduce pH at different levels after 24 h of incubation, while isolates obtained from silage were categorized as low acidifiers (less than 1.5 pH units after 24 h) (Table 2): this result demonstrates that the dairy-related isolates are more adapted to a dairy environment than silage-related isolates, allowing their proper use as starter cultures due to the acidification potential in dairy matrices. Cavanagh et al. [27] showed that non-dairy strains of L. lactis would be unsuitable for use as starters, as they are unable to reach the desired pH. However, because these strains are capable of growing in milk without the use of supplements, they can be considered as non-starter adjunct cultures due to their abilities in producing citrate and diacetyl. Although most of the tested isolates were initially slow acidifiers (after 6 and 12 h), most acid production increased later, after 24 h; at this time, 33.3% of the isolates were categorized as belonging to classes I and II (Table 2). Class III strains with low acidification profiles may contain technological characteristics suitable for adjunct cultures to be used in the production of certain types of cheese [27, 29]. Therefore, the variable acidifying activity indicates that this feature is strain-dependent: based on the recorded genetic profiles, Q1C5 and Q1C10 were highly similar (94.6%, Fig. 1), but presented different acidifying abilities (Table 2). This demonstrates how genetically similar isolates may present different technological characteristics. According to Wouters et al. [41], wild lactococci are usually characterized as less acidifying than commercial strains.
The NaCl tolerance assay revealed that all the bv. diacetylactis isolates were able to grow even at the highest concentrations assessed (Fig. 2). All isolates showed excellent growth in NaCl concentrations up to 4%. Six strains showed low growth at 8% and 10% NaCl. Isolates obtained from silage samples showed a high tolerance to NaCl (up to 10%), and presented a similar growth behavior in all NaCl concentrations. As observed for acidifying potential, growth at NaCl concentrations can be considered as a strains specific-feature: even isolates that shared a high genetic similarity (Fig. 1) presented different growth profiles at different NaCl concentrations (Fig. 2). The ability of a starter culture to adapt and survive in various salt concentrations (including high concentrations) is extremely important during cheese production. Some L. lactis subsp. lactis tested by Perin et al. [30] were able to grow in NaCl at 10% (w/v), while Dal Bello et al. [5] were unable to identify growth of LAB strains inoculated in culture media added with NaCl at concentrations higher than 6% (w/v). Nomura et al. [4] reported that all of the L. lactis subsp. lactis strains grew well in a culture medium with NaCl at 6.5% (w/v), and identified poor growth by some cremoris and bv. diacetylactis strains. The salt tolerance of certain Lactococcus isolates taken from cheese may also reflect an adaptation to the cheese environment (2 to 10% NaCl for different Caciocavallo varieties), and strains isolated from cheeses with the lowest salt content have low salt tolerance [33].
Despite the L. lactis subsp. lactis bv diacetylactis being characterized as promising starter cultures due to their technological features, prior to their potential use by the food or dairy industry, they must be assessed regarding their virulence traits. Lactococcus strains may harbor a number of virulence-related genes, that can confer to them some important pathogenic features (hemolysis, DNAse, lipolysis, among other features), which can pose as a risk for consumers; even not being expressed, such virulence-related genes can be transferred to non-pathogenic strains in the gut and be expressed [29, 42–45]. Associated to the virulence features, the isolates must be assessed according to their antibiotic resistance profiles: pathogenic and resistant strains can represent a challenge for proper treatment, and the genetic elements that confers such resistance can be transferred to other bacteria in the human gut [29, 44, 46]. Finally, LAB are known as capable of producing biogenic amines, important nitrogenous compounds formed due to the decarboxylation of amino acids that can accumulate in fermented dairy products and cause toxicological effects in consumers [30, 44, 47, 48].
Conclusions
Based on the obtained data, we were able to identify L. lactis subsp. lactis bv diacetylactis isolates and to characterize their technological potential, allowing propose their use as starter cultures in the food industry after a deep analysis of their safety for application.
Funding information
CNPq, CAPES (financial code 001), FAPEMIG, and CIRM-BIA for the concession of reference strains.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Luís Augusto Nero, Email: nero@ufv.br.
Antônio Fernandes de Carvalho, Email: antoniofernandes@ufv.br.
References
- 1.Carr FJ, Chill D, Maida N. The lactic acid bacteria: a literature survey. Crit Rev Microbiol. 2002;28(4):281–370. doi: 10.1080/1040-840291046759. [DOI] [PubMed] [Google Scholar]
- 2.Leroy F, De Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol. 2004;15(2):67–78. doi: 10.1016/j.tifs.2003.09.004. [DOI] [Google Scholar]
- 3.Schleifer KH, Kraus J, Dvorak C, Kilpper-Bälz R, Collins MD, Fischer W. Transfer of Streptococcus lactis and related Streptococci to the genus Lactococcus gen. nov. Syst Appl Microbiol. 1985;6(3):183–195. doi: 10.1016/S0723-2020(85)80052-7. [DOI] [Google Scholar]
- 4.Nomura M, Kobayashi M, Narita T, Kimoto-Nira H, Okamoto T. Phenotypic and molecular characterization of Lactococcus lactis from milk and plants. J Appl Microbiol. 2006;101(2):396–405. doi: 10.1111/j.1365-2672.2006.02949.x. [DOI] [PubMed] [Google Scholar]
- 5.Dal Bello B, Cocolin L, Zeppa G, Field D, Cotter PD, Hill C. Technological characterization of bacteriocin producing Lactococcus lactis strains employed to control Listeria monocytogenes in Cottage cheese. Int J Food Microbiol. 2012;153(1–2):58–65. doi: 10.1016/j.ijfoodmicro.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Laroute V, Tormo H, Couderc C, Mercier-Bonin M, Le Bourgeois P, Cocaign-Bousquet M, Daveran-Mingot ML. From genome to phenotype: an integrative approach to evaluate the biodiversity of Lactococcus lactis. Microorganisms. 2017;5(2):27. doi: 10.3390/microorganisms5020027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempler GM, McKay LL. Biochemistry and genetics of citrate utilization in Streptococcus lactis ssp. diacetylactis. J Dairy Sci. 1981;64(7):1527–1539. doi: 10.3168/jds.s0022-0302(81)82721-x. [DOI] [Google Scholar]
- 8.García-Quintáns N, Repizo G, Martín M, Magni C, López P. Activation of the diacetyl/acetoin pathway in Lactococcus lactis subsp. lactis bv. diacetylactis CRL264 by acidic growth. Appl Environ Microbiol. 2008;74(7):1988–1996. doi: 10.1128/AEM.01851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curioni PMG, Bosset JO. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int Dairy J. 2002;12(12):959–984. doi: 10.1016/S0958-6946(02)00124-3. [DOI] [Google Scholar]
- 10.Urbach G. The flavour of milk and dairy products: II. Cheese: Contribution of volatile compounds. Int J Dairy Technol. 1997;50(3):79–89. doi: 10.1111/j.1471-0307.1997.tb01743.x. [DOI] [Google Scholar]
- 11.Smit G, Smit BA, Engels WJM. Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol Rev. 2005;29(3 SPEC. ISS):591–610. doi: 10.1016/j.femsre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Deegan LH, Cotter PD, Hill C, Ross P. Bacteriocins: biological tools for bio-preservation and shelf-life extension. Int Dairy J. 2006;16(9):1058–1071. doi: 10.1016/j.idairyj.2005.10.026. [DOI] [Google Scholar]
- 13.Cotter PD, Ross RP, Hill C. Bacteriocins-a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11(2):95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 14.Delorme C, Godon JJ, Ehrlich SD, Renault P. Mosaic structure of large regions of the Lactococcus lactis subsp. cremoris chromosome. Microbiology. 1994;140(11):3053–3060. doi: 10.1099/13500872-140-11-3053. [DOI] [PubMed] [Google Scholar]
- 15.Beimfohr C, Ludwig W, Schleifer KH. Rapid genotypic differentiation of Lactococcus lactis subspecies and biovar. Syst Appl Microbiol. 1997;20(2):216–221. doi: 10.1016/S0723-2020(97)80068-9. [DOI] [Google Scholar]
- 16.Passerini D, Laroute V, Coddeville M, Le Bourgeois P, Loubière P, Ritzenthaler P, Cocaign-Bousquet M, Daveran-Mingot M-L. New insights into Lactococcus lactis diacetyl- and acetoin-producing strains isolated from diverse origins. Int J Food Microbiol. 2013;160(3):329–336. doi: 10.1016/j.ijfoodmicro.2012.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Pu Z, Dobos M, Limsowtin G (2002) Integrated polymerase chain reaction-based procedures for the detection and identification of species and subspecies of the Gram-positive bacterial genus. J Appl :353–361. [DOI] [PubMed]
- 18.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kempler GM, McKay LL. Improved medium for detection of citrate-fermenting Streptococcus lactis subsp. diacetylactis. Appl Environ Microbiol. 1980;39(4):926–927. doi: 10.1128/AEM.39.4.926-927.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franciosi E, Settanni L, Cavazza A, Poznanski E. Biodiversity and technological potential of wild lactic acid bacteria from raw cows’ milk. Int Dairy J. 2009;19(1):3–11. doi: 10.1016/j.idairyj.2008.07.008. [DOI] [Google Scholar]
- 21.Dal Bello B, Rantsiou K, Bellio A, Zeppa G, Ambrosoli R, Civera T, Cocolin L. Microbial ecology of artisanal products from North West of Italy and antimicrobial activity of the autochthonous populations. LWT Food Sci Technol. 2010;43(7):1151–1159. doi: 10.1016/j.lwt.2010.03.008. [DOI] [Google Scholar]
- 22.Alves MP, Salgado RL, Eller MR, Vidigal PMP, Fernandes de Carvalho A. Characterization of a heat-resistant extracellular protease from Pseudomonas fluorescens 07A shows that low temperature treatments are more effective in deactivating its proteolytic activity. J Dairy Sci. 2016;99(10):7842–7851. doi: 10.3168/jds.2016-11236. [DOI] [PubMed] [Google Scholar]
- 23.Adams DM, Barach JT, Speck ML. Effect of psychrotrophic bacteria from raw milk on milk proteins and stability of milk proteins to ultrahigh temperature treatment. J Dairy Sci. 1976;59(5):823–827. doi: 10.3168/jds.S0022-0302(76)84282-8. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Psoni L, Kotzamanidis C, Yiangou M, Tzanetakis N, Litopoulou-Tzanetaki E. Genotypic and phenotypic diversity of Lactococcus lactis isolates from Batzos, a Greek PDO raw goat milk cheese. Int J Food Microbiol. 2007;114(2):211–220. doi: 10.1016/j.ijfoodmicro.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 26.Kahala M, Mäki M, Lehtovaara A, Tapanainen JM, Katiska R, Juuruskorpi M, Juhola J, Joutsjoki V. Characterization of starter lactic acid bacteria from the Finnish fermented milk product viili. J Appl Microbiol. 2008;105(6):1929–1938. doi: 10.1111/j.1365-2672.2008.03952.x. [DOI] [PubMed] [Google Scholar]
- 27.Cavanagh D, Casey A, Altermann E, Cotter PD, Fitzgerald GF, McAuliffe O. Evaluation of Lactococcus lactis isolates from nondairy sources with potential dairy applications reveals extensive phenotype-genotype disparity and implications for a revised species. Appl Environ Microbiol. 2015;81(12):3961–3972. doi: 10.1128/AEM.04092-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siezen RJ, Bayjanov JR, Felis GE, van der Sijde MR, Starrenburg M, Molenaar D, Wels M, van Hijum SAFT, van Hylckama Vlieg JET. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb Biotechnol. 2011;4(3):383–402. doi: 10.1111/j.1751-7915.2011.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Domingos-Lopes MFP, Stanton C, Ross PR, Dapkevicius MLE, Silva CCG. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017;63:178–190. doi: 10.1016/j.fm.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 30.Perin LM, Belviso S, Dal Bello B, Nero LA, Cocolin L. Technological properties and biogenic amines production by bacteriocinogenic Lactococci and Enterococci strains isolated from raw goat’s milk. J Food Prot. 2017;80(1):151–157. doi: 10.4315/0362-028x.jfp-16-267. [DOI] [PubMed] [Google Scholar]
- 31.Starrenburg M, Hugenholtz J. Citrate fermentation by Lactococcus and Leuconostoc spp. Metab Clin Exp. 1991;57(12):3535–3540. doi: 10.1128/aem.57.12.3535-3540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucey JA, Johnson ME, Horne DS. Invited review: perspectives on the basis of the rheology and texture properties of cheese. J Dairy Sci. 2003;86(9):2725–2743. doi: 10.3168/jds.s0022-0302(03)73869-7. [DOI] [PubMed] [Google Scholar]
- 33.Piraino P, Zotta T, Ricciardi A, McSweeney PLH, Parente E. Acid production, proteolysis, autolytic and inhibitory properties of lactic acid bacteria isolated from pasta filata cheeses: a multivariate screening study. Int Dairy J. 2008;18(1):81–92. doi: 10.1016/j.idairyj.2007.06.002. [DOI] [Google Scholar]
- 34.Herreros MA, Fresno JM, González Prieto MJ, Tornadijo ME. Technological characterization of lactic acid bacteria isolated from Armada cheese (a Spanish goats’ milk cheese) Int Dairy J. 2003;13(6):469–479. doi: 10.1016/S0958-6946(03)00054-2. [DOI] [Google Scholar]
- 35.Liu M, Bayjanov JR, Renckens B, Nauta A, Siezen RJ. The proteolytic system of lactic acid bacteria revisited: a genomic comparison. BMC Genomics. 2010;11(1):5–8. doi: 10.1186/1471-2164-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tulini FL, Hymery N, Haertlé T, Le Blay G, De Martinis ECP. Screening for antimicrobial and proteolytic activities of lactic acid bacteria isolated from cow, buffalo and goat milk and cheeses marketed in the southeast region of Brazil. J Dairy Res. 2016;83(1):115–124. doi: 10.1017/S0022029915000606. [DOI] [PubMed] [Google Scholar]
- 37.Visser S. Proteolytic enzymes and their relation to cheese ripening and flavor: an overview. J Dairy Sci. 1993;76(1):329–350. doi: 10.3168/jds.s0022-0302(93)77354-3. [DOI] [Google Scholar]
- 38.González L, Sacristán N, Arenas R, Fresno JM, Eugenia Tornadijo M. Enzymatic activity of lactic acid bacteria (with antimicrobial properties) isolated from a traditional Spanish cheese. Food Microbiol. 2010;27(5):592–597. doi: 10.1016/j.fm.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Morandi S, Brasca M, Lodi R. Technological, phenotypic and genotypic characterisation of wild lactic acid bacteria involved in the production of Bitto PDO Italian cheese. Dairy Sci Technol. 2011;91(3):341–359. doi: 10.1007/s13594-011-0016-7. [DOI] [Google Scholar]
- 40.Zuljan FA, Mortera P, Alarcón SH, Blancato VS, Espariz M, Magni C. Lactic acid bacteria decarboxylation reactions in cheese. Int Dairy J. 2016;62:53–62. doi: 10.1016/j.idairyj.2016.07.007. [DOI] [Google Scholar]
- 41.Wouters JTM, Ayad EHE, Hugenholtz J, Smit G. Microbes from raw milk for fermented dairy products. Int Dairy J. 2002;12(2–3):91–109. doi: 10.1016/S0958-6946(01)00151-0. [DOI] [Google Scholar]
- 42.Deveau H, Labrie SJ, Chopin MC, Moineau S. Biodiversity and classification of lactococcal phages. Appl Environ Microbiol. 2006;72(6):4338–4346. doi: 10.1128/AEM.02517-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahony J, Murphy J, Van Sinderen D. Lactococcal 936-type phages and dairy fermentation problems: from detection to evolution and prevention. Front Microbiol. 2012;3(SEP):1–9. doi: 10.3389/fmicb.2012.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perin LM, Miranda RO, Todorov SD, Franco BDG d M, Nero LA. Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int J Food Microbiol. 2014;185:121–126. doi: 10.1016/j.ijfoodmicro.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Lemay ML, Tremblay DM, Moineau S. Genome engineering of virulent Lactococcal phages using CRISPR-Cas9. ACS Synth Biol. 2017;6(7):1351–1358. doi: 10.1021/acssynbio.6b00388. [DOI] [PubMed] [Google Scholar]
- 46.Devirgiliis C, Zinno P, Perozzi G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front Microbiol. 2013;4:1–13. doi: 10.3389/fmicb.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buňková L, Buňka F, Hlobilová M, Vaňátková Z, Nováková D, Dráb V. Tyramine production of technological important strains of Lactobacillus, Lactococcus and Streptococcus. Eur Food Res Technol. 2009;229(3):533–538. doi: 10.1007/s00217-009-1075-3. [DOI] [Google Scholar]
- 48.Flasarová R, Pachlová V, Buňková L, Menšíková A, Georgová N, Dráb V, Buňka F. Biogenic amine production by Lactococcus lactis subsp. cremoris strains in the model system of Dutch-type cheese. Food Chem. 2016;194:68–75. doi: 10.1016/j.foodchem.2015.07.069. [DOI] [PubMed] [Google Scholar]