Abstract
Disease risk varies significantly between ethnic groups, however, the clinical significance and implications of these observations are poorly understood. Investigating ethnic differences within the human proteome may shed light on the impact of ancestry on disease risk. We used admixture mapping to explore the impact of genetic ancestry on 237 cardiometabolic biomarkers in 2,216 Latin Americans within the Outcomes Reduction with an Initial Glargine Intervention (ORIGIN) study. We developed a variance component model in order to determine the proportion of variance explained by inter-ancestry differences, and we applied it to the biomarker panel. Multivariable linear regression was used to identify and localize genetic loci affecting biomarker variability between ethnicities. Variance component analysis revealed that 5% of biomarkers were significantly impacted by genetic admixture (p < 0.05/237), including C-peptide, apolipoprotein-E, and intercellular adhesion molecule 1. We also identified 46 regional associations across 40 different biomarkers (p < 1.13 × 10−6). An independent analysis revealed that 34 of these 46 regions were associated at genome-wide significance (p < 5 × 10−8) with their respective biomarker in either Europeans or Latin populations. Additional analyses revealed that an admixture mapping signal associated with increased C-peptide levels was also associated with an increase in diabetes risk (odds ratio [OR] = 6.07 per SD, 95% confidence interval [CI] 1.44 to 25.56, p = 0.01) and surrogate measures of insulin resistance. Our results demonstrate the impact of ancestry on biomarker levels, suggesting that some of the observed differences in disease prevalence have a biological basis, and that reference intervals for those biomarkers should be tailored to ancestry. Specifically, our results point to a strong role of ancestry in insulin resistance and diabetes risk.
Keywords: admixture, biomarker, ancestry, proteome
Introduction
The human proteome plays a principal role in biological processes such as signaling, transport, growth, repair, and defense against infection. These proteins represent intermediate phenotypes, and they are often directly and causally involved in disease pathophysiology. Indeed, many biomarkers are measured clinically and used as non-invasive markers of a patient’s overall health, guiding diagnosis, prognosis, and treatment management.1 However, biomarker profiles have been shown to vary widely between ethnic groups and the clinical significance and implications of these observed differences is poorly understood.2 Furthermore, it is unknown whether these differences correspond to ethnic-specific susceptibility to disease. Disease risk varies significantly between ethnic groups, as well. For instance, Mexican, Latin American, and African populations have a higher risk of type 2 diabetes (T2D) compared to populations with European ancestries.3, 4, 5 This disparity in risk has been hypothesized to be due, at least in part, to genetic and biological factors.6,7
Biomarker differences that exist between populations may also lead to clinical challenges. Consistent differences have been reported for many biomarkers; these include C-reactive protein, vitamin D binding protein, and many circulating adipokines.8, 9, 10, 11, 12 Interpretation of these markers is based on reference intervals which are defined using population values. However, for biomarkers that are markers of disease, this might lead to erroneous diagnosis if ancestry leads to differences in concentrations. For biomarkers that are directly involved in disease progression, this might lead to wrongful evaluation of risk if ancestry leads to increased risk through that mediator. Ideally, these intervals should be determined based on a random sample of healthy individuals from a population similar to the patient. Traditionally, reference intervals have been determined using predominantly European individuals’ intervals, and these do not necessarily extend to other ethnic groups.13
Although differences in levels of biomarkers have been observed between ethnic groups, the reasons for these differences are difficult to determine through classic epidemiological studies. Admixture mapping is a powerful tool, used in genetic epidemiological studies, that may shed light on these observations. Genetic admixture occurs when two or more previously independent populations interbreed, resulting in the introduction of new genetic lineages. This has occurred in Latin Americans, for instance. Latin Americans are an admixed population with Native American, European, and African ancestors. Admixture mapping is applied to recently admixed populations in order to locate disease-causing genetic variants that differ in frequency across ancestral groups.14 The approach is based on the assumption that the frequency of risk alleles varies between populations such that the proportion of ancestry near causal loci will be associated with risk of disease. In this way, differential risk across ancestral groups can be observed at specific genetic loci.15 This approach has been particularly effective in African Americans for identifying novel loci for various diseases.16, 17, 18 Most recently, this technique has been used to reveal novel susceptibility loci in atherosclerosis and albuminuria.19,20
In this study, we used admixture mapping to investigate the impact of ancestry on the human serum proteome by conducting a comprehensive investigation of a multiplex biomarker panel. Specifically, we evaluated the effect of genetic ancestry on 237 serum biomarker concentrations measured in the Latin American population from the ORIGIN (Outcomes Reduction with an Initial Glargine Intervention) trial.21 Although ethnicity has been determined to be a strong predictor of biomarker concentrations, few studies have leveraged the genetic admixture in order to assess the impact of ancestry on biomarker variability that may, in turn, impact their risk of disease. Furthermore, admixture mapping studies offer a unique advantage for identifying genes that confer differential risk between populations because admixture mapping can distinguish biological effects that are due to ancestry at specific loci from individual-level proportions of ancestry, which can be confounded by environmental factors.
Material and Methods
Study Population—ORIGIN
The design and findings of the ORIGIN trial have been described in detail. In brief, 12,537 people who had established cardiovascular risk factors and who also had T2D, impaired glucose tolerance, or impaired fasting glucose were studied. Participants were randomly divided into groups to test two therapies through the use of a factorial design (testing basal insulin glargine versus standard care and omega 3 fatty acid supplements versus placebo); participants were then followed for a median of 6.2 years, watching for cardiovascular events and other health outcomes. As previously described,22 biomarker levels were analyzed in the serum drawn at the beginning of the study from a subset of 8,401 people (66% men; mean age 63.7 years). The analysis was done using a customized human discovery multi-analyte profile (MAP) on the Luminex 100/200 platform, and the biomarkers were selected based on their implications in physiologic processes related to cardiovascular and metabolic diseases. A further subset of 5,078 participants consented to genetic analyses, and 4,147 (1,931 Europeans and 2,216 Latin Americans) passed quality control (QC). Study characteristics were similar across the two groups.
Genotyping
A subset of 5,078 individuals from the ORIGIN study consented to genetic analyses and were genotyped on Illumina’s HumanCore Exome chip. Standard QC measures were used. Single-nucleotide polymorphism (SNPs) were excluded on the basis of low call rate (< 99%), deviation from Hardy-Weinberg (p < 10−6), and low minor allele frequency (MAF) (<0.01 in all ethnic groups). Samples with low call rates (< 99%), sex or ethnicity mismatches, or cryptic relatedness were also removed. We also removed samples from ethnicities with small sample sizes (n < 100). All QC steps were performed using PLINK23 and GCTA.24 After QC, the sample consisted of 4,390 participants and 284,024 SNPs from three ethnic groups (Europeans, Latin Americans, and Africans). Imputation was then performed on the post-QC data in order to predict unobserved genotypes in the study population. Over 30 million SNPs were imputed, allowing for comprehensive coverage of known genetic variants. The 1000 Genomes Project25 was used as the reference panel for ORIGIN imputation, which was performed using the software IMPUTE2.26,27 We removed SNPs imputed with low certainty (info < 0.6, as defined by IMPUTE2).27 For the current report, participants of self-reported Latin American ethnicity comprised the primary analysis group (n = 2,216) and those of European ethnicity were included in the validation and replication analyses (n = 1,931)
Genetic Ancestry Estimation
We used phased, consensus data from the 1000 Genomes Project to create reference panels for Europeans (CEU, FIN, GBR, IBS, and TSI), Africans (ASW, LWK, and YRI), and Asians alleles (CHB, CHS, and JPT, which were used as a proxy for Native American ancestry, as previously described15). (For specific definitions of these population codes, please see Web Resources for a link to the 1000 Genomes Project population codes web page.) Using only genotyped SNPs, we removed ambiguous SNPs and used Beagle28 to phase ORIGIN genotypes. Subsequently, we used RFMix15 to infer the local ancestries of 259,778 SNPs in 2,216 Latin Americans. Probabilities of Asian, European, and African ancestry were derived for each SNP, thus accounting for uncertainty in ancestry ascertainment. Probabilities at each allele ranged from 0 to 1 and were summed at each SNP, representing the dosage of the allele from a given ancestry and ranging from 0 to 2 where, for example, a value of 2 for the European local component at a given SNP would represent both alleles having European ancestry. The procedure has been described in detail elsewhere.15 It is worth clarifying that although local ancestry can be derived for each individual SNP (referred to as “local ancestry components”), each component tags large regions of the genome. Therefore, only a subset of components need to be interrogated in order to fully capture local ancestry variability.
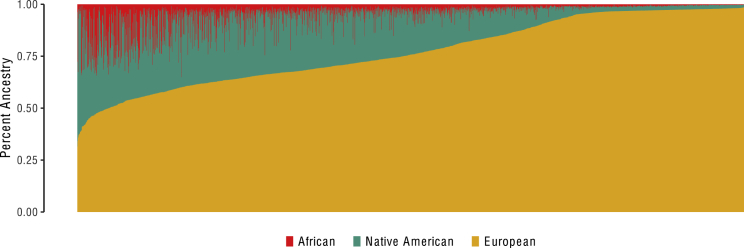
To calculate individual-level ancestries, a set of minimally pruned sites was generated, according to a linkage disequilibrium (LD) correlation matrix based on local SNP European ancestry components, in R (pairwise r2 < 0.95). Specifically, for each chromosome, a square matrix was constructed containing the Pearson’s r2 correlation coefficient between all sites (i.e., pairwise correlation). For example, for any two sites (x and y), the r2 was calculated between the local European components at sitex and sitey. The resulting matrix was pruned agnostically at a threshold of r2 < 0.95. Currently, there is no standard method for pruning local admixture signals, and this threshold was chosen to reduce redundant (identical) associations while retaining as much ancestry information as possible. Pruning using genotype LD (rather than local ancestry LD) is not sensible here because admixture regions are much larger than haplotype blocks across the genome. Therefore, this threshold was selected in an effort to balance over-pruning, which would result in loss of local admixture signals, and under-pruning, which would result in redundant signals. Following pruning, 7,246 local components remained. This set was used for all subsequent analyses. Individual-level (global) ancestry was then obtained for each individual by averaging the ancestry at each of the retained sites (Figure 1). Thus, following this procedure, for each site, each individual had three local ancestry components (one for each of the three ancestral ethnicities) ranging from 0 to 2 and three global ancestry components ranging from 0 to 2, and these represented the average of all locally derived estimates.
Figure 1.
Estimated Admixture Proportion in ORIGIN
Bar graph shows individual ancestry estimates for each of the 2,216 Latin American participants in the Outcomes Reduction with an Initial Glargine Intervention (ORIGIN) study in increasing order of European ancestry estimates. y axis corresponds to the percentages of the genome of each individual originating from each parental population, inferred by the local ancestry estimates.
Genetic Association Models to Determine Contribution of Local Ancestry on Phenotypic Variation
Through the use of simulations, we evaluated the performance of genetic association models in order to capture the phenotypic variance explained by local ancestry. Because associations with global ancestry may represent confounding by environmental or societal factors rather than a true biological difference, we sought to distinguish between local and global effects in order to determine the variance explained according to biological differences (i.e., local ancestry) between ethnic groups. Continuous phenotypes were simulated for each of 2,216 Latin Americans in ORIGIN through the use of the derived local and global ancestry components as predictors. We explored various parameters for their impact on estimated local ancestry variance; these parameters included the effect of non-directional versus directional local effects (i.e., restricting local effects to be positive for a given ancestry in the directional case), the number of causal loci associated with the simulated trait, and the presence and absence of a global ancestry effect. Local directional effects were evaluated to test the models’ ability to distinguish among the many local signals exerting an effect in the same direction versus a single global (confounding) effect. Total trait variance and mean were set at 1 and 0, respectively, in all simulations. For each simulation, a pre-specified set of causal loci ranging from 1 to 10 (1, 2, 3, 5, and 10) were randomly selected from a stringently pruned set of 46 local components (r2 < 0.05) to ensure that independent regions were selected. The pre-defined, unobserved, true local variance ranging from 0 to 0.4 was evenly distributed among the randomly selected causal loci. Similarly, the effect of each global component was standardized and fixed according to a pre-defined overall variance value of either 0 or 0.1. The remaining phenotypic variance was randomly determined. The effect of each locus on the simulated trait was evaluated using adjusted linear models.
Because ancestry, as opposed to genotype data, tends to be highly correlated over longer regions of the chromosome, the number of independent tests estimated is small despite the inclusion of genome-wide ancestry data in the model. Therefore, to determine an appropriate significance level, we performed 10,000 simulations under the null hypothesis, assuming no effect of local ancestry on the simulated trait. For each simulation, a continuous phenotype was derived with no effect of local ancestry and both with and without an effect of global ancestry. Next, each local Asian and African component was tested independently in a linear model adjusted for global Asian and global African components. In other words, for each simulation, 14,492 (7,246 loci times two ethnicities) linear models were tested for an association with the simulated trait. In this way, the reference group was of either non-African or non-Asian ancestry, depending on which component was being evaluated. The lowest p value (pminimum) from the 14,492 independent tests was recorded. We did not identify any difference in distribution of pminimum with and without a global effect. We selected a p value threshold which corresponded to <1% of the pminimum; the result was a significance threshold level of p < 1.13 × 10−6.
For each set of conditions, 100 simulations were completed and both the effect of local ancestry and the effect of global ancestry on trait variance were estimated. We used variance component (VC) models to assess the overall effect of ancestry on the simulated trait through the use of the mmer function in the sommer R package.29 Using local ancestry estimates at the remaining 7,246 sites after pruning (described above), genetic-related matrices (GRMs) were calculated for each ancestry. The local ancestry matrices (2,216 × 7,246) were scaled to have mean of 0 and standard deviation (SD) of 1. Next, the GRM was calculated as the cross-product of the scaled local ancestries. Global Asian ancestry and global African ancestry were each included in the model as fixed effects. Proportions of variance explained by global and local ancestry (both together and separately) were then estimated for each model and compared to the value specified for each simulation. Global ancestry variance was estimated using the regression coefficients from the fixed effect estimates in the VC model. The mmer function provided variance-covariance components for each random effect (i.e., two local ancestry GRMs and residual variance) and were used to estimate local and residual variance accordingly. Total trait variance was estimated as the sum of global, local, and residual variance estimates. Next, we calculated the proportion of variance explained due to local, global, and the sum of local and global ancestry as their respective estimated variance divided by total trait variance. Estimates were recorded for each simulation, and the average (±SD) of each set of conditions was calculated and compared to their unobserved, true, respective values.
We sought to identify the individual loci selected for a causal association with the simulated trait. Specifically, each local ancestry component for both Asian and African ethnicities was independently tested in a linear model with the simulated trait as the dependent variable, adjusted for global Asian and global African components. A forward-selection approach was then used to identify the local components that independently and cumulatively predicted the dependent variable, with a p value for inclusion set at the pre-specified threshold according to simulations under the null (p < 1.13 × 10−6). The minimum p value of each of the 14,492 models representing all local ancestry components for both African and Asian ancestries was evaluated, and if it fell below the threshold for inclusion, the respective local component was added into the predictive model in addition to global African and global Asian components. This process was repeated until no local component association p value fell below 1.13 × 10−6. Because biomarkers with levels falling below the level of quantification in >10% of individuals were analyzed as ordinal variables (resulting in 45 biomarkers transformed into ordinal variables), all simulations were repeated using a simulated ordinal trait to test the models’ ability to perform with a non-continuous dependent variable, and our findings remained consistent (see Supplemental Information).
We then evaluated the proportion of randomly selected causal loci that matched the identified loci based on the forward selection algorithm for a given simulation (i.e., proportion of true causal regions identified). To define a regional association, we used a threshold of r2 > 0.8 with a causal locus. Identified loci with r2 < 0.8 with all randomly selected causal loci were classified as false positives.
Estimation of Effect of Local Ancestry on Serum Biomarkers and Baseline Phenotypes in ORIGIN
The VC model and forward selection process described above were then performed on the 237 measured biomarkers in ORIGIN in an effort to determine the proportion of variance explained by local ancestry for each serum biomarker. A predictive model was constructed for each biomarker according to the following procedure. First, biomarkers were linearly residualized for age and sex. Second, VC models were used to assess the proportion of trait variance explained by local ancestry; these models used global components as fixed effects (as in simulations). Third, linear models were used to test each local component independently for an effect on the residualized biomarker; these models were adjusted for global Asian and global African components. As described above, the minimum p values of all local components were assessed, and any component with a p value less than our inclusion threshold (p < 1.13 × 10−6) was added to the predictive model. This process was then repeated until no p values were less than 1.13 × 10−6. Therefore, for each biomarker, one VC model was used to assess overall local and global variance, and a linear predictive model was constructed which included global African and global Asian components in addition to local components selected from the forward selection algorithm. This forward selection process revealed specific genetic regions which were independently associated with serum biomarkers, residualized for age and sex. Associations were classified as in cis if the identified locus had r2 < 0.8 with any SNP ± 300 Kb for the respective gene.
We sought to further reinforce these local ancestry associations by testing genotype associations in Europeans and Native Latins from the ORIGIN trial (n = 1,931 and 2,216, respectively). For each identified local ancestry association, we implemented the following process. First, an investigation window surrounding the local ancestry signal was created according to pairwise r2 of European local ancestry data. Pairwise r2 was examined both upstream and downstream of the locus of interest until a local ancestry estimate had r2 < 0.8 with the locus of interest to create a window of association. Second, the association of each SNP in this window was tested in ORIGIN Europeans with the respective biomarker through the use of a linear model, adjusted for age, sex, and the first five principal components. Third, the association of each SNP in this window was also tested in ORIGIN Native Latins with the respective biomarker using a linear model, adjusted for age, sex, global ancestry and the corresponding local ancestry components. SNPs with MAF < 0.01 or INFO < 0.6 were removed. Therefore, for each identified local ancestry association, we obtained an estimate of the effect of the local ancestry component on its respective biomarker, and also the effect of SNPs within the derived local ancestry window on the same biomarker, in both ORIGIN Europeans and Native Latin samples. To determine if the local ancestry associations were mainly due to European genotypic associations, we included the effect of the most significant European SNP in the admixture window as a fixed effect in the local ancestry linear model (by weighting the genotype by the beta coefficient obtained in ORIGIN Europeans).
Results
Evaluation of Genetic Association Models Using Simulations
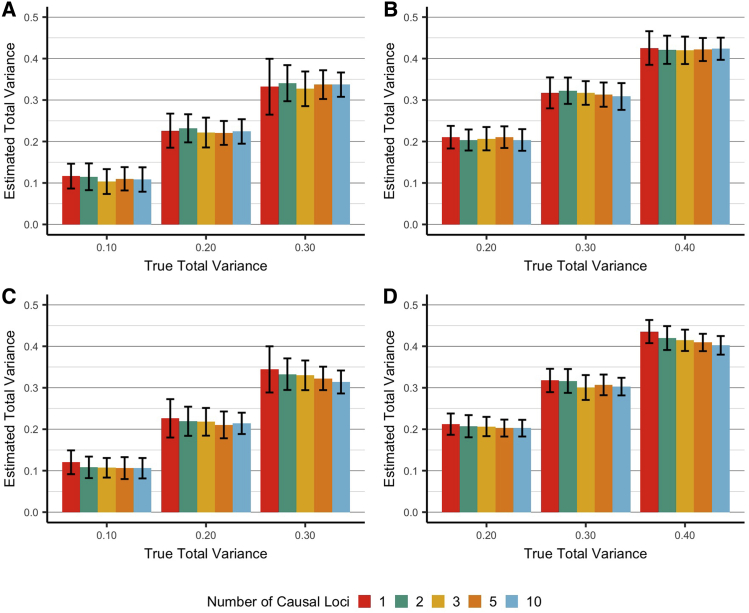
We evaluated the performance of our VC models to estimate the phenotypic variance explained by local admixture associations. In these simulations, we assumed that varying the number of loci (1, 2, 3, 5, or 10) had an ancestry effect on the quantitative trait and that the proportion of variance explained was 0.0, 0.1, 0.2, 0.3, and 0.4, respectively. We then tested conditions with and without a directional condition on the causal ancestry effects (i.e., all effects greater than 0 for a given ancestry) and with and without an effect of global admixture. When a global effect was specified, it was split evenly over the two components (African and Asian), each with a proportion of variance explained of 0.025. Our simulations show that total variance attributed to local and global ancestry can be determined using VC models (Figure 2). Similarly, our simulations show that it is possible to derive unbiased estimates of local variance using VC models (Figure 3). These estimates are stable both with a directional local effect and in the presence of a global effect. However, local estimates were lower in the directional scenarios compared to non-directional. For instance, considering a scenario with 10 causal loci and local variance specified at 0.1, two-way ANOVA revealed significant differences between directional and non-directional simulations (Figure 3A and 3B versus Figure 3C and 3D, p < 5 × 10−16) and no difference between simulations with and without a global effect (Figure 3A and 3C versus Figure 3B and 3D, p = 0.73). This is likely due to the fact that it is difficult for the model to distinguish a global effect from a directional local signal, particularly when many causal loci are present.
Figure 2.
Estimated Proportion of Variance Explained by Local and Global Ancestry under Various Conditions
Average (±SD) estimated local and global variance explained under various simulated conditions. The sum of the true, unobserved, local and global variances was pre-specified. Global variance was set at either 0 (A and C) or 0.1 (B and D), and local variance was set as 0.1, 0.2, and 0.3 as determined by 1, 2, 3, 5, or 10 causal SNPs. The sum of the two variances is shown on the x axis. Each bar represents an average of 100 simulations; error bars show ±SD. (A and B) illustrate simulated conditions with no directional condition, while (C and D) restrict local effects to be greater than 0. (A and C) illustrate simulated conditions with no global effect, and (B and D) each have a pre-specified global effect.
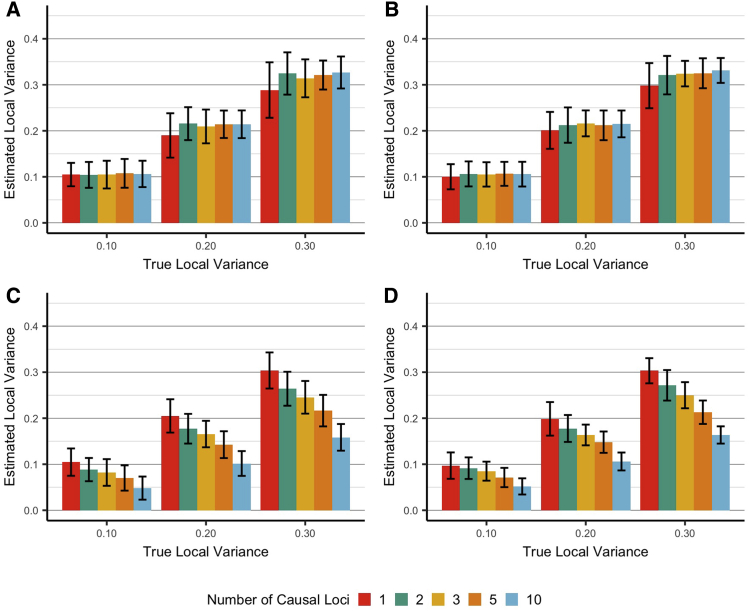
Figure 3.
Estimated Proportion of Variance Explained by Local Ancestry under Various Conditions
Average (±SD) estimated local variance explained under various simulated conditions. True, unobserved, local variances were pre-specified at 0.1, 0.2, and 0.3 (x axis) as determined by 1, 2, 3, 5, or 10 causal SNPs. Each bar represents an average of 100 simulations; error bars show ± SD. (A and B) illustrate simulated conditions with no directional condition, while (C and D) restrict local effects to be greater than 0. (A and C) illustrate simulated conditions with no global effect, and (B and D) each have a pre-specified global effect.
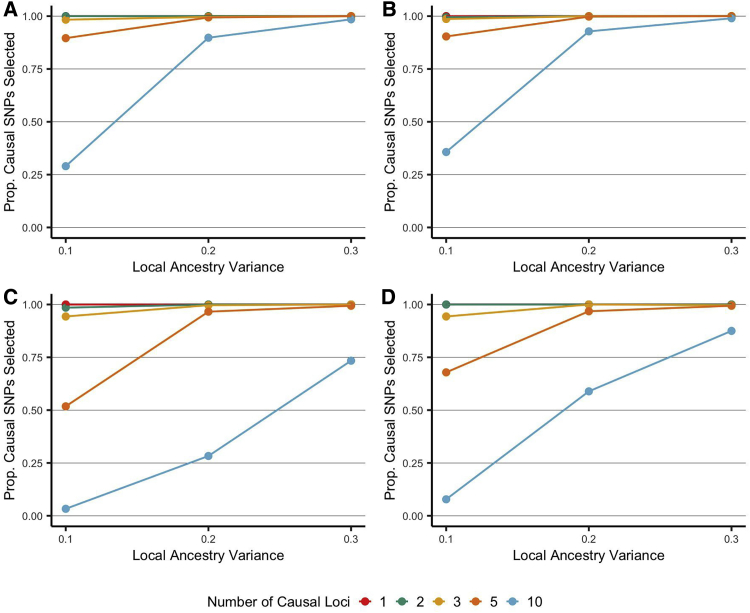
We also sought to determine the ability of the model to select the true, unobserved causal loci. The proportion of causal SNPs selected increased as specified local variance increased (Figure 4), and this did not vary significantly across conditions. When only one SNP was specified as having an effect on the phenotype, the algorithm performed well and identified this locus in >95% of simulations when local variance was greater than 0.05. Conversely, as the number of causal loci increased, the resulting effects were diluted across the randomly selected SNPs, and power to detect individual loci decreased. Consequently, the algorithm was unable to detect all of the true, causal SNPs. This pattern was apparent for all conditions; however, this was strongest in the presence of directional local effect (Figure 4C and 4D). Specifically, a smaller proportion of causal loci were identified on average (e.g., 10 causal loci, local variance specified at 0.05, ANOVA Figure 4A and 4B versus Figure 4C and 4D: p < 5 × 10−16).
Figure 4.
Proportion of Causal SNPs Selected under Various Conditions
Proportion of selected SNPs which were causal or regional, selected by the forward selection algorithm under various simulated conditions. Proportion of causal SNPs (y axis) was calculated as: (number of selected causal SNPs + number of selected regional SNPs) / (number of true, unobserved, causal SNPs). True, unobserved, local variances were pre-specified at 0.1, 0.2, and 0.3 (x axis) as determined by 1, 2, 3, 5 or 10 causal SNPs. (A and B) illustrate simulated conditions with no directional condition, while (C and D) restrict local effects to be greater than 0. (A and C) illustrate simulated conditions with no global effect, and (B and D) each have a pre-specified global effect.
Estimation of Effect of Local Ancestry in ORIGIN
The VC and forward selection models tested through simulations were then applied to the 237 ORIGIN biomarkers. For each biomarker, a model was built, comprised of global Asian and African components in addition to local components selected according to the forward selection algorithm. The proportion of variance attributed to local variance was estimated from the VC model and the individual associated loci (p < 1.13 × 10−06) identified in the linear model were inspected. VC models revealed that 5% (11/237) of biomarkers have a significant proportion of variance explained by both local ancestries after adjusting for multiple hypothesis testing (p < 0.05/237); the proportion of variance explained ranged from 0.11 to 0.24 (see Table 1). Models were also run after regressing out smoking status (yes or no), body mass index (BMI), LDL cholesterol levels, and fasting plasma glucose in addition to age and sex, and the same 11 biomarkers were found to be significant (see Table S2). The global associations and estimated variance were also evaluated as fixed effects from the VC model (Figure 5). We identified 23 and six global African and Asian associations, respectively, representing 12% (29/237) of biomarkers (see Table 2).
Table 1.
Summary of Biomarkers with Significant Proportion of Variation Explained by Local Ancestry in Using VC Analysis (p < 0.05/237)
| Biomarker | Both | African | Asian | |||
|---|---|---|---|---|---|---|
| Proportion Explained and p Value | Proportion Explained(95% CI) | p Value | Proportion Explained(95% CI) | p Value | Proportion Explained(95% CI) | p Value |
| C-peptide | 0.24 (0.16, 0.32) | 1.6 × 10−11 | 0.22 (0.15, 0.29) | 9.9 × 10−12 | 0.02 (−0.02, 0.06) | 0.13 |
| eotaxin-3 | 0.22 (0.14, 0.29) | 5.0 × 10−10 | 0.03 (0.00, 0.07) | 0.02 | 0.19 (0.12, 0.25) | 8.1 × 10−9 |
| clusterin | 0.18 (0.11, 0.25) | 2.2 × 10−8 | 0.16 (0.10, 0.22) | 1.6 × 10−8 | 0.02 (−0.01, 0.06) | 0.10 |
| fatty acid-binding protein liver | 0.13 (0.07, 0.19) | 2.4 × 10−6 | 0.11 (0.06, 0.16) | 1.3 × 10−6 | 0.02 (−0.02, 0.06) | 0.13 |
| intercellular adhesion molecule-1 | 0.14 (0.08, 0.21) | 3.1 × 10−6 | 0.08 (0.03, 0.13) | 0.00018 | 0.06 (0.01, 0.11) | 0.0042 |
| apolipoprotein E | 0.14 (0.07, 0.20) | 4.0 × 10−6 | 0.10 (0.05, 0.15) | 9.4 × 10−6 | 0.03 (−0.01, 0.08) | 0.05 |
| Fas ligand | 0.14 (0.08, 0.20) | 4.6 × 10−6 | 0.08 (0.03, 0.12) | 0.00030 | 0.06 (0.02, 0.11) | 0.0037 |
| alpha-2 macroglobulin | 0.12 (0.06, 0.18) | 1.2 × 10−5 | 0.03 (0.00, 0.06) | 0.038 | 0.09 (0.04, 0.14) | 0.00010 |
| apolipoprotein A-IV | 0.12 (0.06, 0.18) | 3.2 × 10−5 | 0.07 (0.03, 0.12) | 0.00039 | 0.05 (0.00, 0.09) | 0.017 |
| interleukin-2 | 0.12 (0.06, 0.18) | 4.0 × 10−5 | 0.11 (0.06, 0.16) | 1.1 × 10−5 | 0.02 (−0.02, 0.06) | 0.22 |
| paraoxanase-1 | 0.11 (0.05, 0.17) | 8.7 × 10−5 | 0.07 (0.03, 0.12) | 0.00028 | 0.04 (−0.01, 0.08) | 0.048 |
Biomarkers were residualized for age and sex.
CI = confidence interval
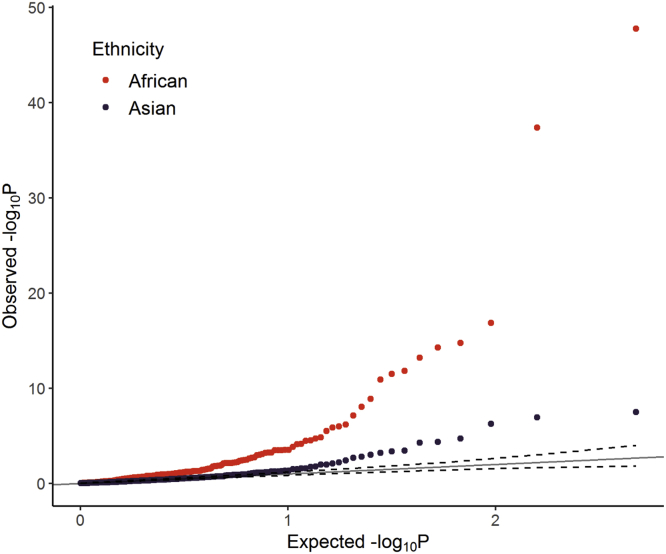
Figure 5.
Global Ancestry QQ Plot
QQ plot of association statistics of the effect of African (red) and Asian (blue) global ancestry on biomarker levels. Variance component models were used to assess the effect of global ancestry, with local ancestry included as random effects. Biomarker levels were first residualized for age and sex.
Table 2.
Summary of Biomarkers with Significant Global Ancestry Association for Either African or Asian Global Ancestry (p < 0.05/237)
| Biomarker | Global African Ancestry | Global Asian Ancestry | ||
|---|---|---|---|---|
| β and p Value | β (95% CI) | p Value | β (95% CI) | p Value |
| Kallikrein 5 | −0.33 (−0.40, −0.25) | <5 × 10−10 | 0.21 (0.02, 0.40) | 0.027 |
| vitronectin | −0.29 (−0.33, −0.25) | <5 × 10−10 | 0.07 (−0.45, 0.59) | 0.79 |
| factor VII | −0.24 (−0.27, −0.21) | <5 × 10−10 | 0.03 (−0.19, 0.25) | 0.81 |
| insulin-like growth factor binding protein 5 | 0.17 (0.13, 0.22) | 1.8 × 10−15 | −0.15 (−0.50, 0.20) | 0.40 |
| immunoglobulin M | 0.24 (0.18, 0.30) | 5.3 × 10−15 | 0.00 (−0.31, 0.31) | 0.98 |
| apolipoprotein B | −0.74 (−0.93, −0.55) | 6.2 × 10−14 | 0.04 (−0.33, 0.40) | 0.84 |
| monocyte chemotactic protein 4 | 0.54 (0.39, 0.69) | 1.6 × 10−12 | 0.08 (0.02, 0.14) | 0.011 |
| interleukin-12 subunit p40 | −0.08 (−0.10, −0.06) | 3.2 × 10−12 | 0.01 (−0.22, 0.24) | 0.95 |
| resistin | 0.34 (0.24, 0.44) | 1.3 × 10−11 | 0.01 (−0.20, 0.22) | 0.95 |
| hepatocyte growth factor receptor | −0.27 (−0.35, −0.18) | 1.3 × 10−9 | 0.10 (−0.34, 0.55) | 0.65 |
| ficolin-3 | −0.13 (−0.17, −0.09) | 9.0 × 10−9 | −0.19 (−0.48, 0.09) | 0.19 |
| protein S100-A4 | 0.55 (0.35, 0.75) | 7.3 × 10−8 | 0.07 (−0.04, 0.17) | 0.22 |
| cortisol | −0.41 (−0.58, −0.25) | 6.5 × 10−7 | −0.13 (−0.38, 0.13) | 0.33 |
| 6Ckine | −0.27 (−0.38, −0.16) | 1.1 × 10−6 | −0.18 (−0.39, 0.02) | 0.080 |
| immunoglobulin E | 0.37 (0.22, 0.52) | 1.4 × 10−6 | 0.45 (0.16, 0.74) | 0.0020 |
| hepatocyte growth factor | −0.46 (−0.65, −0.26) | 3.2 × 10−6 | −0.18 (−0.37, 0.02) | 0.073 |
| ferritin | −0.36 (−0.52, −0.20) | 1.5 × 10−5 | −0.03 (−0.36, 0.30) | 0.86 |
| adrenomedullin | −0.62 (−0.90, −0.33) | 2.0 × 10−5 | −0.23 (−0.54, 0.07) | 0.13 |
| creatine kinase-MB | −0.61 (−0.89, −0.32) | 3.0 × 10−5 | −0.10 (−0.48, 0.28) | 0.61 |
| prostatic acid phosphatase | −0.26 (−0.38, −0.14) | 3.4 × 10−5 | −0.07 (−0.17, 0.02) | 0.13 |
| methylglyoxal | −0.50 (−0.74, −0.25) | 6.8 × 10−5 | −0.05 (−0.54, 0.44) | 0.83 |
| glucose-6-phosphate isomerase | 0.40 (0.20, 0.60) | 8.1 × 10−5 | 0.17 (−0.37, 0.71) | 0.54 |
| sex hormone-binding globulin | 0.26 (0.13, 0.40) | 0.00015 | 0.17 (−0.11, 0.45) | 0.24 |
| mesothelin | 0.25 (−0.01, 0.52) | 0.064 | 0.22 (0.14, 0.29) | 3.2 × 10−8 |
| thrombospondin-1 | 0.00 (−0.24, 0.24) | 0.99 | 0.12 (0.08, 0.17) | 1.21 × 10−7 |
| pulmonary and activation-regulated chemokine | 0.27 (−0.08, 0.62) | 0.13 | 0.25 (0.16, 0.35) | 5.4 × 10−7 |
| T lymphocyte-secreted protein I-309 | 0.23 (0.05, 0.41) | 0.014 | −0.07 (−0.11, −0.04) | 1.9 × 10−5 |
| pigment epithelium derived factor | −0.01 (−0.15, 0.14) | 0.92 | −0.26 (−0.39, −0.14) | 4.6 × 10−5 |
| chemokine CC-4 | 0.00 (−0.09, 0.09) | 0.98 | −0.53 (−0.79, −0.27) | 5.5 × 10−5 |
β per SD increase in global ancestry. Global ancestry is included as fixed effects in the variance component model. Biomarkers were residualized for age and sex.
CI = confidence interval
Using the fixed-effect forward-selection framework, 17% (40/237) of biomarkers were found to have at least one significant local association, and five of these 40 biomarkers overlapped with the 11 associations identified using VC analysis. A total of 46 local components were associated with these biomarkers (i.e., some biomarkers were associated with more than one local component) (see Table S3). Of the 46 local ancestry associations identified, 55% (25/46) were in trans and 45% (21/46) were in cis with the gene encoding the corresponding protein or protein component of the biomarker for which an association was found. Five biomarkers investigated were not direct gene products (e.g., cortisol), and therefore they could not have any cis associations by this definition. One local association identified with Asian ancestry, on chromosome 14, was with one such biomarker, methylglyoxal (included in the 25 trans associations). The number of local associations was similar across ethnicities, with 21 and 25 African and Asian associations, respectively.
Replication in ORIGIN Europeans revealed that 33 of these 46 regions had genotype associations at genome-wide significance with their corresponding biomarkers (p < 5 × 10−8), nine in trans and 24 in cis. Notably, five biomarkers were significantly associated with rs12075, located in the ARCK1 gene which encodes Duffy antigen receptor, which is responsible for the Duffy blood group system. Replication in ORIGIN Native Latin participants revealed an additional cis association at the level of genome-wide significance. Therefore, there were only 34 local associations with no corresponding genome-wide-significant association in either European or Native Latin participants. A summary of local associations and their corresponding genotypic associations in Europeans and Native Latins can be found in Tables S3 and S4. Finally, adjustment for the European SNP association as a fixed effect in the local ancestry linear model resulted in attenuation of 14 of the 46 local ancestry associations (p > 0.05).
Evaluation of the Role of C-Peptide in Disparities in T2D Risk among Ethnic Groups
Our analysis revealed that C-peptide is the biomarker with the most significant involvement of ancestry in determining its levels. Specifically, we found that 24% (95% confidence interval [CI] 16% to 32%, p = 1.7 × 10−11) of the variance of C-peptide is due to local ancestry in Latin Americans, largely due to an effect of African ancestry. We also identified two local ancestry components associated with its levels (p < 1.13 × 10−6) (Figure 6). Our analysis revealed a region on chromosome 9 (with the most significant ancestry association at rs4149261) to have a positive association with C-peptide levels and a region on chromosome 2 (with the most significant ancestry association at rs3769050) to have a negative association with C-peptide (see Table S4). Furthermore, C-peptide has direct medical relevance because of its physiological importance as a marker of insulin secretion, and also as a clinical biomarker used in the recently refined classification of adult-onset diabetes.30 Because diabetes risk is also well known to vary among ethnicities, we sought to explore the roles of C-peptide and genetic ancestry in the context of T2D in order to further elucidate this relationship.
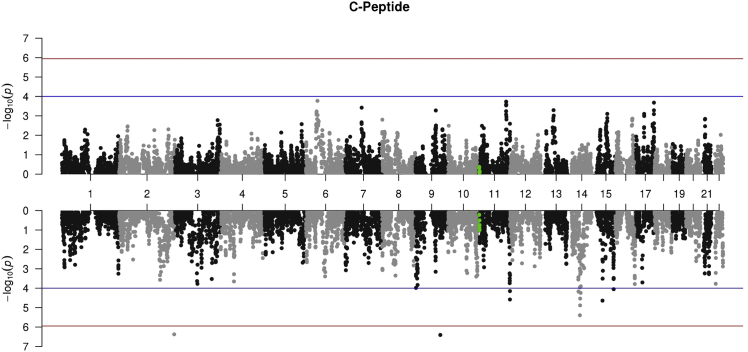
Figure 6.
Manhattan Plot of Admixture Mapping of C-Peptide Protein
Mirror Manhattan plot of the association between local Asian ancestry (top) and local African ancestry (bottom) with C-peptide, made using the additive estimated model as determined by the forward selection algorithm. In other words, each point represents the association between a single local component and C-peptide, after residualizing for age and sex, adjusted for global African and Asian ancestry and the two significant SNPs falling below (for African) or above (for Asian) the selected threshold (shown in red), based on experiment-wide adjusted p value (1.13 × 10−6). Negative log 10 p values are plotted against each local component’s respective position on each chromosome. The blue line corresponds to nominal significance of p = 1 × 10−4. Local components in LD (r2 > 0.8) with any SNP within 300KB of the gene encoding c-peptide protein are shown in green.
First, we sought to determine the impact of including a glycemic-related weighted genetic risk score (GRS) as a fixed effect in the VC model in order to test whether local admixture associations could be explained by known genetic associations from large genome-wide association study (GWAS) meta-analyses. Using estimates from public consortia, we tested a GRS for T2D, HbA1C, fasting glucose, fasting insulin, and 2 h glucose.31,32 The effect of local ancestry remained significant in all models (data not shown). Second, we evaluated the genomic region surrounding the two local ancestry components that were found to be associated with C-peptide levels for association with glycemic traits. Specifically, African local ancestry components on chromosomes 2 and 9 (Figure 6) were shown to be associated with C-peptide levels. Using admixture LD, we derived a local ancestry window (r2 < 0.8) for each local ancestry component and looked to see whether SNPs in these windows were associated with T2D and glycemic traits in DIAGRAM or MAGIC databases.31,32 After adjusting for multiple hypothesis testing, we found no significant associations. Third, we tested the effects of the two local ancestry components associated with C-peptide for an effect on baseline T2D prevalence and insulin resistance in ORIGIN. Insulin resistance was measured as defined previously, using the insulin res in the ORIGIN trial to achieve normoglycemia.33 Models assessing local ancestry associations demonstrated that the African local estimate associated with increased C-peptide at rs4149261 was also associated with increased risk of T2D (OR = 6.07 per SD, 95% CI 1.44 to 25.56, p = 0.01) and median dose of insulin used (per kg of fat-free mass, log transformed) (β = 0.54 per SD, 95% CI 0.16 to 0.92, p = 0.005). However, African local estimates at rs3769050 were not associated with T2D or insulin resistance (p > 0.05).
Discussion
Marked differences in biomarker profiles between populations have been investigated and previously reported.2 However, these observations have not been fully elucidated, and causes may include genetic, lifestyle, or socio-economic factors. In this report, we sought to explore the impact of genetic ancestry on the human proteome and the implications of this for health and disease. We first developed a model to investigate phenotypic variation among admixed individuals. Through simulations, we show that the proportion of variance due to local admixture can be estimated using a VC model, and a forward selection model can be used to reveal causal loci associated with biomarker levels. Using these models, we found that local ancestry affects at least 19% (46/237) of biomarkers with 5% of biomarkers having more than 10% of phenotypic variance explained by local ancestry in Latin Americans with dysglycemia. Additionally, 12% of biomarkers had significant global associations, however, global associations may be confounded by lifestyle or socio-economic factors and might not necessarily represent bona fide biological effects. Local associations, conversely, represent true biological effects and implicate genomic regions involved in phenotypic variance.
Genetic ancestry was found to have a particularly strong influence on C-peptide, and this influence was almost entirely due to an effect of African ancestry. C-peptide is a well-established clinical biomarker which can be used to distinguish type 1 and 2 diabetes.34 Indeed, diabetes is well known to exhibit differential risk patterns among ethnic groups, and this is consistent with these findings.3, 4, 5 We identified two African local components that had an effect on C-peptide. Further examination of these local ancestry components revealed that the region at rs4149261, which was associated with increased C-peptide levels, was also associated with an increased risk of diabetes and insulin resistance. These results validate the use of biomarkers as intermediate endpoints to study the effect of genetic ancestry on disease risk. Because C-peptide is a strong marker of insulin secretion, these findings point to the role of insulin resistance rather than insulin deficiency to explain the inter-ancestry difference in diabetes risk. While both local components showed that increased levels of C-peptide are associated with increased risk of T2D, the ancestry-specific effects on C-peptide levels were inconsistent. In other words, African ancestry increased C-peptide levels at one locus and decreased C-peptide levels at another locus. Therefore, more studies are needed to further resolve the disparity in diabetes risk among ethnic groups.
The genetics of biomarker concentrations have been extensively investigated in the context of GWAS.35 Numerous loci have been identified for many biomarkers, and these loci have also been linked to disease; this suggests causal relationships and potential drug targets. However, few studies have leveraged genetic admixture as a complimentary approach for discovering novel chromosomal regions that impact biomarker levels. Our analysis revealed 46 regions linked to biomarker concentration. Specifically, we found an association between local Asian ancestry and ACE levels, and further genotypic mapping suggests this is an effect at the ACE locus. ACE has a well-established role in regulating blood pressure and is also used to diagnose sarcoidosis. Response to ACE inhibitors has been shown to differ among ethnic groups, raising the possibility that current dosage guidelines may not be applicable to non-European ethnicities.36, 37, 38 Additionally, we identified five biomarkers that are associated with rs12075 within the Duffy antigen receptor gene, which encodes for the glycosylated membrane protein and is a non-specific receptor for several cytokines. This gene exhibits known genetic admixture, and variation in this gene are responsible for the Duffy blood group system.39 The association of the ARCK1 variant with multiple protein levels replicates previous findings, substantiating a potential role for Duffy antigen receptor for chemokines (DARC) in the regulation of serum cytokines.40 Finally, 32 of these 46 regional local associations remained significant after we adjusted for the most significant European GWAS signal in the region. These findings point to a polygenic model whereby the local ancestry associations are capturing large genomic regions which harbor many genetic variants that each confer a very small effect on biomarker concentrations, and some of these regions might be monomorphic in one or more ethnicities.
Understanding the impact of genetics on biomarker profiles also has clinical implications. Predictive thresholds for each specific ethnic group are necessary for accurate risk stratification. Otherwise, there is potential for misclassification of risk and inappropriate use of pharmacotherapies. Notably, after multiple hypothesis testing, we found that 5% of biomarkers are affected by local ancestry, and 30% showed nominal significance (p < 0.05) ranging from a 0.05 to a 0.31 proportion of variation due to an effect of ancestry. These findings suggest that these biomarkers harbor true biological inter-ancestry differences in concentration that are genetically determined. These differences may lead to differences in disease risk and clinical diagnosis. For instance, we found local associations with clinically relevant biomarkers, including vitamin-D-binding protein, apolipoprotein-E, and vascular endothelial growth factor. These results are consistent with previous reports and demonstrate that specific genetic polymorphisms may partially explain the observed differences in concentrations between populations.41,42 These findings may have implications for the interpretation of clinical markers across different ethnic groups.
A few limitations are worth mentioning. First, for the 11 biomarkers for which we found significant effects of local ancestry, we did not identify a specific genetic locus contributing to the variation of six of these biomarkers. These results are consistent with the polygenic model of inheritance, which is hypothesized to underlie many complex traits. According to this model, a large number of loci of small effect sizes together explain the variation of a single trait, such as a biomarker. If hundreds of genetic variants contribute to the observed differences between Asian and African ancestry for a single biomarker, relative to European ancestry, then the proportion of Asian and African ancestry in Latins will act as a proxy for the overall contribution of variants. However, identification of any specific variant will require an appropriately large sample size. Indeed, our simulations have shown that even with 10 loci, the power to detect local associations was very weak, particularly in the presence of directional associations. Second, we identified a local association for 17% (40/237) of biomarkers, however, after multiple hypothesis testing, the VC models showed only 5% of biomarkers to have a significant effect of ancestry. These results suggest that power was limited in our VC analysis compared to the linear model, and larger studies using VC analysis to identify additional markers with an effect of ancestry are needed. Third, pruning was performed based on European ancestry, which does not necessarily represent the genetic architecture of the Asian and/or African ancestral components. Finally, we were not able to identify a significant, corresponding genotype association in either Europeans or Native Latins for every local association identified. This could be because multiple causal variants account for the local association, and we were underpowered for this, or because the causal variant was not well tagged in our study. Likewise, in the Native Latin GWAS, the causal variant(s) could be perfectly correlated with ancestry, and therefore impossible to distinguish from local ancestry itself. It is also worth noting that we did not have access to an African or an Asian cohort, so we could not assess these genotypic relationships in these ancestries.
Genetically admixed populations provide a powerful model for exploring the contribution of genetics to differences in biomarker concentrations among populations. Studying Latin Americans within the framework of a large, international study, we provide evidence for an effect of genetic ancestry on biomarker variability. Our results show that ancestry has a role in the concentration of at least 5% of biomarkers, although this is likely a lower boundary. This finding has many implications, namely that differences in disease prevalence likely have biological bases in many cases, and that use of reference intervals for those biomarkers should be tailored to ancestry. These results highlight the need for specific cutoff values and prognostic measures to be determined for each ethnicity and implemented accordingly. Finally, we also show that some loci appear to have pleiotropic ancestry effects and therefore appear to be of particular importance. Because serum proteins are frequently dysregulated in disease, identification of factors that determine protein variability is a clinical priority. These findings shed light on the contribution of ancestry in disease and pave the way for better informed, ethnicity-specific, defined cut-offs. Further research will be needed in order to identify specific factors responsible for these differences and to gain a better understanding of underlying biological mechanisms.
Declaration of Interests
H.C.G. has received consulting fees from Sanofi, Novo Nordisk, Lilly, AstraZeneca, Boehringer Ingelheim, and GlaxoSmith-Kline and support for research or continuing education through his institution from Sanofi, Lilly, Takeda, Novo Nordisk, Boehringer Ingelheim, and AstraZeneca. G.P. has received consulting fees from Sanofi, Bristol Myers Squibb, Lexicomp, and Amgen and support for research through his institution from Sanofi. Dr. Hess is an employee of Sanofi.
Acknowledgments
The ORIGIN trial and biomarker project were supported by Sanofi and the Canadian Institutes of Health Research (CIHR; award number 125794). H.C.G. received support from the Population Health Institute Chair in Diabetes Research and Care; G.P. received support from the Canada Research Chair in Genetic and Molecular Epidemiology and the CISCO Professorship in Integrated Health Systems; and Z.K. received support from the Swiss National Science Foundation.
Published: February 13, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.01.016.
Web Resources
1000 Genomes Project, https://www.genome.gov/27528684/1000-genomes-project
1000 Genomes Project population codes, https://www.internationalgenome.org/category/population/
Supplemental Data
References
- 1.Biomarkers Definitions Working Group Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Tahmasebi H., Trajcevski K., Higgins V., Adeli K. Influence of ethnicity on population reference values for biochemical markers. Crit. Rev. Clin. Lab. Sci. 2018;55:359–375. doi: 10.1080/10408363.2018.1476455. [DOI] [PubMed] [Google Scholar]
- 3.Florez J.C., Price A.L., Campbell D., Riba L., Parra M.V., Yu F., Duque C., Saxena R., Gallego N., Tello-Ruiz M. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52:1528–1536. doi: 10.1007/s00125-009-1412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spanakis E.K., Golden S.H. Race/ethnic difference in diabetes and diabetic complications. Curr. Diab. Rep. 2013;13:814–823. doi: 10.1007/s11892-013-0421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowler W.C., Pettitt D.J., Saad M.F., Bennett P.H. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab. Rev. 1990;6:1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 6.Hara K., Fujita H., Johnson T.A., Yamauchi T., Yasuda K., Horikoshi M., Peng C., Hu C., Ma R.C.W., Imamura M., DIAGRAM consortium Genome-wide association study identifies three novel loci for type 2 diabetes. Hum. Mol. Genet. 2014;23:239–246. doi: 10.1093/hmg/ddt399. [DOI] [PubMed] [Google Scholar]
- 7.Williams A.L., Jacobs S.B.R., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P., Aguilar-Salinas C.A., SIGMA Type 2 Diabetes Consortium Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gijsberts C.M., den Ruijter H.M., Asselbergs F.W., Chan M.Y., de Kleijn D.P.V., Hoefer I.E. Biomarkers of Coronary Artery Disease Differ Between Asians and Caucasians in the General Population. Glob. Heart. 2015;10:301–311.e11. doi: 10.1016/j.gheart.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Morimoto Y., Conroy S.M., Ollberding N.J., Kim Y., Lim U., Cooney R.V., Franke A.A., Wilkens L.R., Hernandez B.Y., Goodman M.T. Ethnic differences in serum adipokine and C-reactive protein levels: the multiethnic cohort. Int. J. Obes. 2014;38:1416–1422. doi: 10.1038/ijo.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan U.I., Wang D., Sowers M.R., Mancuso P., Everson-Rose S.A., Scherer P.E., Wildman R.P. Race-ethnic differences in adipokine levels: the Study of Women’s Health Across the Nation (SWAN) Metabolism. 2012;61:1261–1269. doi: 10.1016/j.metabol.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Talib H.J., Ponnapakkam T., Gensure R., Cohen H.W., Coupey S.M. Treatment of Vitamin D Deficiency in Predominantly Hispanic and Black Adolescents: A Randomized Clinical Trial. J. Pediatr. 2016;170 doi: 10.1016/j.jpeds.2015.11.025. 266–72.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielson C.M., Jones K.S., Bouillon R., Chun R.F., Jacobs J., Wang Y., Hewison M., Adams J.S., Swanson C.M., Lee C.G., Osteoporotic Fractures in Men (MrOS) Research Group Role of Assay Type in Determining Free 25-Hydroxyvitamin D Levels in Diverse Populations. N. Engl. J. Med. 2016;374:1695–1696. doi: 10.1056/NEJMc1513502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim E., Miyamura J., Chen J.J. Racial/Ethnic-Specific Reference Intervals for Common Laboratory Tests: A Comparison among Asians, Blacks, Hispanics, and White. Hawaii J. Med. Public Health. 2015;74:302–310. [PMC free article] [PubMed] [Google Scholar]
- 14.Sankararaman S., Sridhar S., Kimmel G., Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu X., Luke A., Cooper R.S., Quertermous T., Hanis C., Mosley T., Gu C.C., Tang H., Rao D.C., Risch N., Weder A. Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet. 2005;37:177–181. doi: 10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 17.Freedman M.L., Haiman C.A., Patterson N., McDonald G.J., Tandon A., Waliszewska A., Penney K., Steen R.G., Ardlie K., John E.M. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc. Natl. Acad. Sci. USA. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown L.A., Sofer T., Stilp A.M., Baier L.J., Kramer H.J., Masindova I., Levy D., Hanson R.L., Moncrieft A.E., Redline S. Admixture Mapping Identifies an Amerindian Ancestry Locus Associated with Albuminuria in Hispanics in the United States. J. Am. Soc. Nephrol. 2017;28:2211–2220. doi: 10.1681/ASN.2016091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shendre A., Wiener H., Irvin M.R., Zhi D., Limdi N.A., Overton E.T., Wassel C.L., Divers J., Rotter J.I., Post W.S., Shrestha S. Admixture Mapping of Subclinical Atherosclerosis and Subsequent Clinical Events Among African Americans in 2 Large Cohort Studies. Circ Cardiovasc Genet. 2017;10:e001569. doi: 10.1161/CIRCGENETICS.116.001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstein H., Yusuf S., Riddle M.C., Ryden L., Bosch J., ORIGIN Trial Investigators Rationale, design, and baseline characteristics for a large international trial of cardiovascular disease prevention in people with dysglycemia: the ORIGIN Trial (Outcome Reduction with an Initial Glargine Intervention) Am. Heart J. 2008;155:26–32, 32.e1–32.e6. doi: 10.1016/j.ahj.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Gerstein H.C., Paré G., McQueen M.J., Haenel H., Lee S.F., Pogue J., Maggioni A.P., Yusuf S., Hess S., Outcome Reduction With Initial Glargine Intervention Trial Investigators Identifying novel biomarkers for cardiovascular events or death in people with dysglycemia. Circulation. 2015;132:2297–2304. doi: 10.1161/CIRCULATIONAHA.115.015744. [DOI] [PubMed] [Google Scholar]
- 23.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Browning S.R., Browning B.L. Haplotype phasing: existing methods and new developments. Nat. Rev. Genet. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Covarrubias-Pazaran G. Genome-Assisted prediction of quantitative traits using the r package sommer. PLoS ONE. 2016;11:e0156744. doi: 10.1371/journal.pone.0156744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahlqvist E., Storm P., Käräjämäki A., Martinell M., Dorkhan M., Carlsson A., Vikman P., Prasad R.B., Aly D.M., Almgren P. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018;6:361–369. doi: 10.1016/S2213-8587(18)30051-2. [DOI] [PubMed] [Google Scholar]
- 31.Scott R.A., Scott L.J., Mägi R., Marullo L., Gaulton K.J., Kaakinen M., Pervjakova N., Pers T.H., Johnson A.D., Eicher J.D., DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott R.A., Lagou V., Welch R.P., Wheeler E., Montasser M.E., Luan J., Mägi R., Strawbridge R.J., Rehnberg E., Gustafsson S., DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat. Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerstein H.C., Ferrannini E., Riddle M.C., Yusuf S., ORIGIN Trial Investigators Insulin resistance and cardiovascular outcomes in the ORIGIN trial. Diabetes Obes. Metab. 2018;20:564–570. doi: 10.1111/dom.13112. [DOI] [PubMed] [Google Scholar]
- 34.Jones A.G., Hattersley A.T. The clinical utility of C-peptide measurement in the care of patients with diabetes. Diabet. Med. 2013;30:803–817. doi: 10.1111/dme.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun B.B., Maranville J.C., Peters J.E., Stacey D., Staley J.R., Blackshaw J., Burgess S., Jiang T., Paige E., Surendran P. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferdinand K.C., Armani A.M. The management of hypertension in African Americans. Crit. Pathw. Cardiol. 2007;6:67–71. doi: 10.1097/HPC.0b013e318053da59. [DOI] [PubMed] [Google Scholar]
- 37.Cohn J.N., Julius S., Neutel J., Weber M., Turlapaty P., Shen Y., Dong V., Batchelor A., Guo W., Lagast H. Clinical experience with perindopril in African-American hypertensive patients: a large United States community trial. Am. J. Hypertens. 2004;17:134–138. doi: 10.1016/j.amjhyper.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 38.ALLHAT Officers and Coordinators for the ALLHAT Collaborative Research Group. The Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial Major outcomes in high-risk hypertensive patients randomized to angiotensin-converting enzyme inhibitor or calcium channel blocker vs diuretic: The Antihypertensive and Lipid-Lowering Treatment to Prevent Heart Attack Trial (ALLHAT) JAMA. 2002;288:2981–2997. doi: 10.1001/jama.288.23.2981. [DOI] [PubMed] [Google Scholar]
- 39.Howes R.E., Patil A.P., Piel F.B., Nyangiri O.A., Kabaria C.W., Gething P.W., Zimmerman P.A., Barnadas C., Beall C.M., Gebremedhin A. The global distribution of the Duffy blood group. Nat. Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voruganti V.S., Laston S., Haack K., Mehta N.R., Smith C.W., Cole S.A., Butte N.F., Comuzzie A.G. Genome-wide association replicates the association of Duffy antigen receptor for chemokines (DARC) polymorphisms with serum monocyte chemoattractant protein-1 (MCP-1) levels in Hispanic children. Cytokine. 2012;60:634–638. doi: 10.1016/j.cyto.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powe C.E., Evans M.K., Wenger J., Zonderman A.B., Berg A.H., Nalls M., Tamez H., Zhang D., Bhan I., Karumanchi S.A. Vitamin D-binding protein and vitamin D status of black Americans and white Americans. N. Engl. J. Med. 2013;369:1991–2000. doi: 10.1056/NEJMoa1306357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braithwaite V.S., Jones K.S., Schoenmakers I., Silver M., Prentice A., Hennig B.J. Vitamin D binding protein genotype is associated with plasma 25OHD concentration in West African children. Bone. 2015;74:166–170. doi: 10.1016/j.bone.2014.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.